Abstract
Demonstrated successes of protein design and engineering suggest significant potential to produce diverse protein architectures and assemblies beyond those found in nature. Here, we describe a new class of synthetic protein architecture through the successful design and atomic structures of water-soluble cross-β proteins. The cross-β motif is formed from the lamination of successive β-sheet layers, and it is abundantly observed in the core of insoluble amyloid fibrils associated with protein-misfolding diseases. Despite its prominence, cross-β has been designed only in the context of insoluble aggregates of peptides or proteins. Cross-β’s recalcitrance to protein engineering and conspicuous absence among the known atomic structures of natural proteins thus makes it a challenging target for design in a water-soluble form. Through comparative analysis of the cross-β structures of fibril-forming peptides, we identified rows of hydrophobic residues (“ladders”) running across β-strands of each β-sheet layer as a minimal component of the cross-β motif. Grafting a single ladder of hydrophobic residues designed from the Alzheimer’s amyloid-β peptide onto a large β-sheet protein formed a dimeric protein with a cross-β architecture that remained water-soluble, as revealed by solution analysis and x-ray crystal structures. These results demonstrate that the cross-β motif is a stable architecture in water-soluble polypeptides and can be readily designed. Our results provide a new route for accessing the cross-β structure and expanding the scope of protein design.
Keywords: beta-sheet, OspA, protein design, self-assembly, fibril
The cross-β motif is a major class of polypeptide architecture common to a wide range of insoluble protein assemblies (1). While it is prominently known as the underlying structure of the amyloid fibrils associated with protein-misfolding diseases (2), the cross-β architecture also comprises robust materials such as spider silk and bacterial biofilms (3). Cross-β’s versatility and remarkable stability therefore make it an attractive architecture for protein and nanomaterial design (4).
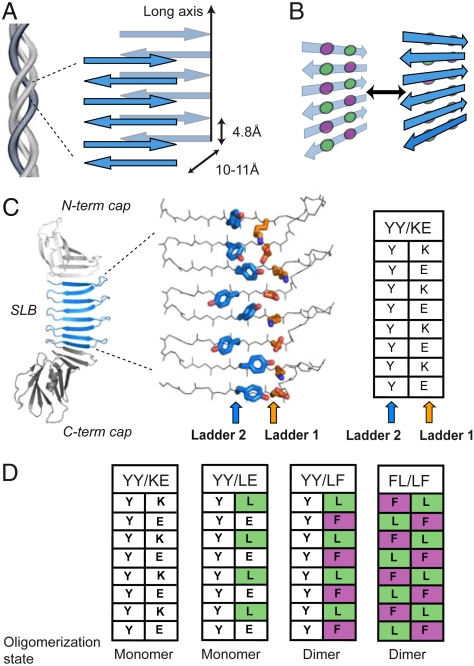
The rapidly growing repertoire of structures of the cross-β spine (5–10) demonstrates that the fundamental unit of cross-β is laminated β-sheets whose strands run perpendicular to the long-axis of the protein assembly (1) (Fig. 1A). This β-rich composition produces the characteristic 4.8 Å and 10–11 Å reflections observed in x-ray diffraction experiments of fibrils, which are attributed to the strand spacing within and between β-sheet layers, respectively (Fig. 1A) (1, 11). Though a regular pattern of backbone hydrogen bonds provides the framework for cross-β, amino acid side chains play a key role in the formation and stabilization of the laminated architecture. The periodicity of peptide units inherent in cross-β self-assemblies gives rise to a repeating pattern of side chains (“ladders”) running across β-strands within each β-sheet layer (Fig. 1B). These ladders control the β-sheet-forming propensity and surface properties of the self-assembly as well as direct the formation of dry and wet interfaces between the β-sheet layers (5, 6). The ladders on the dry face are well packed and form tightly interdigitated interactions termed “steric zippers”, whereas the wet face is hydrated and more loosely packed (5, 6). Designed short peptides that recapitulate insoluble cross-β self-assemblies have further defined general characteristics of fibril-forming sequences beyond those associated with diseases (12, 13).
Fig. 1.
The architecture of cross-β fibrils and PSAMs. (A) Diagram of cross-β architecture formed from the lamination of β-sheets. The long-axis and typical distances between intra- and intermolecular β-strands are shown. Though antiparallel β-sheets are shown, cross-β can be formed from either antiparallel or parallel β-sheets. (B) Schematic drawing depicting the concept of cross-strand ladders and the formation of a cross-β dimer from β-sheets containing hydrophobic ladders. Circles denote amino acid side chains that form cross-strand ladders consisting of two amino acid types (colored differently). (C) Left, the PSAM scaffold is shown as a cartoon, with the single-layer β-sheet segment (SLB) colored blue and the N- and C-terminal globular domains colored gray. The side chains of the two cross-strand ladders used as hosts are shown as stick models and colored by element. The carbon atoms of Ladders 1 and 2 are shown in orange and blue, respectively. The table summarizes the amino acid identities of the ladders. (D) Amino acid identities of PSAMs used in this work presented in the table format as in C. Their amino acid sequences are presented in Fig. S1.
Although cross-β structures are abundant in insoluble protein deposits and thus comprise an important class of insoluble protein architecture, there are few examples of cross-β interactions among the tens of thousands of atomic structures of natural water-soluble proteins. Though early protein and amyloid research focused on analyses of these natural polypeptides, advances in our understanding of molecular recognition and evolution in conjunction with recombinant DNA technologies have now enabled the generation of “synthetic” proteins that do not exist in nature. Design, production, and characterization of synthetic proteins test our fundamental understanding of proteins as well as expand the universe of protein architecture and function (14–16). A robust design strategy should be applicable to cross-β investigation. However, the scarcity of well-characterized, water-soluble cross-β motifs has made it difficult to understand how this architecture may be designed outside the context of self-assemblies of short peptides (3), for which substantial computational design has been performed (17–20). To date, no water-soluble cross-β structures have been successfully produced. Attempts to graft segments of amyloid-forming peptides into water-soluble proteins have resulted in insoluble fibril-like assemblies (21, 22) or structurally ill-characterized materials (23). Several previous studies which grafted segments of amyloid-forming peptides into globular proteins have produced fibril-like structures through either domain swapping or intermolecular association of amyloidogenic sequences, but unfortunately either the atomic structure of these assemblies could not be validated or they have demonstrated architecture which is inconsistent with that of cross-β (22–26). As such, one might consider the cross-β architecture to be incompatible with requirements for water-soluble polypeptides.
Adapting the cross-β motif to a water-soluble form would be an important achievement in expanding the repertoire of available protein architectures. The ability to generate water-soluble cross-β proteins would not only further advance our understanding of factors governing cross-β formation in natural systems, but would also broaden the scope of protein design and engineering. Here we report the successful design and atomic structures of a unique class of synthetic water-soluble proteins built to form the cross-β architecture. Our results reveal surprisingly simple requirements for the cross-β structure and suggest a unique direction in protein design.
Results and Discussion
Cross-β Design Using Minimalist Motifs.
The rapid growth in the number of structures of fibril-forming peptides obtained by x-ray crystallography and solid-state NMR spectroscopy provides a wealth of common characteristics that could be applied towards the rational design of the cross-β motif in globular proteins (5–9, 27, 28). We envisioned that a water-soluble cross-β architecture could be designed by mimicking structural features of cross-β while controlling the degree of self-assembly. We identified the “peptide self-assembly mimic” (PSAM) system, which captures a segment of a peptide self-assembly within a water-soluble and monomeric protein, as a promising scaffold for this approach (29) (Fig. 1C). The PSAM scaffold employed in this work is a variant of Borrelia burgdorferi Outer surface protein A (OspA) containing three additional copies of a β-hairpin segment derived from the OspA single-layer β-sheet (SLB) (29). The PSAM scaffold, previously referred to as “OspA+3bh” (29), thus contains a large SLB comprised of four identical 23-residue β-hairpins that are capped by the N- and C-terminal globular domains of OspA (Fig. S1). Because the PSAM is a single polypeptide, its sequence can be precisely and easily manipulated while maintaining both β-strand register and orientation (30). Importantly, we have recently produced a PSAM containing a particularly flat β-sheet (termed “YY/KE”; Fig. 1C and Fig. S1) suitable for cross-β design in this work (30).
Our strategy for generating water-soluble cross-β was to identify and graft minimal architectural features from known cross-β assemblies into the PSAMs. Hydrophobic ladders are commonly found in many amyloid fibril models and may play an important role in generating a dry interface (31, 32), and were thus selected as the basic design principle. We recapitulated the hydrophobic ladder motif by introducing mutations into adjacent host ladders located on the same face of the PSAM β-sheet (denoted as Ladder 1 and Ladder 2; Fig. 1C). The ladders were built using alternating Leu and Phe residues (Fig. 1B, D; Fig. S1). This ladder motif would be present in an antiparallel assembly of the Leu-X-Phe or Phe-X-Leu sequence (where X is any amino acid) (33). The Leu-X-Phe sequence has a high propensity for fibrillization (12) and forms a hydrophobic ladder in the amyloid-β (Aβ) core region (the amino acid sequence, LVFFA) (8, 31, 34). This Aβ segment can form the core of either parallel or antiparallel assemblies depending on its flanking sequences (33). A Leu-Phe ladder is also present in fibrils of the human islet amyloid polypeptide (32). We initially designed a PSAM mutant containing a single Leu-Phe ladder to determine if this minimal motif was sufficient for generating cross-β (termed “YY/LF”; Fig. 1D). To gauge the effects of having two neighboring hydrophobic ladders, we also produced a mutant in which the Leu-Phe pattern was introduced into both of the host ladders (“FL/LF”; Fig. 1D and Fig. S1).
Designed Cross-β Proteins are Soluble and Stable.
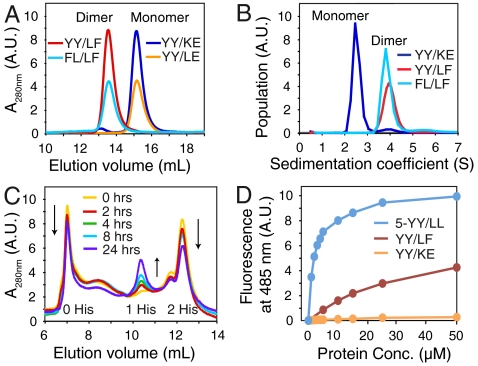
The PSAMs containing hydrophobic ladders (YY/LF and FL/LF) were expressed in E. coli as soluble proteins, and, unlike the starting YY/KE PSAM that is predominantly monomeric, they formed dimers as monitored by gel filtration chromatography (Fig. 2A) and analytical ultracentrifugation (Fig. 2B). This dimerization is consistent with the formation of a cross-β architecture through lamination of linear stretches of hydrophobic residues on the PSAMs. In contrast, a mutant in which only the Leu residues were introduced at every other ladder position (“YY/LE”; Fig. 1D and Fig. S1) was monomeric (Fig. 2A), suggesting the importance of a contiguous hydrophobic ladder in promoting β-sheet lamination. Urea denaturation experiments further showed the mutants were thermodynamically stable (Fig. S2).
Fig. 2.
Dimer formation of cross-β PSAMs. (A) Size-exclusion chromatograms of PSAMs. Proteins are identified in the figure and the positions corresponding to the monomeric and dimeric species are marked. (B) Sedimentation velocity analytical ultracentrifugation. The relative populations of YY/KE, YY/LF and FL/LF are plotted as a function of sedimentation coefficient, S. The monomer and dimer peaks are labeled. (C) Time-dependent subunit exchange monitored via cation exchange chromatography. An equimolar mixture of YY/LF dimers containing either zero or two His-tags was analyzed over a 24 h incubation period. Peaks corresponding to dimers containing zero and two His-tags (starting materials) and that for a dimer species containing one His-tag (exchange product) are marked. (D) ThT binding to the YY/LF cross-β PSAM. Fluorescence emission at 485 nm of 10 μM ThT is plotted as a function of PSAM concentration. The 5-YY/LL and YY/KE PSAMs are included as positive and negative controls.
To investigate dimerization kinetics, we monitored the time-dependent exchange of subunits between PSAM complexes. We prepared two samples of the YY/LF PSAM with either an N-terminal His-tag fusion (two His-tags per dimer) or with the His-tag cleaved (zero His-tags per dimer). Using cation exchange chromatography, we successfully separated the zero and two His-tag species at opposite ends of a salt gradient (Fig. 2C). By combining equal amounts of these samples and incubating the mixture between 0–24 h, we observed an incremental depletion of the zero and two His-tag dimer peaks and the growth of a new peak (shown by size-exclusion chromatography to also be a dimer) midway between them. These data are consistent with the formation of a dimer species containing one His-tag resulting from subunit exchange (i.e. dissociation and reassociation) of PSAM subunits. The PSAM subunits exchanged with a half-life of approximately 1 d, demonstrating the kinetic stability of the dimers (Fig. 2C).
We also tested whether YY/LF and FL/LF could form a heterodimer. Mixing equal amounts of the two proteins in urea and refolding them resulted in the formation of the expected 1∶2∶1 ratio of the YY/LF homodimer: heterodimer: FL/LF homodimer (Fig. S3). These results suggest that dimerization is not highly sensitive to the amino acid identity of the hydrophobic ladders. The importance of hydrophobic interactions has also been observed in the Aβ peptide, where a number of hydrophobic substitutions maintain its ability to form fibrils (35).
Thioflavin-T Binding to Cross-β PSAMs.
We have recently designed a monomeric PSAM containing hydrophobic ladders composed of Tyr and Leu across five β-strands (5-YY/LL) that binds the amyloid dye Thioflavin-T (ThT) with high affinity. Its crystal structure and molecular dynamics simulations strongly suggest that ThT binds across the β-strands of the SLB within a shallow groove formed by the designed ladders (36, 37). Because of the sequence similarity of the 5-YY/LL hydrophobic ladders to those in YY/LF and FL/LF, we investigated ThT binding to these dimeric PSAMs. Interestingly, although the dimeric PSAMs bound to ThT, they exhibited much weaker affinity than the monomeric 5-Y/LL PSAM (Fig. 2D). As described below, the weak ThT binding of the dimeric PSAMs, despite the presence of aromatic/hydrophobic ladders seemingly capable of ThT binding, can be attributed to the sequestration of the ladders within the dimerization interface.
X-ray Crystal Structures of Cross-β PSAMs.
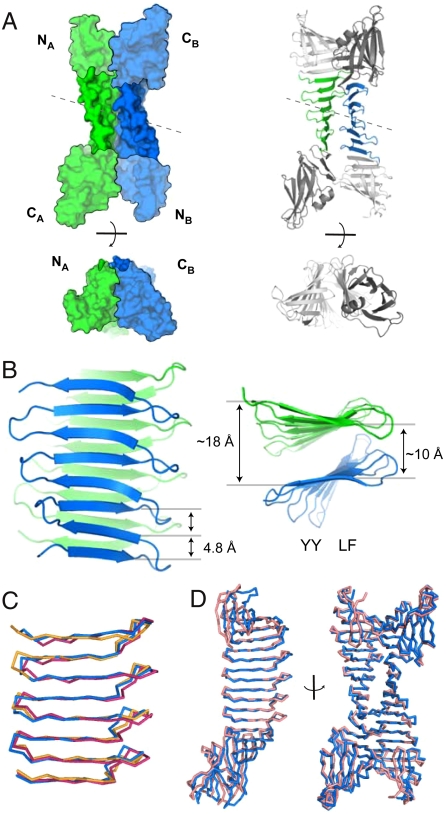
The crystal structures of the YY/LF and FL/LF dimers were determined at 1.65 and 2.50 Å, respectively (statistics are given in Table S1). Because of the weak but significant ThT binding, we performed crystal screens of the dimeric PSAMs both in the presence and absence of ThT. Interestingly, though YY/LF crystallized in conditions with and without ThT, only crystals grown in the presence of the dye were of diffraction quality. However, no unambiguous electron density for ThT was observed in YY/LF. In contrast, FL/LF crystallized only in the absence of ThT.
The structures of YY/LF and FL/LF revealed laminated eight-stranded β-sheets exhibiting features characteristic of the cross-β motif (Fig. 3). In both, the asymmetric unit contains two conformationally similar PSAM molecules bound head-to-tail (the Cα root mean squared deviations (RMSDs)) between PSAM subunits in each asymmetric unit are 1.4 Å for YY/LF and 0.7 Å for FL/LF), and PSAM dimerization was mediated by mutual interactions of the designed hydrophobic ladders. The long axes of the two sheets, defined as in fibrils (Fig. 1A), are aligned in a nearly parallel manner (Fig. 3B). The peptide backbones of the two β-sheets are separated by 10 Å at their closest point, consistent with a typical cross-β architecture (2), and 18 Å at the farthest (Fig. 3B). Consequently, the planes of the two β-sheets have a relative pitch of ∼30° (Fig. 3B, Fig. 4C, D). The two β-sheet layers are staggered, positioning β-strands on each side of the dimer interface between the strands on the opposite face, as seen in cross-β spine structures (5, 6) and amyloid-β models (8) (Fig. 4C).
Fig. 3.
The x-ray crystal structures of cross-β PSAMs. (A) The overall structure of the YY/LF PSAM dimer is shown in surface and cartoon representations, in two orthogonal views. Molecule A is in green and molecule B in blue. The N- and C-terminal domains are indicated. The two molecules are related by pseudo-two-fold symmetry (dashed line). (B) The arrangement of the laminated β-sheet segment of YY/LF PSAM. Residues 118–209 are shown in orthogonal orientations, with the same coloring scheme as in A. The Cα-Cα distances across the β-sheets and the positions of the YY and LF ladders are indicated. (C) Comparisons of the backbone conformations of the central β-sheet regions of the YY/KE (Orange), YY/LF (Blue), FL/LF (Magenta) PSAMs. (D) Comparisons of the cross-β PSAM dimers, showing alignments of molecule A (Left) and an orthogonal view of the entire dimer complex (Right). Proteins are shown as Cα traces, with YY/LF in blue and FL/LF in pink.
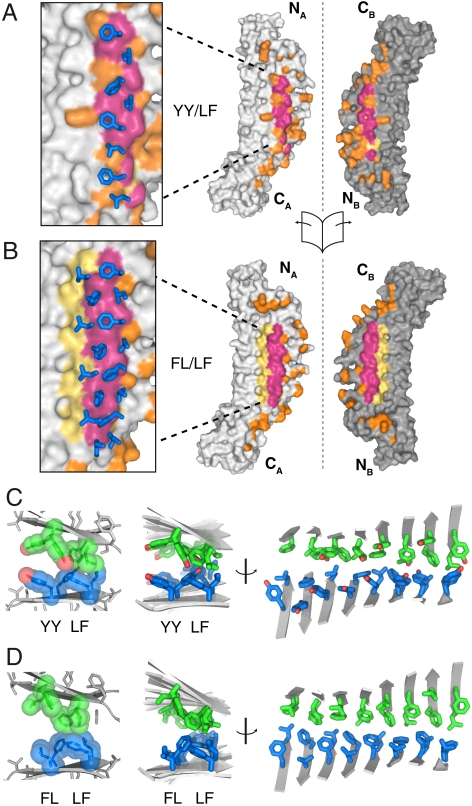
Fig. 4.
The interface structures of synthetic cross-β. (A) The dimer interface of the YY/LF PSAM dimer. The dimer has been opened like a book along the axis indicated, with molecule A on the right and molecule B on the left. The red coloration indicates surfaces in the LF ladder that are buried in the interface (defined as atoms within 5 Å of the adjacent molecule). The yellow surfaces are for those atoms in the LF ladder that are not buried in the interface. All other contacts are colored orange. The insert shows how the hydrophobic ladder from subunit B (shown as stick models) overlays on the subunit A surface. (B) The dimer interface of the FL/LF PSAM dimer. The coloring scheme is the same as in A, but the red and yellow surfaces are for atoms in both hydrophobic ladders. (C) The packing of the cross-β segment of YY/LF. At left, the side chains of the YY and LF ladders are shown as spheres. For clarity, only two adjacent β-strands from molecules A and B are shown. At Center and Right are shown orthogonal representations of all side chains of the YY and LF ladders. The β-strands are shown as cartoons. The loop regions have been omitted for clarity. Side-chain carbons are colored green and blue for molecule A and molecule B, respectively. (D) The packing of the cross-β segment of FL/LF depicted in the same manner as in C.
YY/LF and FL/LF are structurally similar, with a Cα-Cα RMSD of 2.4 Å for the overall dimeric complex (Fig. 2D, right), 2.0 Å for the underlying PSAM subunits (Fig. 2D, left), and 1.6 Å for the central β-sheets (Fig. 2C). The fact that YY/LF and FL/LF share similar solution properties and have nearly identical structures suggests that the addition of ThT is not required for stabilizing the designed architecture. Importantly, the site analogous to the ThT binding site of the monomeric 5-YY/LL PSAM is sterically occluded in the YY/LF and FL/LF structures due to tight packing of the hydrophobic ladders. These results are in agreement with molecular dynamics simulations indicating that ThT rarely binds between laminated β-sheets but rather preferentially binds onto exposed surfaces (37).
In both cross-β PSAM structures, the residues in the hydrophobic ladders form a dry interface dominated by intermolecular hydrophobic interactions. Over two-thirds of the contacts between the laminated β-sheets (∼895 Å2 and ∼710 Å2 buried surface area per monomer in the cross-β regions of YY/LF and FL/LF, respectively, Table 1) are nonpolar in composition (Fig. 4A). The hydrophobic ladder residues pack both across and within strands to form a largely continuous hydrophobic surface on each monomer (Fig. S4). Although FL/LF contains two hydrophobic ladders, the Leu-Phe ladder of Ladder 2 was partially exposed to solvent (Fig. 4B, D), as was the equivalent ladder consisting of Tyr in YY/LF (Fig. 4A, C). The inability of the second hydrophobic ladder to form a dry interface appears to be due to steric clashes of the capping domains (Fig. 4A, B). The capping domains in both structures make numerous intersubunit contacts (∼450–550 Å2), and they exhibit a localized hinge-like bending relative to their conformations in monomeric PSAMs (Fig. S5 and Movie S1). Despite these deformations, the secondary structures of the cross-β PSAMs remain effectively unchanged (Fig. 3). The fact that PSAMs lacking the hydrophobic ladder mutations do not form dimers suggests that the contacts involving the capping domains are primarily opportunistic and artifacts of bringing the SLBs into close proximity. Thus, the intermolecular interactions of the hydrophobic ladders occurred even at the expense of the distortion of the scaffold, suggesting that these cross-β structures are energetically favorable.
Table 1.
Interface composition of cross-β PSAMs
| Surface Area Buried (Å2) * |
Nonpolar Contacts |
|
| YY/LF † | 1323 | 66% |
| YY/LF (SLB) ‡ | 894 | 71% |
| FL/LF † | 1264 | 68% |
| FL/LF (SLB) ‡ | 712 | 77% |
*Surface area burial is reported for a single subunit, and is derived from the average value of the two subunits comprising each cross-β PSAM.
†Buried surface area for the entire PSAM.
‡Buried surface area only for the single-layer β-sheet segment (residues 118–209).
β-Sheet lamination in the PSAMs occurs essentially parallel to the hydrophobic cross-strand ladders, causing steric clashes of the N- and C-terminal capping domains (Fig. 3A). The majority of these clashes would have been avoided by laminating diagonally (with perpendicular strands instead of parallel strands), as is often observed in the case of β-sandwich proteins. However, diagonal lamination would incorporate a large number of charged and polar residues into the hydrophobic interface. These results suggest that hydrophobic burial in the PSAM dimers is a particularly favorable mode of β-sheet lamination, and underscores how the parallel orientation of β-sheet layers in cross-β maximizes the number of repetitive, productive chemical contacts in a self-assembly.
The fact that both PSAMs exhibit the same head-to-tail dimerization suggests that this is an energetically preferred orientation (Fig. 3A). In this arrangement, homotrophic interactions between the two hydrophobic ladders (Ladder 1-Ladder 1 and Ladder 2-Ladder 2) were formed, consistent with Class 8 of Eisenberg’s nomenclature for amyloid fibril orientations (6). The opposite, head-to-head arrangement of the PSAMs would have led to heterotrophic ladder interactions (Ladder 1-Ladder 2). Although the YY/LF PSAM would produce distinctly different dimer interfaces between the two arrangements, the similarity of Ladder 1 and Ladder 2 in FL/LF would produce effectively identical β-sheet interfaces in either arrangement. It is therefore unlikely that the amino acid sequence of the ladders solely dictated the orientation of the PSAMs. Instead, the crystal structures suggest that the observed head-to-tail dimerization is favored because it creates fewer steric conflicts among the capping domains. In contrast, the hypothetical head-to-head orientation would bring the larger C-terminal domains of two subunits in close proximity (there are ∼130 residues in the C-terminal domain versus ∼90 residues in the N-terminal domain), thus leading to severe steric conflicts between the underlying PSAMs. These results indicate that cross-β architectures can be controlled by altering regions flanking the laminated β-sheet interface.
Comparison of Soluble Cross-β and Amyloid Fibril Models.
The cross-β PSAM structures represent the largest segments of cross-β architecture determined to date. It should be noted that cross-β structures of self-assemblies of short peptides determined with x-ray crystallography or solid-state NMR spectroscopy inherently assume that all self-assembling subunits have an identical structure (5–9). Furthermore, infinite β-sheet lamination is a prerequisite for crystallizing small self-assembling peptides (5–7). Thus, these PSAMs offer a unique opportunity to characterize cross-β structures with fewer constraints imposed by experimental limitations. The cross-β PSAMs have 6–8° twist per β-strand (Fig. 2B), similar to those found for computationally optimized cross-β spine structures (38) but smaller than those estimated from micrographs of amyloid fibrils (∼15°) (2). Additionally, the four β-hairpin repeats constituting the cross-β PSAMs exhibit slightly different conformations with pairwise Cα RMSD values of ∼0.4–1.2 Å, providing a rare insight into structural heterogeneity within a cross-β assembly. Such subtle conformational heterogeneity can lead to differences in morphology when propagated over a large number of peptide units (29).
Role of Hydrophobic Ladders in Cross-β Assembly.
The ability of the PSAM system to precisely replace amino acid side chains without significant changes in the underlying β-sheet framework has allowed us to dissect the contributions of a number of normally overlapping factors and to directly address the role of amino acid ladders in the cross-β architecture. These results demonstrate that hydrophobic residues linearly aligned across β-strands are a particularly favorable force in driving β-sheet lamination and may play a central role in generating ordered self-assemblies and protein oligomers. Because a mutation in a peptide subunit is propagated throughout a peptide self-assembly, a single hydrophilic-to-hydrophobic mutation (such as the Glu-to-Phe mutation in the transition from the monomeric YY/LE to the dimeric YY/LF; Fig. 1D) can lead to the formation of a fully hydrophobic ladder and drastically change the overall surface properties of the assembled β-sheet, thus tipping the conformational equilibrium towards the formation of cross-β (13, 39). The capacity of even a short hydrophobic ladder to drive β-sheet lamination suggests that a peptide containing multiple hydrophobic residues may form a variety of cross-β structures by employing distinct hydrophobic ladder configurations. Such alternative modes can explain the structural polymorphs of amyloid fibrils that are observed for the same amino acid sequence (32, 40–42).
The use of hydrophobic cross-strand ladders on a single face of the SLB allowed us to create a highly soluble “self-sealing” architecture with a well-defined orientation (43). These characteristics were essential to designing a cross-β architecture free of the aggregation tendencies present in uncontrolled or infinitely extending modes of self-assembly typically associated with cross-β. The fact that the PSAM mutants were either predominantly monomeric or predominantly dimeric is notable in the engineering of protein assemblies, as previous attempts to generate protein assemblies by introducing point mutations have often produced complexes with substantial monomer-oligomer equilibria (44). In contrast, because the PSAMs contain multiple copies of a self-assembly-like β-strand unit, a mutation in the underlying unit drastically changes the overall chemical and structural properties of the β-sheet. These results reinforce how dramatically a point mutation in a small peptide unit can affect its capability to self-assemble (39) and help rationalize the abundance of polypeptides that can be converted into amyloid fibrils by small sequence perturbations. However, we emphasize that our findings do not imply that hydrophobic residues are generally sufficient for fibril formation. In the case of true fibril-forming peptides, the entropic loss incurred upon fibrillization is much greater than in our system where the β-sheets of each PSAM fold independently prior to lamination.
Scarcity of Hydrophobic Ladders in Natural Proteins.
Though hydrophobic ladders with a repeating binary pattern (e.g. LF) are commonly found in cross-β assemblies formed from short peptides, we have not identified this motif in natural water-soluble proteins. Most β-sheets in natural, water-soluble proteins contain four or fewer β-strands and often exhibit considerable levels of distortion (45, 46), rendering them unsuitable scaffolds for long cross-strand hydrophobic ladders. The closest analogs that we found are rows of hydrophobic residues in the interface of β-helix proteins (47, 48) and other β-sheet oligomers (49). However, these ladders do not comprise an alternating pattern of two amino acid types, but rather they are punctuated with polar amino acids that likely enhance oligomerization specificity (50). Because a long hydrophobic ladder can lead to strong interactions between β-sheet layers as demonstrated here, it is probable that there is a strong selective pressure against proteins containing rows of surface-exposed hydrophobic residues in nature.
The ability of amyloid fibrils to be formed by hundreds of sequence-unrelated peptides suggests that the energetic factors underlying cross-β’s stability are heavily opportunistic or promiscuous in nature. This hypothesis is supported by the degenerate pairing of hydrophobic residues in the cross-β PSAM dimer interface and by our observation that the PSAMs can form asymmetric interfaces from two distinct subunits between dimers (e.g. the heterodimer formed by YY/LF and FL/LF). The stable but nonspecific interactions between dry, hydrophobic β-sheet interfaces may lead to uncontrolled self-assembly of β-sheets containing these motifs, further rationalizing why hydrophobic ladders are so infrequently observed in natural proteins. In the case of functional amyloids, tight regulation and cellular localization appears to be key to sequestering the potentially nonspecific and self-templating nature of the cross-β architecture (3, 51, 52).
Conclusions
This work has expanded the architectural repertoire of water-soluble proteins and demonstrated that the cross-β architecture is inherently stable and surprisingly easily designed. Because water-soluble cross-β proteins can be studied using a variety of quantitative biophysical and structural methods, they should provide an excellent model with which to probe the structural and thermodynamic consequences of mutations associated with protein-misfolding diseases and to investigate interactions of fibrils with small organic molecules (36). The design concept described here may be further employed to produce a variety of cross-β proteins and other precisely defined nanomaterials.
Materials and Methods
Protein Production.
Mutagenesis, expression, purification, and urea denaturation of PSAMs were performed as described previously (53). Except for ThT binding measurements, all data were obtained using PSAM mutants constructed from the “sm1” PSAM scaffold, which contains mutations in the N- and C-terminal globular domains of OspA that increase crystallization efficiency without perturbing global structure or the SLB region (54). An N-terminal His-tag was cleaved prior to all measurements, unless otherwise stated.
Size-Exclusion Chromatography and Ultracentrifugation.
Proteins were analyzed using a Superdex 200 column (Amersham) in 10 mM phosphate buffer, pH 6.0 with 150 mM NaCl. Sedimentation velocity measurements were performed using a XL-A ultracentrifuge (Beckman Coulter). The protein concentrations were adjusted so that absorbance at 280 nanometers (nm) was 0.4. Experiments were performed at 40,000 rpm and 20 °C. Data were analyzed using the SEDFIT program (55).
Stability Measurements.
Urea-induced unfolding of the PSAMs was monitored using elipticity at 235 nm and tryptophan (Trp) fluorescence emission at 290 nm at 30 °C as previously established (53).
Dimer Exchange Kinetics Measurements.
Two samples of the YY/LF PSAM either with or without an N-terminal His-tag were prepared. They were combined in an equimolar ratio, and after incubation at 25 °C, the samples were analyzed using cation exchange chromatography (SP-Sepharose, Amersham) in 20 mM sodium acetate buffer, pH 5.0 with a sodium chloride gradient.
Structure Analysis.
Crystallization and structure determination are described in Table S1. Solvent accessible surface areas were determined using CNS (56) and their chemical compositions were analyzed using home-made scripts. The twist angles between β-hairpins were determined as described (29).
Supplementary Material
Acknowledgments.
We thank S. Meredith and T. Sosnick for critical reading of the manuscript. This work was supported by the National Institutes of Health Grants R01-GM057215 (to S.K.) and T90-DK070076 (to M.B.), and the National Science Foundation Grant CMMI-0709079 (to S.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0912654107/DCSupplemental.
References
- 1.Nelson R, Eisenberg D. Recent atomic models of amyloid fibril structure. Curr Opin Struct Biol. 2006;16:260–265. doi: 10.1016/j.sbi.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 2.Sunde M, et al. Common core structure of amyloid fibrils by synchrotron x-ray diffraction. J Mol Biol. 1997;273:729–739. doi: 10.1006/jmbi.1997.1348. [DOI] [PubMed] [Google Scholar]
- 3.Otzen D, Nielsen PH. We find them here, we find them there: Functional bacterial amyloid. Cell Mol Life Sci. 2008;65:910–927. doi: 10.1007/s00018-007-7404-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Knowles TP, et al. Role of intermolecular forces in defining material properties of protein nanofibrils. Science. 2007;318:1900–1903. doi: 10.1126/science.1150057. [DOI] [PubMed] [Google Scholar]
- 5.Nelson R, et al. Structure of the cross-beta spine of amyloid-like fibrils. Nature. 2005;435:773–778. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sawaya MR, et al. Atomic structures of amyloid cross-beta spines reveal varied steric zippers. Nature. 2007;447:453–457. doi: 10.1038/nature05695. [DOI] [PubMed] [Google Scholar]
- 7.Makin OS, Atkins E, Sikorski P, Johansson J, Serpell LC. Molecular basis for amyloid fibril formation and stability. Proc Natl Acad Sci U S A. 2005;102:315–320. doi: 10.1073/pnas.0406847102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luhrs T, et al. 3D structure of Alzheimer’s amyloid-beta(1–42) fibrils. Proc Natl Acad Sci U S A. 2005;102:17342–17347. doi: 10.1073/pnas.0506723102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wasmer C, et al. Amyloid fibrils of the HET-s(218–289) prion form a beta solenoid with a triangular hydrophobic core. Science. 2008;319:1523–1526. doi: 10.1126/science.1151839. [DOI] [PubMed] [Google Scholar]
- 10.Petkova AT, Yau WM, Tycko R. Experimental constraints on quaternary structure in Alzheimer’s beta-amyloid fibrils. Biochemistry. 2006;45:498–512. doi: 10.1021/bi051952q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Makin OS, Serpell LC. Structures for amyloid fibrils. Febs J . 2005;272:5950–5961. doi: 10.1111/j.1742-4658.2005.05025.x. [DOI] [PubMed] [Google Scholar]
- 12.Lopez De La Paz M, Serrano L. Sequence determinants of amyloid fibril formation. Proc Natl Acad Sci U S A. 2004;101:87–92. doi: 10.1073/pnas.2634884100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiti F, Stefani M, Taddei N, Ramponi G, Dobson CM. Rationalization of the effects of mutations on peptide and protein aggregation rates. Nature. 2003;424:805–808. doi: 10.1038/nature01891. [DOI] [PubMed] [Google Scholar]
- 14.Kuhlman B, et al. Design of a novel globular protein fold with atomic-level accuracy. Science. 2003;302:1364–1368. doi: 10.1126/science.1089427. [DOI] [PubMed] [Google Scholar]
- 15.Yeates TO, Padilla JE. Designing supramolecular protein assemblies. Curr Opin Struct Biol. 2002;12:464–470. doi: 10.1016/s0959-440x(02)00350-0. [DOI] [PubMed] [Google Scholar]
- 16.Riechmann L, Winter G. Novel folded protein domains generated by combinatorial shuffling of polypeptide segments. Proc Natl Acad Sci U S A. 2000;97:10068–10073. doi: 10.1073/pnas.170145497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ivanova MI, Thompson MJ, Eisenberg D. A systematic screen of beta(2)-microglobulin and insulin for amyloid-like segments. Proc Natl Acad Sci U S A. 2006;103:4079–4082. doi: 10.1073/pnas.0511298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kammerer RA, et al. Exploring amyloid formation by a de novo design. Proc Natl Acad Sci U S A. 2004;101:4435–4440. doi: 10.1073/pnas.0306786101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsai CJ, et al. Principles of nanostructure design with protein building blocks. Proteins. 2007;68:1–12. doi: 10.1002/prot.21413. [DOI] [PubMed] [Google Scholar]
- 20.Lopez De La Paz M, et al. De novo designed peptide-based amyloid fibrils. Proc Natl Acad Sci U S A. 2002;99:16052–16057. doi: 10.1073/pnas.252340199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chiti F, Dobson CM. Amyloid formation by globular proteins under native conditions. Nat Chem Biol. 2009;5:15–22. doi: 10.1038/nchembio.131. [DOI] [PubMed] [Google Scholar]
- 22.Sambashivan S, Liu Y, Sawaya MR, Gingery M, Eisenberg D. Amyloid-like fibrils of ribonuclease A with three-dimensional domain-swapped and native-like structure. Nature. 2005;437:266–269. doi: 10.1038/nature03916. [DOI] [PubMed] [Google Scholar]
- 23.Otzen DE, Miron S, Akke M, Oliveberg M. Transient aggregation and stable dimerization induced by introducing an Alzheimer sequence into a water-soluble protein. Biochemistry. 2004;43:12964–12978. doi: 10.1021/bi048509k. [DOI] [PubMed] [Google Scholar]
- 24.Guo Z, Eisenberg D. The structure of a fibril-forming sequence, NNQQNY, in the context of a globular fold. Protein Sci. 2008;17:1617–1623. doi: 10.1110/ps.036368.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamasaki M, Li W, Johnson DJ, Huntington JA. Crystal structure of a stable dimer reveals the molecular basis of serpin polymerization. Nature. 2008;455:1255–1258. doi: 10.1038/nature07394. [DOI] [PubMed] [Google Scholar]
- 26.Otzen DE, Kristensen O, Oliveberg M. Designed protein tetramer zipped together with a hydrophobic Alzheimer homology: A structural clue to amyloid assembly. Proc Natl Acad Sci U S A. 2000;97:9907–9912. doi: 10.1073/pnas.160086297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wiltzius JJ, et al. Atomic structure of the cross-β spine of Islet Amyloid Polypeptide (Amylin) Protein Sci. 2008;17:1467–1474. doi: 10.1110/ps.036509.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee SW, et al. Steric zipper of the amyloid fibrils formed by residues 109–122 of the Syrian hamster prion protein. J Mol Biol. 2008;378:1142–1154. doi: 10.1016/j.jmb.2008.03.035. [DOI] [PubMed] [Google Scholar]
- 29.Makabe K, et al. Atomic structures of peptide self-assembly mimics. Proc Natl Acad Sci U S A. 2006;103:17753–17758. doi: 10.1073/pnas.0606690103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Biancalana M, Makabe K, Koide A, Koide S. Aromatic cross-strand ladders control the structure and stability of beta-rich peptide self-assembly mimics. J Mol Biol. 2008;383:205–213. doi: 10.1016/j.jmb.2008.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petkova AT, et al. Solid state NMR reveals a pH-dependent antiparallel beta-sheet registry in fibrils formed by a beta-amyloid peptide. J Mol Biol. 2004;335:247–260. doi: 10.1016/j.jmb.2003.10.044. [DOI] [PubMed] [Google Scholar]
- 32.Madine J, et al. Structural insights into the polymorphism of amyloid-like fibrils formed by region 20–29 of amylin revealed by solid-state NMR and x-ray fiber diffraction. J Am Chem Soc. 2008;130:14990–15001. doi: 10.1021/ja802483d. [DOI] [PubMed] [Google Scholar]
- 33.Gordon DJ, Balbach JJ, Tycko R, Meredith SC. Increasing the amphiphilicity of an amyloidogenic peptide changes the beta-sheet structure in the fibrils from antiparallel to parallel. Biophys J. 2004;86:428–434. doi: 10.1016/S0006-3495(04)74119-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoyer W, Gronwall C, Jonsson A, Stahl S, Hard T. Stabilization of a beta-hairpin in monomeric Alzheimer’s amyloid-beta peptide inhibits amyloid formation. Proc Natl Acad Sci U S A. 2008;105:5099–5104. doi: 10.1073/pnas.0711731105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim W, Hecht MH. Generic hydrophobic residues are sufficient to promote aggregation of the Alzheimer’s Abeta42 peptide. Proc Natl Acad Sci U S A. 2006;103:15824–15829. doi: 10.1073/pnas.0605629103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Biancalana M, Makabe K, Koide A, Koide S. Molecular Mechanism of Thioflavin-T Binding to the Surface of beta-Rich Peptide Self-Assemblies. J Mol Biol. 2009;385:1052–1063. doi: 10.1016/j.jmb.2008.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu C, et al. The binding of thioflavin T and its neutral analog BTA-1 to protofibrils of the Alzheimer’s disease Abeta(16–22) peptide probed by molecular dynamics simulations. J Mol Biol. 2008;384:718–729. doi: 10.1016/j.jmb.2008.09.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Esposito L, Pedone C, Vitagliano L. Molecular dynamics analyses of cross-beta-spine steric zipper models: Beta-sheet twisting and aggregation. Proc Natl Acad Sci U S A. 2006;103:11533–11538. doi: 10.1073/pnas.0602345103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Esteras-Chopo A, Serrano L, Lopez De La Paz M. The amyloid stretch hypothesis: Recruiting proteins toward the dark side. Proc Natl Acad Sci U S A. 2005;102:16672–16677. doi: 10.1073/pnas.0505905102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Petkova AT, et al. Self-propagating, molecular-level polymorphism in Alzheimer’s beta-amyloid fibrils. Science. 2005;307:262–265. doi: 10.1126/science.1105850. [DOI] [PubMed] [Google Scholar]
- 41.Wiltzius JJ, et al. Molecular mechanisms for protein-encoded inheritance. Nat Struct Mol Biol. 2009;16:973–978. doi: 10.1038/nsmb.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paravastu AK, Leapman RD, Yau WM, Tycko R. Molecular structural basis for polymorphism in Alzheimer’s beta-amyloid fibrils. Proc Natl Acad Sci U S A. 2008;105:18349–18354. doi: 10.1073/pnas.0806270105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vendruscolo M, Dobson CM. Chemical biology: More charges against aggregation. Nature. 2007;449:555. doi: 10.1038/449555a. [DOI] [PubMed] [Google Scholar]
- 44.Grueninger D, et al. Designed protein-protein association. Science. 2008;319:206–209. doi: 10.1126/science.1150421. [DOI] [PubMed] [Google Scholar]
- 45.Chothia C. Conformation of twisted beta-pleated sheets in proteins. J Mol Biol. 1973;75:295–302. doi: 10.1016/0022-2836(73)90022-3. [DOI] [PubMed] [Google Scholar]
- 46.Richardson JS, Richardson DC. Natural beta-sheet proteins use negative design to avoid edge-to-edge aggregation. Proc Natl Acad Sci U S A. 2002;99:2754–2759. doi: 10.1073/pnas.052706099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Steinbacher S, et al. Crystal structure of phage P22 tailspike protein complexed with Salmonella sp. O-antigen receptors. Proc Natl Acad Sci U S A. 1996;93:10584–10588. doi: 10.1073/pnas.93.20.10584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Angkawidjaja C, et al. Crystal structure of a family I.3 lipase from Pseudomonas sp. MIS38 in a closed conformation. FEBS Lett. 2007;581:5060–5064. doi: 10.1016/j.febslet.2007.09.048. [DOI] [PubMed] [Google Scholar]
- 49.Yeo HJ, et al. Structural basis for host recognition by the Haemophilus influenzae Hia autotransporter. EMBO J. 2004;23:1245–1256. doi: 10.1038/sj.emboj.7600142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Harbury PB, Zhang T, Kim PS, Alber T. A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science. 1993;262:1401–1407. doi: 10.1126/science.8248779. [DOI] [PubMed] [Google Scholar]
- 51.Fowler DM, Koulov AV, Balch WE, Kelly JW. Functional amyloid—from bacteria to humans. Trends Biochem Sci. 2007;32:217–224. doi: 10.1016/j.tibs.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 52.Fowler DM, et al. Functional amyloid formation within mammalian tissue. PLoS Biol. 2006;4:100–107. doi: 10.1371/journal.pbio.0040006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yan S, et al. Conformational heterogeneity of an equilibrium folding intermediate quantified and mapped by scanning mutagenesis. J Mol Biol. 2004;338:811–825. doi: 10.1016/j.jmb.2004.02.063. [DOI] [PubMed] [Google Scholar]
- 54.Makabe K, Tereshko V, Gawlak G, Yan S, Koide S. Atomic-resolution crystal structure of Borrelia burgdorferi outer surface protein A via surface engineering. Protein Sci. 2006;15:1907–1914. doi: 10.1110/ps.062246706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brunger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.