Abstract
A severe coagulopathy is a life-threatening complication of acute promyelocytic leukemia (APL) and is ascribable mainly to the excessive levels of tissue factor (TF) in APL cells regulated in response to the promyelocytic leukemia/retinoic acid receptor α (PML/RARα) fusion protein. The underlying molecular mechanisms for this regulation remain ill-defined. With U937-PR9 cell lines stably expressing luciferase reporter gene under the control of different mutants of the TF promoter, both luciferase and ChIP data allowed the localization of the PML/RARα-responsive sequence in a previously undefined region of the TF promoter at position −230 to −242 devoid of known mammalian transcription factor binding sites. Within this sequence a GAGC motif (−235 to −238) was shown to be crucial because deletion or mutation of these nucleotides impaired both PML/RARα interaction and promoter transactivation. However, EMSA results showed that PML/RARα did not bind to DNA probes encompassing the −230 to −242 sequences, precluding a direct DNA association. Mutational experiments further suggest that the activator protein 1 (AP-1) sites of the TF promoter are dispensable for PML/RARα regulation. This study shows that PML/RARα transactivates the TF promoter through an indirect interaction with an element composed of a GAGC motif and the flanking nucleotides, independent of AP-1 binding.
Keywords: transcription, regulation, GAGC motif, acute promyelocytic leukemia, coagulopathy
Thrombohemorrhagic complications are frequent in patients with hematological malignancies (1), and hemorrhage is a major cause of morbidity and mortality in patients with acute leukemia (2, 3). In particular in acute promyelocytic leukemia (APL), up to 90% of patients present with hemorrhagic complications at diagnosis, and fatal hemorrhage develops in ≈10–20% of these patients (4, 5). The bleeding diathesis is thought to result from disseminated intravascular coagulation triggered by the procoagulant activities such as tissue factor (TF) produced by APL leukemic cells (6). Treatment of APL patients with all-trans retinoic acid (ATRA) or arsenic trioxide produces a high rate of complete remission, together with a down-regulation of TF expression which usually precedes a rapid resolution of the coagulopathy (7 –9).
TF, a 47-kDa membrane-bound glycoprotein that initiates the blood coagulation cascade (10), is constitutively expressed on the surface of most nonvascular cells. Inducible expression of TF triggers intravascular clotting activity associated with various diseases such as sepsis, atherosclerosis, and cancer (3). Constitutive expression of the TF gene is maintained mainly by transcription factor Sp1, whereas the inducible TF expression appears to be regulated via transcription factors activator protein 1 (AP-1), NF-κB, and early growth response factor 1 (11 –13). Nevertheless, knowledge regarding the mode and the mechanisms for the regulation of TF gene in response to promyelocytic leukemia/retinoic acid receptor α (PML/RARα) remains incomplete.
APL is a unique subtype of acute myeloid leukemia characterized by the generation of the PML/RARα fusion gene as a result of a reciprocal chromosome translocation with breakpoints within the RARα gene and the PML gene. In hematopoietic precursor cells, PML/RARα disrupts RARα function in a dominant negative manner by recruiting the nuclear corepressor-histone deacetylase (HDAC) complex with a higher affinity and thus induces differentiation blockade of hematopoietic precursors at the promyelocytic stage (14). The PML moiety of the fusion protein also is implicated in transcription regulation by interacting with important molecules such as retinoblastoma protein through blocking its interaction with HDAC (15) or Daxx regulated by sumoylation at K160 site (16). However, the regulatory mechanisms for PML/RARα in TF expression in APL cells remain largely unknown.
Previous work (17) has established that the TF promoter is constitutively active in the promyelocytic NB4 cells and undergoes a down-regulation with ATRA treatment dependent on its proximal −383 to +121 bp. Thus it is possible that PML/RARα might regulate the expression of TF through an interaction with the TF promoter in a region proximal to −383. However, it is still unclear whether PML/RARα can bind to TF promoter and which nucleotides of the promoter are involved in PML/RARα–TF promoter interaction. More detailed analysis of the precise molecular mechanisms for the regulation of TF expression by PML/RARα will facilitate the efforts to access the molecular etiology of the coagulopathy in APL and to improve understanding of the PML/RARα action.
ATRA can reduce the level of PML/RARα in NB4 cells, but whether it interferes directly with the expression of TF without the involvement of PML/RARα is yet unknown. The monoblastic cell line U937 constitutively expresses TF, and this expression is markedly increased when the cells are transfected with PML/RARα fusion gene (17), suggesting that the fusion protein is involved in the regulation of excessive TF expression in U937 cells. U937-PR9 is a subclone of U937 cells stably transfected with the PML/RARα cDNA under the control of a zinc-inducible promoter and thus expresses PML/RARα only in the presence of zinc (18). Application of this cell model allows investigation of the bona fide roles of PML/RARα in regulation of TF expression by switching the PML/RARα gene transcription on and off.
In this study, we examined whether the expression level of PML/RARα per se regulates the TF promoter in U937-PR9 cells and defined the molecular interaction that is involved in this process. In addition, the use of the recombinant mutation strategies in luciferase reporter and ChIP assays allowed us to map the nucleotide sequences in the TF promoter critical for mediating PML/RARα interaction upon transactivation of the promoter.
Results
Interaction of PML/RARα with the TF Promoter During Up-Regulation of TF Expression.
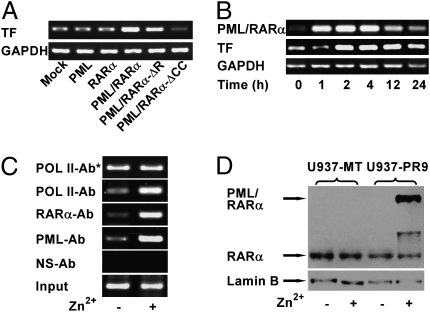
Equilibrated by pRL-SV40 activity, transfection of wild-type PML/RARα in U937 cells enhanced the TF gene transcription in contrast to a significant inhibition caused by the PML/RARαΔCC mutant. Interestingly, PML/RARαΔR still was able to elicit a substantial increase of TF gene transcription (Fig. 1A). Fig. 1B shows a sustained increase of TF gene transcription in U937-PR9 cells in response to zinc induction. ChIP experiments revealed that the genomic DNA fragments containing the TF promoter precipitated by both anti-PML and anti-RARα antibodies were strikingly enhanced in the presence of ZnSO4 (Fig. 1C), clearly demonstrating that the zinc-inducible PML/RARα, as evidenced by Fig. 1D, is able to interact with the TF promoter. In luciferase experiments, the U937-PR9 cells bearing the full-length TF promoter (−2174) showed increased luminescence in response to zinc induction (Fig. 2A) along with the PML/RARα expression (Fig. 2B). These data indicate that PML/RARα transactivates the TF promoter in U937-PR9 cells.
Fig. 1.
Stimulatory effect of PML/RARα on the transcription of the TF gene in U937 cells through an interaction with the TF promoter. (A) pSG5 eukaryocytic expression vectors encoding wild-type PML, RARα, and PML/RARα or its mutant forms, PML/RARαΔR and PML/RARαΔCC, were transfected into U937 cells. Total RNAs were isolated from the transfectants, TF transcription was determined by RT-PCR, and the samples were normalized based on GAPDH expression. (B) Total RNAs were isolated from U937-PR9 cells treated with 100 μM of ZnSO4. The transcription of the TF and genes was examined by RT-PCR which was normalized by GAPDH expression. (C) Sonicated chromatin fragments of U937-PR9 cells were immunoprecipitated with antibodies specific for RARα (RARα-Ab), PML (PML-Ab), RNA polymerase II (POL II-Ab and POL II-Ab*), or a nonspecific antibody (NS-Ab). PCR was carried out with primers corresponding to the genomic TF promoter sequences except for the POL II-Ab–precipitated sample (POL II-Ab*), in which primers corresponding to GAPDH were applied. (D) Nuclear proteins extracted from U937-PR9 or U937-MT cells induced with or without ZnSO4 were electrophoresed with 8% polyacrylamide gels and blotted with a polyclonal anti-RARα antibody. Lamin B served as a control.
Fig. 2.
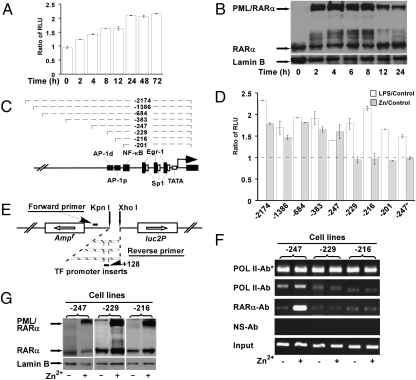
Critical role of the sequences between −229 and −247 of the TF promoter in its transactivation by PML/RARα in U937-PR9 cells. (A) U937-PR9 cells stably expressing the luciferase gene under the control of full-length TF promoter (−2174) were incubated with ZnSO4 for the indicated period. Luciferase data are expressed as the mean and SD of triplicate tests. (B) Nuclear proteins of the cells were analyzed by Western blotting with an anti-RARα antibody. (C) Schematic presentation of the TF promoter shows the putative transcription factor binding sites and the different truncation mutations used in this study. Numbering is indicated with respect to the transcriptional start site (bent arrow). (D) Luciferase analysis (mean and SD of triplicate tests) for the truncated TF promoters was performed; −247* represents U937-MT cells expressing TF promoter truncated at −247. (E) Designing strategies for the luciferase vectors and the exogenous TF promoter-specific primers are shown schematically. (F) ChIP assays were performed as described in the legend of Fig. 1. (G) Protein expression of PML/RARα and RARα in stable sublines (−247, −229, and −216) was examined by Western blotting.
Localization of the PML/RARα-Responsive Sequences Within the TF Promoter.
U937-PR9 cells stably expressing luciferase reporter gene under the control of 5′ flanking regions of TF promoter were tested (Fig. 2C). In the presence of ZnSO4 luciferase activities were increased in U937-PR9 cells bearing the TF promoters truncated at or more distal to the −247 position, whereas the truncations at −229, −216, and −201 resulted in a failure of response to PML/RARα (Fig. 2D), suggesting that the sequence −247 cagctccgcgctcggtgg−230 in the TF promoter serves as the responsive region during PML/RARα-induced activation. In contrast, luciferase expression controlled by the −247-truncated TF promoter in U937-MT cells remained unaltered (Fig. 2D; −247*), indicating that ZnSO4 alone may not transactivate the TF promoter.
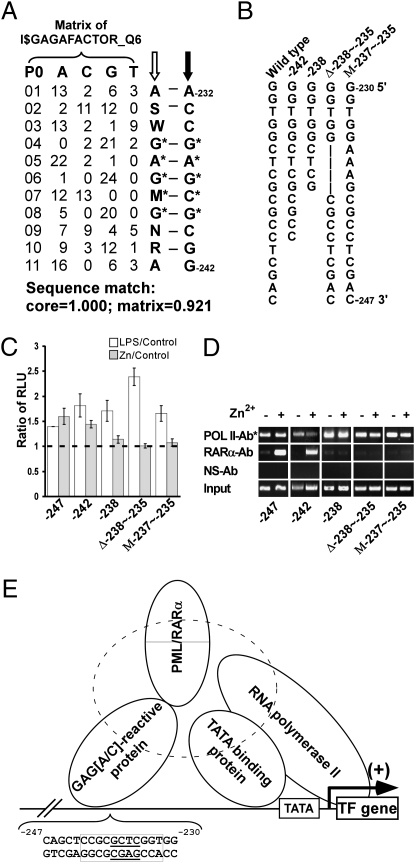
With a modified ChIP protocol (Fig. 2E) that allows exclusive amplification of the plasmid-delivered TF promoters, an unequivocal enrichment of the TF promoter sequence was achieved in anti-RARα antibody–precipitated materials from cells expressing truncated promoter at −247 treated with ZnSO4 but not in those from −229 and −216 mutants (Fig. 2F). These ChIP data are consistent with the luciferase activities in these promoter mutants. The expression of PML/RARα in these cells was monitored in parallel experiments (Fig. 2G). Alignment data (Fig. 3A) further showed that the sequence −247cagctccgcgctcggtgg−230 contains a motif (−242 to −232) highly homologous to the Drosophila GAGA motif with a GCTC core from −238 to −235. In an attempt to identify the critical nucleotides within this region, several mutations, including truncations at −242 or −238 and deletion or substitution mutations, were introduced into the −247 luciferase reporter vector (Fig. 3B). After stable transfection, luciferase experiments showed that the −242-truncated TF promoter was still transactivated in response to PML/RARα, but the −238 truncation was not (Fig. 3C). Furthermore, deletion of GCTC (GAGC, complementary strain) sequence (Δ−238∼−235) or replacement of the nucleotides CTC with AAA (M−237∼−235) shifted the promoter from active to inactive status (Fig. 3C). In line with the promoter activity, ChIP results (Fig. 3D) demonstrated a failure of the mutant TF promoters with truncation at −238 and deletion or substitution mutations of the GAGC sequence in interacting with PML/RARα. In Western blot, all cell lines exhibited a comparable level of PML/RARα expression in response to zinc induction (Fig. S1).
Fig. 3.
Identification of the region within TF promoter pivotal for its transactivation regulated by the interaction with PML/RARα. (A) The sequence homology between the nucleotides −232 to −242 of the TF promoter (noncoding strand) and the consensus sequence of the matrix I$GAGAFACTOR_Q6 corresponding to the putative Drosophila melanogaster GAGA factor reactive element. The matrix I$GAGAFACTOR_Q6 was originated from TRANSFAC MATRIX TABLE (Release 9.2). The P0 column is the position index of the matrix. The A, C, G, and T columns indicate the occurrence of nucleotide at the corresponding positions. The consensus sequence of I$GAGAFACTOR_Q6 using IUPAC alphabets (S = C/G; W = A/T; M = A/C; N = A/C/G/T; R = A/G) is indicated by a hollow arrow, and the noncoding strand of the TF promoter is indicated by a solid arrow. The asterisked nucleotides constitute the core of the consensus sequence. (B) Schematic presentation of the TF promoter shows the mutations in TF promoter used in this study. U937-PR9 cells expressing the TF promoter containing truncations at −242 or at −238, a deletion from −235 to −238 (Δ−238∼−235) or a mutation from −235 to −237 (M−237∼−235) were established. Luciferase (C) and ChIP (D) data of these mutants are shown. (E) A schematic diagram that delineates a proposed model for the interaction of PML/RARα with the TF promoter. PML/RARα forms a complex through the coiled-coil domain of the PML moiety with the intermediary/partner protein(s) which interact with the−232∼−242 nucleotides (boxed) of the TF promoter with a crucial GAGC core (underlined). This interaction may influence the chromatin remodeling that alters the RNA polymerase II-related transcription initiation complex and thus the transactivation of the promoter.
Interaction of PML/RARα with the Regulatory Region of TF Promoter in an Indirect Manner.
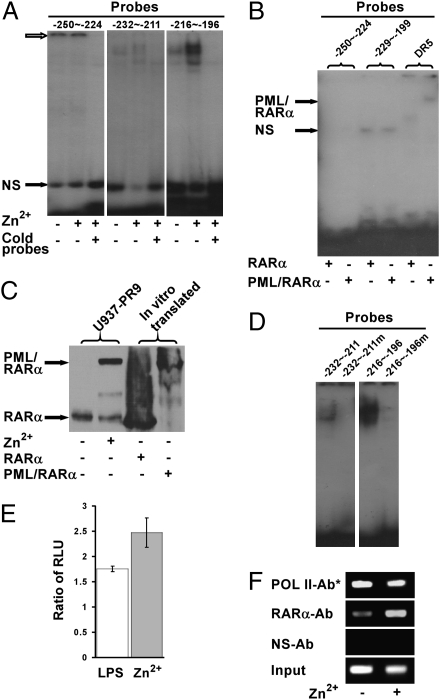
Given the evidence showing that PML/RARα interacts with the TF promoter via a region proximal to the −247 position, we chose to explore further the molecular basis for this interaction. In EMSA, nuclear extracts of the U937-PR9 cells with or without zinc induction showed no band shift with the −250∼−224 probe, but the substantial signals in the sampling wells suggested the formation of large complexes unable to enter the gels (Fig. 4A Left). On the other hand, the −232∼−211 and −216∼−196 probes yielded bands with U937-PR9 nuclear extracts (Fig. 4A Middle and Right). “Super shift” with antibodies specific for c-Jun and JunD (Fig. S2) confirmed the specificity of AP-1-proximal (AP-1p) for the shift band.
Fig. 4.
Effect of the mutated AP-1 sites on the regulation of the TF promoter by PML/RARα. EMSA analysis for the interaction of PML/RARα with the TF promoter was performed using radiolabeled DNA probes spanning from position −250 to −196 of the TF promoter. (A) Nuclear extracts from U937-PR9 cells were incubated with −250∼−224, −232∼−211, and −216∼−196 probes. The upper (hollow) arrow indicates the position of sample wells. (B) The in vitro translated PML/RARα or RARα proteins were incubated with the −250∼−224 and −229∼−199 probes. The DR5 probe containing a putative RARE was used as a positive control; NS indicates nonspecific bands. (C) Nuclear extracts from U937-PR9 cells or in vitro translated PML/RARα or RARα proteins were electrophoresed and blotted with an anti-RARα antibody. (D) EMSA was performed with probes containing wild-type or mutated AP-1d (−232∼−211 or −232∼−211m) and AP-1p (−216∼−196 or −216∼−196m) sites. (E) U937-PR9 cells stably expressing luciferase gene under the control of TF promoter with simultaneously mutated AP-1d and AP-1p sites were assessed for luciferase activity. (F) ChIP assays were carried out with the same cells per the foregoing procedures.
Regulatory factors are known to interact with promoters either directly by binding to DNA or indirectly by forming complexes with partner molecule(s), and ChIP assays reveal the interaction of proteins with promoters regardless which mode is involved. To clarify whether PML/RARα can bind directly to the regulatory regions of TF promoter, in vitro translated PML/RARα and RARα proteins were employed in EMSA. As depicted in Fig. 4B, the −250∼−224 probe containing the PML/RARα regulatory motif did not bind either PML/RARα or wild-type RARα, nor did the −229∼−199 probe encompassing both AP-1 distal (AP-1d) and AP-1p sites, indicating that neither PML/RARα nor RARα directly binds to the TF promoter. In contrast to the −250∼−224 and −229∼−199 probes, the DR5 oligonucleotide containing a canonical retinoic acid response element (RARE) did bind PML/RARα. The expression of PML/RARα or wild-type RARα protein in the EMSA samples was monitored by Western blot (Fig. 4C). The EMSA results invoke a model whereby PML/RARα interacts with the TF promoter in an indirect manner.
Dispensability of the AP-1 Sites for PML/RARα-Induced Transactivation of TF Promoter in U937-PR9 Cells.
To test whether the PML/RARα-mediated regulation of TF expression relies on the AP-1 sites, mutations were introduced into the −232∼−211 and −216∼−196 probes to remove their ability to bind AP-1 proteins. Indeed, both probes failed to bind the proteins, in contrast to the wild-type probes that did produce shift bands (Fig. 4D). The same AP-1p and AP-1d mutations also were introduced simultaneously into the −247 luciferase reporter vector (M-AP-1dp) and transfected into the U937-PR9 cells. Fig. 4E shows that the TF promoter with mutated AP-1 sites still could induce an enhancement of luminescence of sufficient magnitude to account for an unimpaired potential of the promoter. In ChIP analysis, as expected, a significant enrichment was achieved with anti-RARα antibody that parallels the luciferase results (Fig. 4F). Data indicate that mutation of the AP-1 sites did not impair PML/RARα-mediated TF promoter transactivation and further suggest that the AP-1 sites should be dispensable for this regulation.
Discussion
The present work provides insights into the molecular basis for the regulation of the TF expression by PML/RARα. First, PML/RARα up-regulates TF expression through interactions with the TF promoter, most likely using the protein–protein interaction domain of the PML moiety. Second, the −230∼−242 sequence of the TF promoter, or more accurately the −235∼−238 sequence and the flanks, are critical for the interaction with PML/RARα and the promoter transactivation. Third, PML/RARα does not bind directly to the TF promoter but most likely forms a complex with it through intermediary/partner molecules. Last, TF promoter with mutated AP-1 sites remains fully active in response to PML/RARα in either the promoter transactivation or in the fusion protein interaction. It is likely, therefore, that the unique regulatory pathway elucidated in this study may contribute to the specific regulation of the aberrant TF expression and its pathological consequences in APL.
Even based on the less stringent criteria (19), the human TF promoter contains no potential PML/RARα binding site, providing an additional argument that PML/RARα interacts with the TF promoter through an indirect mechanism. This perception is reinforced by the observations that the PML/RARαΔR does not reduce the TF gene transcription significantly (Fig. 1A) and that the oligonucleotide probes encompassing the PML/RARα-interacting sequences do not recognize PML/RARα (Fig. 4 A and B). The ΔCC version of PML/RARα was much less efficient in inducing transactivation (Fig. 1A), suggesting that the protein–protein interaction through PML, possibly including the PML/RARα homodimerization, is critical for PML/RARα in regulating the transcription. These observations establish the importance of the indirect mode by which PML/RARα induces the transcription of target genes via protein–protein interaction with intermediary protein(s) but not direct DNA binding. Our data are consistent with a recently published study that described a gain-of-function for PML/RARα in transactivating the inhibitor of differentiation-1 and -2 promoters following ATRA treatment through an indirect interaction mediated by the Sp1 transcription factor (20), despite the agonist-independent feature of the PML/RARα-induced TF promoter transactivation. Métivier and coworkers (21) proposed a mode for the transcription of an estrogen receptor-α target gene in which as many as 46 transcription factors associate with the promoter to assemble a transcriptionally productive complex. In our study the ChIP assays confirmed the interaction of PML/RARα with the TF promoter (Figs. 1C, 2 F, 3D, and 4F), whereas the EMSA results ruled out the possibility of direct binding and suggested the formation of large complexes (Fig. 4A Left). Thus, present data point to a mechanism in which PML/RARα transactivates the TF promoter by forming complexes mediated by partner proteins binding to the promoter.
In luciferase assays PML/RARα induced a moderate but reproducible TF promoter transactivation in U937-PR9 cells indicating that, in line with the previous report (22), the extent of transactivation does not necessarily reflect the functional importance of the regulation. Moreover, the magnitude of the promoter transactivation in this work corroborates the results in a published study, in which U937-PR9 cells stably transfected with native promoter constructs that usually lead to moderate levels of transactivation (23) also were used (17). The conventional ChIP protocols cannot localize the target sequences precisely and are not applicable to the recombinant mutational analysis. In stably transfected cells the exogenous constructs are integrated into the genome that can be immunoprecipitated in ChIP (24). In our experiments the chromosomal integration of the TF promoter reporter constructs into the host-cell genome resulted in different activity states because of the different interactions between the promoters and PML/RARα. Thus, ChIP with PML/RARα for the exogenous TF promoters (Fig. 2E) in these cells revealed the PML/RARα interaction with the promoters paralleling the reporter activity.
w?>Previous evidence suggested that liganded RARα is able to repress AP-1–mediated transcription (25). In contrast, PML/RARα stimulates AP-1–dependent transcription in the presence of ligand, and, notably, unliganded PML/RARα inhibits AP-1–dependent transcription (26). The present study shows that the TF promoter was transactivated by PML/RARα without the induction of ligand. These data led us to hypothesize that there is a signaling pathway other than the AP-1–dependent pathway and prompted us to examine the role of the AP-1 sites in PML/RARα-regulated TF promoter transactivation. As expected, mutated AP-1 sites resulted in a loss of AP-1 protein binding (Fig. 4D). Interestingly, these mutations did not render the TF promoter incapable of responding to PML/RARα either in transactivation or in molecular interaction in ChIP (Fig. 4 E and F). Our results thus preclude a scenario in which the AP-1 sites play an indispensable role in PML/RARα-regulated TF expression and define a regulatory pathway with which PML/RARα per se transactivates the TF promoter.
The nucleotides −230 to −242 of the TF prompter contain no putative mammalian transcription factor binding site. However, computational analysis of this sequence shows a high homology to the putative Drosophila melanogaster GAGA motif (Fig. 3A). In Drosophila the GAGA-binding factor (GAF) is required for the expression of a wide range of genes through activating the RNA polymerase II transcription (27). The very limited information concerning the GAGA element in mammals indicates that the GAGA motif also might be involved in gene transcription regulation. Mutation of the GAGA sites on the rat CD44 promoter resulted in a decrease of the promoter transactivation in COS-7 cells (28). Introduction of the anti-Drosophila GAF antibody into the mouse embryos caused a suppression of the transcriptional activation of the hsp70 promoter (29), indicating that the murine GAF might be structurally related to the Drosophila factors. The present study also suggests that the motif with a GAGA(C) core in human TF promoter is essential to PML/RARα-mediated transactivation (Fig. 3 C and D) and provides a plausible explanation for the roles of the hypothetical mammalian GAF. Our proposed model, illustrated in Fig. 3E, underscores the complexity of the mechanism of action of PML/RARα in TF up-regulation and highlights the need for further identification of the PML/RARα binding partners to understand fully the molecular basis for PML/RARα-regulated TF expression.
Materials and Methods
Cell Lines.
The U937-PR9 cell line expressing PML/RARα under the control of the zinc-inducible MT promoter and the mock-transfected cell line U937-MT were kindly provided by P. G. Pelicci (Perugia University, Perugia, Italy).
Plasmids.
The plasmids pSG5-PML, pSG5-RARα, and pSG5-PML/RARα have been described previously (30). The PML/RARα mutants encoding PML/RARαΔR (with a deleted RARα DNA binding domain) or PML/RARαΔCC (lacking the coiled-coil protein-protein interaction domain of PML) were created by using the SauI and BstE2 sites of RARα and the BssHII sites of PML, respectively. For construction of the luciferase reporter vectors, a series of the sequences with different 5′ flanking regions of the TF promoter were PCR-amplified from human genomic DNA with primers as indicated (Table S1) and inserted into the multicloning site of the pGL4.15 (Luc2P/Hygro) plasmid (Promega) (Fig. 2E). Plasmids M-AP-1dp in which both AP-1d and AP-1p sites were mutated (Δ−238∼−235, M−237∼−235) were made by site-directed mutagenesis (Stratagene) based on the plasmid pGL4-247 (Fig. 3B). The correctness of all of the mutant plasmids was confirmed by DNA sequencing.
Cell Transfection.
U937 cells were electroporated with the plasmid constructs cotransfected with the pRL-SV40 vector (Promega) to calibrate the transfection efficiency. To establish the stable cell lines, transfected U937-PR9 cells were selected with hygromycin B (Calbiochem). Cells were subcloned and selected by increased luciferase activity in response to lipopolysaccharide (LPS) (Sigma) (17). In this study, data reported for each stable cell line represent at least three clones.
RT-PCR.
The cDNAs were synthesized from total RNA extracted from the cells. PCR was performed with primers for PML/RARα, TF, and GAPDH (Table S2).
Luciferase Assays.
Cells (2 × 105) treated with LPS or ZnSO4 for 8 h were lysed with passive lysis buffer (Promega), and untreated cells were used as control. Firefly luciferase activity was measured, and all experiments were performed in triplicate on three independent occasions.
Nuclear Extracts and in Vitro Protein Translation.
Nuclear extracts were prepared as described previously (31). PML/RARα and RARα proteins were synthesized in vitro with pSG5-RARα and pSG5-PML/RARα as DNA templates according to the manufacturer’s protocol (Promega).
Western Blot Analysis.
PML/RARα, RARα, and lamin B were detected with antibodies against RARα (Santa Cruz) and lamin B (Santa Cruz).
EMSA.
The oligonucleotide probes spanning the TF promoter regulatory regions (−250 to −196) and a control DR5-type RARE probe (31) were synthesized (Table S3). EMSA was performed as described in manufacturer’s protocol (Amersham). Probes end-labeled with [γ-32P] dATP (Amersham) were incubated with 10 μg nuclear extracts or 5 μg protein produced by in vitro translation. The protein–oligonucleotide complex was separated on 4.8% nondenaturing polyacrylamide gels.
ChIP.
ChIP was performed using the ChIP-IT kit (Active Motif) according to manufacturer’s specifications. Cells were crosslinked with 1% formaldehyde and lysed by SDS lysing buffer. Cross-linked chromatin was sonicated to fragments with an average size of 400–600 bp and immunoprecipitated with antibodies against RARα or PML (Santa Cruz) and preimmune serum (Active Motif). The PCR primers included forward primer 5′-cgtctctcaaggataagtaa-3′, based on the sequence of pGL4.15 vector, and reverse primer 5′-−169cctcccggtaggaaact−185-3′, compatible with TF promoter sequences (Fig. 2E).
Supplementary Material
Acknowledgments
The authors thank Dr. Zhenyi Wang for thoughtful advice and helpful suggestions, Dr. Hongli Wang for helpful discussion, Dr. Baiwei Gu and Dr. Ruihua Zhao for their efforts in preliminary studies, and Yuanjing Lu, Tao Zhen, Zheng Ruan, Dr. Xiaoyu Su, and Dr. Ping Wang for technical assistance. This work was supported by Grants 30871107 and 30710103905 from the National Natural Science Foundation of China, 2006AA02A245 and 2009CB825607 from the Ministry of Science and Technology of China, and 09410706800 from the Shanghai Municipal Commission for Science and Technology. It also was supported in part by Grants 863, 2006AA 02A405 from the National High Tech Program for Biotechnology, 973, 2010CB529200 from the Chinese National Key Basic Research Project, Y0201 from the Key Discipline program of Shanghai Municipal Education Commission, 30821063 from the Innovation Group of the National Natural Science Foundation of China, and 06DZ2202, 07DZ05908, and 08DZ2200100 from the Shanghai Municipal Commission for Science and Technology. N.K. was supported in part by a grant from the Fondation BNP-Paribas in Shanghai, China.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0915006107/DCSupplemental.
References
- 1.Chong BH, Lee SH. Management of thromboembolism in hematologic malignancies. Semin Thromb Hemost. 2007;33:435–448. doi: 10.1055/s-2007-976179. [DOI] [PubMed] [Google Scholar]
- 2.Rickles FR, et al. Bleeding and thrombosis in acute leukemia: What does the future of therapy look like? Thromb Res. 2007;120(Suppl 2):S99–S106. doi: 10.1016/S0049-3848(07)70137-8. [DOI] [PubMed] [Google Scholar]
- 3.Tallman MS, Kwaan HC. Intravascular clotting activation and bleeding in patients with hematologic malignancies. Rev Clin Exp Hematol. 2004;8:E1. [PubMed] [Google Scholar]
- 4.Kwaan HC, Wang J, Boggio LN. Abnormalities in hemostasis in acute promyelocytic leukemia. Hematol Oncol. 2002;20:33–41. doi: 10.1002/hon.687. [DOI] [PubMed] [Google Scholar]
- 5.Zhou GB, Zhang J, Wang ZY, Chen SJ, Chen Z. Treatment of acute promyelocytic leukaemia with all-trans retinoic acid and arsenic trioxide: A paradigm of synergistic molecular targeting therapy. Philos Trans R Soc Lond B Biol Sci. 2007;362:959–971. doi: 10.1098/rstb.2007.2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arbuthnot C, Wilde JT. Haemostatic problems in acute promyelocytic leukaemia. Blood Rev. 2006;20:289–297. doi: 10.1016/j.blre.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 7.Barbui T, Finazzi G, Falanga A. The impact of all-trans-retinoic acid on the coagulopathy of acute promyelocytic leukemia. Blood. 1998;91:3093–3102. [PubMed] [Google Scholar]
- 8.Zhao W, et al. Effects of all-trans-retinoic acid and arsenic trioxide on the hemostatic disturbance associated with acute promyelocytic leukemia. Thromb Res. 2001;102:197–204. doi: 10.1016/s0049-3848(01)00233-x. [DOI] [PubMed] [Google Scholar]
- 9.Zhu J, et al. Tissue factors on acute promyelocytic leukemia and endothelial cells are differently regulated by retinoic acid, arsenic trioxide and chemotherapeutic agents. Leukemia. 1999;13:1062–1070. doi: 10.1038/sj.leu.2401448. [DOI] [PubMed] [Google Scholar]
- 10.Mackman N. Role of tissue factor in hemostasis, thrombosis, and vascular development. Arterioscler Thromb Vasc Biol. 2004;24:1015–1022. doi: 10.1161/01.ATV.0000130465.23430.74. [DOI] [PubMed] [Google Scholar]
- 11.Guha M, et al. Lipopolysaccharide activation of the MEK-ERK1/2 pathway in human monocytic cells mediates tissue factor and tumor necrosis factor alpha expression by inducing Elk-1 phosphorylation and Egr-1 expression. Blood. 2001;98:1429–1439. doi: 10.1182/blood.v98.5.1429. [DOI] [PubMed] [Google Scholar]
- 12.Krikun G, et al. Regulation of tissue factor gene expression in human endometrium by transcription factors Sp1 and Sp3. Mol Endocrinol. 2000;14:393–400. doi: 10.1210/mend.14.3.0430. [DOI] [PubMed] [Google Scholar]
- 13.Mackman N. Regulation of the tissue factor gene. Thromb Haemost. 1997;78:747–754. [PubMed] [Google Scholar]
- 14.Grignani F, et al. Fusion proteins of the retinoic acid receptor-α recruit histone deacetylase in promyelocytic leukaemia. Nature. 1998;391:815–818. doi: 10.1038/35901. [DOI] [PubMed] [Google Scholar]
- 15.Khan MM, et al. PML-RARalpha alleviates the transcriptional repression mediated by tumor suppressor Rb. J Biol Chem. 2001;276:43491–43494. doi: 10.1074/jbc.C100532200. [DOI] [PubMed] [Google Scholar]
- 16.Zhu J, et al. A sumoylation site in PML/RARA is essential for leukemic transformation. Cancer Cell. 2005;7:143–153. doi: 10.1016/j.ccr.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 17.Tenno T, Oberg F, Mackman N, Nilsson K, Siegbahn A. PML/RARalpha plays a role for basal activity and retinoid-induced repression of the tissue factor promoter in acute promyelocytic leukemia cells. Thromb Haemost. 2003;90:930–939. doi: 10.1160/TH03-02-0087. [DOI] [PubMed] [Google Scholar]
- 18.Nervi C, et al. Caspases mediate retinoic acid-induced degradation of the acute promyelocytic leukemia PML/RARalpha fusion protein. Blood. 1998;92:2244–2251. [PubMed] [Google Scholar]
- 19.Kamashev D, Vitoux D, De Thé H. PML-RARA-RXR oligomers mediate retinoid and rexinoid/cAMP cross-talk in acute promyelocytic leukemia cell differentiation. J Exp Med. 2004;199:1163–1174. doi: 10.1084/jem.20032226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Wageningen S, et al. Gene transactivation without direct DNA binding defines a novel gain-of-function for PML-RARalpha. Blood. 2008;111:1634–1643. doi: 10.1182/blood-2007-04-081125. [DOI] [PubMed] [Google Scholar]
- 21.Métivier R, et al. Estrogen receptor-α directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell. 2003;115:751–763. doi: 10.1016/s0092-8674(03)00934-6. [DOI] [PubMed] [Google Scholar]
- 22.Nishizawa M, Kataoka K, Vogt PK. MafA has strong cell transforming ability but is a weak transactivator. Oncogene. 2003;22:7882–7890. doi: 10.1038/sj.onc.1206526. [DOI] [PubMed] [Google Scholar]
- 23.Tenen DG, Hromas R, Licht JD, Zhang DE. Transcription factors, normal myeloid development, and leukemia. Blood. 1997;90:489–519. [PubMed] [Google Scholar]
- 24.Hebbar PB, Archer TK. Altered histone H1 stoichiometry and an absence of nucleosome positioning on transfected DNA. J Biol Chem. 2008;283:4595–4601. doi: 10.1074/jbc.M709121200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nicholson RC, et al. Negative regulation of the rat stromelysin gene promoter by retinoic acid is mediated by an AP1 binding site. EMBO J. 1990;9:4443–4454. doi: 10.1002/j.1460-2075.1990.tb07895.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doucas V, Brockes JP, Yaniv M, de Thé H, Dejean A. The PML-retinoic acid receptor α translocation converts the receptor from an inhibitor to a retinoic acid-dependent activator of transcription factor AP-1. Proc Natl Acad Sci USA. 1993;90:9345–9349. doi: 10.1073/pnas.90.20.9345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Croston GE, Kerrigan LA, Lira LM, Marshak DR, Kadonaga JT. Sequence-specific antirepression of histone H1-mediated inhibition of basal RNA polymerase II transcription. Science. 1991;251:643–649. doi: 10.1126/science.1899487. [DOI] [PubMed] [Google Scholar]
- 28.Kim SW, et al. A novel mechanism of thyroid hormone-dependent negative regulation by thyroid hormone receptor, nuclear receptor corepressor (NCoR), and GAGA-binding factor on the rat cD44 promoter. J Biol Chem. 2005;280:14545–14555. doi: 10.1074/jbc.M411517200. [DOI] [PubMed] [Google Scholar]
- 29.Bevilacqua A, Fiorenza MT, Mangia F. A developmentally regulated GAGA box-binding factor and Sp1 are required for transcription of the hsp70.1 gene at the onset of mouse zygotic genome activation. Development. 2000;127:1541–1551. doi: 10.1242/dev.127.7.1541. [DOI] [PubMed] [Google Scholar]
- 30.Zhu Q, et al. Synergic effects of arsenic trioxide and cAMP during acute promyelocytic leukemia cell maturation subtends a novel signaling cross-talk. Blood. 2002;99:1014–1022. [PubMed] [Google Scholar]
- 31.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.