Abstract
Thioredoxins (Trxs) are small ubiquitous regulatory disulfide proteins. Plants have an unusually complex complement of Trxs composed of six well-defined types (Trxs f, m, x, y, h, and o) that reside in different cell compartments and function in an array of processes. The extraplastidic h type consists of multiple members that in general have resisted isolation of a specific phenotype. In analyzing mutant lines in Arabidopsis thaliana, we identified a phenotype of dwarf plants with short roots and small yellowish leaves for AtTrx h9 (henceforth, Trx h9), a member of the Arabidopsis Trx h family. Trx h9 was found to be associated with the plasma membrane and to move from cell to cell. Controls conducted in conjunction with the localization of Trx h9 uncovered another h-type Trx in mitochondria (Trx h2) and a Trx in plastids earlier described as a cytosolic form in tomato. Analysis of Trx h9 revealed a 17-amino acid N-terminal extension in which the second Gly (Gly2) and fourth cysteine (Cys4) were highly conserved. Mutagenesis experiments demonstrated that Gly2 was required for membrane binding, possibly via myristoylation. Both Gly2 and Cys4 were needed for movement, the latter seemingly for protein structure and palmitoylation. A three-dimensional model was consistent with these predictions as well as with earlier evidence showing that a poplar ortholog is reduced by a glutaredoxin rather than NADP-thioredoxin reductase. In demonstrating the membrane location and intercellular mobility of Trx h9, the present results extend the known boundaries of Trx and suggest a role in cell-to-cell communication.
Keywords: protein movement, redox regulation, signal transduction, thioredoxin h, myristoylation–palmitoylation
Current evidence indicates that redox state is one of the factors determining the fate and growth of cells during the development of multicellular organisms. The ubiquitous disulfide regulatory protein thioredoxin (Trx) appears to play a role in linking redox to this process (1, 2). Trxs belong to a complex family of regulatory proteins consisting of at least six distinct types in plants, f, m, x, y, h, and o (Fig. S1A). Trxs reside in different cell compartments and function in an array of processes. Whereas the role of chloroplast Trxs is relatively clear, our understanding of the function of the multiple members of the extraplastidic h type is limited. Moreover, mutant phenotypes of h-type Trxs are scant; a loss-of-function mutant has been described in only one case: Trx h5 was found to be required for response of Arabidopsis to fungal infection (3, 4).
In exploring the function of the h type of Trxs, we elected to focus on the member of this group whose ortholog, Trx h9, was recently found to be important in the germination of cereal seeds (5). As Arabidopsis appeared ideally suited for this purpose, we sought a phenotype for a mutant defective in this Trx. Our efforts have been successful: We now report the identification of a phenotype for a null, loss-of-function mutation in Trx h9. The trx h9 mutant, showed chlorotic leaves and impaired growth and development. In contrast to other Trxs tested as controls, Trx h9 was found to be associated with the plasma membrane and, surprisingly, was able to move from cell to cell. By constructing structural models, we found that Arabidopsis Trx h9 may be reduced with NADPH by glutaredoxin (Grx) and glutathione (GSH) rather than the usual NADP-thioredoxin reductase (NTR) enzyme as shown for its ortholog from poplar (6). Our findings suggest that Trx h9 plays a role in plant growth, possibly by allowing cells to relay redox information to one another.
Results and Discussion
Arabidopsis Trx h9.
Sequence analysis revealed that Trx h9 is evolutionarily conserved and shares high sequence identity with its orthologs in different species. For example, we found that 63% of the Trx h9 protein sequence is identical to that of its rice counterpart (Os01g0168200; GenBank accession no. NP_001042127), whereas only 46% is identical with Trx h1, the most closely related paralog in Arabidopsis. Based on an N-terminal extension characterized by a conserved Gly at position 2 (Gly2) and a cysteine at position 4 (Cys4), Trx h9 and its orthologs form a new branch of h-type Trxs clearly separate from other members of the group (Figs. S1 and S2A). These features suggest that this region may endow Trx h9 with unique functions and properties. The microarray data revealed that Trx h9 is expressed in a tissue-specific manner (Fig. S2B), although RT-PCR analyses showed that Trx h9 is expressed throughout the plant (Fig. S2C), consistent with results reported by Reichheld et al. (7). Expression was strongest in the epidermal cells in the elongation zone of the roots, stomata, stamen, and pollen (Fig. S2 B and C and Table S1).
Recessive Loss-of-Function Mutation in Trx h9 Impairs Growth and Development.
Genetic screening of T-DNA insertions and subsequent sequencing revealed that Salk line 086660 contained a T-DNA insertion in the second exon of the Trx h9 coding sequence between the 135th and 136th nucleotide (Fig. S2D). RT-PCR results further indicated that homozygous Salk_086660 plants were null loss-of-function mutants of Trx h9 (Fig. S2E). There was no detectable trx h9 RNA in plants from two individual T3 homozygous mutants of Salk_086660 (Salk_08660-3 and Salk_08660-9).
Phenotypic analyses revealed that loss of Trx h9 function seriously impaired growth and development. When cultured on Murashiga-Skoog (MS) medium without sucrose, mutant plants ceased growth after germination (Fig. S3A). Roots and leaves of the homozygous mutants were significantly shorter and smaller than wild-type counterparts grown on MS medium with 1% sucrose (Fig. 1 A and B). Root tips of 7-day-old mutant seedlings grown on MS medium with 1% sucrose had a shortened apical meristem and, cells were more compact, in keeping with noticeably shorter roots (Fig. 1B; Fig. S3B). When grown in soil, mutant plants were dwarf with small yellowish leaves (Fig. 1C; Fig. S3C). Mesophyll cells from mutant leaves were smaller and irregularly shaped, with fewer chloroplasts versus wild type (Fig. 1D). Consistent with lower chloroplast numbers, mature rosette leaves of mutant plants contained on a relative basis ca 50% of the chlorophyll of wild type based on HPLC analysis (1477 vs. 760.7 for Chl a and 804.9 vs. 406.4 for Chl b in wild type and ath9 mutant, respectively). Heterozygous and F1 plants from a backcross to Salk_086660 yielded a wild-type plant, suggesting that Salk_086660 contains a recessive Trx h9 mutation.
Fig. 1.
Phenotypic and complementation analysis of trx h9 mutation in Salk_08660 plants. Seven-day-old Arabidopsis seedlings (A) and root tips (B) grown on MS medium plus 1.0% sucrose (left to right: wild-type Arabidopsis Col-0 plant, homozygous trx h9 mutant, and 35S::Trx h9 in trx h9 background). Root tips in B were fixed [methanol:acetic acid (3:1) buffer at 4°C overnight], stained with DAPI for 20 min, and viewed immediately under a fluorescence microscope. (C) Thirty-five-day-old Arabidopsis grown in soil. (D) Leaves from 35-day-old Arabidopsis grown in soil and viewed under a fluorescence microscope. Order in all panels is the same as (A). [Scale bars, 1 cm (A and C); 50 μm (B and D).]
At this point, we deemed it desirable to characterize a novel Arabidopsis chloroplast Trx because the loss-of function Trx h9 seems to affect chloroplasts. For this purpose, we selected a previously unreported Arabidopsis Trx [called “putative thioredoxin” (GenBank accession no. NP_187329; TAIR: At3G06730) in the Arabidopsis Information Resource database] that was predicted to be in plastids based on the use of the ChloroP 1.1 Server (http://www.cbs.dtu.dk/services/ChloroP) and to be closely related to plastid Trx y (Fig. S1A). We provisionally designated this protein Trx p, due to its putative and plastidic nature. The Trx p ortholog in tomato, CITRX (Cf-9-interacting thioredoxin), was earlier shown to function in disease resistance (8). It was thus of interest to deermine loss-of-function phenotypes of Trx p by studying a T-DNA insertion line, Salk_028162, in which a T-DNA was inserted at a position 161 bp upstream of the first intron of the Trxp p gene (Fig. S1B). This insertion resulted in a null knockout in the homozygous mutant plants (trx p) (Fig. S1C) which appeared completely albino, ceased growth, and died shortly after germination (Fig. S1D). Plastids in trx p plants lacked visible internal membrane structures, including stromal and granal thylakoids as well as starch granules (Fig. S1F). There were, however, many densely stained globular structures, possibly plastoglobuli of degenerated thylakoid lipids (9) (Fig. S1G). Trx p thus may play a role in plastid development in addition to its role in plant disease (8).
35S::Trx h9 Construct Fully Rescued the Loss-of-Function Phenotypes of Trx h9 Mutant.
In further characterization of its function, the full-length Trx h9 gene driven by the 35S promoter was introduced into homozygous Salk_086660 mutant plants. Sixty individual transgenic 35S::Trx h9 lines in the trx h9 mutant background were analyzed in the T1 generation. Complete rescue of the mutant phenotypes was observed in 44 lines (77.33%), yielding plants indistinguishable from wild-type Arabidopsis (Col-0) (Fig. 1 A–D). These results confirmed that the mutant phenotypes were due to the loss-of-function mutation in Trx h9.
Trx h9 Is a Plasma Membrane Protein.
To obtain insight into its function, we determined the subcellular localization of Trx h9. We fused green fluorescence protein (GFP, a 27-kDa resident of the cytosol) in-frame to the full-length Trx h9 at the C terminus. Localization of the Trx h9-GFP fusion protein was assayed in both transient and stable transformation systems.
Before examining Trx h9, we tested GFP fusions with Arabidopsis proteins targeted to different organelles using the isolated onion epidermal cell layer for the transient expression assay. These studies revealed that free GFP (a cytosolic protein) was present throughout the cell (Fig. S4A). Moreover, when cells expressing free GFP were plasmolyzed, GFP remained in the cytosol (Fig. S4B), not in the Hechtian strands (see below) or cell wall, as revealed by DAPI staining (Fig. S4C). By contrast, when fused to a nuclear localization signal (NLS), GFP accumulated, as expected, in the nucleus (Fig. S4D). In short, GFP fused to control proteins was localized as predicted.
We also examined the location of Trx p that was predicted to be in plastids by the PSORT program (http://psort.ims.u-tokyo.ac.jp/form.html). Using transient and stable transformations with GFP fusions, we localized Arabidopsis Trx p in plastids (Fig. 2 D and E). In parallel transient expressions, the Trx p counterpart (GenBank accession no. BG299573) and Trx f (GenBank accession no. AK250725) from barley (Hordeum vulgare) were identically localized. This finding contrasts with the report that the Trx p ortholog from tomato, CITRX, is localized in the cytosol (8). Further work is needed to resolve this discrepancy.
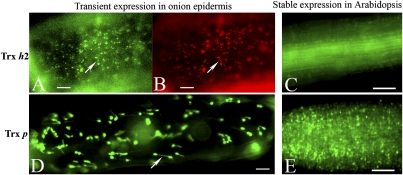
Fig. 2.
Subcellular localization of Trx h2 and Trx p in onion epidermal and transgenic Arabidopsis cells using GFP tagging. (A–C) Trx h2 (At5g39950); (D and E) Trx p (At3G06730). (A, B, and D) Transient expression in onion epidermal cells. (C and E) Stable expression in transgenic Arabidopsis root cells. (A, C, D, and E) Visualization of GFP. (B) MitoTracker orange. [Scale bars, 10 μm (A, B, and D); 50 μm (C and E).] In A and B, arrows point to the same mitochondrion, and in D to the long stromule.
We also examined the localization of Trx h2, which, as anticipated, was in the cytosol. However, unexpectedly, it was also observed in mitochondria in the onion transient expression system. This location was seen with both the onion transient expression assay, where GFP (Fig. 2A) was colocalized using MitoTrack orange, a specific dye for mitochondria (Invitrogen) (Fig. 2B), and in stably transformed Arabidopsis (Fig. 2C). In addition to providing evidence for a mitochondrial h-type Trx in Arabidopsis, these results complement earlier work with castor seed (10) and poplar (11).
Cellular localization of Trx h9 was examined against this background. When transiently expressed in onion cells, Trx h9 fused to GFP localized to the cell periphery—that is, the plasma membrane, cell wall, or both (Fig. 3A). To localize the protein more accurately, onion cells were plasmolyzed after being transformed with Trx h9-GFP. Under these conditions, protoplasts pull away from the cell wall, becoming spherical and leaving large numbers of thin plasma membrane bridges, known as Hechtian strands, firmly anchored to the cell wall (12). Trx h9-GFP displayed a pattern consistent with its location in the plasma membrane (Fig. 3B, Hechtian strands marked with arrow). Similar results were obtained with the barley Trx h-like protein (GenBank accession no. AAN63616), an ortholog that shares 65% sequence identity with Trx h9.
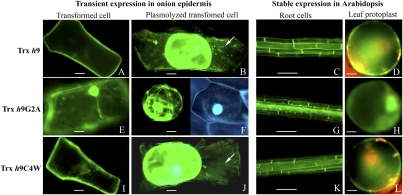
Fig. 3.
Subcellular localization of wild-type and mutated Trx h9 in onion epidermal and transgenic Arabidopsis cells using GFP tagging. (A–D) Trx h9. (E–H) Trx h9G2A. (I–L) Trx h9C4W. A and B, E and F, and I and J show transient expression of wild-type and mutated Trx h9-GFP in onion epidermal cells. C and D, G and H, and K and L show stable expression patterns of wild-type and mutated Trx h9-GFP in Arabidopsis. A, E, and I are unplasmolyzed transformed epidermal cells; B, F, and J are plasmolyzed, transformed epidermal cells. C, G, and K are stably transformed root cells; D, H, and L, are stably transformed mesophyll protoplasts. Image in F viewed for GFP (Left) and DAPI (Right). [Scale bars, 50 μm (C, G, and K); 10 μm (remainder).] In B and J, arrows point to Hechtian strands.
Results obtained with Trx h9-GFP stably expressed in Arabidopsis were in agreement with the transient expression assays. GFP from Trx h9-GFP fusion proteins in transgenic Arabidopsis seedlings was observed in the plasma membrane and/or cell wall of root cells (Fig. 3C). Analyses of protoplasts from plants stably expressing Trx h9-GFP revealed that fluorescence remained in the plasma membrane of spherical protoplasts after cell walls were removed (Fig. 3D). Further, western blots using a GFP antibody demonstrated that the Trx h9-GFP fusion protein was prevalent in the insoluble, not the soluble, fraction extracted from transgenic plants stably expressing the protein (Fig. S5). This pattern contrasted with that of Trx h2, which localized to both the cytosol and mitochondria (Fig. 2 A–C). Overall, these results suggest that Trx h9 is a plasma membrane protein.
Trx h9 May Be Myristoylated and Palmitoylated.
Although there are exceptions for mitochondria and endoplasmic reticulum, plant h-type Trxs are generally considered to be soluble, cytosolic proteins due to the absence of protein-sorting signals and transmembrane domains (10, 13, 14). However, other factors are known to affect membrane-binding properties of proteins through either co- or posttranslational addition of a variety of lipids, such as myristyl (C14), farnesyl (C15), palmityl (C16), and geranylgeranyl (C20) groups. Covalent linkage of these moieties to a protein can affect its membrane-binding properties (15). Trx h9 possesses a conserved N-terminal Gly (Gly2) and Cys (Cys4) suggestive of lipid modification. N-myristoylation refers to the cotranslational and irreversible addition of myristate at the N-terminal Gly of a protein via amide linkage (16, 17). Palmitoylation, on the other hand, represents a reversible posttranslational attachment of a palmityl group to specific Cys residues through thioester linkage (15, 18). Proteins with Cys residues adjacent to or near an N-myristoylated Gly residue, like Trx h9, may be sequentially palmitoylated (17).
By using a specific N-terminal prediction program, Myristoylator (http://ca.expasy.org/tools/myristoylator), we analyzed Arabidopsis Trxs for potential myristoylation. Trx h9 (At3g08710), Trx h2 (At5g39950), Trx h7 (At1G59730), and Trx h8 (At1G69880) were found to have high confidence scores for myristoylation (respective S values of 0.9872, 0.9864, 0.9890, and 0.9901). The S score is based on the average responses of 25 artificial neural network predictions, which are defined as S = positive minus negative; 0.85 < S < 1 indicates a high confidence prediction. As the only h-type representative with the necessary third Cys (Cys4) at the N terminus, Trx h9 was subjected to palmitoylation prediction using the CSS-PALM program (http://csspalm.biocuckoo.org). Perhaps not surprisingly, the N-terminal conserved Cys at position 4 (Cys4) of Trx h9 was positive for palmitoylation with a score (S) of 4.852 (S ≥ 1.0 indicates high confidence). Thus, whereas Trxs h2, h7, and h8 may be myristoylated, Trx h9 appears to be the only member of the Trx h group capable of undergoing dual lipid modification (myristoylation and palmitoylation). Furthermore, based on these analyses, we found that other plant Trxs (f, m, x, y, and o) would also not undergo lipid modification. The role of Gly (Gly2) in Trxs h2, h7, and h8 is yet to be determined.
Mutation of Gly2, Not Cys4, in Conserved N-Terminal Extension Abolishes Trx h9 Membrane Localization.
The Gly2 and Cys4 residues in the N-terminal extension of Trx h9 are highly conserved with its orthologs (Fig. S2A). Gly2 and Cys4 are canonical sites, respectively, for N-myristoylation and palmitoylation (15 –17). Further, Cys4 is required for catalysis of the poplar ortholog of Trx h9, PtTrxh4 (19). It was therefore of interest to determine the importance of Gly2 and Cys4 in Trx h9. To this end, the two amino acids were mutated to Ala2 (Trx h9G2A) and Trp4 (Trx h9C4W), respectively. Subcellular localization of the mutated GFP proteins was determined in both transient and stable transformants.
Mutation of Gly2 abolished membrane localization, as seen with transient expression (Fig. 3E), such that Trx h9 showed typical cytosolic localization similar to GFP alone (Fig. S4A). Moreover, unlike Trx h9-GFP, Trx h9G2A-GFP fusion was not seen in the plasma membrane but rather was distributed in the cytosol, as visualized by GFP and confirmed by DAPI staining (Fig. 3F). In contrast to Gly2, mutation of Cys4 did not affect localization of Trx h9, which, like the wild type, was observed in the plasma membrane (Fig. 3 I and J). Results with stably transformed Arabidopsis were consistent with transient expression assays of the mutants in both Gly2 (Fig. 3 G and H) and Cys4 (Fig. 3 K and L). Localization results led to the conclusion that association of Trx h9 with the plasma membrane is possibly linked to N-myristoylation modification at Gly2 but is independent of Cys4.
Trx h9 Can Move from Cell to Cell.
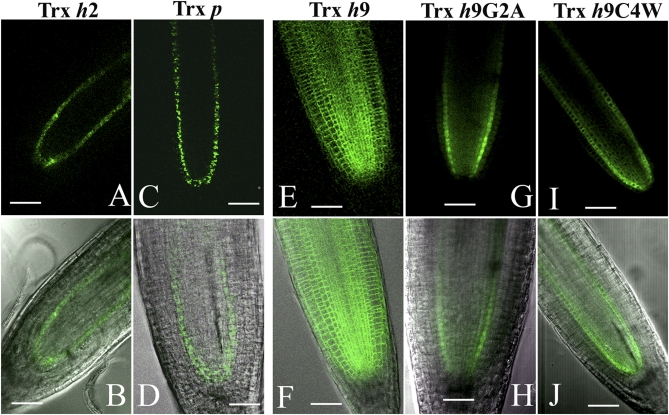
That Trx h9 is membrane-anchored and has the potential for palmitoylation suggested a unique function relative to its Trx counterparts. As palmitoylation has been associated with protein movement in animal cells, we sought evidence of a related function in plants (18, 20). We therefore tested the possible movement of Trx h9 using the Arabidopsis tissue-specific promoter SCARECROW (pSCR), which directs downstream gene expression specifically in the single endodermal cell layer of the root (21). Before examining expression of Trx h9, we confirmed promoter specificity with two other Trxs (Trx h2 and Trx p), their full-length protein-coding sequences fused to GFP, pSCR::Trx h2-GFP and pSCR::Trx p-GFP, respectively. Confocal imaging confirmed that both Trx-GFP fusions (pSCR::Trx h2-GFP and pSCR::Trx p-GFP) showed the expected expression in the single endodermal layer of the root tip (Fig. 4 A, B and C, D). By contrast, in plants transformed with pSCR::Trx h9-GFP, GFP was seen throughout the root (Fig. 4 E and F). These results suggest that, unlike other Trxs examined to date, Trx h9 can move from cell to cell, that is, migrate from its original expression site in endodermal cells to other cell layers of the root (Fig. 4 E and F). This type of movement differs from that described for truncated human Trx (Trx80) and NaTrxh from Nicotiana alata that cross the cell boundary during secretion (22, 23). It is also different from that for 13-kDa rice cytosolic h-type Trx, RPP13-1, that moves from companion cells through plasmodesmata into adjacent sieve elements in mature phloem (24). It is noted that the apparent absence of Trx h9 movement in isolated epidermal onion tissue described above suggests the requirement for a condition provided only by the living plant.
Fig. 4.
Movement analysis of Trx h9 protein in root tips of 7-day-old Arabidopsis (Col-0) seedlings using Arabidopsis SCARECROW promoter (pSCR) and GFP tag. (A and B) Trx h2 (At5g39950). (C and D) Trx p (At3G06730). (E and F) Trx h9. (G and H) Trx h9G2A. (I and J) Trx h9C4W. (Scale bars, 50 μm.)
Trx h9 Movement Requires Both Gly2 and Cys4.
As observed for its membrane localization (Fig. 3 E and F), Trx h9 movement was abolished by replacing Gly2 with alanine (Fig. 4 G and H). However, unlike localization, which was not altered by mutagenizing Cys4 (Fig. 3 I–L), movement of Trx h9 was also abolished by converting Cys4 to tryptophan (Fig. 4 I and J). Palmitoylation occurs in membranes, and thus cytosolic proteins must interact with membranes to be palmitoylated (17, 18). Gly2 may, therefore, be required for membrane localization of Trx h9 and for subsequent palmitoylation of Cys4. Palmitoylated proteins, which favor specific protein-protein interactions, modulate the activity of signaling cascades (18). Although Cys4 was not required for membrane localization of Trx h9, the dynamically palmitoylated Cys4 may enhance its membrane association and facilitate its movement by regulating its activities and interactive properties. Taken together, these results suggest that the two conserved amino acids in the N-terminal extension of Trx h9 are essential for its unusual properties: Gly2 for membrane anchoring and both Gly2 and Cys4 for mobility. Interestingly, Cys4 of Trx h9 seems to be the target not only for movement but also for its reduction by glutaredoxin, as suggested by results obtained with the ortholog from poplar (19). However, mechanisms as to how these two reactions involving the same thiol group are regulated with respect to each other remain to be determined. Mutation analysis in terms of movement and palmitoylation of Cys57 in the catalytic site of Trx h9 would provide insight into this question.
Trx h9 May Dock via Interaction of N-Terminal Cys4 with Catalytic Cys57.
Sequence analysis reveals that Trx h9 has 70% identity with a Trx from poplar (PtTrxh4), the 3D structure of which has been determined (19). Using PtTrx h4 and other close homologs as templates for comparative modeling, we determined a predicted 3D structure for Trx h9 by applying I-TASSER simulation (25) (Fig. 5A) that matched nearly perfectly with Zits templates (Fig. 5B) (SI Materials and Methods). Analysis of its structure revealed that, as discussed by Koh et al. (19), the N-terminal extension appears to act like an arm appended to the main body of Trx h9 with the potential to be a protein docking site (binding pocket)—through possible interaction of the N-terminal Cys (Cys4) with Cys57 in the classical catalytic site (C57GPC60) (Fig. 5A).This docking site could confer specific binding properties to Trx h9 in its interaction with other proteins in a manner possibly modulated by palmitoylation of Cys4. In addition, the C terminus of Trx h9 could form a smaller binding pocket in which Ser136 is located at the inside surface. The reported phosphorylation of Ser136 in response to sucrose (26) could alter binding properties of the pocket. An analysis of the evolutionary conservation of its surface amino acids using ConSurf (http://consurf.tau.ac.il) provided further evidence that the N-terminal extension of Trx h9, especially Gly2, appears to have functional significance (Fig. 5C).
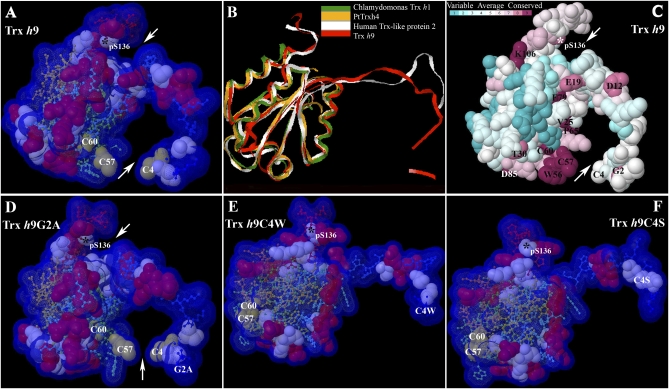
Fig. 5.
Three-dimensional models and conserved residue prediction for Trx h9. (A) Three-dimensional model of Trx h9. (B) Superimposition of 3D model of Trx h9 (red) and the top three templates of Trx h9: C. reinhardtii Trx h1 (green; PDB ID code 1ep7A); poplar PtTrxh4 (yellow; PDB ID code 3d21); and human Trx-like protein 2 (white; PDB ID code 2diyA), using 3d-SS (3-Dimensional Structural Superposition) service. (C) Conserved residue analysis of Trx h9. Residue conservation from variable to conserved is shown in green to dark red, respectively. (D–F) Three-dimensional model of Trx h9G2A, Trx h9C4W, and Trx h9C4S, respectively. Arrows point to potential docking sites of Trx h9 in A, C, and D. Cys, Ser, and positively charged residues are shown in off-yellow, light blue, and red, respectively, as 100% of van der Waals. The remainder of residues from N to C terminus are shown in blue to red as 20% of van der Waals in A and D, and E and F. All atoms are coupled with Solvent-Accessible Surface (VDW + 1.4 Å) in A and D–F. Asterisks indicate phosphorylated Ser at position 136 (pS136) at the C terminus of Trx h9. Conserved amino acids with single-letter abbreviations are indicated at their numbered position in C.
To assess the contribution of the conserved N-terminal Gly2 and Cys4 to structure, we constructed 3D models for three Trx h9 mutants—Trx h9G2A, Trx h9C4W, and Trx h9C4S—using the I-TASSER method. Gly2 was replaced by Ala in Trx h9G2A and Cys4 by both Trp in Trx h9C4W and Ser in Trx h9C4S. Mutation of Gly2 had relatively little effect on the 3D structure; the N-terminal extension appeared still to be able to form a binding pocket (Fig. 5D). However, replacing Cys4 with either Trp or Ser dramatically altered the structure (Fig. 5 E and F). The N-terminal extension appeared to move away from the main body of the molecule, seemingly leading to loss of the binding pocket and the ability of Cys4 to interact with catalytic Cys57.
Grx, Not NTR, Fits in the Potential Binding Pocket of Trx h9.
Computational docking methods used to predict protein-protein interactions can help define parameters valuable to understanding biochemical mechanisms. When analyzed in this manner, Trx h9 appeared not to interact with NADP-thioredoxin reductase (NTR), like other h-type Trxs such as h1 (Fig. S6 A vs. B). Rather, its predicted structure indicated that it might be preferentially reduced by the GSH/Grx system, as for the poplar ortholog of Trx h9, PtTrxh4 (6) (Fig. S6 D and E) in an interaction dependent on Cys4 but not Gly2 (Fig. S6 H and I and F and G and SI Text).
Conclusion
An h-type Trx h9, was found to bind to the plasma membrane despite lacking a transmembrane domain. Experiments with the SCARECROW promoter revealed that Trx h9 was mobile, with the capability to move from cell to cell. Two amino acids in its N-terminal extension appeared to be responsible for these unique properties. Gly2 was required for association with the plasma membrane (possibly for myristoylation), and both Gly2 and Cys4 were essential for mobility (the latter seemingly for structure and palmitoylation). A modeling analysis indicated that Trx h9 is preferably reduced by GSH and Grx, similar to its poplar ortholog, rather than by NTR, as described for other h-type Trxs. A T-DNA insertion mutation revealed that Trx h9 was required for growth and development. Trx h9 thus appears to resemble Trx h3 (27) in bridging the Grx/Trx interface in relaying information to maintain cellular redox balance. It seems possible that Trx h9 may be required for redox signaling. Finally, control experiments uncovered a Trx h (h2) residing in mitochondria and a plastid Trx (Trx p) previously identified described as cytosolic (8). It will be of interest to see how Trx h9 contributes to plant growth and development, including the germination of seeds, and how Trx h2 and Trx p function in their respective organelles.
Materials and Methods
Expression Analysis and Screening for T-DNA Insertion Mutants.
Expression was analyzed by quantitative RT-PCR (SI Materials and Methods). Lines with a putative T-DNA insertion in Trx h9, Salk_086660, or in Trx p, Salk_028162, were obtained from the Arabidopsis Biological Resource Center. T-DNA insertions and expression of the relevant genes in the Salk lines were screened by PCR sequencing and RT-PCR, respectively (SI Materials and Methods).
Constructs.
Full-length coding sequence of the Trxs and an ∼2.6-kb fragment immediately upstream of the SCARECROW coding sequence were amplified by PCR from genomic DNA of Arabidopsis Col-0 plants using appropriate primers (Table S2). EcoRI and BamHI cloning sites and mutated sequences were incorporated into the 5′ forward primers (Table S2, indicated by underline). PCR products from the Trxs were cloned into pEZS-NL vector at the EcoRI and BamHI sites for transient transformation. For stable transformation, a NotI fragment, with each cloned Trx gene linked to the 35S promoter and OCS 3′, was removed from pEZS-NL and inserted into binary vector pART27. The ∼2.6-kb fragment from Scarecrow (21) replaced the 35S promoter in pEZS-NL at NotI and EcoRI sites (Table S2). The NotI fragment with pSCR promoter::Trx-GFP was transferred to pART27.
Transient Expression Assays Using Particle Bombardment of Isolated Onion Epidermal Cell Layer.
Plasmids with the 35S::Trx-GFP fusion were individually bombarded into onion epidermal cells (28), which were examined for GFP (see below). After 20–24 h postparticle bombardment, onion epidermal cells were stained with DAPI (29). Cells were plasmolyzed by incubating 10 min in each of 0.25, 0.50, and 0.75 M sucrose.
Plant Transformation, Selection, Protoplast Generation, and Fluorescence Visualization.
Trx constructs in pART27 were introduced into Agrobacterium tumefaciens GV3101, using the freeze–thaw method (30), and the transformed Agrobacterium was then introduced into Arabidopsis via the floral-dip method (31). T1 plants were selected on MS medium containing 50 μg/L kanamycin. Arabidopsis plants were grown in soil in the greenhouse or on MS medium with 1.0% sucrose (16 h light/8 h dark cycle, 23 °C). Protoplasts were generated from transgenic leaves by treating 30 min with 1.0% cellulase, 0.5% pectinase, 1.0 mg/mL BSA, 0.048% PVP, 0.5 M sucrose, 1.0 mM CaCl2 (pH 5.5), and used immediately to examine for GFP. Fluorescence was visualized using a Leica DM LB fluorescence microscope or a Zeiss LSM 510 confocal microscope with 488 nm/530 nm excitation/emission light for GFP, 364/470 for DAPI, and 543/576 for MitoTracker orange.
Prediction of 3D Structure of Trx h9.
The three-dimensional structure of Trx h9 was predicted using I-TASSER (threading/assembly/refinement), that is, Zhang service (25), available at http://zhang.bioinformatics.ku.edu/I-TASSER. Superimposition analysis of the 3D models of Trx h9 and its templates [human Trx-like protein 2 (PDB ID code 2diyA), PtTrxh4 (PDB ID code 3d21A), and Trx h1 from Chlamydomonas reinhardtii (PDB ID code 1ep7A)] was done using 3-Dimensional Structural Superposition (3d-SS) service (http://cluster.physics.iisc.ernet.in/3dss/severalinput.html) (32). Conserved amino acids at the protein surface were determined using ConSurf (33) (http://consurf.tau.ac.il/overview.html).
Supplementary Material
Acknowledgments
The authors thank Drs. S. Ruzin and D. Schichnes of the UCB Biological Imaging Facility for assistance, S. Cutler and D. Ehrhardt for pEZS-NL, Catherine Lie, SuFey Ong, and Khanh Nguyen for laboratory assistance, and Z. Li for chlorophyll analysis. L.M. was partially supported by the Agricultural and Environmental Chemistry Graduate Program, P.G.L. by the US Department of Agriculture Cooperative Extension, and B.B.B. by the Agricultural Experiment Station, University of California.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0913759107/DCSupplemental.
References
- 1.Foyer CH, Noctor G. Redox homeostasis and antioxidant signaling: A metabolic interface between stress perception and physiological responses. Plant Cell. 2005;17:1866–1875. doi: 10.1105/tpc.105.033589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fujino G, Noguchi T, Takeda K, Ichijo H. Thioredoxin and protein kinases in redox signaling. Semin Cancer Biol. 2006;16:427–435. doi: 10.1016/j.semcancer.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 3.Sweat TA, Wolpert TJ. Thioredoxin h5 is required for victorin sensitivity mediated by a CC-NBS-LRR gene in Arabidopsis . Plant Cell. 2007;19:673–687. doi: 10.1105/tpc.106.047563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tada Y, et al. Plant immunity requires conformational changes [corrected] of NPR1 via S-nitrosylation and thioredoxins. Science. 2008;321:952–956. doi: 10.1126/science.1156970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li YC, et al. The level of expression of thioredoxin is linked to fundamental properties and applications of wheat seeds. Mol Plant. 2009;2:430–441. doi: 10.1093/mp/ssp025. [DOI] [PubMed] [Google Scholar]
- 6.Gelhaye E, Rouhier N, Jacquot JP. Evidence for a subgroup of thioredoxin h that requires GSH/Grx for its reduction. FEBS Lett. 2003;555:443–448. doi: 10.1016/s0014-5793(03)01301-2. [DOI] [PubMed] [Google Scholar]
- 7.Reichheld J-P, Mestres-Ortega D, Laloi C, Meyer Y. The multigenic family of thioredoxin h in Arabidopsis thaliana: Specific expression and stress response. Plant Physiol Biochem. 2002;40:685–690. [Google Scholar]
- 8.Rivas S, et al. CITRX thioredoxin interacts with the tomato Cf-9 resistance protein and negatively regulates defence. EMBO J. 2004;23:2156–2165. doi: 10.1038/sj.emboj.7600224. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9.Lichtenthaler HK. Die Plastoglobuli von Spinat, ihre Grösse, Isolierung und Lipochinonzusammensetzung. Protoplasma. 1969;68:65–77. [Google Scholar]
- 10.Marcus F, et al. Plant thioredoxin h: An animal-like thioredoxin occurring in multiple cell compartments. Arch Biochem Biophys. 1991;287:195–198. doi: 10.1016/0003-9861(91)90406-9. [DOI] [PubMed] [Google Scholar]
- 11.Gelhaye E, et al. A specific form of thioredoxin h occurs in plant mitochondria and regulates the alternative oxidase. Proc Natl Acad Sci USA. 2004;101:14545–14550. doi: 10.1073/pnas.0405282101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lang-Pauluzzi I, Gunning BES. A plasmolytic cycle: The fate of cytoskeletal elements. Protoplasma. 2000;212:174–185. [Google Scholar]
- 13.Buchanan BB, Balmer Y. Redox regulation: A broadening horizon. Annu Rev Plant Biol. 2005;56:187–220. doi: 10.1146/annurev.arplant.56.032604.144246. [DOI] [PubMed] [Google Scholar]
- 14.Gelhaye E, Rouhier N, Navrot N, Jacquot JP. The plant thioredoxin system. Cell Mol Life Sci. 2005;62:24–35. doi: 10.1007/s00018-004-4296-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Casey PJ. Protein lipidation in cell signaling. Science. 1995;268:221–225. doi: 10.1126/science.7716512. [DOI] [PubMed] [Google Scholar]
- 16.Maurer-Stroh S, Eisenhaber B, Eisenhaber F. N-terminal N-myristoylation of proteins: Refinement of the sequence motif and its taxon-specific differences. J Mol Biol. 2002;317:523–540. doi: 10.1006/jmbi.2002.5425. [DOI] [PubMed] [Google Scholar]
- 17.Resh MD. Fatty acylation of proteins: New insights into membrane targeting of myristoylated and palmitoylated proteins. Biochim Biophys Acta. 1999;1451:1–16. doi: 10.1016/s0167-4889(99)00075-0. [DOI] [PubMed] [Google Scholar]
- 18.Smotrys JE, Linder ME. Palmitoylation of intracellular signaling proteins: Regulation and function. Annu Rev Biochem. 2004;73:559–587. doi: 10.1146/annurev.biochem.73.011303.073954. [DOI] [PubMed] [Google Scholar]
- 19.Koh CS, et al. An atypical catalytic mechanism involving three cysteines of thioredoxin. J Biol Chem. 2008;283:23062–23072. doi: 10.1074/jbc.M802093200. [DOI] [PubMed] [Google Scholar]
- 20.Zhou B, Liu L, Reddivari M, Zhang XA. The palmitoylation of metastasis suppressor KAI1/CD82 is important for its motility- and invasiveness-inhibitory activity. Cancer Res. 2004;64:7455–7463. doi: 10.1158/0008-5472.CAN-04-1574. [DOI] [PubMed] [Google Scholar]
- 21.Malamy JE, Benfey PN. Analysis of SCARECROW expression using a rapid system for assessing transgene expression in Arabidopsis roots. Plant J. 1997;12:957–963. doi: 10.1046/j.1365-313x.1997.12040957.x. [DOI] [PubMed] [Google Scholar]
- 22.Juárez-Díaz JA, et al. A novel thioredoxin h is secreted in Nicotiana alata and reduces S-RNase in vitro. J Biol Chem. 2006;281:3418–3424. doi: 10.1074/jbc.M511687200. [DOI] [PubMed] [Google Scholar]
- 23.Pekkari K, et al. Truncated thioredoxin (Trx80) exerts unique mitogenic cytokine effects via a mechanism independent of thiol oxido-reductase activity. FEBS Lett. 2003;539:143–148. doi: 10.1016/s0014-5793(03)00214-x. [DOI] [PubMed] [Google Scholar]
- 24.Ishiwatari Y, et al. Rice phloem thioredoxin h has the capacity to mediate its own cell-to-cell transport through plasmodesmata. Planta. 1998;205:12–22. doi: 10.1007/s004250050291. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Niittylä T, Fuglsang AT, Palmgren MG, Frommer WB, Schulze WX. Temporal analysis of sucrose-induced phosphorylation changes in plasma membrane proteins of Arabidopsis . Mol Cell Proteomics. 2007;6:1711–1726. doi: 10.1074/mcp.M700164-MCP200. [DOI] [PubMed] [Google Scholar]
- 27.Reichheld J-P, et al. Inactivation of thioredoxin reductases reveals a complex interplay between thioredoxin and glutathione pathways in Arabidopsis development. Plant Cell. 2007;19:1851–1865. doi: 10.1105/tpc.107.050849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scott A, Wyatt S, Tsou PL, Robertson D, Allen NS. Model system for plant cell biology: GFP imaging in living onion epidermal cells. Biotechniques. 1999;26:1125–1132. 1128–1132. doi: 10.2144/99266st04. [DOI] [PubMed] [Google Scholar]
- 29.Khar A, Mitchison JM. Observations on ultracentrifuging wild-type and mutant (cdc2.33) cells of Schizosaccharomyces pombe . J Cell Sci. 1989;92:345–348. doi: 10.1242/jcs.92.3.345. [DOI] [PubMed] [Google Scholar]
- 30.Höfgen R, Willmitzer L. Storage of competent cells for Agrobacterium transfor-mation. Nucleic Acids Res. 1988;16:9877. doi: 10.1093/nar/16.20.9877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana . Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 32.Russell RB, Breed J, Barton GJ. Conservation analysis and secondary structure prediction of the SH2 family of phosphotyrosine binding domains. FEBS Lett. 1992;304:15–20. doi: 10.1016/0014-5793(92)80579-6. [DOI] [PubMed] [Google Scholar]
- 33.Landau M, et al. ConSurf: The projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 2005;33(Web Server issue):W299–W302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.