Abstract
Argonaute (AGO) proteins interact with distinct classes of small RNAs to direct multiple regulatory outcomes. In many organisms, including plants, fungi, and nematodes, cellular RNA-dependent RNA polymerases (RdRPs) use AGO targets as templates for amplification of silencing signals. Here, we show that distinct RdRPs function sequentially to produce small RNAs that target endogenous loci in Caenorhabditis elegans. We show that DCR-1, the RdRP RRF-3, and the dsRNA-binding protein RDE-4 are required for the biogenesis of 26-nt small RNAs with a 5′ guanine (26G-RNAs) and that 26G-RNAs engage the Piwi-clade AGO, ERGO-1. Our findings support a model in which targeting by ERGO-1 recruits a second RdRP (RRF-1 or EGO-1), which in turn transcribes 22G-RNAs that interact with worm-specific AGOs (WAGOs) to direct gene silencing. ERGO-1 targets exhibit a nonrandom distribution in the genome and appear to include many gene duplications, suggesting that this pathway may control overexpression resulting from gene expansion.
Keywords: RNAi, 22G-RNAs, 26G-RNAs, ERI pathway, gene duplication
RNA interference (RNAi) is a mechanism of gene regulation directed by Argonaute (AGO) proteins in conjunction with their sequence-specific small RNA cofactors. A multitude of distinct AGO-mediated regulatory modules have been identified in plants, fungi, and animals (1). In all of these pathways, base pair interactions between the small RNA and a target molecule provide specificity, whereas the AGO protein, which contains a conserved nuclease domain, can direct silencing through target cleavage or through recruitment of transcriptional or posttranscriptional regulators.
AGO pathways can be triggered by natural or exogenous sources of double-stranded (ds)RNA. The Dicer family of RNase III-related enzymes processes dsRNA into small RNAs of 20–25 nucleotides (nt) in length (2). For example, micro (mi)RNAs are processed from genomically encoded hairpins and mediate AGO-dependent silencing at the posttranscriptional level (3 –5). Short interfering (si)RNAs are processed from both endogenous and exogenous sources of dsRNA and direct AGO-dependent cleavage of target mRNAs (2). siRNAs processed from primary dsRNA sources (for example, hairpins or convergent transcripts) are referred to as primary siRNAs. In fungi, plants, and nematodes, RNA-dependent RNA polymerases (RdRPs) are required for the amplification of silencing signals. siRNAs that are processed from dsRNA generated by RdRP are referred to as “secondary siRNAs.” In Caenorhabditis elegans, secondary siRNAs appear to be directly synthesized by RdRP, independently of DCR-1, and are loaded onto AGOs (6 –8).
In C. elegans, two RdRPs, RRF-1 and EGO-1, are required for the biogenesis of an abundant class of endogenous small RNAs called 22G-RNAs (9), which are predominantly 22 nt in length and contain a triphosphorylated 5′ guanine. Interestingly, 22G-RNAs are antisense to more than 50% of annotated genes (9). Two major 22G-RNA systems exist in C. elegans: those that interact with the AGO CSR-1 and those that interact with the expanded family of worm-specific AGO (WAGO) proteins (9, 10). The CSR-1/22G-RNA system is required for the proper organization of holocentric chromosomes and is essential for chromosome segregation. The WAGO/22G-RNA system provides surveillance against transposable elements and aberrant endogenous transcripts (the endo-RNAi pathway) and is also required for the response to foreign dsRNA (the exo-RNAi pathway).
The ERI endo-RNAi pathway was defined by mutations that result in an enhanced exo-RNAi response (11, 12). The Eri phenotype appears to reflect relaxed competition for limiting RNAi factors that are also required for the response to exogenous dsRNA (12, 13). The RdRP RRF-3 was identified as an Eri mutant (14) and as a physical interactor with Dicer and other proteins defined genetically as Eri factors (12). On the basis of these and other studies, it was proposed that RRF-3 produces endogenous dsRNA that is processed by Dicer and loaded onto the AGO, ERGO-1, for which loss-of-function also results in an Eri phenotype (12, 13). WAGOs were identified as possible secondary AGOs in the ERI pathway (13).
Here, we show that the ERI pathway is indeed a two-step AGO pathway and that the pathway also involves two separate rounds of RdRP-mediated small RNA biogenesis. We show that, in embryos, ERGO-1 interacts with the previously described 26-nt small RNAs with a 5′ guanine (26G-RNAs) (15). Furthermore, components of the ERI complex, including RRF-3, DCR-1, and the dsRNA-binding protein RDE-4, are required for the biogenesis of both 26G- and 22G-RNAs on Eri targets. In contrast, RRF-1 and WAGOs are required for the accumulation of 22G- but not 26G-RNAs. Hence, we propose that 26G-RNAs are the primary small RNAs in the ERI pathway that drive the downstream production of 22G-RNAs. Many ERGO-1 26G-RNA targets appear to be ancient duplications, suggesting that the function of this pathway may be to buffer the expression of rapidly expanding gene families.
Results
ERGO-1 Interacts with 26G-RNAs.
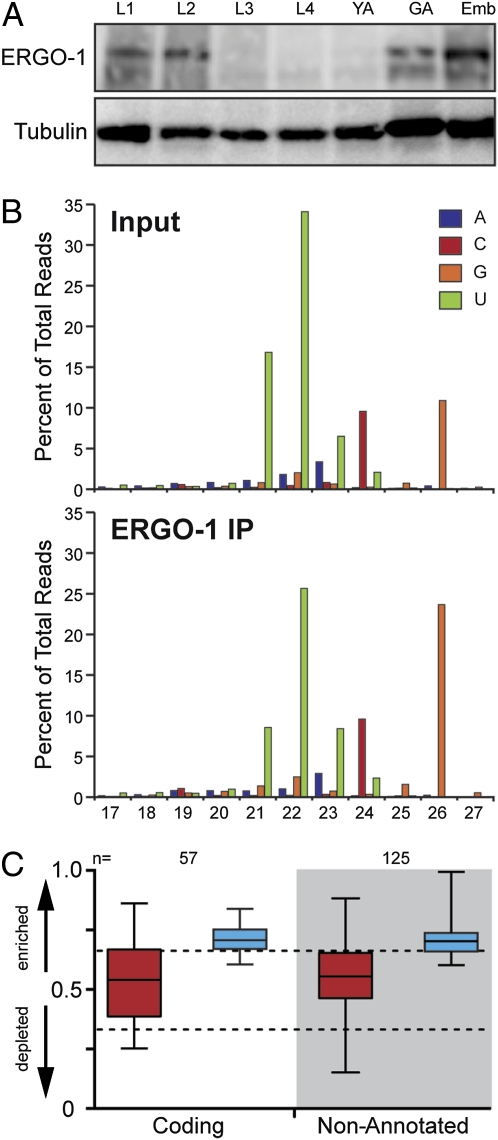
Previous work identified ERGO-1 as an AGO that functions in the ERI endo-siRNA pathway (13). To identify small RNAs that interact with ERGO-1, we deep sequenced small RNAs prepared from ERGO-1 immunoprecipitation (IP) and input samples. Developmental expression studies indicated that ERGO-1 was primarily expressed in embryos and was virtually absent from L3 and L4 larvae and young adults (lacking embryos) (Fig. 1A and Fig. S1). Therefore, ERGO-1 IP experiments were performed using embryo lysates. Analyses of both size and first-nucleotide distribution of reads revealed that 26G-RNAs were enriched ≈2.2-fold in the ERGO-1 IP sample over the input library (Fig. 1B). Although 21- to 23-nt small RNAs with a 5′ uracil (5′U), including miRNAs and 21U-RNAs, were cloned at high levels, they were not enriched in the ERGO-1 IP sample (Fig. 1B). The modest enrichment of 26G-RNAs and the high background of 21U-RNAs and miRNAs is consistent with the low efficiency of ERGO-1 IP (Fig. S1). However, the interaction between ERGO-1 and a representative 26G-RNA appears to be specific, because both the expression of the 26G-RNA and its interaction with ERGO-1 were abrogated in an ergo-1 null mutant (Fig. S1).
Fig. 1.
ERGO-1 interacts with 26G-RNAs in embryos. (A) Expression profile of ERGO-1 protein. (B) Length and first nucleotide distribution of genome-matching reads in Input and ERGO-1 IP small RNA libraries. (C) Enrichment or depletion of small RNAs derived from 26G-RNA genes (white area) or nonannotated clusters (gray area) in the ERGO-1 IP. Small RNAs were separated by read length into 26 nt (blue) and <26 nt (red). Values approaching 1 indicate enrichment of small RNA; values approaching 0 indicate depletion. Relative enrichment was calculated as ratio of IP/(IP + wild-type). “n” loci with at least 10 reads per million total reads (not including structural) in either the wild-type or the mutant sample were analyzed. The top and bottom of each box represent the 75th and 25th percentiles, respectively. The horizontal line within each box represents the median value. Dotted lines denote 2-fold enrichment (Upper) and twofold depletion (Lower).
ERGO-1-associated 26G-RNAs mapped to genes (23%), pseudogenes (8%), and nonannotated loci (64%) in proportions similar to those observed for the 26G-RNA species present in our non-IP dataset (Fig. S1). Comparing these two datasets, 26G-RNAs targeting a set of 57 genes were enriched by ≥1.5-fold in the ERGO-1 IP dataset (Fig. 1C and Table S1). When we examined each 5′G size population independently, we observed varying degrees of enrichment in the ERGO-1 IP. However, of the reads that matched 26G-RNA targets, only 24G–28G reads were enriched twofold (Fig. S2). The frequency distribution of 24G–28G reads was consistent with these populations being derived from 26G-RNAs (Fig. S2). Greater than 90% of the enriched 24G and 25G reads represented 3′ truncations of 26G-RNAs, whereas the 27G and 28G reads appeared to represent terminal transferase products (10). Small RNA species, ranging from 17 to 23 nt, including 22G-RNAs (9), were also present, and in some cases enriched, in the IP dataset. The significance of this enrichment is not clear. However, because such reads were present at very low levels, were derived from loci that are not targeted by 26G-RNAs, and were not dependent on ERGO-1 (Fig. 1 and Fig. S2). Therefore, although we cannot rule out specific interactions with other small RNA species, the above data strongly support the direct association between ERGO-1 and 26G-RNAs that are abundant during embyrogenesis.
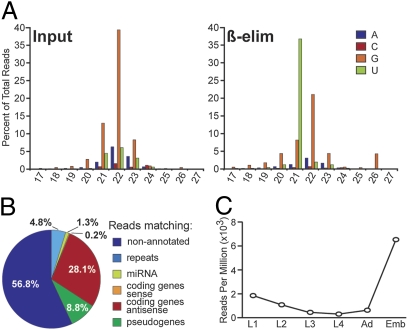
26G-RNAs were previously shown to be 5′ monophosphorylated small RNAs with a 3′ modification that is resistant to oxidation by periodate (β-elimination) (15). To determine whether the ERGO-1 interacting 26G-RNAs correspond to those identified by Ruby et al. (15), small RNAs purified from adult animals containing embryos were oxidized with periodate (β-eliminated) before cloning and deep sequencing. A second library was prepared in parallel from untreated small RNAs as a control and both libraries were prepared using a method compatible with cloning mono- or triphosphorylated small RNAs, i.e., 22G-RNAs (9).
Of 2.77 million genome-matching reads in the untreated sample, 0.5% corresponded to potential 26G-RNAs and 2.9% to 21U-RNAs, whereas the majority of reads represented 22G-RNAs (Fig. 2A). In the β-eliminated sample, the 26G and 21U species were enriched 8.6- and 12.3-fold, constituting 4.3 and 35.8% of the 5.68 million genome-matching reads, respectively. In both samples, ∼40% of 26G-RNA reads mapped antisense to coding genes (30%) or pseudogenes (10%). More than half (∼56%) of the reads mapped to genomic loci lacking any annotation (Fig. 2B), similar to the assignment of ERGO-1 interacting 26G-RNAs. We identified 49 genes with antisense 26G-RNAs that were enriched in the β-eliminated sample over the untreated control (Table S1), 48 of which were enriched at least 1.5-fold in the ERGO-1 IP (Fig. S2). Analyzing deep-sequencing data across developmental time points (16), we observed that 26G-RNAs derived from these 48 genes were most abundant during embryogenesis and decrease dramatically during larval development (Fig. 2C).
Fig. 2.
26G-RNAs cloned after β-elimination. (A) Plot of the first nucleotide composition and length of small RNA reads that were sequenced in Input and β-eliminated (β-elim) samples. (B) Pie chart indicating the assignment of genome-matching 26-nt reads according to genome annotation. (C) Expression profile of ERGO-1–dependent 26G-RNAs during development. Twenty-six-nucleotide reads targeting 48 ERGO-1–dependent loci were extracted from deep-sequencing data generated by Batista et al. (16). Plot shows reads per million in each developmental stage.
Identification of Nonannotated 26G-RNA Loci.
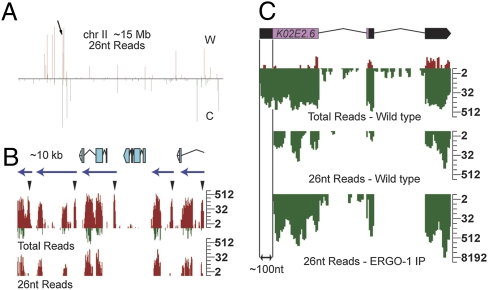
The majority of 26G-RNA reads were derived from unique nonannotated genomic sequences. Despite the lack of annotation, these 26G-RNAs were in clusters and oriented on one strand as though antisense to an expressed transcript (Fig. 3A). To further characterize these nonannotated genomic loci, 26G-RNA reads matching genome annotations were removed from the data and each chromosome strand was scanned using a 500-nt window to build and annotate 26G-RNA clusters (Methods). This analysis defined 147 genomic loci with a 26G-RNA density of at least 10 reads per million (rpm) in our dataset (Table S2). These clusters are much more extensive than recently reported clusters (17) and appear to encompass complete transcription units that have not been annotated. Analysis of the ERGO-1 IP data revealed that 126 of these loci were enriched above a threshold of 1.5-fold in the IP relative to input (Fig. 1C). By visual inspection, ∼17 nonannotated 26G-RNA clusters appeared to extend from, or were very close to, annotated 26G-RNA genes, raising the possibility that these clusters may target incompletely annotated transcripts (9).
Fig. 3.
26G-RNA clusters are targeted by 22G-RNAs. (A) Density profile of 26-nt reads along chromosome (chr) II, which is ∼15 Mb. C, Crick; W, Watson. Arrow indicates location of 26G-RNA cluster shown in B. (B) Density profile of small RNAs targeting an ∼10-kb cluster in adults with embryos (gravid). Twenty-six-nucleotide read density shown in the Lower graph represents 26G-RNAs. “Total reads” shown in Upper graph include both 26G- and 22G-RNA reads. Several peaks within this cluster lack 26G-RNA reads (arrowheads). The majority of reads from this cluster are on the Watson strand (red). Reads that map to the Crick strand (green) in this cluster are associated with 26G-RNA reads only. The blue arrows above density profiles predict transcription units based on the observed 26G-/22G-RNA patterns at ERGO-1 targets. The annotated gene predictions within this ∼10-kb interval are illustrated above the density plots. Log2 scales are shown (Right). (C) Density profiles of small RNAs targeting K02E2.6 in wild-type adult and ERGO-1 IP datasets. Gene structure is shown at the top. Reads matching the Watson strand (red) are sense reads. Reads matching the Crick strand (green) are antisense. “Total Reads” include 26G- (“26-nt Reads”) and 22G-RNAs. 26G-RNAs are excluded from ∼100 nt of the 5′-UTR of K02E2.6. A log2 scale is shown (Right).
In total, 26G-RNAs targeting annotated and nonannotated loci defined a set of ≈180 26G-target loci with read densities >10 rpm. The genomic location of 26G-RNA targets appeared to be nonrandom. The most abundant 26G-RNA loci tended to map within 5 Mb of the chromosome ends (Fig. 3A and Fig. S3). This pattern was strikingly different from the entire set of 22G-RNA loci, which were more evenly distributed throughout each chromosome (9, 10). Interestingly, we observed groups of 26G-RNA targets that were oriented in tandem in the genome. For example, an ∼10-kb region of chromosome II exhibited 5 different 26G-RNA loci (Fig. 3B) that appear to be homologous, tandemly repeated units, suggesting that they are duplications that have diverged in sequence.
Essentially all 26G-RNA targets were also targeted by 22G-RNAs (Fig. 3 and Tables S3 and S4) (9). In fact, 22G-RNAs were several orders of magnitude more abundant than 26G-RNAs at many loci in gravid adult samples. Interestingly, 26G-RNAs but not 22G-RNAs were excluded from the first ∼100 nt at the 5′ end of target genes (Fig. 3B). For example, 26G-RNAs mapped with similar density to both the exons and the 3′-UTR of K02E2.6, but were absent from the 5′-UTR (Fig. 3B). In contrast, the entire K02E2.6 transcript was targeted by 22G-RNAs, including the 5′-UTR. Sense reads were almost exclusively derived from regions targeted by 26G-RNAs and rarely derived from the regions targeted only by 22G-RNAs. This differential small RNA pattern was also observed at virtually all 26G-RNA target loci and helped us to define 26G-RNA loci (Methods). Surprisingly, 22G-RNAs corresponding to more than half of the 26G-RNA loci were enriched in the somatic tissues of adult animals (Table S3), where 26G-RNAs were relatively depleted (Fig. 2C). Taken together, these observations are consistent with the idea that targeting by 26G-RNAs drives the secondary biogenesis of 22G-RNAs both in the embryo and in subsequent developmental stages.
26G-RNA Biogenesis Is Dependent on Components of the ERI Pathway.
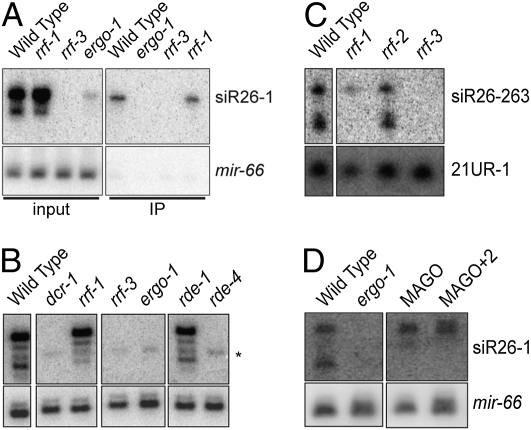
Having demonstrated that ERGO-1 interacts with 26G-RNAs, we next examined what other factors influence the expression of small RNAs targeting the ERGO-1 26G-RNA loci. Consistent with our deep-sequencing data, Northern blot analysis using a probe for siR26-1 (15), targeting C40A11.10, revealed association of this 26G-RNA with the ERGO-1 IP complex in embryo lysates (Fig. 4A). We found that siR26-1 expression was abrogated by mutations in the ERI pathway, including ergo-1, rrf-3, rde-4, and a viable, Eri allele of dcr-1(mg375Eri) (18) (Fig. 4 A and B). The requirement for rde-4 in the biogenesis of 26G-RNAs was independent of its role in the exo-RNAi pathway, as rde-1 was not required for 26G-RNA biogenesis (Fig. 4B).
Fig. 4.
Genetic requirements for 26G-RNA expression. (A) Northern blot of siR26-1 in wild-type and mutant embryos. Input (Left) and ERGO-1 IP (Right) samples are indicated. Loading control: mir-66. (B) Northern blot of siR26-1 in RNAi mutant (adult). The membrane was first hybridized to a mir-66 probe as a loading control, which could not be removed completely, and is indicated by the asterisk (*). All lanes shown were from the same membrane and exposure. Only relevant lanes are shown. (C) Northern blot of siR26-263 in the indicated RdRP mutants (adult). Loading control: 21U-1. All lanes were from the same membrane and exposure. Only the relevant lanes are shown. (D) Northern blot of siR26-1 in mutants with multiple WAGO deletions (MAGO and MAGO+2) (9, 13). Loading control: mir-66. Adults were used. All lanes were from the same membrane and exposure. Only the relevant lanes are shown.
Among the four C. elegans RdRP genes, only rrf-3 was required for the expression of 26G-RNAs as determined by Northern blot analysis (Fig. 4 A–C and Fig. S4). 26G-RNA expression was unaltered in both rrf-1 and rrf-2 mutants (Fig. 4 A–C and Fig. S4). Although most 26G-RNAs were not detected in ego-1 mutants (Fig. S4), this could reflect the fact that ego-1 mutants are sterile and thus lack embryos, which is the stage when ERGO-1-dependent 26G-RNAs are most abundantly expressed (Fig. 2C). As expected, the expression of siR26-1 was unaffected in mutants lacking WAGO-1 and other WAGO-class Argonautes that are required for, and interact with, 22G-RNAs (Fig. 4D). Taken together, these findings indicate that 26G-RNA accumulation is dependent on components of the ERI pathway, but independent of several components of the exo-RNAi and 22G-RNA pathways.
ERI Pathway Stimulates 22G-RNA Accumulation.
As described above, ERGO-1-dependent 26G-RNA loci were also targeted by 22G-RNAs. Some of our probes (siR26-1 and siR26-263) detected both 26G-RNAs and a 22-nt RNA species, whereas the K02E2.6 probe detected only 22G-RNAs (Fig. 4 and Fig. S4) (9). The ERI-pathway genes rrf-3, ergo-1, rde-4, and dcr-1 were all required for expression of both the 26G- and 22G-RNA species at these target loci (Fig. 4B and Fig. S4). In contrast, rrf-1 and several wago Argonautes assayed were not required for 26G-RNA expression at these targets, but were required for 22G-RNA expression (Fig. 4 C and D) (9). ERGO-1 still interacted with 26G-RNAs in rrf-1 mutant embryos (Fig. 4A). Together, these data suggest that the expression of ERGO-1 26G-RNAs is required for the RRF-1-dependent biogenesis of 22G-RNAs at these loci.
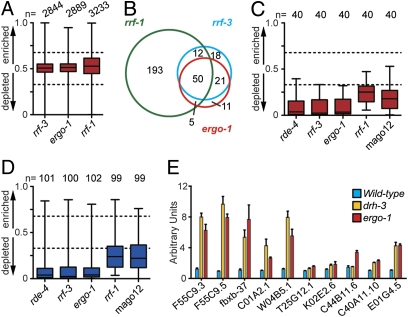
To examine the requirements of RRF-3, ERGO-1, and RRF-1 for 22G-RNA biogenesis on a genome scale, small RNAs were cloned and deep sequenced from rrf-3, ergo-1, and rrf-1 mutants. Compared to our wild-type dataset (9), the overall distribution of small RNA classes was largely unaffected in each mutant (Fig. S5) and 22G-RNAs targeting annotated genes were unaffected as a whole (Fig. 5A). However, ergo-1 and rrf-3 mutants were depleted of 22G-RNAs targeting 87 and 101 genes, respectively, and were highly overlapping with 71 genes in common (Fig. 5B and Table S4).
Fig. 5.
The biogenesis of 22G-RNAs targeting 26G-RNA loci is dependent on the ERI pathway. (A) Overall enrichment or depletion of 22G-RNAs targeting n genes in each mutant, as described in Fig. 1C. (B) Venn comparison of genes depleted of 22G-RNAs (≥2-fold) in indicated mutants (loci below the lower dashed line in A). (C) Enrichment or depletion of 22G-RNAs derived from n 26G target genes, as described in Fig. 1C. (D) Enrichment or depletion of 22G-RNAs derived from n 26G nonannotated clusters, as described in Fig. 1C. (E) qRT-PCR analysis of 26G target genes expression in drh-3 (yellow) and ergo-1 (red) mutants and wild type (blue). Error bars represent the standard deviation of the mean.
Within the set of 48 annotated genes targeted by 26G-RNAs, only 40 exhibited 22G-RNA levels that satisfied the rigorous criteria of 25 rpm in wild-type or mutant samples. All 40 of these genes were depleted of 22G-RNAs in both rrf-3 and ergo-1 mutants (Fig. 5C). Of the ∼100 nonannotated clusters that satisfy the 25-rpm cutoff, 90 were depleted of 22G-RNAs in both ergo-1 and rrf-3 mutants (Fig. 5D). Consistent with our Northern data demonstrating a role for RDE-4 in 26G-RNA biogenesis, virtually all of the 26G-RNA loci (38 annotated genes and 92 nonannotated clusters) were depleted of 22G-RNAs in an rde-4 mutant RNA sample (Fig. 5 C and D) (9). Together, these data demonstrate that the ERI endo-siRNA pathway is required for the expression of both 26G-RNAs and 22G-RNAs at these ERGO-1 target loci.
RRF-1 and WAGOs Are Required for 22G-RNAs in the ERI Pathway.
In the rrf-1 mutant, ∼260 genes were depleted of 22G-RNAs (Table S4), suggesting that RRF-1 is more broadly required for 22G-RNA biogenesis. Of the 71 loci depleted of 22G-RNAs in both rrf-3 and ergo-1 mutants, 62% were also RRF-1 dependent (Fig. 5B). More significantly, 31 (78%) 26G-RNA target genes were depleted of 22G-RNAs in the rrf-1 mutant dataset, all of which were depleted of 22G-RNAs in rrf-3 and ergo-1 mutants (Fig. 5C). Of the genes that were not depleted of 22G-RNAs in the rrf-1 mutant and that satisfy our 22G-RNA criteria for analysis, all but one were dependent on the RdRP EGO-1 (10). The remaining target was not depleted of 22G-RNAs in either rrf-1 or ego-1, suggesting that RRF-1 and EGO-1 are redundant for 22G-RNAs targeting this gene. In addition, 76 nonannotated clusters were depleted of 22G-RNAs in the rrf-1 mutant (Fig. 5D), of which 74 were also dependent on rrf-3, ergo-1, and rde-4.
Finally, we examined deep-sequencing data from a mutant bearing deletions in all 12 wago genes, MAGO12 (9). 22G-RNAs targeting 35 (88%) 26G-RNA target genes and 71 nonannotated clusters were depleted in the MAGO12 mutant (Fig. 5 C and D). Thus, the biogenesis of 22G-RNAs at ERGO-1 target loci is dependent on rrf-1 and WAGOs.
WAGO/22G-RNA pathways have been shown to silence their targets (9), and previous reports have indicated that the ERI pathway is an endogenous-silencing pathway (12, 18). To look for silencing of the ERI targets, we used quantitative PCR after reverse transcription (qRT-PCR). Indeed, ERGO-1 26G-RNA targets were up-regulated in drh-3 and ergo-1 mutants (Fig. 5E), both of which are required for the expression of 22G-RNAs targeting these loci (Figs. 4 and 5) (9).
Discussion
Here, we have analyzed the genetic and small-RNA components of the ERI endogenous-RNAi pathway. We have identified Dicer-dependent 26-nt RNA species (26G-RNAs) as cofactors of the ERI pathway Argonaute ERGO-1. Components of the ERI complex, including DCR-1, the RdRP RRF-3, and the dsRNA-binding protein RDE-4, are required for the biogenesis of both 26G-RNAs and 22G-RNAs on ERI targets. A second RdRP, RRF-1, and additional WAGO Argonautes are required for the accumulation of 22G-RNAs (but not 26G-RNAs). These findings support a two-step model in which 26G-RNAs function upstream in the ERI pathway and drive the downstream production of 22G-RNAs (Fig. 6).
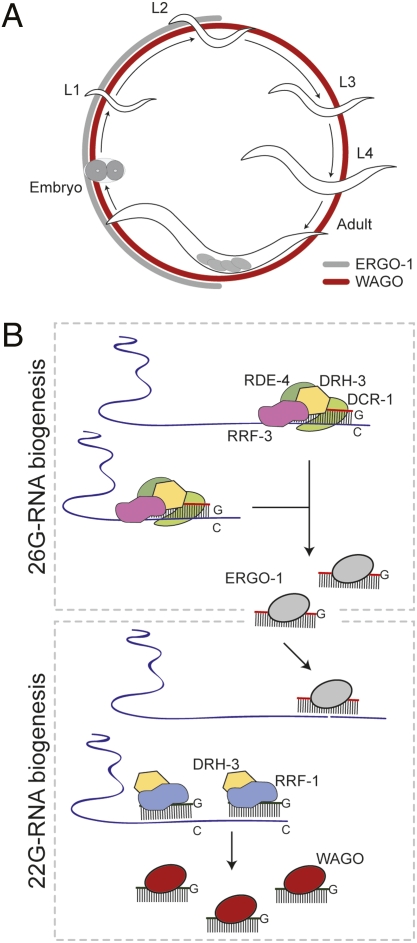
Fig. 6.
Model for 26G-/22G-RNA biogenesis. (A) ERGO-1/26G-RNA (gray) and WAGO/22G-RNA (red) expression during the C. elegans life cycle: embryo, L1–L4 larval stages, and adult with embryos. (B) Processive 26G-RNA biogenesis by the ERI complex and ERGO-1 loading. Targeting by ERGO-1 results in recruitment of the 22G-RNA RdRP machinery and WAGO loading.
During embryogenesis, 26G-RNAs drive the biogenesis of 22G-RNAs that persist into later developmental stages, when ERGO-1/26G-RNAs are present at very low levels (Fig. 6). Thus, 22G-RNA expression may function to maintain silencing through a self-sustaining amplification loop in the absence of further 26G-RNA expression. This possibility is also consistent with the long-lasting silencing observed in response to exogenous dsRNA (19), which involves a distinct upstream AGO but shares with the ERI pathway the RRF-1-dependent secondary 22G-RNA/WAGO pathway (9, 13).
In some cases, ERI pathway-dependent 22G-RNAs were derived from loci that were not significantly targeted by 26G-RNAs. For example, the well-characterized X-cluster is an ERI-dependent 22G-RNA locus (9, 12, 20, 21), but is not targeted by 26G-RNAs above the 10-rpm cutoff. However, the X-cluster shares a region of significant nucleotide identity with a 26G-RNA-producing locus on chromosome 1 [linkage group (LG)I]. The region of identity in the LGI locus is targeted almost exclusively by 22G-RNAs. These and other similar findings are consistent with a transitive biogenesis of 22G-RNAs downstream of 26G-RNA targeting (22).
The dependence of 26G-RNAs on components of the DCR-1/ERI complex is consistent with a concerted mechanism of biogenesis (12, 23, 24). RNA duplexes generated by RRF-3 could be processed by DCR-1 to generate duplex siRNAs that are loaded into ERGO-1. However, several observations are not in agreement with 26G-RNA biogenesis via a direct, DCR-1-mediated cleavage of RRF-3-generated RNA duplexes. As noted by Ruby et al. (15), the 5′G bias of 26G-RNAs is more consistent with direct synthesis by RdRP and does not fit well with the thermodynamic rules thought to govern the loading of Dicer products into AGO complexes (25, 26). In addition, although sense small RNAs were cloned from 26G-RNA loci, their length and position with respect to the corresponding 26G-RNAs are not consistent with Dicer processing. Although some phasing of 26G-RNAs was apparent (when the most abundant species were considered), for most loci we observed a highly overlapping distribution of 26G-RNAs. Rather than the phasing one might expect from Dicer-mediated processing, these findings are most consistent with cycles of 26-mer synthesis by RdRP initiating at multiple sites along the target mRNA. Further biochemical and genetic analyses will be necessary to understand the role of Dicer in the ERI pathway.
ergo-1 mutants are viable and exhibit no overt phenotypes other than an enhanced sensitivity to exogenous RNAi. Indeed, ergo-1 mutants do not display the Him or male-specific, temperature-sensitive sterile phenotypes associated with other Eri mutants (18). Instead, these male-specific functions depend on spermatogenesis-expressed 26G-RNAs that engage two partially redundant AGOs, ALG-3 and ALG-4 (27). ergo-1 may function to regulate the exo-RNAi pathway in somatic tissues and/or may have other as yet undetected biological functions. Most ERGO-1 targets are not recognizable as genes and often reside in clusters of what appear to represent ancient duplications. Therefore, it is conceivable that the ERGO-1 pathway may function to buffer against deleterious affects arising from expression of these duplicated noncoding sequences. Whereas the specific biological function of the ERGO-1 pathway remains unclear, it provides a striking example of interdependence and competition between AGO systems and points to the complexity and rapidly evolving nature of AGO/small-RNA networks.
Methods
Worm Culture and Strains.
C. elegans culture and genetics were essentially as described (28). The Bristol N2 strain was used as the wild-type control. Alleles used are listed by chromosome: LGI, sago-2(tm894), ppw-1(tm914), ppw-2(tm1120), avr-14(ad1302), rrf-1(pk1417), ego-1(om97), hT2[qIs48](I;III), C04F12.1(tm1637), rrf-2(pk2040); LGII, rrf-3(pk1426), C06A1.4(tm887), F58G1.1(tm1019); LGIII, dcr-1(mg375Eri), rde-4(ne337); LGIV, M03D4.6(tm1144); and LGV, ergo-1(tm1860), sago-1(tm1195), rde-1(ne300), avr-15(ad1051), glc-1(pk54).
Generation of ERGO-1 Antibodies.
A C-terminal ERGO-1-specific peptide (CEVNKDMNVNEKLEGMTFV) was coupled to KLH and used to immunize four rabbits (Capralogics).
ERGO-1 Immunoprecipitation.
ERGO-1 antiserum (∼500 μL) was incubated with ∼50 mg of embryo lysate. Immune complexes were precipitated with Protein A Sepharose beads (GE Healthcare) and washed with cold lysis buffer. RNA was extracted from the immune complexes using TRI Reagent (MRC). See SI Text for additional details.
Western Blot Analysis.
Proteins immobilized on Hybond-C Extra membrane (GE Healthcare) were probed with anti-ERGO-1 (1:3,000) and anti-tubulin (1:5,000). HRP-conjugated secondary antibodies were used at 1:5,000. See SI Text for additional details.
Small RNA Purification and Cloning.
Extraction of total RNA and enrichment for small RNA < ∼200 nt were as described (9).
Oxidation of Small RNA.
Small RNA (∼20 μg) was oxidized using 0.2 M NaIO4 (periodate) in 60 μL of 0.3 M borate buffer (pH 8.6) for 10 min at room temperature. Excess NaIO4 was destroyed by adding 2 μL of glycerol and incubating for 10 min at room temperature. Oxidized RNA was desalted using a Bio-spin 6 column (Bio-Rad) and precipitated with 4 vol of ethanol.
Small RNA Cloning and Sequencing.
RNAs (18 to 30 nt) were gel purified and cloned using a 5′ ligation-dependent protocol as described (9). Some small RNA samples were pretreated with calf-intestine phosphatase and polynucleotide kinase (rrf-3, ergo-1, and rrf-1) or tobacco acid pyrophosphatase (β-elimination and input) to make the 5′ ends of 22G-RNAs available for ligation (9). cDNA libraries were sequenced by the University of Massachusetts (Worcester, MA) Deep Sequencing Core, using an Illumina Genome Analyzer II.
Data Analysis.
Small RNA sequences were processed and mapped to the C. elegans genome (Wormbase release WS192) as well as Repbase (13.07), using custom Perl scripts (Perl 5.8.6) as described (9). Clusters were generated from 26G-RNAs that matched nonannotated genome sequences, using the ERGO-1 IP dataset. Details are provided in SI Text.
Northern Blot Analysis.
Small RNA Northern blots were performed as described (9). Starfire probe sequences are provided in Table S5.
Real-Time PCR.
qRT-PCR was performed as described (9). cDNA was generated using 5 μg of total RNA, random hexamers, and SuperScript III Reverse Transcriptase (Invitrogen). The expression level of each target RNA was normalized to gpd-2 or act-3. Primer sequences are provided in Table S5.
Supplementary Material
Acknowledgments
The authors thank members of the Mello Laboratory for helpful comments on the manuscript, E. Kittler and the University of Massachusetts Deep Sequencing Core for processing Illumina samples, S. Kennedy for sharing the dcr-1(mg375Eri) allele, and the Caenorhabditis Genetics Center for providing strains. P.J.B. is supported by SFRH/BD/11803/2003 from Fundação para Ciência e Tecnologia, Portugal. J.M.C. is a Howard Hughes Medical Institute fellow of the Life Sciences Research Foundation. E.M.Y. is a Damon Runyon Fellow supported by the Damon Runyon Cancer Research Foundation (DRG-1983-08). C.C.M. is a Howard Hughes Medical Institute Investigator. This work was supported by the Canadian Institute of Health Research and Fonds de la Recherche en Santé du Québec (T.F.D.), by Ruth L. Kirschstein National Research Service Award GM63348 (to D.C.), and by Grant R01GM58800 (to C.C.M.) from the National Institute of General Medical Sciences.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession nos. GSE18714).
This article contains supporting information online at www.pnas.org/cgi/content/full/0911908107/DCSupplemental.
References
- 1.Chapman EJ, Carrington JC. Specialization and evolution of endogenous small RNA pathways. Nat Rev Genet. 2007;8:884–896. doi: 10.1038/nrg2179. [DOI] [PubMed] [Google Scholar]
- 2.Ghildiyal M, Zamore PD. Small silencing RNAs: An expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grishok A, et al. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- 4.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 5.Hutvágner G, et al. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 6.Aoki K, Moriguchi H, Yoshioka T, Okawa K, Tabara H. In vitro analyses of the production and activity of secondary small interfering RNAs in C. elegans. . EMBO J. 2007;26:5007–5019. doi: 10.1038/sj.emboj.7601910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sijen T, Steiner FA, Thijssen KL, Plasterk RH. Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science. 2007;315:244–247. doi: 10.1126/science.1136699. [DOI] [PubMed] [Google Scholar]
- 8.Pak J, Fire A. Distinct populations of primary and secondary effectors during RNAi in C. elegans . Science. 2007;315:241–244. doi: 10.1126/science.1132839. [DOI] [PubMed] [Google Scholar]
- 9.Gu W, et al. Distinct Argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Mol Cell. 2009;36:231–244. doi: 10.1016/j.molcel.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Claycomb JM, et al. The Argonaute CSR-1 and its 22G-RNA cofactors are required for holocentric chromosome segregation. Cell. 2009;139:123–134. doi: 10.1016/j.cell.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kennedy S, Wang D, Ruvkun G. A conserved siRNA-degrading RNase negatively regulates RNA interference in C. elegans . Nature. 2004;427:645–649. doi: 10.1038/nature02302. [DOI] [PubMed] [Google Scholar]
- 12.Duchaine TF, et al. Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell. 2006;124:343–354. doi: 10.1016/j.cell.2005.11.036. [DOI] [PubMed] [Google Scholar]
- 13.Yigit E, et al. Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell. 2006;127:747–757. doi: 10.1016/j.cell.2006.09.033. [DOI] [PubMed] [Google Scholar]
- 14.Simmer F, et al. Loss of the putative RNA-directed RNA polymerase RRF-3 makes C. elegans hypersensitive to RNAi. Curr Biol. 2002;12:1317–1319. doi: 10.1016/s0960-9822(02)01041-2. [DOI] [PubMed] [Google Scholar]
- 15.Ruby JG, et al. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans . Cell. 2006;127:1193–1207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- 16.Batista PJ, et al. PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans . Mol Cell. 2008;31:67–78. doi: 10.1016/j.molcel.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stoeckius M, et al. Large-scale sorting of C. elegans embryos reveals the dynamics of small RNA expression. Nat Methods. 2009;6:745–751. doi: 10.1038/nmeth.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pavelec DM, Lachowiec J, Duchaine TF, Smith HE, Kennedy S. Requirement for the ERI/DICER complex in endogenous RNA interference and sperm development in Caenorhabditis elegans . Genetics. 2009;183:1283–1295. doi: 10.1534/genetics.109.108134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grishok A, Tabara H, Mello CC. Genetic requirements for inheritance of RNAi in C. elegans . Science. 2000;287:2494–2497. doi: 10.1126/science.287.5462.2494. [DOI] [PubMed] [Google Scholar]
- 20.Ambros V, Lee RC, Lavanway A, Williams PT, Jewell D. MicroRNAs and other tiny endogenous RNAs in C. elegans . Curr Biol. 2003;13:807–818. doi: 10.1016/s0960-9822(03)00287-2. [DOI] [PubMed] [Google Scholar]
- 21.Lee RC, Hammell CM, Ambros V. Interacting endogenous and exogenous RNAi pathways in Caenorhabditis elegans . RNA. 2006;12:589–597. doi: 10.1261/rna.2231506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sijen T, et al. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell. 2001;107:465–476. doi: 10.1016/s0092-8674(01)00576-1. [DOI] [PubMed] [Google Scholar]
- 23.Colmenares SU, Buker SM, Buhler M, Dlakić M, Moazed D. Coupling of double-stranded RNA synthesis and siRNA generation in fission yeast RNAi. Mol Cell. 2007;27:449–461. doi: 10.1016/j.molcel.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 24.Lee SR, Collins K. Physical and functional coupling of RNA-dependent RNA polymerase and Dicer in the biogenesis of endogenous siRNAs. Nat Struct Mol Biol. 2007;14:604–610. doi: 10.1038/nsmb1262. [DOI] [PubMed] [Google Scholar]
- 25.Schwarz DS, et al. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 26.Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 27.Conine C, et al. The Argonautes ALG-3 and ALG-4 are required for spermatogenesis-specific 26G-RNAs and for thermotolerant sperm in C. elegans . Proc Natl Acad Scil. 2010 doi: 10.1073/pnas.0911685107. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brenner S. The genetics of Caenorhabditis elegans . Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.