Abstract
The type I lissencephaly gene product LIS1, a key regulator of cytoplasmic dynein, is critical for cell proliferation, survival, and neuronal migration. However, little is known about the regulation of LIS1. Here, we identify a previously uncharacterized mammalian homolog of Aspergillus NudC, NudCL2 (NudC-like protein 2), as a regulator of LIS1. NudCL2 is localized to the centrosome in interphase, and spindle poles and kinetochores during mitosis, a pattern similar to the localization of LIS1 and cytoplasmic dynein. Depletion of NudCL2 destabilized LIS1 and led to phenotypes resembling those of either dynein or LIS1 deficiency. NudCL2 complexed with and enhanced the interaction between LIS1 and Hsp90. Either disruption of the LIS1-Hsp90 interaction with the C terminus of NudCL2 or inhibition of Hsp90 chaperone function by geldanamycin decreased LIS1 stability. Thus, our results suggest that NudCL2 regulates the LIS1/dynein pathway by stabilizing LIS1 with Hsp90 chaperone. This represents a hitherto undescribed mechanism of the LIS1/dynein regulation in mammalian cells.
Keywords: cochaperone, heat shock protein 90, lissencephaly 1, migration, p23
L is1 was identified as a causative gene for classic lissencephaly, a developmental brain abnormality characterized by defects in neuronal positioning (1). LIS1 has been reported to be required for neuronal migration and cell proliferation and survival (2– 4). LIS1 dynamically colocalizes with cytoplasmic dynein at centrosome, mitotic spindles, kinetochores, and cell cortex to execute various biological processes (5– 9). LIS1 directly binds to multiple sites within dynein heavy chain, including the stem domain and the AAA1 domain (ATPase associated with diverse cellular activities), which is the site for ATP hydrolysis (9). Purified recombinant LIS1 is shown to increase the microtubule-stimulated ATPase activity of the dynein motor in vitro (10). Recent studies indicate that LIS1 is able to suppress the motility of dynein on microtubules (11). These results clearly suggest that LIS1 serves as a key regulator of the cytoplasmic dynein complex; however, the regulation of LIS1 itself remains largely unknown.
A number of studies in the filamentous fungus Aspergillus nidulans have demonstrated that the genes in the nuclear distribution (nud) pathway are homologs to components or regulators of the cytoplasmic dynein/dynactin complex (12). The NudA, NudG, and NudI genes encode cytoplasmic dynein heavy, light, and intermediate chain, respectively (12). The proteins encoded by nudM and nudK are p150Glued and actin-related protein 1, components of the dynactin complex (12, 13). The NudF gene encodes NudF protein, an Aspergillus ortholog of LIS1 (14).
In A. nidulans, a nudC mutation greatly reduces the protein level of NudF at the nonpermissive temperature, and extra copies of the NudF gene can suppress the nudC mutation (14). Moreover, all of the suppressors of nudC mutation reverse its temperature-sensitive phenotype by restoring the protein level of NudF (15). These data indicate that nudC may be upstream of nudF in A. nidulans. In mammals, we and other groups have found that NudC associates with LIS1 and the cytoplasmic dynein complex and influences the dynein’s function (16, 17); however, there is no report supporting a direct role of NudC in the regulation of LIS1. Recently, we showed that a mammalian NudC-like protein, NudCL, is required specifically for the stabilization of dynein intermediate chain, but not LIS1 (18). Thus, whether a NudC homolog could be responsible for the regulation of LIS1 in mammals is not yet clear.
In this communication, we identified a previously undescribed mammalian homolog of Aspergillus NudC, NudCL2 (NudC-like protein 2), which shares significant homology with human NudC and NudCL. NudCL2 appears to be a regulator of LIS1 and influences the LIS1/dynein pathway by stabilizing LIS1 with Hsp90 chaperone, which represents another mechanism of the LIS1/dynein regulation in mammals.
Results
Identification of NudCL2.
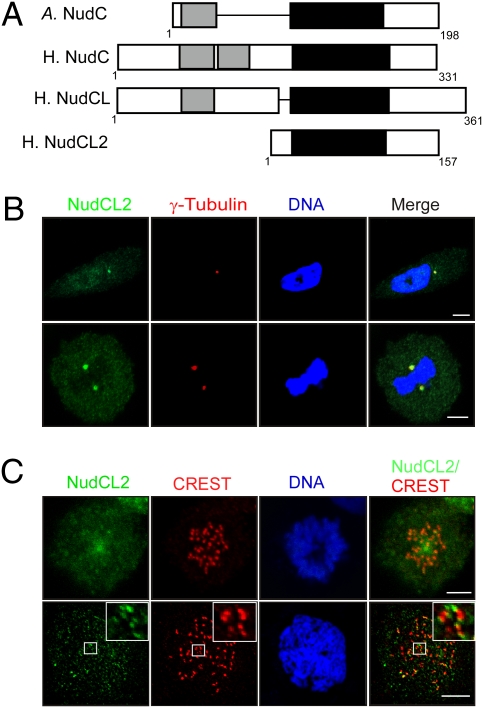
To discover other regulators of LIS1, we used tBLASTn program to search unidentified mammalian homologs of Aspergillus nudC and detected a previously uncharacterized human sequence (GenBank release no. NM_145266) homologous to Aspergillus nudC. We used RT-PCR to clone this cDNA with total RNA isolated from HeLa cells (Fig. S1A). Sequencing analyses showed that the cDNA encoded a deduced protein of 157 amino acid residues (Fig. S1B). This protein shares significant homology with Aspergillus NudC, human NudC, and NudCL (Fig. S1C); therefore, we designated it NudCL2. Bioinformatics analysis revealed that human NudCL2 contained a conserved p23_NudC_like domain (cd06467, the conserved domain database of National Center for Biotechnology Information), which is also present in human NudC, NudCL, and Aspergillus NudC (Fig. 1A). BLAST searches for proteins homologous to human NudCL2 in other species showed that putative orthologs of NudCL2 are highly conserved in bovine, chicken, chimpanzee, mouse, rat, zebrafish, and human (Fig. S1D).
Fig. 1.
Characterization of NudCL2. (A) Schematic comparison of human NudCL2, NudCL, and NudC, and Aspergillus NudC. Light gray bars indicate coiled-coil domains and black filled bars show p23_NudC_like domains. (A) Aspergillus; H., Human. (B and C) Subcellular localization of NudCL2. HeLa cells grown on cover slides were subjected to immunofluorescence analyses with the indicated antibodies. DNA was visualized by DAPI. (Scale bar, 10 μm.) The centrosome and mitotic spindle poles were determined by staining with anti-γ tubulin antibody (B). The mitotic kinetochores were identified by immunofluorescence with CREST autoimmune serum (C). A mitotic chromosome spread was performed to confirm the kinetochore localization of NudCL2 (Lower). High magnifications are shown in Insets.
To explore the expression pattern of NudCL2, we raised two rabbit polyclonal antibodies against purified NudCL2 protein (anti-NudCL2 antibody) and a synthetic peptide of NudCL2 (anti-NudCL2 peptide antibody), respectively (Figs. S2A and S3A). After affinity purification, each of them specifically recognized NudCL2 protein (Figs. S2B and S3B). Immunofluorescence analysis revealed that NudCL2 was clearly colocalized with γ-tubulin in addition to punctate distribution throughout the cell in interphase (Fig. 1B, Upper), indicating that NudCL2 localizes to the centrosome. During M phase, NudCL2 was accumulated at spindle poles (Fig. 1B, Lower) and kinetochores that were stained with CREST autoimmune serum (Fig. 1C, Upper). To confirm the kinetochore localization of NudCL2, we used the mitotic chromosome spread method and found that NudCL2 was detected at the outer region of the kinetochores relative to the signal of CREST, which is known to recognize the inner region of the kinetochores (Fig. 1C, Lower). These data suggest that NudCL2 is localized to the centrosome in interphase, spindle poles, and kinetochores during mitosis, a pattern similar to the localization of LIS1 and cytoplasmic dynein (5, 6, 8, 19–21).
Down-Regulation of LIS1 in NudCL2-Depleted Cells.
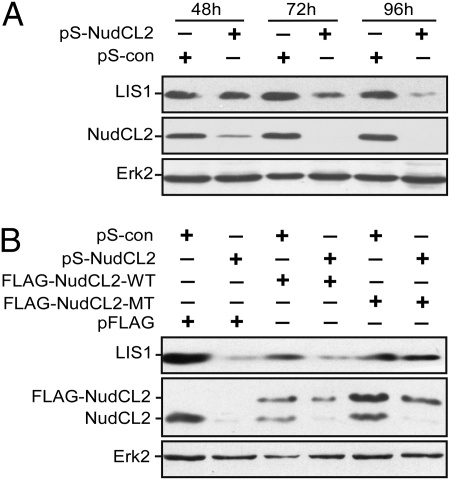
In Aspergillus, nudC has been reported to influence the protein level of NudF, an Aspergillus ortholog of LIS1 (14). Here, we determined whether NudCL2, a mammalian homolog of Aspergillus NudC, affects the stability of LIS1 in mammals. Vector-based RNAi was used to deplete endogenous NudCL2 by constructing an RNAi vector, pSilencer-NudCL2 (pS-NudCL2). Western blotting showed that the level of NudCL2 in cells transfected with pS-NudCL2 was greatly reduced from 48 h to 96 h posttransfection compared to that transfected with pSilencer vector (pS-con), whereas the level of extracellular signal-regulated kinase 2 (ERK2) remained unchanged (Fig. 2A). Meanwhile, NudCL2 depletion resulted in a significant down-regulation of LIS1 from 72 h to 96 h posttransfection (Fig. 2A). To exclude the potential off-target effects of NudCL2 RNAi, we performed an RNAi rescue experiment and found that ectopic expression of RNAi-resistant NudCL2 efficiently restored the protein levels of LIS1 (Fig. 2B). Taken together, these data suggest that NudCL2 may play a role in regulating the stability of LIS1.
Fig. 2.
Down-regulation of LIS1 in cells depleted of NudCL2. HeLa cells were cotransfected with the indicated vectors and pBABE-puro. At 24 h after transfection, puromycin was added to enrich the transfection-positive cells. (A) NudCL2 depletion leads to reduced LIS1. The cells harvested at various times were subjected to immunoblotting analyses. (B) Ectopic expression of RNAi-resistant NudCL2 partially reverses the down-regulation of LIS1 caused by NudCL2 depletion. RNAi-resistant NudCL2 mutant vector (pFLAG-NudCL2-MT) was constructed by the silent mutation of three nucleic acids in the RNAi targeting region of NudCL2. At 72 h posttransfection, the cells were subjected to Western analysis. Erk2 was used as a loading control.
NudCL2 Acts as a Regulator of LIS1 in the LIS1/Dynein Pathway.
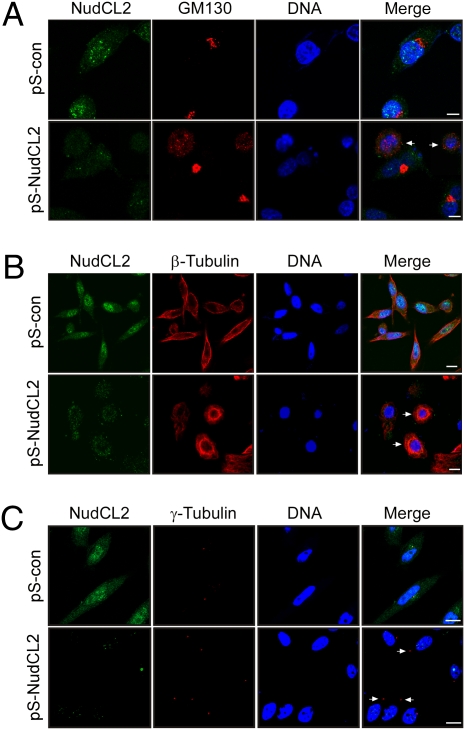
To elucidate the function of NudCL2, we investigated cellular phenotypes in NudCL2-depleted cells using immunofluorescence analysis. The results showed that depletion of NudCL2 significantly resulted in the dispersion of Golgi apparatus (Fig. 3A and Fig. S4A), which was often found in cells with defects in LIS1 and cytoplasmic dynein (8, 22). Emerging studies have demonstrated that down-regulation of either LIS1 or dynein induces perinuclear accumulation of microtubules and overexpression of LIS1 leads to peripheral accumulation of microtubules (7, 8, 23). Here, we found that NudCL2 depletion caused a marked perinuclear accumulation of microtubules (Fig. 3B and Fig. S4B). Both LIS1 and dynein were reported to be required for the coupling of the centrosome and nucleus (7, 8, 23, 24). The distance between the centrosome and nucleus in more than 90% of HeLa cells is less than 3 μm (Fig. S5). However, the cells with enlarged distance between the centrosome and nucleus (>3 μm) was significantly increased in NudCL2-depleted cells as compared with that in control cells (Fig. 3C and Fig. S4C), indicating defects in the coupling of the centrosome and nucleus in NudCL2-depleted cells. To eliminate the possibility of NudCL2 depletion-induced phenotypes by off-target effects of RNAi, we designed another NudCL2 RNAi vector (pS-NudCL2-1) with a targeting region of NudCL2 mRNA different from that of pS-NudCL2. The results from depletion of NudCL2 with pS-NudCL2-1 showed similar phenotypes to that with pS-NudCL2 (Fig. S6). These data clearly reveal that NudCL2 depletion results in the phenotypes that are frequently detected in cells with the deficiency of either LIS1 or dynein, implying that NudCL2 may regulate the function of the LIS1/dynein pathway.
Fig. 3.
Cellular phenotypes induced by depletion of NudCL2. HeLa cells were transfected with either pS-con or pS-NudCL2 and pBABE-puro. At 24 h after transfection, puromycin was added to select the transfection-positive cells. The cells were subjected to immunofluorescence analyses with the indicated antibodies at 72 h posttransfection. DNA was stained with DAPI. (Scale bar, 10 μm.) (A) The cells indicated with arrows show the dispersion of Golgi apparatus visualized by staining with anti-GM130 antibody. (B) The perinuclear microtubule enrichment is detected in cells with arrows. (C) The cells with arrows exhibit a significant increase in the distance between the centrosome and nucleus (>3 μm).
To further confirm whether NudCL2 regulates LIS1 function, we performed the reciprocal rescue assay in NudCL2-depleted cells by overexpression of LIS1 or in LIS1-depleted cells by ectopic expression of NudCL2 (Fig. S7). The data showed that overexpression of LIS1 partially reversed the phenotypes of NudCL2-depleted cells, including dispersion of Golgi apparatus, perinuclear accumulation of microtubules, and uncoupling of the centrosome and nucleus. However, ectopic expression of NudCL2 in LIS1-depleted cells failed to counteract the phenotypes caused by LIS1 knockdown. These results indicate that NudCL2 may be an upstream regulator of LIS1 in the LIS1/dynein pathway.
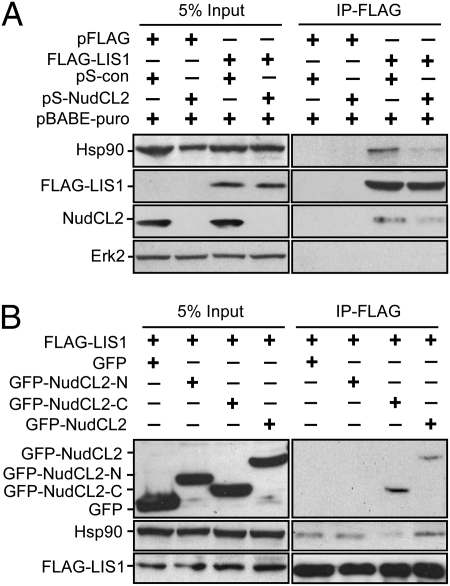
NudCL2 Forms a Complex with LIS1 and Hsp90.
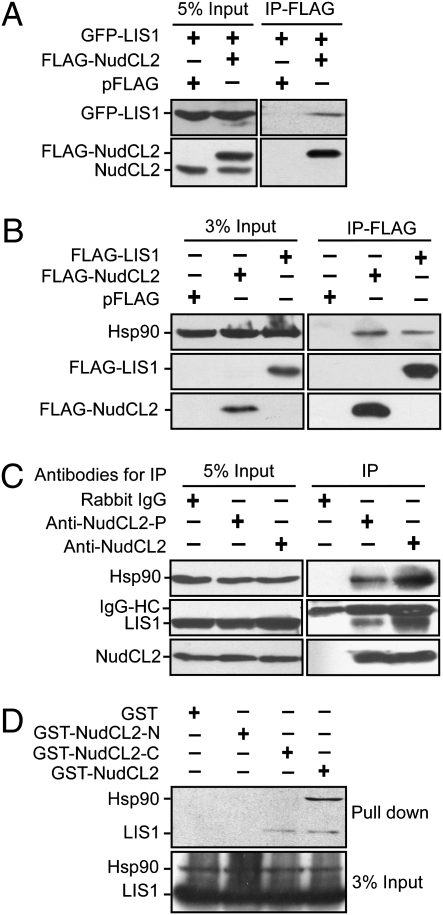
To explore the mechanism of LIS1 regulation by NudCL2, we tested whether NudCL2 binds to LIS1. GST pull-down assays revealed that purified NudCL2 associated with LIS1 in vitro (Fig. S8A). To examine the interaction between NudCL2 and LIS1 in vivo, we cotransfected pFLAG-NudCL2 and pGFP-LIS1 vectors into HeLa cells. The proteins immunoprecipitated with anti-FLAG antibody-coupled beads were subjected to Western analysis and the data showed that NudCL2 associated with LIS1 in vivo (Fig. 4A). Further data revealed that endogenous NudCL2 was able to bind to endogenous LIS1 in mammalian cells (Fig. 4C).
Fig. 4.
The interaction among NudCL2, LIS1 and Hsp90. (A and B) HeLa cells transfected with the indicated vectors were subjected to coimmunoprecipitation analyses with anti-FLAG antibody-coupled beads at 48 h posttransfection. Immunoblot analyses were performed with the antibodies as shown. NudCL2 was found to bind to LIS1 in vivo (A). Both NudCL2 and LIS1 coimmunoprecipitated with endogenous Hsp90 in vivo (B). (C) Three aliquots of lysates from HeLa cells were subjected to immunoprecipitation experiments with anti-rabbit IgG, anti-NudCL2 antibody (anti-NudCL2) and anti-NudCL2 peptide antibody (anti-NudCL2-P), respectively. Immunoprecipitated proteins were then analyzed by Western blotting with the indicated antibodies. IgG-HC, heavy chain of IgG. (D) The association of NudCL2 fragments with Hsp90 and LIS1 in vitro. Purified GST, GST-NudCL2-N, GST-NudCL2-C, and GST-NudCL2 proteins were incubated with the lysates of HeLa cells, respectively. Either LIS1 or Hsp90 that interacts with NudCL2 fragments was detected by Western blotting with the antibodies as shown.
Because NudCL2 has a conserved p23_like domain that may be associated with molecular chaperone Hsp90 (25, 26), we investigated if NudCL2 associate with Hsp90. GST pull-down and immunoprecipitation experiments with anti-FLAG, NudCL2 or NudCL2-peptide antibodies revealed that NudCL2 interacted with Hsp90 in vitro and in vivo (Fig. S8B and Fig. 4 B–C). To identify which domain of NudCL2 is responsible for binding to LIS1 and Hsp90, we purified fragments of NudCL2 protein, GST-NudCL2-N (N terminus of NudCL2, 1–108aa) that contains the p23_like domain (9–101aa), and GST-NudCL2-C (C terminus of NudCL2, 109–157aa). GST pull-down assays showed that NudCL2-C, but not NudCL2-N, associated with LIS1 in vitro (Fig. 4D). Immunoprecipitation experiment confirmed the interaction between NudCL2-C with LIS1 in vivo (Fig. 5B). Furthermore, only full-length NudCL2, but not N or C terminus of NudCL2, was able to bind to Hsp90 (Fig. 4D), suggesting that the p23_like domain in NudCL2 may be essential, but not sufficient, for binding to Hsp90. Because NudCL2 associates with LIS1 and Hsp90, we were interested in the association of LIS1 with Hsp90. The data revealed that LIS1 interacted with Hsp90 in vitro (Fig. S8C) and in vivo (Figs. 4B and 5). Further results showed that endogenous LIS1 was able to interact with endogenous Hsp90 and NudCL2 (Fig. S9). Taken together, these results suggest that NudCL2, LIS1, and Hsp90 may form a biochemical complex in mammalian cells.
Fig. 5.
NudCL2 regulates the interaction between LIS1 and Hsp90 in vivo. HeLa cells were transfected with the indicated vectors and subjected to coimmunoprecipitation analysis with anti-FLAG antibody-coupled beads at 72 h posttransfection. Western analyses were performed with the antibodies as shown. (A) Depletion of NudCL2 inhibits the association of LIS1 with Hsp90 in vivo. At 24 h after transfection, puromycin was added to enrich the transfection-positive cells. (B) Overexpression of the C terminus of NudCL2 that associates with LIS1 interferes with the interaction between Hsp90 and LIS1 in vivo.
NudCL2 Facilitates the Interaction Between Hsp90 and LIS1.
Considering some proteins containing the p23_like domain may enhance the binding of Hsp90 to its client proteins (27), we determined whether NudCL2 increases the association of Hsp90 with LIS1. Depletion of NudCL2 significantly decreased the interaction between LIS1 and Hsp90 in vivo (Fig. 5A). Furthermore, overexpression of NudCL2-C that binds to LIS1, but not Hsp90 (Fig. 4D), obviously inhibited the association of Hsp90 with LIS1 in vivo (Fig. 5B). The above data suggest that NudCL2 may be important for the interaction between Hsp90 with LIS1.
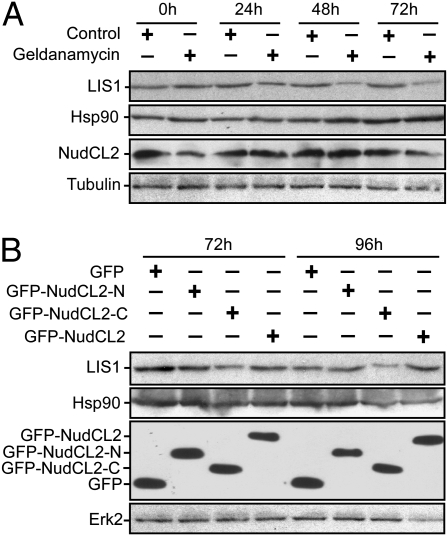
NudCL2 Regulates LIS1 Stability with Hsp90.
For the interaction between Hsp90 chaperone and LIS1, we investigated whether Hsp90 chaperoning function plays a role in the regulation of LIS1. Inhibition of Hsp90 function by geldanamycin, a specific inhibitor of Hsp90 ATPase activity (28), significantly reduced the protein level of LIS1 (Fig. 6A), indicating that Hsp90 may influence LIS1 stability by its chaperoning function.
Fig. 6.
Hsp90 is involved in the regulation of LIS1 stability by NudCL2. (A) Inhibition of Hsp90 chaperone function reduces LIS1 stability. HeLa cells treated with geldanamycin were collected at the indicated times and subjected to immunoblotting analyses with the antibodies as shown. Tubulin was used as a loading control. (B) Overexpression of the C terminus of NudCL2 that inhibits the interaction between Hsp90 and LIS1 decreases the stability of LIS1. HeLa cells transfected with the indicated vectors were harvested at various times and subjected to Western analyses. Erk2 was used as a loading control.
Because NudCL2 is important for LIS1 stability and promotes the interaction between Hsp90 and LIS1, we assess the possibility that NudCL2 may modulate LIS1 stability by facilitating the association of Hsp90 with LIS1. The data showed that ectopic expression of NudCL2-C that interferes with the interaction between LIS1 and Hsp90, was able to decrease the level of LIS1 (Fig. 6B), suggesting that NudCL2 may stabilize LIS1 with Hsp90 chaperone by facilitating their interaction.
Discussion
In this study, we have identified NudCL2 as a regulator of LIS1 and found that NudCL2 forms a complex with LIS1 and Hsp90. NudCL2 modulates the LIS1/dynein pathway by stabilizing LIS1 with Hsp90 chaperone, which represents a previously uncharacterized mechanism of LIS1 regulation.
Regulation of LIS1.
Even though the nud pathway in A. nidulans are evolutionarily conserved with the LIS1/dynein pathway in vertebrates, the functions of mammalian homologs of the nud genes have evolved to be more complicated and diverse. For example, nudE was identified as a multicopy suppressor of a mutation in the NudF gene in A. nidulans (29), whereas the functional relationship between LIS1 and mammalian homologs of NudE (NDEL1 and NDE1) is perplexing. The abnormalities of microtubule organization and Golgi morphology in Lis1-null mouse embryonic fibroblasts and the neuronal migration defects in granule neurons of Lis1-null mice are effectively counteracted by exogenous expression of LIS1, but not NDEL1 and NDE1 (7). However, similar defects in Ndel1-null neurons are reversed completely by exogenous expression of NDEL1, partially by NDE1 expression and moderately by LIS1 expression. Furthermore, overexpression of LIS1 is able to partially alleviate the Ndel1 RNAi effect but fails to counteract the defects caused by dynein RNAi (23). These reports suggest that NDEL1 may act upstream of LIS1 to regulate dynein function in mammals, which is distinct from that in the nud pathway in A. nidulans.
LIS1 has been also identified as the noncatalytic β subunit of platelet-activating factor acetyl-hydrolase complex 1B (PAF-AH 1B) (30). Studies of structural biology have shown that LIS1 binds to the catalytic subunit of PAF-AH 1B via the highly conserved top surface of the LIS1 β-propellers (31). This surface also includes sites of mutations causing lissencephaly and overlaps with a hypothetical dynein binding region. NDEL1 has been demonstrated to compete with the catalytic subunit of PAF-AH 1B complex for LIS1 binding to the dynein complex (31), suggesting that NDEL1 may be involved in the molecular switch of LIS1 for either the dynein or PAF-AH 1B complex.
NudCL2 Stabilizes LIS1 with Hsp90.
Hsp90 is a highly conserved molecular chaperone essential for activating many signaling proteins in eukaryotic cells (32). The Hsp90 cochaperone p23 appears to directly interact with ATP-bound form of Hsp90 and promote the maturation of client proteins (26). p23 is also found to stabilize the client protein-Hsp90 complex in vivo (33). Even though the proteins containing the p23_like domain, the core-folding motif of p23 protein may have diverse functions, most of them seem to associate with Hsp90 (25, 26). For example, Sgt1p, a conserved protein with the p23_like domain, has been found to act as an adaptor protein that recruits specific client proteins to Hsp90 complex by binding to both Hsp90 and its client proteins (27). Here, we found that NudCL2 with the p23_like domain not only directly bound to Hsp90 and LIS1 but also enhanced the association of Hsp90 with LIS1. Either interruption of the LIS1-Hsp90 interaction with the C terminus of NudCL2 or inhibition of Hsp90 chaperone function by geldanamycin led to reduce LIS1 stability, suggesting that NudCL2 may act as an adaptor protein to facilitate the interaction between Hsp90 and its potential client protein, LIS1.
Regulation of the LIS1/Dynein Pathway by the NudC Family.
Genetic screening in A. nidulans has identified a number of genes in the nud pathway that appear to be components or regulators of the dynein/dynactin complex (12). The mammalian homologs of nudE and nudF were studied more extensively than that of nudC even though the nudC null mutant has more severe defects than any other nud mutants in A. nidulans (34). In mammals, there are at least three homologs to Aspergillus NudC: NudC, NudCL, and NudCL2. We and other groups have found that NudC is involved in cell cycle progression via phosphorylation by Polo-like kinase 1, a key mitotic kinase (16, 17, 35). NudC is shown to associate with the LIS1/dynein complex (36); however, the function of this interaction was still uncharacterized. Recently, we provide evidence that NudCL also interacts with the LIS1/dynein complex and is involved in the stability of dynein intermediate chain (18). There is no direct data supporting a role of either NudC or NudCL in the regulation of LIS1 in mammals even though nudC appears to act upstream of nudF in A. nidulans. Here, we have identified NudCL2 as a regulator of LIS1, likely by stabilizing LIS1 with Hsp90 chaperone. These data suggest that mammalian members of the NudC family may play diverse roles in the regulation of the LIS1/dynein pathway (Fig. S10). Further study is clearly needed to explore the mechanisms by which the NudC family members modulate the LIS1/dynein pathway and their physiological significance.
Materials and Methods
Molecular Cloning.
To identify mammalian homologs of Aspergillus nudC, the protein sequence of Aspergillus NudC was applied to search in the databases in National Center for Biotechnology Information with tBLASTn program. A previously uncharacterized human sequence (GenBank release no. NM_145266) that encoded a deduced protein of 157 amino acid residues (GenBank release no. NP_660309) was detected. One pair of primers, 5′-TTatgtcggccccgtttgagga-3′ and 5′-Aatgcaggaaaaaaagcagtta-3′, was designed to clone NudCL2 by RT-PCR with total RNA extracted from HeLa cells. Three independent clones of NudCL2 cDNA were isolated and sequenced.
Construction of Plasmids.
Full-length human NudCL2 was subcloned into pFLAG (pCMV-Tag 2C, Stratagene), pGFP (pEGFP-C1; Clontech), and pGST (pGEX-5×-1; Amersham Pharmacia) vectors, respectively. The vectors pGST-NudCL2-N, pGST-NudCL2-C, pGFP-NudCL2-N, and pGFP-NudCL2-C were also separately constructed. pFLAG-LIS1 and pGST-LIS1 were made by PCR using pGFP-LIS1 (kindly provided by Dr. X. L. Zhu, Shanghai Institute of Biochemistry and Cell Biology, Shanghai, China) as a template. To construct the plasmids for RNAi, oligos containing complementary hairpin sequences were synthesized and cloned into pS vector (pSilencer 2.1-U6; Ambion). The targeted sequences of NudCL2 and LIS1 genes are 5′-accttgagaaataactgct-3’ and 5′-CGGACAAGTAGAATAAATG-3′, respectively. PCR was also used to construct RNAi-resistant NudCL2 mutant vector by the silent mutation of three nucleic acids in the RNAi targeting region of NudCL2 gene (accttgaaaagtgactgct). All of these constructs were confirmed by DNA sequencing.
Cell Culture, Transfection, and Drug Treatment.
HeLa cells were maintained in DMEM (Invitrogen) containing 10% FBS (Invitrogen) in an atmosphere containing 5% CO2. HeLa cells were transfected by GenePORTER transfection reagent (Gene Therapy Systems) with various vectors as indicated in the text. Geldanamycin (InvivoGenTM) were stored at −20 °C as a stock solution of 1.78 mM in DMSO. Cells were incubated with geldanamycin or DMSO for various periods as described in the text.
Preparation of GST-Fused Proteins and Antibody.
Recombinant GST-fused proteins generated in Escherichia coli DH5α were purified by incubation with glutathione-agarose beads followed by elution with glutathione elution buffer. Rabbit polyclonal anti-NudCL2 and anti-NudCL2 peptide antibodies, which were generated by using purified GST-NudCL2 and NudCL2 peptide (GAEISGNYTKGGPDFSNLEK, 138–157aa) as antigens, respectively, were affinity-purified for multiple analyses (Proteintech).
Immunofluorescence Staining.
HeLa cells grown on coverslips were fixed with cold methanol and then stained with anti-GM130 (BD Biosciences), β-tubulin, γ-tubulin (Sigma), human CREST autoimmune serum (Antibodies, Inc.) or NudCL2 antibodies, followed by incubation with either Cy3- or FITC-conjugated anti-mouse, anti-rabbit or anti-human Ig secondary antibody (Jackson ImmunoResearch Laboratories). DNA was stained with DAPI (Sigma). The mounted coverslips were analyzed by confocal fluorescence microscopy (FV1000; Olympus). The distance between the centrosome and nucleus was defined as the shortest distance from the centrosome to the edge of nucleus.
Chromosome Spread Preparation.
Chromosome spread was prepared as previously described (37). HeLa cells grown on slides were swollen in a hypotonic solution (75 mM KCl), followed by centrifugation and fixation, and then were subjected to immunofluorescence analysis.
GST Pull-Down Assay.
GST pull-down assays were performed as described previously (17). To detect the association among NudCL2, LIS1, and Hsp90, the blots were probed with the antibodies as indicated in the text.
Immunoprecipitation and Western Blotting.
Immunoprecipitation for FLAG-tagged proteins was performed as described previously (38). In brief, the cells transfected with the indicated vectors were subjected to coimmunoprecipitation analysis with anti-FLAG antibody-coupled beads (Sigma). Endogenous NudCL2 was immunoprecipitated with either anti-NudCL2 antibody or anti-NudCL2-peptdie antibody. Western analyses were performed with anti-Erk2, GFP, Hsp90α (Santa Cruz), FLAG, β-tubulin, LIS1 (Sigma), and NudCL2 antibodies, respectively. Either Erk2 or β-tubulin was used as a loading control.
Supplementary Material
Acknowledgments
We thank Drs. Raymond L. Erikson (Harvard University, Cambridge, MA), Xueliang Zhu, and Dangsheng Li (Shanghai Institute of Biochemistry and Cell Biology, Shanghai, China) for their technical assistance and critical comments on the manuscript. This work was supported by grants from the Natural Scientific Foundation of China (30671070 and 30771107), Ministry of Science and Technology of China (2007CB914500), Ministry of Education of China (NCET-06-0530) and Natural Scientific Foundation of Zhejiang Province, China (R205291, Y206103, and 2007R10G2010103).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0914307107/DCSupplemental.
References
- 1.Reiner O, et al. Isolation of a Miller-Dieker lissencephaly gene containing G protein beta-subunit-like repeats. Nature. 1993;364:717–721. doi: 10.1038/364717a0. [DOI] [PubMed] [Google Scholar]
- 2.Wynshaw-Boris A, Gambello MJ. LIS1 and dynein motor function in neuronal migration and development. Genes Dev. 2001;15:639–651. doi: 10.1101/gad.886801. [DOI] [PubMed] [Google Scholar]
- 3.Wynshaw-Boris A. Lissencephaly and LIS1: Insights into the molecular mechanisms of neuronal migration and development. Clin Genet. 2007;72:296–304. doi: 10.1111/j.1399-0004.2007.00888.x. [DOI] [PubMed] [Google Scholar]
- 4.Vallee RB, Tsai JW. The cellular roles of the lissencephaly gene LIS1, and what they tell us about brain development. Genes Dev. 2006;20:1384–1393. doi: 10.1101/gad.1417206. [DOI] [PubMed] [Google Scholar]
- 5.Faulkner NE, et al. A role for the lissencephaly gene LIS1 in mitosis and cytoplasmic dynein function. Nat Cell Biol. 2000;2:784–791. doi: 10.1038/35041020. [DOI] [PubMed] [Google Scholar]
- 6.Feng Y, et al. LIS1 regulates CNS lamination by interacting with mNudE, a central component of the centrosome. Neuron. 2000;28:665–679. doi: 10.1016/s0896-6273(00)00145-8. [DOI] [PubMed] [Google Scholar]
- 7.Sasaki S, et al. Complete loss of Ndel1 results in neuronal migration defects and early embryonic lethality. Mol Cell Biol. 2005;25:7812–7827. doi: 10.1128/MCB.25.17.7812-7827.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith DS, et al. Regulation of cytoplasmic dynein behaviour and microtubule organization by mammalian Lis1. Nat Cell Biol. 2000;2:767–775. doi: 10.1038/35041000. [DOI] [PubMed] [Google Scholar]
- 9.Tai CY, Dujardin DL, Faulkner NE, Vallee RB. Role of dynein, dynactin, and CLIP-170 interactions in LIS1 kinetochore function. J Cell Biol. 2002;156:959–968. doi: 10.1083/jcb.200109046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mesngon MT, et al. Regulation of cytoplasmic dynein ATPase by Lis1. J Neurosci. 2006;26:2132–2139. doi: 10.1523/JNEUROSCI.5095-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamada M, et al. LIS1 and NDEL1 coordinate the plus-end-directed transport of cytoplasmic dynein. EMBO J. 2008;27:2471–2483. doi: 10.1038/emboj.2008.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morris NR. Nuclear migration. From fungi to the mammalian brain. J Cell Biol. 2000;148:1097–1101. doi: 10.1083/jcb.148.6.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang J, Li S, Fischer R, Xiang X. Accumulation of cytoplasmic dynein and dynactin at microtubule plus ends in Aspergillus nidulans is kinesin dependent. Mol Biol Cell. 2003;14:1479–1488. doi: 10.1091/mbc.E02-08-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiang X, Osmani AH, Osmani SA, Xin M, Morris NR. NudF, a nuclear migration gene in Aspergillus nidulans, is similar to the human LIS-1 gene required for neuronal migration. Mol Biol Cell. 1995;6:297–310. doi: 10.1091/mbc.6.3.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chiu YH, Morris NR. Extragenic suppressors of nudC3, a mutation that blocks nuclear migration in Aspergillus nidulans . Genetics. 1995;141:453–464. doi: 10.1093/genetics/141.2.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aumais JP, et al. Role for NudC, a dynein-associated nuclear movement protein, in mitosis and cytokinesis. J Cell Sci. 2003;116:1991–2003. doi: 10.1242/jcs.00412. [DOI] [PubMed] [Google Scholar]
- 17.Zhou T, Aumais JP, Liu X, Yu-Lee LY, Erikson RL. A role for Plk1 phosphorylation of NudC in cytokinesis. Dev Cell. 2003;5:127–138. doi: 10.1016/s1534-5807(03)00186-2. [DOI] [PubMed] [Google Scholar]
- 18.Zhou T, Zimmerman W, Liu X, Erikson RL. A mammalian NudC-like protein essential for dynein stability and cell viability. Proc Natl Acad Sci USA. 2006;103:9039–9044. doi: 10.1073/pnas.0602916103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pfarr CM, et al. Cytoplasmic dynein is localized to kinetochores during mitosis. Nature. 1990;345:263–265. doi: 10.1038/345263a0. [DOI] [PubMed] [Google Scholar]
- 20.Steuer ER, Wordeman L, Schroer TA, Sheetz MP. Localization of cytoplasmic dynein to mitotic spindles and kinetochores. Nature. 1990;345:266–268. doi: 10.1038/345266a0. [DOI] [PubMed] [Google Scholar]
- 21.Sasaki S, et al. A LIS1/NUDEL/cytoplasmic dynein heavy chain complex in the developing and adult nervous system. Neuron. 2000;28:681–696. doi: 10.1016/s0896-6273(00)00146-x. [DOI] [PubMed] [Google Scholar]
- 22.Harada A, et al. Golgi vesiculation and lysosome dispersion in cells lacking cytoplasmic dynein. J Cell Biol. 1998;141:51–59. doi: 10.1083/jcb.141.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shu T, et al. Ndel1 operates in a common pathway with LIS1 and cytoplasmic dynein to regulate cortical neuronal positioning. Neuron. 2004;44:263–277. doi: 10.1016/j.neuron.2004.09.030. [DOI] [PubMed] [Google Scholar]
- 24.Tanaka T, et al. Lis1 and doublecortin function with dynein to mediate coupling of the nucleus to the centrosome in neuronal migration. J Cell Biol. 2004;165:709–721. doi: 10.1083/jcb.200309025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garcia-Ranea JA, Mirey G, Camonis J, Valencia A. p23 and HSP20/alpha-crystallin proteins define a conserved sequence domain present in other eukaryotic protein families. FEBS Lett. 2002;529:162–167. doi: 10.1016/s0014-5793(02)03321-5. [DOI] [PubMed] [Google Scholar]
- 26.Felts SJ, Toft DO. p23, a simple protein with complex activities. Cell Stress Chaperones. 2003;8:108–113. doi: 10.1379/1466-1268(2003)008<0108:paspwc>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Catlett MG, Kaplan KB. Sgt1p is a unique co-chaperone that acts as a client adaptor to link Hsp90 to Skp1p. J Biol Chem. 2006;281:33739–33748. doi: 10.1074/jbc.M603847200. [DOI] [PubMed] [Google Scholar]
- 28.Buchner J. Hsp90 & Co.—a holding for folding. Trends Biochem Sci. 1999;24:136–141. doi: 10.1016/s0968-0004(99)01373-0. [DOI] [PubMed] [Google Scholar]
- 29.Efimov VP, Morris NR. The LIS1-related NUDF protein of Aspergillus nidulans interacts with the coiled-coil domain of the NUDE/RO11 protein. J Cell Biol. 2000;150:681–688. doi: 10.1083/jcb.150.3.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hattori M, Adachi H, Tsujimoto M, Arai H, Inoue K. Miller-Dieker lissencephaly gene encodes a subunit of brain platelet-activating factor acetylhydrolase [corrected] Nature. 1994;370:216–218. doi: 10.1038/370216a0. [DOI] [PubMed] [Google Scholar]
- 31.Tarricone C, et al. Coupling PAF signaling to dynein regulation: structure of LIS1 in complex with PAF-acetylhydrolase. Neuron. 2004;44:809–821. doi: 10.1016/j.neuron.2004.11.019. [DOI] [PubMed] [Google Scholar]
- 32.Pearl LH, Prodromou C. Structure and mechanism of the Hsp90 molecular chaperone machinery. Annu Rev Biochem. 2006;75:271–294. doi: 10.1146/annurev.biochem.75.103004.142738. [DOI] [PubMed] [Google Scholar]
- 33.Morishima Y, et al. The hsp90 cochaperone p23 is the limiting component of the multiprotein hsp90/hsp70-based chaperone system in vivo where it acts to stabilize the client protein: hsp90 complex. J Biol Chem. 2003;278:48754–48763. doi: 10.1074/jbc.M309814200. [DOI] [PubMed] [Google Scholar]
- 34.Chiu YH, Xiang X, Dawe AL, Morris NR. Deletion of nudC, a nuclear migration gene of Aspergillus nidulans, causes morphological and cell wall abnormalities and is lethal. Mol Biol Cell. 1997;8:1735–1749. doi: 10.1091/mbc.8.9.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nishino M, et al. NudC is required for Plk1 targeting to the kinetochore and chromosome congression. Curr Biol. 2006;16:1414–1421. doi: 10.1016/j.cub.2006.05.052. [DOI] [PubMed] [Google Scholar]
- 36.Aumais JP, et al. NudC associates with Lis1 and the dynein motor at the leading pole of neurons. J Neurosci. 2001;21(RC187):1–7. doi: 10.1523/JNEUROSCI.21-24-j0002.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liang Y, et al. Nudel modulates kinetochore association and function of cytoplasmic dynein in M phase. Mol Biol Cell. 2007;18:2656–2666. doi: 10.1091/mbc.E06-04-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yan X, et al. Human Nudel and NudE as regulators of cytoplasmic dynein in poleward protein transport along the mitotic spindle. Mol Cell Biol. 2003;23:1239–1250. doi: 10.1128/MCB.23.4.1239-1250.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.