Abstract
Regulated activation of Ras by receptor tyrosine kinases (RTK) constitutes a key transduction step in signaling processes that control an array of fundamental cellular functions including proliferation, differentiation, and survival. The principle mechanism by which Ras is activated down stream of RTKs involves the stimulation of guanine nucleotide exchange by the ubiquitous guanine nucleotide exchange factor Son of sevenless (Sos). In resting conditions, Sos activity is constrained by intramolecular interactions that maintain the protein in an autoinhibited conformation. Structural, biochemical, and genetic studies have implicated the histone domain (Sos-H), which comprises the most N-terminal region of Sos, in the regulation of Sos autoinhibition. However, the molecular underpinnings of this regulatory function are not well understood. In the present study we demonstrate that Sos-H possesses in vitro and in vivo membrane binding activity that is mediated, in part, by the interactions between a cluster of basic residues and phosphatidic acid. This interaction is required for Sos-dependent activation of Ras following EGF stimulation. The inducible association of Sos-H with membranes contributes to the catalytic activity of Sos by forcing the domain to adopt a conformation that destabilizes the autoinhibitory state. Thus, Sos-H plays a critical role in governing the catalytic output of Sos through the coupling of membrane recruitment to the release of autoinhibition.
Keywords: Ras, guanine nucleotide exchange factor, Noonan syndrome, phosphatidic acid
Ras proteins are essential transducers of signals that originate at the cell surface following the ligand-dependent stimulation of receptor tyrosine kinases (RTK). The activation of Ras by RTK proceeds through the inducible conversion of Ras-GDP to Ras-GTP by the multidomain guanine nucleotide exchange factor Son of sevenless (Sos) (Fig. 1A). Structural and biochemical studies have assigned specific functions to the different domains of Sos in regulating guanine nucleotide exchange on Ras. The catalytic region consisting of the Rem and Cdc25 domains harbors two Ras-binding sites, one for Ras-GDP and the other for Ras-GTP (1). The Ras-GDP binding site (also referred to as the catalytic site) mediates nucleotide dissociation, and the Ras-GTP binding site (also referred to as the allosteric site) promotes the release of Sos from an inactive autoinhibited state imposed by the intramolecular interaction between the DH domain and the Rem segment of the catalytic domain (Fig. 1A). The allosteric site has also been implicated in the membrane recruitment of Sos along with the proline-rich C-terminal region and the PH domain (2–4). The N-terminus of Sos (residues 1–198) contains a segment with sequence and structural similarity to histones and accordingly has been termed the histone domain (Sos-H) (5). The only known function ascribed to date to this domain is in the maintenance of the autoinhibited conformation of Sos through the in cis binding to a helical linker that connects the DH-PH module of Sos to the catalytic domain (3, 6–8) (Fig. 1A). Analysis of the surface electrostatic potential of Sos-H revealed positively charged regions and it has been proposed that these regions may mediate the association with negatively charged membrane components (7). In the present study we have sought to determine the existence and significance of Sos-H membrane interactions.
Fig. 1.
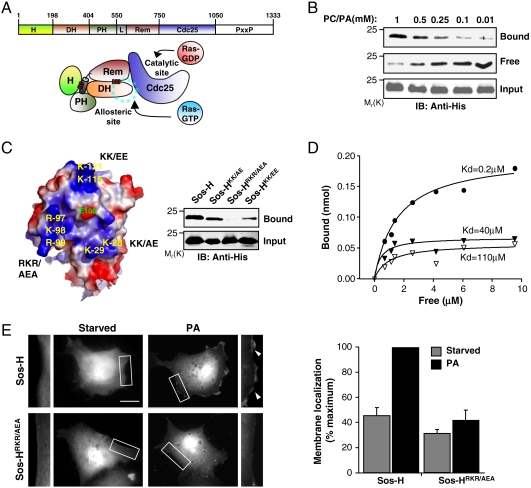
The histone domain of Sos (Sos-H) binds to membrane lipids. (A) The domain arrangement of Sos. H, Histone; DH, Dbl homology; PH, Pleckstrin homology; L, helical Linker; REM, Ras exchange motif; CDC25, yeast CDC25 homology; and PxxP, proline-rich region. Below, a cartoon depiction of the relative orientation of the various domains of Sos in the autoinhibited state. The red bars indicate intramolecular interactions. The blue dotted line circle indicates the binding site for allosteric Ras, which is occluded by the DH domain. (B) His-tagged Sos-H (0.5 μM) was mixed with the indicated concentrations of lipid vesicles comprised PC∶PA (90∶10). Vesicles were pelleted and the amount of Sos-H in the pellet (Bound) or in the supernatant (Free) was detected by immunoblotting (IB). (C) Surface electrostatic potential map (blue, positive; red, negative) showing the 3 basic patches in Sos-H and the residues mutated (see Materials and Methods). Right, Sos-H constructs containing the indicated mutations were mixed with 1 mM PC∶PA vesicles (90∶10) and binding was determined as in B. (D) Increasing amounts of Sos-H were incubated with 1 mM PC∶PA (closed circles) or PC lipid vesicles (open triangles) and Sos-HRKR/AEA with PC∶PA vesicles (closed triangles). Kd was determined as previously described (2). Data presented in panels B–D are representative of three independent experiments. (E) COS-1 cells were transfected with GFP-tagged Sos-H and membrane localization scored in either starved cells or upon incubation with PA (100 μM) for 20 min. The image represents a single slice of 0.25 μm from a serial Z-section of the cell. The boxed areas are enlarged in the left and right adjacent images with arrowheads showing the relative increase in fluorescence intensity at the membrane. Results (bar graph) are mean ± s.d. of three independent experiments with at least 100 expressing cells counted for each condition (see Materials and Methods). Scale bar: 10 μm.
Results and Discussion
The Histone Domain Displays Lipid-Dependent Membrane Binding Activity.
An initial unbiased screen for potential interactions between Sos-H and phospholipids indicated preferential binding to phosphatidic acid (PA) (Fig. S1A). To validate this observation, we used a lipid vesicle-sedimentation assay in which the partitioning of purified Sos-H between the vesicle-containing pellet (bound) and the supernatant (free) is assessed post ultracentrifugation. As illustrated in Fig. 1B, Sos-H bound to PA-containing vesicles in a concentration-dependent manner, with detectable binding observed at a concentration of 0.01 mM PA. In contrast, no binding was observed to vesicles composed of phosphatidylcholine only or to vesicles containing phosphatidylserine, another acidic phospholipid (Fig. S1B). Consistent with the observation of Gureasko et al. (9), Sos-H also bound to PIP2-containing vesicles (Fig. S1B). Because the binding of Sos-H to PA was significantly higher relative to PIP2 binding, we have focused our analysis on the Sos-H/PA interactions. The modulation of Sos function by interactions with membrane components other than PA, i.e. PIP2, is the subject of the accompanying study by Gureasko et al (9). Whether or not these interactions play a redundant or complementary role with that of PA remains to be determined.
The solvent exposed surface of Sos-H contains three prominent basic patches surrounding a negatively charged E108 residue (Fig. 1C, Left). Significantly, all of these patches and E108 are conserved across Sos sequences (Fig. S1D). To assess the individual contribution of these patches to PA-binding, lysine and arginine residues within each patch were mutated. As illustrated in Fig. 1C Right , each set of mutations compromised PA-binding. However, the Sos-H mutant in which AEA residues were substituted for 97RKR99 (Sos-HRKR/AEA) displayed the most pronounced reduction in binding activity indicating that this basic cluster serves as a major PA-binding determinant. To substantiate this conclusion, the binding affinities of PA for Sos-H and Sos-HRKR/AEA were measured. Sos-H bound to PA-containing vesicles with a Kd of 0.2 μM, nearly 600 times stronger than PC-containing vesicles (Kd = 110 μM) (Fig. 1D). By comparison, the binding of Sos-HRKR/AEA to PA-containing vesicles was 200-fold weaker (Kd = 40 μM). Of note, the binding of Sos-HRKR/AEA to PIP2 was not altered relative to Sos-H suggesting that PA and PIP2 interact at distinct sites (Fig. S1C).
To test whether PA-binding could play a role in the membrane association of Sos-H in vivo, GFP-fusion constructs of Sos-H and Sos-HRKR/AEA were transiently transfected into COS-1 cells and their subcellular distribution analyzed by fluorescence microscopy. In unstimulated cells, Sos-H displayed a basal level of membrane localization plausibly due to charge-mediated binding to membrane phospholipids (Fig. 1E). The extent of this constitutive membrane association was dependent on the levels of ectopically expressed constructs making it difficult to assess its physiological relevance. The addition of membrane-permeable PA to serum-deprived cells stimulated the translocation of Sos-H, but not Sos-HRKR/AEA, from the cytoplasm to the membrane as evident by a rim of fluorescence at the cell periphery (Fig. 1E). Similar results were obtained when serum-deprived cells were stimulated with EGF (Fig. S1E). Because PA can stimulate PIP2 production by activating PI5K (10) it is possible that the observed Sos-H recruitment is mediated, at least in part, by PIP2. However, because the RKR to AEA substitution on Sos-H does not affect PIP2 binding (Fig. S1C), our findings are most consistent with the interpretation that the PA-induced membrane localization of Sos-H results from the preferential binding of PA to the RKR motif. Previously, we have demonstrated that PLD2-generated PA recruits Sos to the membrane via the PH domain (2). To determine whether PLD2 is also required to promote the translocation of Sos-H to the membrane, RNA interference was used to knock-down PLD2. Suppression of PLD2 expression impaired the ability of Sos-H to translocate to the plasma membrane in response to EGF stimulation (Fig. S2). Together, these data indicate that the affinity of Sos-H to the plasma membrane can be modulated by growth factor-dependent activation of PLD2 that in turn stimulates an increase in the levels of the Sos-H ligands PA and PIP2.
Sos-H-Mediated Membrane Interaction Modulates Sos Activity.
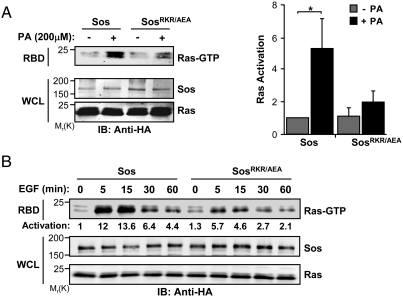
To assess the functional relevance of the interaction of Sos-H with PA, COS-1 cells were cotransfected with full-length wild-type (WT) Sos or SosRKR/AEA and HRas expression vectors, and the levels of activated RAS were monitored using the Raf1 RAS-binding domain (RBD) pull-down assay. The relative expression levels of the constructs were adjusted to ensure no contribution from endogenous Sos (Fig. S3). As illustrated in Fig. 2A, the addition of membrane-permeable PA enhanced the activity of WT Sos. In contrast, SosRKR/AEA was refractory to the stimulatory effect of PA addition. Similarly, SosRKR/AEA was appreciably compromised in its ability to mediate EGF-induced Ras activation, (Fig. 2B) indicating a role for Sos-H/PA interactions in regulating the catalytic output of Sos. The remaining Ras activation displayed by SosRKR/AEA likely reflects the contribution of other recruitment/activation mechanisms that are engaged following EGF stimulation. These may include C-terminal-mediated interactions with Grb2, PH-mediated interactions with PA and PIP2, and Sos-H-mediated interactions with PIP2.
Fig. 2.
The interaction between Sos-H and PA is required for Sos activation. (A, B) COS-1 cells were cotransfected with HA-tagged Ras and HA-tagged Sos constructs. Cells were serum starved and then stimulated with PA for 15 min (A) or 10 nM EGF for the indicated intervals (B). Ras activation (Ras-GTP) was measured by the RBD pull-down assay as described in Materials and Methods. Results are representative of three independent experiments. Error bars are ± s.d.∗, P < 0.05.
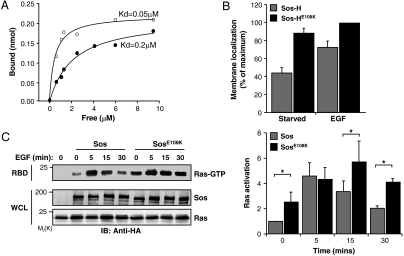
Sos-H is the site for the Noonan syndrome-causing mutation E108K (8). The location of E108 is adjacent to the PA-binding motif, and the charge reversal caused by the E → K mutation would generate a contiguous patch of basic residues that could potentiate phospholipid binding (Fig. 1C, Left). Consistent with this idea, the apparent affinity of Sos-H containing an E108K substitution (Sos-HE108K) to PA was 4-fold higher in comparison to Sos-H (Fig. 3A). The enhanced phospholipid binding potential conferred by the E108K mutation is further substantiated by the observation that Sos-HE108K displayed a constitutive membrane association (Fig. 3B). Furthermore, in the context of full-length Sos, the E108K mutation led to an enhanced basal and prolonged EGF-induced Ras activation relative to WT Sos (Fig. 3C). These observations point to a critical role for E108 in preventing the promiscuous binding of Sos-H to the membrane and the ensuing Sos activation. The predicted failure to execute this function in Noonan syndrome patients harboring the E108K mutation is consistent with the unchecked Ras signaling that is characteristic of these patients (11).
Fig. 3.
The Noonan syndrome mutation E108K activates Sos by increasing PA-binding and membrane association. (A) Sos-H (closed circles) and Sos-HE108K (open circles) were incubated with 1 mM PA-containing lipid vesicles and affinities calculated as in Fig. 1D. (B) GFP-tagged Sos-H or Sos-HE108K were transfected into COS-1 cells and membrane localization quantified as in Fig. 1E. Results are mean ± s.d. of three independent experiments. (C) COS-1 cells were cotransfected with HA-tagged Ras and either HA-tagged Sos or SosE108K. Cells were serum starved and then stimulated with 10 nM EGF for the indicated intervals and RBD pull-down performed as in Fig. 2. Results are mean ± s.d. of three independent experiments. ∗,P < 0.05.
Sos-H-Membrane Interaction Activates Sos Through a Conformational Switch.
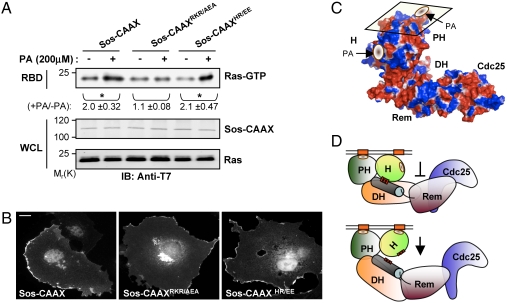
Recently, we have found that the PH domain of Sos (Sos-PH) binds to PA and that the interaction is critical for the ligand-induced membrane recruitment of Sos and hence for Sos-mediated Ras activation (2). By analogy, the binding of PA to Sos-H could serve a membrane-targeting function. Alternatively, PA-binding to Sos-H could contribute to Sos activity through another mechanism that is independent of membrane targeting. To distinguish between these possibilities, a Sos construct that is constitutively targeted to the membrane was generated by replacing the C-terminal region (residues 1050–1333) of Sos with a CAAX box (Sos-CAAX) containing sequence derived from the C-terminal region of HRas (12). The addition of PA to COS-1 cells expressing Sos-CAAX led to an increase in Sos-mediated Ras activation indicating that PA has a Sos-activating function that is distinct from its membrane-targeting function (Fig. 4A). Significantly, this activating function is strictly dependent on the binding of PA to Sos-H as it was selectively abolished by mutation of residues that are critical for PA-binding to Sos-H (Sos-CAAXRKR/AEA) but not Sos-PH (Sos-CAAXHR/EE) (Fig. 4A). All constructs displayed a similar subcellular localization pattern with pronounced membrane association (Fig. 4B). Together, these data suggest that the two PA-binding activities of Sos may have complementary functions; the Sos-PH–PA interaction mediates the targeting of Sos to the membrane and the Sos-H–PA interaction activates Sos at the membrane. The molecular details of this activation process remain to be established. However, because the PA-binding motif on Sos-H lay perpendicular to the plane of the membrane (Fig. 4C), its association with the membrane would necessitate an upward swivel movement, which in turn would result in the disruption of the interaction of the histone domain with the helical linker. As a consequence, the autoinhibited conformation would be destabilized permitting Sos activation through the allosteric site (Fig. 4D). Moreover, this conclusion is in agreement with the structural and biochemical analyses of Sos regulation by the histone domain (9). A similar mechanism for the enhancement of Sos activity has been proposed for the Noonan syndrome-associated Sos activating mutation R552G that interferes with the formation of ion pair interactions that are critical for the intramolecular association of Sos-H with the helical linker (3, 8, 13). Thus, membrane binding of Sos-H may serve to alleviate the constraining effect of this domain on Sos activity.
Fig. 4.
Activation of Sos by the Sos-H–PA interaction is independent of membrane targeting. (A) COS-1 cells were cotransfected with T7-tagged Ras and the indicated T7-tagged Sos-CAAX constructs. Cells were serum starved and then stimulated with PA for 15 min. RBD pull-down was performed as in Fig. 2. Activation was measured as the relative increase of Ras-GTP levels upon PA stimulation. Numerical values represent the mean ± s.d. of three independent experiments. ∗,P < 0.05. (B) COS-1 cells were transfected with the indicated T7-tagged Sos-CAAX constructs and processed for indirect immunofluorescence. Images were acquired as in Fig. 1E. Scale bar: 20 μm. (C) Surface electrostatic potential map (blue, positive; red, negative) of Sos oriented at the membrane (parallelogram) showing the PA-binding motifs on the Sos-H and Sos-PH domains (see Materials and Methods). (D) Model of Sos activation by Sos-H–PA interaction. (Upper) Membrane recruitment via the binding of the PH domain to PA does not relieve the autoinhibition but positions the Sos-H in proximity to PA-containing membrane regions. (Lower) Sos-H–PA interaction causes an upward rotation of the Sos-H leading to the destabilization of the histone–linker interaction.
Conclusions
The process of Sos-mediated Ras activation involves multiple regulatory steps that need to be tightly coordinated to ensure an appropriate biological output. We have shown that the binding of phospholipids to Sos-PH (2, 3) and Sos-H [this work and (9)] controls the membrane-targeting and allosteric modulation of Sos, respectively. Together, these findings suggest a mechanism by which the two principal modalities for regulating Sos activity, membrane recruitment and release of autoinhibition, can be coupled. Thus, membrane-remodeling events involving a localized increase in the levels of phospholipids could have profound effects on the dynamics of Ras activation by Sos. Currently, the two known phospholipids implicated in the targeting and activation of Sos are PA and PIP2. PA, which is generated predominantly by PLD2, can stimulate the synthesis of PIP2 by activating PI5K (10, 14). PIP2 in turn acts as a positive regulator of PLD2 (15). This feedback relationship may serve to coordinate the activation of Sos in specific membrane domains that can support the coincident production of PA and PIP2.
Materials and Methods
Protein Preparation.
Sos-H (residues 1–191) of human Sos1 was expressed and purified as described earlier (5).
General Reagents.
1,2-Dilauroyl-sn-glycero-3-phosphate, 1-palmitoyl-2- oleoyl-sn-glycero-3-phosphate, 1-palmitoyl-2-oleoyl-sn-glycero-3- phosphocholine, 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine, and phosphatidylinositol-4,5-bisphosphate (PIP2) (swine brain) were from Avanti Polar Lipids. EGF was from Invitrogen and Ni-NTA resin was from Pierce.
Antibodies.
Primary antibody sources were T7, Novagen; HA and Sos1 (C-terminus), Upstate; His, Abgent; Sos1 (N-terminus), BD Biosciences. Secondary antibody sources were Alexa Fluor 680-conjugated antimouse, Invitrogen; IRDye 800-conjugated goat antirabbit, Rockland.
Cell Culture and Transfection.
COS-1 cells were cultured in DMEM (Invitrogen) supplemented with 5% FBS (Invitrogen). Cells were maintained in 5% CO2 at 37 °C. Transient transfections were performed with FuGENE 6 (Roche) according to the manufacturer’s instructions.
Plasmids.
The following plasmids were previously described: HA tagged WT human Sos1 (amino acids 1–1333) (16) and Sos-CAAX (amino acids 1–1049) (12); His-Sos-H (amino acids 1–191) (5); and GST-Raf1-RBD (17). GFP-tagged Sos-H was generated by cloning the sequence corresponding to Sos amino acids 1–200 into pEGFPC3 mammalian expression vector (Clontech). Sos mutants were generated by site directed mutagenesis and verified by DNA sequencing.
Lipid-Binding Assays.
The expression and purification of Sos-H and the generation of lipid vesicles were described previously (5, 18). The lipid binding experiments and Kd determination were done as previously described (2).
Ras Activation Assay.
The levels of Ras-GTP were determined by the GST-RBD pull-down assay, as previously described (17). Levels of Ras-GTP/Ras were determined using the Odyssey system (LiCor).
Fluorescence Microscopy.
Transfected cells grown on cover slips were treated, fixed, and processed for indirect immunofluorescence and direct fluorescence microscopy as described previously (2). Cells were scored as displaying membrane localization when the GFP signal was detected in > 50% of the cell periphery. Results were expressed as percentage of the total number of cells scored and are normalized to the maximal value obtained for each experiment.
Structural Models and Sequence Alignment.
PyMol (DeLano Scientific) was used with default parameters to generate surface electrostatic potential maps using the Histone domain (PDB code 1Q9C), DH-PH-Rem-Cdc25 (PDB code 1XD4) and the docked structure described in (5). Alignment of Sos sequences was done by MEGA4.0 (19) using accession numbers Q07889, Q07890 (Homo sapiens Sos1 and Sos2), Q62245 (Mus musculus Sos1), P26675 (Drosophila melanogaster), and NP_504235 (Caenorhabditis elegans).
Supplementary Material
Acknowledgments.
We thank Dr. Laura Taylor for help with data analysis and manuscript preparation and members of the Bar-Sagi laboratory for input and helpful discussions. This work was supported by National Institutes of Health Grant GM078266 (D.B.-S.).
Footnotes
The authors declare no conflict of interest.
See companion article on page 3430.
This article contains supporting information online at www.pnas.org/cgi/content/full/0914315107/DCSupplemental.
References
- 1.Margarit SM, et al. Structural evidence for feedback activation by Ras.GTP of the Ras-specific nucleotide exchange factor SOS. Cell. 2003;112:685–695. doi: 10.1016/s0092-8674(03)00149-1. [DOI] [PubMed] [Google Scholar]
- 2.Zhao C, Du G, Skowronek K, Frohman MA, Bar-Sagi D. Phospholipase D2-generated phosphatidic acid couples EGFR stimulation to Ras activation by Sos. Nat Cell Biol. 2007;9:706–712. doi: 10.1038/ncb1594. [DOI] [PubMed] [Google Scholar]
- 3.Gureasko J, et al. Membrane-dependent signal integration by the Ras activator Son of sevenless. Nat Struct Mol Biol. 2008;15:452–461. doi: 10.1038/nsmb.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gale NW, Kaplan S, Lowenstein EJ, Schlessinger J, Bar-Sagi D. Grb2 mediates the EGF-dependent activation of guanine nucleotide exchange on Ras. Nature. 1993;363:88–92. doi: 10.1038/363088a0. [DOI] [PubMed] [Google Scholar]
- 5.Sondermann H, Soisson SM, Bar-Sagi D, Kuriyan J. Tandem histone folds in the structure of the N-terminal segment of the Ras activator Son of sevenless. Structure. 2003;11:1583–1593. doi: 10.1016/j.str.2003.10.015. [DOI] [PubMed] [Google Scholar]
- 6.Jorge R, et al. HSos1 contains a new amino-terminal regulatory motif with specific binding affinity for its pleckstrin homology domain. J Biol Chem. 2002;277:44171–44179. doi: 10.1074/jbc.M204423200. [DOI] [PubMed] [Google Scholar]
- 7.Sondermann H, Nagar B, Bar-Sagi D, Kuriyan J. Computational docking and solution x-ray scattering predict a membrane-interacting role for the histone domain of the Ras activator Son of sevenless. Proc Natl Acad Sci USA. 2005;102:16632–16637. doi: 10.1073/pnas.0508315102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tartaglia M, et al. Gain-of-function SOS1 mutations cause a distinctive form of Noonan syndrome. Nat Genet. 2007;39:75–79. doi: 10.1038/ng1939. [DOI] [PubMed] [Google Scholar]
- 9.Gureasko J, et al. The role of the histone domain in the autoinhibition and activation of the Ras activator Son of Sevenless (accompanying paper) 2010 doi: 10.1073/pnas.0913915107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Foster DA, Xu L. Phospholipase D in cell proliferation and cancer. Mol Cancer Res. 2003;1:789–800. [PubMed] [Google Scholar]
- 11.Tartaglia M, Gelb BD. Noonan syndrome and related disorders: Genetics and pathogenesis. Annu Rev Genomics Hum Genet. 2005;6:45–68. doi: 10.1146/annurev.genom.6.080604.162305. [DOI] [PubMed] [Google Scholar]
- 12.Aronheim A, et al. Membrane targeting of the nucleotide exchange factor Sos is sufficient for activating the Ras signaling pathway. Cell. 1994;78:949–961. doi: 10.1016/0092-8674(94)90271-2. [DOI] [PubMed] [Google Scholar]
- 13.Roberts AE, et al. Germline gain-of-function mutations in SOS1 cause Noonan syndrome. Nat Genet. 2007;39:70–74. doi: 10.1038/ng1926. [DOI] [PubMed] [Google Scholar]
- 14.Jenkins GH, Fisette PL, Anderson RA. Type I phosphatidylinositol 4-phosphate 5-kinase isoforms are specifically stimulated by phosphatidic acid. J Biol Chem. 1994;269:11547–11554. [PubMed] [Google Scholar]
- 15.Hodgkin MN, et al. Phospholipase D regulation and localisation is dependent upon a phosphatidylinositol 4,5-biphosphate-specific PH domain. Curr Biol. 2000;10:43–46. doi: 10.1016/s0960-9822(99)00264-x. [DOI] [PubMed] [Google Scholar]
- 16.Corbalan-Garcia S, Margarit SM, Galron D, Yang SS, Bar-Sagi D. Regulation of Sos activity by intramolecular interactions. Mol Cell Biol. 1998;18:880–886. doi: 10.1128/mcb.18.2.880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boykevisch S, et al. Regulation of Ras signaling dynamics by Sos-mediated positive feedback. Curr Biol. 2006;16:2173–2179. doi: 10.1016/j.cub.2006.09.033. [DOI] [PubMed] [Google Scholar]
- 18.Chen RH, Corbalan-Garcia S, Bar-Sagi D. The role of the PH domain in the signal-dependent membrane targeting of Sos. EMBO J. 1997;16:1351–1359. doi: 10.1093/emboj/16.6.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.