Abstract
Approximately 3,500 mammalian genes are predicted to be secreted or single-pass transmembrane proteins. The function of the majority of these genes is still unknown, and a number of the encoded proteins might find use as new therapeutic agents themselves or as targets for small molecule or antibody drug development. To analyze the physiological activities of the extracellular proteome, we developed a large-scale, high-throughput protein expression, purification, and screening platform. For this study, the complete human extracellular proteome was analyzed and prioritized based on genome-wide disease association studies to select 529 initial target genes. These genes were cloned into three expression vectors as native sequences and as N-terminal and C-terminal Fc fusions to create an initial collection of 806 purified secreted proteins. To determine its utility, this library was screened in an OCT4-based cellular assay to identify regulators of human embryonic stem-cell self-renewal. We found that the pigment epithelium-derived factor can promote long-term pluripotent growth of human embryonic stem cells without bFGF or TGFβ/Activin/Nodal ligand supplementation. Our results further indicate that activation of the pigment epithelium-derived factor receptor-Erk1/2 signaling pathway by the pigment epithelium-derived factor is sufficient to maintain the self-renewal of pluripotent human embryonic stem cells. These experiments illustrate the potential for discovering novel biological functions by directly screening protein diversity in cell-based phenotypic or reporter assays.
Keywords: secreted proteins, human embryonic stem cells, pigment epithelium-derived factor, automated protein expression, high throughput
Many physiological processes are regulated by secreted proteins, making the extracellular proteome a rich source of molecules for understanding basic pathways involved in normal physiological function, development, and human disease. Although many examples of bioactive secreted proteins have been identified, recent technologic advances allow us to directly screen this proteomic diversity for new factors influencing important biological pathways. Efforts to survey the extracellular proteome using conditioned media from transiently transfected cell lines have shown that novel factors can be identified in a prospective screening approach. Lin et al. (1) evaluated ∼3,400 genes encompassing predicted secreted proteins or extracellular domains of single-pass transmembrane proteins. Each gene of interest was expressed by transient transfection using liposomal reagents in 293 T cells, and relative quantitation was determined by ELISA using an affinity tag. Conditioned media from this set was used to screen for biological activity in various assays. However, such screens are complicated by secondary metabolites, the release of cytoplasmic proteins by cell lysis, and variable and undefined levels of extracellular proteins. To overcome these challenges, we have created a collection of purified and physically characterized extracellular proteins that can be screened in a dose-dependent manner under defined conditions. An initial set of 529 distinct secreted-protein candidates was prioritized from the total mouse and human extracellular proteome by genome-wide association studies and genetic mapping of quantitative trait loci (QTL) from mouse genetic studies. This gene set was expressed in multiple expression vectors to provide native and Fc-fusion proteins, and it provided 806 proteins for use in cell-based phenotypic and reporter screens.
To show the utility of this approach for identifying protein factors with novel functions, we performed a high-content imaging screen for secreted proteins that regulate human embryonic stem-cell self-renewal. Human embryonic stem cells (hESCs), derived from the inner cell mass (ICM) of the blastocyst, have the capacity for long-term undifferentiated growth in culture and the theoretical potential to differentiate into all somatic cell types (2). These hESCs offer not only a model system for human development but also a potentially unlimited source of graft material for transplantation-based therapies. Mouse ESCs (mESCs) can be maintained in an undifferentiated state with leukemia inhibitory factor (LIF). In contrast, the maintenance of undifferentiated hESCs does not require LIF and the LIF/Stat3 signaling pathway, suggesting that there is a different molecular requirement for pluripotence in human cells (3). Recent studies have shown that feeder-fibroblast conditioned medium (CM) (4), high concentrations of basic FGF (bFGF; 100 ng/mL) (5), and combinations of bFGF with Noggin (6) or TGF-β/activin/Nodal signaling molecules (7, 8) can support long-term culture of hESCs grown on an extracellular matrix (ECM)-coated surface (e.g., Matrigel) under feeder-free conditions. To identify additional regulators of hESC self-renewal, we screened the 806 purified extracellular proteins for their ability to maintain expression of the transcription factor OCT4, which is only expressed in undifferentiated hESCs (9). We found that pigment epithelium-derived factor (PEDF) promotes long-term hESC self-renewal and maintains their pluripotency in the absence of bFGF or TGFβ/Activin/Nodal ligand supplementation. This study not only shows that PEDF is sufficient to support hESC self-renewal and pluripotency, but it also shows the utility in screening purified proteins from the extracellular proteome for biologic activities.
Results and Discussion
Selection of Candidate Extracellular Genes.
Predicted secreted gene lists were compiled from previous large-scale efforts including the secreted protein discovery initiative (SPDI) (10) and the web-based secreted protein database SPD (11), and they were also gathered using the sequence-based supervised signal peptide-prediction algorithms SignalP (12) and Phobius (13). The most abundant sources of false-positive predictions from all sources were identified and removed, including, among others, genes whose proteins localize to mitochondria and those predicted to encode multipass transmembrane proteins by tied mixture hidden Markov model (TMHMM) algorithm (12). Existing Gene Ontology-traceable author-statement annotations to extracellular localization categories were added. Existing high-quality, annotated UniProt entries were translated into Entrez Gene identifiers and combined with the list. All assorted identifiers were unified to Entrez Gene entities, and for initial targeting efforts, representative exemplar proteins for every gene were selected. The predicted secreted gene list contains 3,530 unique human proteins encoded by 2,881 genes. Large numbers (>1,500, currently) of putative secreted proteins are encoded by model transcripts no longer supported by EntrezGene, and these were excluded from the targeted secreted protein set. Also excluded were those that, according to Gene Ontology, are only associated with the mitochondrion.
Single-pass transmembrane genes were identified by existing Gene Ontology categories for cell-surface/membrane localization and combined with a filtering step by Phobius to include both the signal peptide and a single transmembrane helix, which provided the prediction for the extracellular domain boundaries. This resulted in 1,718 unique proteins encoded by 1,162 genes, of which 615 proteins encoded by 400 genes were removed owing to their previous inclusion in the compiled predicted secreted-proteins list. The net set of secreted and single-pass transmenbrane protein-encoding genes, therefore, currently contains 4,642 unique human proteins encoded by 3,641 unique genes (Table S1), all of which are currently supported by Entrez Gene. This compares with previous studies based on a combination of literature reports and experimental analysis of the human-plasma proteome that predicted 1,175 distinct gene products (14), which were expanded in other studies through sequence analysis to ∼2,200 genes (1, 15).
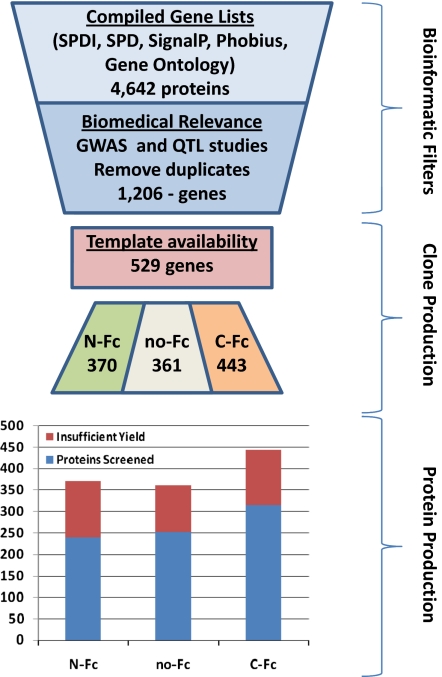
Although a collection of all secreted proteins is currently being assembled, we based our initial collection on candidate genes with additional biologic associations based on both hypothesis-driven and genetic methods. Five filters based on genetic studies were applied to candidate secreted proteins and the extracellular domains (ECD) of single-pass transmembrane proteins. First, a genome-wide association (GWA) scan of 18 traits related to type 2 diabetes (T2D) was used to select 1,058 candidate genes (16, 17). Second, our QTL analysis of inbred mouse strains (18, 19) for 40 metabolic traits added 763 candidate genes. Third, a comparative analysis between the two preceding human and mouse QTL studies identified 129 additional candidate genes in overlapping synthetic loci at slightly relaxed thresholds. Fourth, an integrative analysis across four T2D GWA studies (17) was used to identify 10 candidate genes. Finally, expression QTL analysis of inbred mouse strains was used to screen for genes in 70 Gene Ontology categories or Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways related to metabolic function (20), adding 316 genes to the gene list. After removing duplicates, a total of 1,206 unique genes were identified from the five filters above, and of these, 529 were predicted by the above analysis to be secreted or single-pass transmembrane and were chosen to be candidates for our initial target collection. A summary of the target selection, cloning, and protein-expression process is provided (Fig. 1).
Fig. 1.
Flowchart of target selection through protein production. An initial bioinformatic evaluation identified 4,642 predicted extracellular targets. Filtering for biomedical relevance reduced this set to 1,206, 529 of which were selected for production based on template availability. A total of 1,174 clones were processed, yielding a final protein collection of 806 purified proteins (69%), which met threshold criteria.
Expression, Purification, and Characterization of Extracellular Proteins.
A previous study has shown that interrogation of the extracellular proteome through the use of conditioned media is a useful approach to finding novel biological activities (1). That study reported median protein concentrations in CM of 20 ng/mL and an average of 75 ng/mL. However, these levels are nearly 1,000-fold less that the average concentration of the purified-protein collection reported here. It is reasonable to predict that the specific activity of some proteins would fall below the limit of detection from CM production. The use of purified proteins for biological screens has several other advantages. Purifying the proteins separates them from transfection reagents, secondary metabolites, proteases, and intra- and extracellular proteins produced by the production cell line that can interfere with interpretation of results from subsequent activity screens. Typically, purified proteins are also available in much higher and defined concentrations than in conditioned media. This allows dose-dependent activity testing and confirmation studies using identical materials in secondary follow-up assays. In addition, the purified proteins can be characterized for endotoxin and other contaminants, posttranslational modifications, and heterogeneity. Purified proteins can also be readily evaluated in combination; however, such a combinatorial matrix is too large for a primary screen but would be useful for evaluating synergistic or antagonistic effects.
The primary disadvantage to a purified-protein approach is the logistical difficulty in processing thousands of expression constructs. A previous report by Battle et al. (15) described a 4-year effort to produce 2,200 proteins that were subsequently screened for bioactivity. In a manner analogous to the current study, the proteins were expressed by transient transfection in mammalian cells (HEK293-EBNA) to take advantage of proper posttranslation modification and a highly efficient secretory pathway. The authors describe a combination of manual and automated tasks requiring significant human resources to produce the protein collection. To overcome the logistical difficulty of a purified-protein approach, we developed custom robotics, Protein Expression and Purification Platform (PEPP), to perform mammalian cell culture, transient transfection, protein expression, harvest, and purification in a fully automated manner. A more detailed description of the PEPP platform and its application to the mouse and human extracellular proteome is provided in SI Materials and Methods.
A mammalian expression system was used to increase the likelihood for correct folding and posttranslational modifications of the extracellular proteins. Transient transfection in human embryonic kidney (HEK293 Freestyle) cells was performed at a volume of 35 mL followed by affinity purification, protein characterization, and dispensation into multiple single-use aliquots. The average yield of protein and the relative success rate for target expression on PEPP were similar to the Battle et al. (15) study, despite full automation and reduced culture volumes (35 mL by PEPP versus 100- or 500-mL cultures). Our reagent collection provides a consistent, characterized, and concentrated protein pool (averaging over 100 μg per protein) for screening in biologically relevant functional assays. It is also noteworthy that, unlike the HEK Freestyle cells that are cultured in serum/protein-free media, the HEK293-EBNA cells used in the previous study are cultured in 1–2% FCS. A single-step affinity purification, using an undisclosed tag at the carboxy-terminus, is unlikely to remove all contaminants from CM, which can potentially confound downstream-activity assessments.
To facilitate uniform and high-level protein processing and secretion, a single IgK secretion leader was used rather than the various native secretion leaders. This leader was selected based on its capacity for high-level expression and secretion of extracellular domains, consistent and efficient protein processing, and cloning ease. Fusion to the Ig Fc domain can provide enhanced expression and stability for recombinant proteins. However, in some cases, such a fusion may interfere with biological activity. Therefore, each target gene was cloned in three forms, one with an N-terminal Fc fusion (N-Fc), one a C-terminal Fc fusion (C-Fc), and one lacking an Fc fusion (no-Fc). All forms were expressed using a CMV promoter and were fused to a C-terminal His-FLAG sequence to facilitate protein purification and detection. A collection of 1,174 expression constructs representing 529 of the prioritized secreted and extracellular domains of single-pass transmembrane genes was assembled for protein production. For most secreted proteins, the complete ORF minus the native signal peptide was subcloned into the three fusion vector variants. For all type I and type II single-pass transmembrane proteins, only the predicted extracellular domain of the ORF was subcloned, thereby permitting IgK-directed extracellular secretion and purification in the absence of producer cell lysis. In previous studies, the native signal peptide and a single carboxy-terminus tag were used for expression of genes of interest. We found that expression of targets with a single signal peptide, yet multiple versions of fusion tags, increased the success of expression and production levels and the ability to generate an active form of the protein.
We selected a set of 48 targets as a proof-of-concept set. This POC48 group contained several well-characterized secreted proteins with biological activity and several extracellular domains and proteins randomly selected from the larger priority target set representing a broad spectrum of protein properties. To validate that proteins could be isolated in active form from this approach, we evaluated the POC48 in several cell-based assays. For example, PCSK9 was identified within the POC48 set as affecting LDL uptake. This protein functions by binding the LDL receptor and blocking its uptake (21, 22). Not all forms of PCSK9 were equally active. Fusion of the Fc to the N terminus interferes with proper folding and processing. The C-terminal Fc fusion in this case appeared more active than the non-Fc. Likewise, in a glucagon-like peptide-1 (GLP1) secretion assay, both gastric inhibitory polypeptide (GIP) and Glucagon were identified as positive effectors. Only the mature GIP constructs were active, whereas the full-length protein was not. Mature Glucagon was active regardless of the tag, whereas the full-length protein was generally inactive. The exception was full-length Glucagon without an Fc fusion, which showed moderate activity in the assay. Inspection of the capillary-electrophoresis results indicated that additional processing to a mature form was occurring with this construct but not with the other Fc fusions. For the larger collection used in this study, there is no easy test to determine whether or not the expression or activity of any individual target may be compromised as a protein fusion, and so a negative result is ambiguous. These results emphasize the value in evaluating multiple variants of each target.
Processing of the prioritized extracellular proteome collection of 1,174 expression constructs was performed over several weeks using the PEPP platform shown in Fig. S1. Approximately 200–400 secreted proteins were expressed and affinity purified each week. A protein concentration of >20 μg/mL, with endotoxin levels <15 EU/mL, was set as a minimal standard; 806 of 1,174 expression clones met this threshold, providing an average of 100–150 μg of protein in 1.5 mL volume per construct with virtually all samples below endotoxin detection limits. In a significant number of cases, proteins resolved by capillary electrophoresis as fairly diffuse bands, likely because of glycosylation and other posttranslational modifications. In most cases, mass-spectral data were not taken because of the large degree of posttranslational modification and microheterogeneity of glycosylation. As an example, the purified proteins for the three constructs of PEDF as resolved by capillary electrophoresis are shown in Fig. S2. All proteins resolved as bands consistent with expected molecular mass for denatured reduced samples and were shown to be N-glycosylated, as shown by a shift in mobility after treatment with the deglycosidase, PNGase-F.
The final protein set of 806 proteins was assembled and dispensed in single-use aliquots for cell-based screening. The 806 proteins were distributed as 30% no-Fc, 30% N-Fc, and 40% C-Fc (Table S2). The success rate of producing protein that met minimal standards was similar for the three types of expression constructs (∼70%). Of the 529 target genes attempted, 443 (84%) yielded sufficient protein with at least one construct to allow screening for biological activity; the majority (55%) of genes was represented by at least two construct forms in the purified protein set.
Primary Screen for Proteins Promoting hESC Self-Renewal.
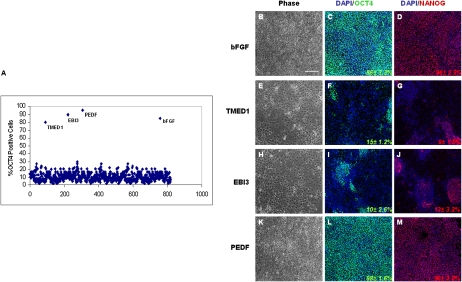
A recently reported high-throughput hESCs self-renewal assay (23) was adapted to screen the collection of 806 purified proteins in 80-μl assay volumes in a 384-well format. This assay measures OCT4, a transcription factor expressed in undifferentiated hESCs (2). Briefly, hESC lines H9 (WA-09) and H1 (WA-01) grown on Matrigel coated plates under feeder-free culture conditions were dissociated into a single-cell suspension and plated in 384-well plates with unconditioned medium (UM) lacking bFGF or TGFβ/Activin/Nodal ligands. hESCs were allowed to attach for 24 h and then, were treated with individual purified proteins. After a 7-day incubation, hESCs were then fixed and stained with antibodies for OCT4. Continued exposure to 100 ng/mL bFGF until day 7 resulted in high OCT4 levels defined as the positive control (promoting self-renewal), whereas bFGF withdrawal at 7 days resulted in differentiation of the hESCs. From the screen, we found that transmembrane emp24 protein-transport domain-containing 1 (TMED1), Epstein–Barr virus-induced 3 (EBI3), and PEDF at 100 ng/mL could individually maintain and increase the number of OCT4 positive cells similar to 100 ng/mL bFGF (Fig. 2A). All three expression constructs for the primary screening hits PEDF1, TMED1, and EBI3, and for the positive controls, bFGF and BMP4 were contained within the protein-screening set, although concentrations of each varied (Table S3). The activities showed for PEDF1 and TMED1 were only associated with the no-Fc fusion proteins and not for the N-Fc or C-Fc fusions, whereas that of EBI3 was elicited only by the N-Fc fusion form, underscoring the need to produce multiple-tag variants of each protein target to enhance the likelihood of avoiding tag-dependent false-negative activities.
Fig. 2.
Human ESC self-renewal assay. (A) Percentage of OCT4 positive hESCs after treatment with 806 different proteins. (B–M) Representative images showing hESC-H9 cell morphology and OCT4 and NANOG expression after growing hESC-H9 cells in either bFGF, TEMD1, EBI3, or PEDF for five passages in UM medium. Values are the mean ± SD for three measurements. (Scale bar, 100 μm.)
TMED1 has been reported to interact with interleukin-1 receptor-like 1 (IL1RL1), but it lacks any similarity to other IL1RL1 ligands (24). EBI3 was first identified by its induced expression in B lymphocytes in response to Epstein–Barr virus infection (25). It encodes a secreted glycoprotein belonging to the hematopoietin receptor family and forms heterodimers with a 28-kDa protein to form interleukin 27 (IL-27) (26). IL-27 regulates T cell and inflammatory responses, in part, by activating the Jak/STAT pathway of CD4+ T cells (27). PEDF is an extracellular multifunctional protein belonging to the serpin superfamily with documented neurotrophic, antiangiogenic, and antitumorigenic properties (28).
Long-Term Growth in Secondary hESC Self-Renewal Assays.
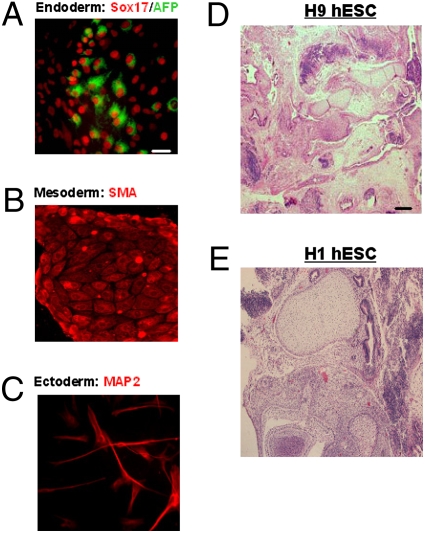
hESCs grown in UM containing either 100 ng/mL TMED1, EBI3, or PEDF seemed morphologically indistinguishable from self-renewing hESCs grown in mouse embryonic fibroblast (MEF) feeder-cell CM after three passages. However, by passage five, hESC cultures grown in the presence of TMED1 and EBI3 lost their typical tight, undifferentiated morphology with concordant loss of OCT4 and NANOG expression (9) (Fig. 2 E–J). The observed reduction of OCT4 expression in hESCs treated with EBI3 or TMED1 by passage five indicates a positive effect on proliferation rather than on hESC self-renewal. In contrast, hESCs grown in the presence PEDF or the bFGF control retained their undifferentiated morphology and maintained OCT4 and NANOG expression. Analysis of undifferentiated stem-cell markers in hESCs grown as long as 15 passages in the presence of PEDF indicated that the cells continued to maintain high levels (>90%) of OCT4, alkaline phosphatase, Tra-1–81, and SSEA-4 (Fig. S3 A–P). After 17 passages, hESCs grown with PEDF remained karyotypically normal (Fig. S4 A and B) and maintained their ability to differentiate into mesodermal (SMA), ectodermal (MAP2), and endodermal (Sox17/AFP) lineages (Fig. 3 A–C). To test in vivo whether or not hESCs remain pluripotent after long-term culture with PEDF (17 passages), we injected 104 undifferentiated hESCs grown with PEDF into SCID mice. Two months after injection, we found that all of the mice injected developed teratomas (Fig. 3 D and E), consistent with the documented association between self-renewal and pluripotency in teratoma formation in this context (2). These results indicate that PEDF promotes hESC self-renewal and is sufficient to maintain their pluripotency.
Fig. 3.
Analysis of hESC cell pluripotency after long-term culture (17 passages) with PEDF. (A–C) H9 cells cultured in vitro were induced to form embryoid bodies, and the derived cells were stained with different differentiation markers: endoderm (AFP and Sox17), mesoderm (SMA), or ectoderm (MAP2). (Scale bar, 100 μm). (D and E) H&E images of teratomas that formed in SCID mice after injection of 1 × 104 hESCs (H9 or H1 cells) that were grown with PEDF for 17 passages. (Scale bar, 50 μm.)
Expression and Requirement for the PEDF Receptor for hESC Self-Renewal.
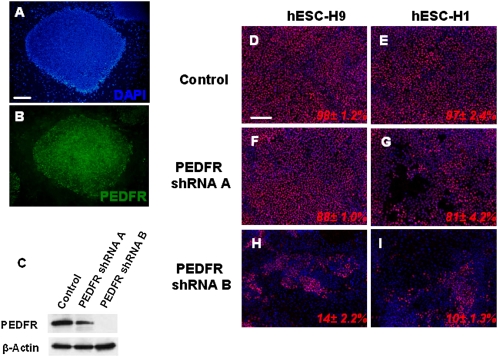
Becerra and coworkers (29) recently identified 80-kDa PLA2/nutrin/patatin-like phospholipase domain-containing 2 (PNPLA2) as a PEDF receptor (PEDFR). By immunofluorescence, we found that undifferentiated hESCs expressed PEDFR (Fig. 4 A and B), suggesting that PEDF may act through this receptor to support hESCs self-renewal. To assess the functional role of PEDFR signaling in hESC self-renewal, the PEDFR mRNA was knocked down using lentivirally delivered PEDFR-targeting shRNAs. PEDFR shRNA-B showed the greatest reduction (>80%) in PEDFR protein levels (Fig. 4C). Treatment of hESCs with PEDFR shRNA-B led to a dramatic change into a differentiated cell morphology and caused hESCs to lose OCT4 expression (Fig. 4 H and I). These results indicate that the PEDFR signaling pathway is important for promoting hESC self-renewal and pluripotency.
Fig. 4.
Expression and effects of PEDF receptor knockdown in hESCs. (A and B) Undifferentiated hESC-H9 cells grown with UM medium plus PEDF (100 ng/mL) were fixed and stained for the PEDF receptor. (C) Western blot analysis of PEDFR protein expression 72 hours after hESC-H9 cells grown in UM medium plus 100 ng/mL PEDF were treated with control or with PEDFR shRNA lenti-particles A or B. (D–I) Analysis of OCT4 expression 72 hours hESC-H9 and H1 cells were treated with control or PEDFR shRNA lenti-particles A or B. Values are the mean ± SD for three measurements. (Scale bar, 100 μm.)
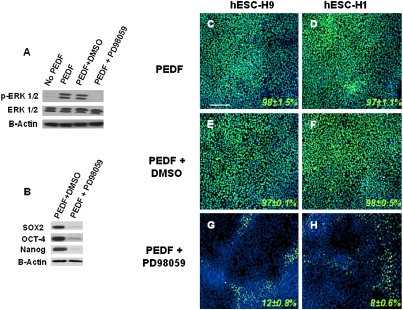
There have been recent reports indicating that PEDF can activate extracellular signal-regulated kinases (ERK1/2). ERK1/2 phosphorylation by bFGF has been recently reported to be essential for hESC self-renewal and pluripotency (30). Consistent with this observation, PEDF was able to stimulate ERK1/2 phosphorylation in hESCs (Fig. 5A), and this phosphorylation could be inhibited by treatment with 10 μM ERK1/2 inhibitor PD98059 (Fig. 5A). Additionally, inhibition of ERK1/2 phoshorylation in hESCs treated with PD98059 in the presence of PEDF caused hESCs to lose OCT4, NANOG, and SOX2 expression (Fig. 5B) and to adopt a differentiated morphology (Fig. 5 G and H). These results suggest that the effects of PEDF on hESC self-renewal are mediated by ERK1/2 signaling.
Fig. 5.
Activation of the ERK1/2 signaling pathway by PEDF supports hESC self-renewal and pluripotency. (A) hES-H9 cells were serum starved for 16 hours and then treated with PEDF (100 ng/mL) or PEDF (100 ng/mL) plus ERK1/2 inhibitor PD98059 (10 μM) for 15 minutes. Cytosolic extracts (30 μg) were subjected to immunoblotting with anti-phospho ERK Ab (pERK; Top) or with anti-general ERK Ab (ERK; Bottom). (B) Analysis of stem cell–associated protein expression 7 days after hESCs were treated with PEDF (100 ng/mL) or PEDF (100 ng/mL) plus PD98059 (10 μM) in UM medium. (C–H) Immunofluorescence analysis of OCT4 expression in hES-H1 and H9 cells treated with PEDF (100 ng/mL), PEDF (100 ng/mL) plus DMSO, or PEDF (100 ng/mL) plus PD98059 (10 μM) in UM medium for 7 days. Values are the mean ± SD for three measurements. (Scale bar, 100 μm.)
PEDF’s effects on stem-cell self-renewal are not restricted to ESCs. For example, PEDF secretion by components of the murine-brain subventricular zone secrete have been shown to promote self-renewal of adult neural stem cells (NSCs) (31), suggesting that PEDF may have a general role in promoting stem-cell self-renewal. We are currently evaluating the effects of PEDF in other stem-cell populations. The above in vitro and in vivo examination of serially passaged hESCs indicates that PEDF can support long-term self-renewal of hESCs in the absence of bFGF or Activin A. Under such conditions, hESCs maintain their characteristic compact-colony morphology (Fig. 2K) and hESC-specific markers (Fig. S3 A–P). More importantly, such long-term, PEDF-expanded hESCs maintain a normal karyotype (Fig. S4 A and B) and full developmental potential as determined by in vitro directed differentiation to the three primary germ-layer derivatives (Fig. 3 A–C) and in vivo complex teratoma formation (Fig. 3 D and E).
Conclusions.
The extracellular proteome represents a rich source of potential therapeutic targets and biology. Beyond the diversity of primary sequence, protein processing and other posttranslational-processing events add to the complexity of the extracellular proteome. Large chemical libraries of over one million compounds are typically screened to find effectors of biological pathways. By contrast, the entire secreted proteome consists of only a few thousand proteins, most of which have one or more biological functions. Screening of cognate pathways with purified secreted proteins will identify new effector proteins and new roles for existing ones. Phenotypic assays such as stem-cell differentiation, cellular proliferation, apoptosis, and cell migration afford the opportunity to interrogate many pathways simultaneously. This collection and an expanded collection encompassing all extracellular genes will undoubtedly identify many biological activities.
Materials and Methods
The hESC lines H9 (WA-09) and H1 (WA-01) were used for this study. hESCs were cultured on mitotically inactivated MEFs as described previously (2); this was followed by feeder-free growth on Matrigel in MEF-CM (4) before 384-well plating. For 384-well plating, cells were harvested after Accutase dissociation for 20 min at 37 °C. At that stage, single-cell suspensions could be obtained without further mechanical dissociation, and dissociated cells displayed high levels of viability (>95% based on trypan exclusion). hESCs were plated at 2,000 cells per well from a stirred single-cell suspension in UM medium [80% DMEM/F-12 and 20% knockout serum replacement supplemented with 1 mM L-glutamine and 1% nonessential amino acids (Invitrogen; 0.1 mM β-mercaptoethanol) using a Multidrop Dispenser (Thermo)].
For details on the large-scale, high-throughput PEPP and other methodology used for immunocytochemistry, immunoblotting, teratoma formation, and PEDFR shRNA knockdown, see SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Paul Anderson, Laura Pratt, Paul Calvin, Dan Sipes, Elisabeth Gardiner, Elena Rodriguez, Yingyao Zhou, Regina Gorski, Jennifer York, Jiadong Zhou, and Dong-In Koo for their efforts in establishing this platform to screen the extracellular proteome. This work was supported by the California Institute for Regenerative Medicine. Cloning efforts were supported in part by National Institutes of Health Grant U54 GM074898 (Joint Center for Structural Genomics) from the National Institute of General Medical Sciences.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0914019107/DCSupplemental.
References
- 1.Lin H, et al. Discovery of a cytokine and its receptor by functional screening of the extracellular proteome. Science. 2008;320:807–811. doi: 10.1126/science.1154370. [DOI] [PubMed] [Google Scholar]
- 2.Thomson JA, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 3.Dahéron L, et al. LIF/STAT3 signaling fails to maintain self-renewal of human embryonic stem cells. Stem Cells. 2004;22:770–778. doi: 10.1634/stemcells.22-5-770. [DOI] [PubMed] [Google Scholar]
- 4.Xu C, et al. Feeder-free growth of undifferentiated human embryonic stem cells. Nat Biotechnol. 2001;19:971–974. doi: 10.1038/nbt1001-971. [DOI] [PubMed] [Google Scholar]
- 5.Levenstein ME, et al. Basic fibroblast growth factor support of human embryonic stem cell self-renewal. Stem Cells. 2006;24:568–574. doi: 10.1634/stemcells.2005-0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu RH, et al. Basic FGF and suppression of BMP signaling sustain undifferentiated proliferation of human ES cells. Nat Methods. 2005;2:185–190. doi: 10.1038/nmeth744. [DOI] [PubMed] [Google Scholar]
- 7.Vallier L, Alexander M, Pedersen RA. Activin/Nodal and FGF pathways cooperate to maintain pluripotency of human embryonic stem cells. J Cell Sci. 2005;118:4495–4509. doi: 10.1242/jcs.02553. [DOI] [PubMed] [Google Scholar]
- 8.James D, Levine AJ, Besser D, Hemmati-Brivanlou A. TGFbeta/activin/nodal signaling is necessary for the maintenance of pluripotency in human embryonic stem cells. Development. 2005;132:1273–1282. doi: 10.1242/dev.01706. [DOI] [PubMed] [Google Scholar]
- 9.Nichols J, et al. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–391. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- 10.Clark HF, et al. The secreted protein discovery initiative (SPDI), a large-scale effort to identify novel human secreted and transmembrane proteins: A bioinformatics assessment. Genome Res. 2003;13:2265–2270. doi: 10.1101/gr.1293003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Y, et al. SPD—a web-based secreted protein database. Nucleic Acids Res. 2005;33:D169–D173. doi: 10.1093/nar/gki093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc. 2007;2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- 13.Käll L, Krogh A, Sonnhammer EL. A combined transmembrane topology and signal peptide prediction method. J Mol Biol. 2004;338:1027–1036. doi: 10.1016/j.jmb.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 14.Anderson NL, et al. The human plasma proteome: A nonredundant list developed by combination of four separate sources. Mol Cell Proteomics. 2004;3:311–326. doi: 10.1074/mcp.M300127-MCP200. [DOI] [PubMed] [Google Scholar]
- 15.Battle T, Antonsson B, Feger G, Besson D. A high-throughput mammalian protein expression, purification, aliquoting and storage pipeline to assemble a library of the human secretome. Comb Chem High Throughput Screen. 2006;9:639–649. doi: 10.2174/138620706778700143. [DOI] [PubMed] [Google Scholar]
- 16.Diabetes Genetics Initiative of Broad Institute of Harvard and MIT, Lund University, and Novartis Institutes of BioMedical Research et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 17.Zeggini E, et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316:1336–1341. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McClurg P, et al. Genomewide association analysis in diverse inbred mice: Power and population structure. Genetics. 2007;176:675–683. doi: 10.1534/genetics.106.066241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pletcher MT, et al. Use of a dense single nucleotide polymorphism map for in silico mapping in the mouse. PLoS Biol. 2004;2:2159–2169. doi: 10.1371/journal.pbio.0020393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu Q, et al. Crystal structure of an ADP-ribosylated protein with a cytidine deaminase-like fold, but unknown function (TM1506), from Thermotoga maritima at 2.70 A resolution. Proteins. 2008;71:1546–1552. doi: 10.1002/prot.21992. [DOI] [PubMed] [Google Scholar]
- 21.Benjannet S, et al. NARC-1/PCSK9 and its natural mutants: Zymogen cleavage and effects on the low density lipoprotein (LDL) receptor and LDL cholesterol. J Biol Chem. 2004;279:48865–48875. doi: 10.1074/jbc.M409699200. [DOI] [PubMed] [Google Scholar]
- 22.Maxwell KN, Breslow JL. Adenoviral-mediated expression of Pcsk9 in mice results in a low-density lipoprotein receptor knockout phenotype. Proc Natl Acad Sci USA. 2004;101:7100–7105. doi: 10.1073/pnas.0402133101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Desbordes SC, et al. High-throughput screening assay for the identification of compounds regulating self-renewal and differentiation in human embryonic stem cells. Cell Stem Cell. 2008;2:602–612. doi: 10.1016/j.stem.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gayle MA, et al. Cloning of a putative ligand for the T1/ST2 receptor. J Biol Chem. 1996;271:5784–5789. doi: 10.1074/jbc.271.10.5784. [DOI] [PubMed] [Google Scholar]
- 25.Devergne O, et al. A novel interleukin-12 p40-related protein induced by latent Epstein-Barr virus infection in B lymphocytes. J Virol. 1996;70:1143–1153. doi: 10.1128/jvi.70.2.1143-1153.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pflanz S, et al. IL-27, a heterodimeric cytokine composed of EBI3 and p28 protein, induces proliferation of naive CD4(+) T cells. Immunity. 2002;16:779–790. doi: 10.1016/s1074-7613(02)00324-2. [DOI] [PubMed] [Google Scholar]
- 27.Lucas S, Ghilardi N, Li J, de Sauvage FJ. IL-27 regulates IL-12 responsiveness of naive CD4+ T cells through Stat1-dependent and -independent mechanisms. Proc Natl Acad Sci USA. 2003;100:15047–15052. doi: 10.1073/pnas.2536517100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tombran-Tink J, Barnstable CJ. PEDF: A multifaceted neurotrophic factor. Nat Rev Neurosci. 2003;4:628–636. doi: 10.1038/nrn1176. [DOI] [PubMed] [Google Scholar]
- 29.Alberdi E, Aymerich MS, Becerra SP. Binding of pigment epithelium-derived factor (PEDF) to retinoblastoma cells and cerebellar granule neurons. Evidence for a PEDF receptor. J Biol Chem. 1999;274:31605–31612. doi: 10.1074/jbc.274.44.31605. [DOI] [PubMed] [Google Scholar]
- 30.Li J, et al. MEK/ERK signaling contributes to the maintenance of human embryonic stem cell self-renewal. Differentiation. 2007;75:299–307. doi: 10.1111/j.1432-0436.2006.00143.x. [DOI] [PubMed] [Google Scholar]
- 31.Ramírez-Castillejo C, et al. Pigment epithelium-derived factor is a niche signal for neural stem cell renewal. Nat Neurosci. 2006;9:331–339. doi: 10.1038/nn1657. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.