Abstract
Plant steroid hormones, brassinosteroids (BRs), regulate essential growth and developmental processes. BRs signal through membrane-localized receptor BRI1 and several other signaling components to regulate the BES1 and BZR1 family transcription factors, which in turn control the expression of hundreds of target genes. However, knowledge about the transcriptional mechanisms by which BES1/BZR1 regulate gene expression is limited. By a forward genetic approach, we have discovered that Arabidopsis thaliana Interact-With-Spt6 (AtIWS1), an evolutionarily conserved protein implicated in RNA polymerase II (RNAPII) postrecruitment and transcriptional elongation processes, is required for BR-induced gene expression. Loss-of-function mutations in AtIWS1 lead to overall dwarfism in Arabidopsis, reduced BR response, genome-wide decrease in BR-induced gene expression, and hypersensitivity to a transcription elongation inhibitor. Moreover, AtIWS1 interacts with BES1 both in vitro and in vivo. Chromatin immunoprecipitation experiments demonstrated that the presence of AtIWS1 is enriched in transcribed as well as promoter regions of the target genes under BR-induced conditions. Our results suggest that AtIWS1 is recruited to target genes by BES1 to promote gene expression during transcription elongation process. Recent genomic studies have indicated that a large number of genes could be regulated at steps after RNAPII recruitment; however, the mechanisms for such regulation have not been well established. The study therefore not only establishes an important role for AtIWS1 in plant steroid–induced gene expression but also suggests an exciting possibility that IWS1 protein can function as a target for pathway-specific activators, thereby providing a unique mechanism for the control of gene expression.
Keywords: plant growth, transcription regulation, transcription elongation, RNA polymerase II, Spt6
Plant steroid hormones, brassinosteroids (BRs), regulate diverse plant growth and developmental processes such as cell elongation, vascular development, photomorphogenesis, reproduction, and stress responses (1, 2). Unlike animal steroid hormones that bind to nuclear receptors to directly regulate expression of target genes, BRs function through membrane-localized receptor BRI1 and several other components to regulate the protein levels and activities of BES1 and BZR1 family transcription factors (3 –8), which have six members in Arabidopsis. Although knockout of BES1 alone does not give obvious phenotype, knockdown of BES1 and BZR1 by RNAi leads to a semidwarf phenotype, suggesting that this family of proteins function redundantly in BR signaling (9). BES1 and BZR1 have atypical basic helix–loop–helix (bHLH) DNA binding domain and bind to E-box (CANNTG) and/or BRRE (BR Response Element, CGTGT/CG) to regulate BR target gene expression (9, 10). In addition, BES1 interacts with other transcription factors such as BIM1 and AtMYB30 as well as putative histone demethylases ELF6 and REF6 (9, 11, 12). Consistent with the important roles of BES1 in mediating BR signaling, the gain-of-function mutant bes1-D, in which BES1 protein accumulates to high level, displays constitutive BR response phenotypes, including excessive cell elongation, resistance to the BR biosynthesis inhibitor brassinazole (BRZ), and increased expression of BR-induced genes (7). Despite the emerging findings, knowledge about the transcriptional mechanisms by which BES1 and BZR1 regulate gene expression is still limited.
IWS1/SPN1 protein was firstly identified in yeast (13, 14). It was reported to be involved in postrecruitment of RNAPII-mediated transcription and required for the expression of several inducible genes (14). It was also copurified with RNAPII and several transcription elongation factors, including Spt6, Spt4/Spt5, and TFIIS (13, 15, 16). Recent studies indicated that the yeast IWS1/SPN1 recruited Spt6 and the SWI/SNF chromatin remodeling complex during transcriptional activation (17). Human IWS1 has been shown to facilitate mRNA export and to coordinate Spt6 and the histone methyltransferase HYBP/Setd2 to regulate histone modifications (18, 19). IWS1 appears to be essential, as depletion/reduction of IWS1 gene results in lethality in yeast (14) and significant inhibition of human cell proliferation in vitro (20). However, the biological function of IWS1/SPN1 and mechanism of its action remain largely elusive.
In this study, we found that AtIWS1, the yeast and human counterparts of which are implicated in gene expression processes after RNAPII recruitment (14, 17 –19, 21), interacts with transcription factor BES1 and is required for BR-induced gene expression. Our results therefore establish an important role for AtIWS1 in plant steroid–induced gene expression and provide insights into the functions of this conserved protein across eukaryotes as well as into the mechanism of transcriptional regulation.
Results
seb1 Suppresses bes1-D Phenotype and Has Reduced BR Response.
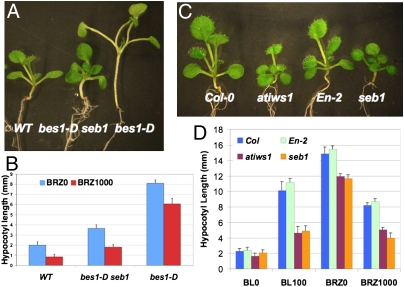
In an attempt to identify BES1 downstream signaling components, we screened for suppressors of bes1-D gain-of-function mutant. One of the suppressors, bes1-D seb1, was selected for further studies because of its strong suppression of the bes1-D phenotype (Fig. 1). seb1 is a single recessive mutation, as the F1 seedlings of bes1-D seb1 and bes1-D cross displayed bes1-D phenotype and F2 segregated suppressors showed a 3:1 ratio. Whereas bes1-D seedlings displayed excessive stem elongation, bes1-D seb1 seedlings had reduced stem elongation (Fig. 1A). Unlike bes1-D, which was resistant to BRZ inhibition in hypocotyl elongation assays, the bes1-D seb1 double mutant restored BRZ sensitivity, suggesting that this suppressor gene is required for the bes1-D phenotype (Fig. 1B).
Fig. 1.
seb1 Suppresses bes1-D phenotype and has reduced BR response. (A) Two-week-old light-grown seedlings. (B) Hypocotyl elongation assay with 2-week-old light-grown seedlings in the absence or presence of 1000 nM BRZ. (C) Two-week-old light-growing seedlings of Col-0, T-DNA insertion line atiws1 (SALK_056238) in Col-0 background, En-2, and seb1 single mutant in En-2 background. (D) Hypocotyl elongation assays with 2-week-old light-grown seedlings (0 and 100 nM BL), or 5-day-old dark-growing seedlings (0 or 1,000 nM BRZ). A total of 15–20 seedlings were used for each line for each condition and the difference between mutants and wild-type were significant, as measured by Student’s t test (P < 0.05).
We next cloned the SEB1 gene by map based cloning (Fig. S1A). The seb1 mutation was first narrowed down to a 60-kb region by a set of flanking markers. Five candidate genes in the interval were then sequenced. A “G” to “A” mutation in Arabidopsis gene At1g32130 was identified, which created an early stop codon in the middle of the gene (Fig. S1B). Expression of a genomic clone of At1g32130 in the bes1-D seb1 mutant complemented the dwarf phenotype (Fig. S1C), indicating that the identified mutation is responsible for the seb1 phenotype. At1g32130 and its close homolog At4g19000 (44% protein sequence identity) have not previously been characterized in plants, and the C-terminal domain shows high homology to C-termini of IWS1 proteins found in other species, including yeast and human beings (Fig. S1D). We therefore named these two Arabidopsis genes AtIWS1 and AtIWS2, respectively. The seb1 mutant by itself also shows a dwarf phenotype compared to En-2 wild-type (Fig. 1C). In addition, we obtained a mutant line, atiws1, which contains a T-DNA insertion in the third exon of the AtIWS1 gene, most likely resulting in a null allele (Fig. S2). The atiws1 in Col-0 background and the seb1 in En-2 background, both null alleles, have similar semidwarf phenotypes with reduced stem elongation in both seedling and adult plants (Fig. 1C and Fig. S3). Whereas AtIWS1 has high levels of expression in both seedling and adult plants, AtIWS2 has barely detectable expression in general (http://bbc.botany.utoronto.ca/efp/cgi-bin/efpWeb.cgi). Consistent with the gene expression data, the T-DNA knockout line atiws2 shows no visible growth phenotype (Fig. S4). Furthermore, the double knockout line atiws1atiws2 and seb1atiws2 do not enhance the atiws1/seb1 semidwarf phenotype (Fig. S4). We therefore conclude that AtIWS1 is the active member in this gene family in BR signaling.
The seb1 and atiws1 mutants not only have overall dwarf phenotypes, but also exhibit altered BR responses in the hypocotyl elongation assays (Fig. 1D). Under light, the hypocotyl lengths of the seb1 and atiws1 mutants were slightly shorter than the wild-type (WT) controls in the absence of BL; however, BL-induced hypocotyl elongation was greatly reduced in both mutants compared with WT. Similarly, in the dark, whereas mutant seedlings were ∼20% shorter than WT without BRZ treatment, they were 50% shorter when treated with 1 μM BRZ, suggesting that the mutants are hypersensitive to reduced BR levels. These results demonstrated that AtIWS1 is required for BR-induced cell elongation.
BR-Induced Gene Expression Is Decreased in seb1 Mutant.
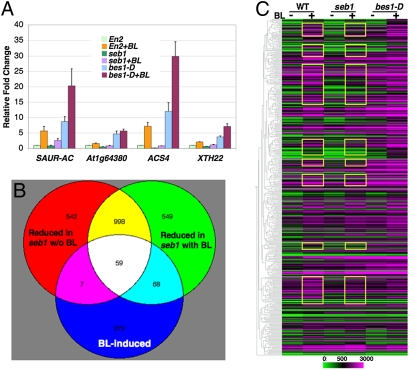
To understand the molecular basis for impaired BR response and growth phenotype of the seb1 mutant, we first tested the expression of several known BR target genes using quantitative RT-PCR. The expression of all four genes tested were significantly reduced in the seb1 mutant and increased in the bes1-D mutant, especially under BL-induced conditions (Fig. 2A). To gain a global view of how many BR-regulated genes are affected in the mutants, we performed microarray analyses using WT, seb1, and bes1-D seedlings (all in En-2 background) with or without BL treatments. These experiments identified 3138 and 3824 differentially expressed genes in seb1 and bes1-D, respectively, compared with WT (Fig. S5). The substantial overlap (1437 genes) between AtIWS1- and BES1-regulated genes suggests that AtIWS1 plays important roles in the BR signaling pathway. To understand this further, we analyzed in greater depth the genes that were induced by BL and were reduced in seb1 mutant (Fig. 2 B and C). Of the 406 genes that were induced by BL, ≈70% were up-regulated in bes1-D (Fig. 2C and Table S1). Comparison of the seb1 and WT gene expression profiles indicated that a large fraction of the BL-responsive genes were also regulated by AtIWS1: 31% (127/406) were reduced in seb1 in the presence of BL, whereas 16% (66 / 406) were reduced in seb1 in the absence of BL (Fig. 2B and Table S1–Table S3). Most of the BL-induced genes that were reduced in seb1 (59/66) were also included in the 127 genes reduced in seb1 in the presence of BL (Fig. 2B). The same conclusion can be drawn from the gene clustering analysis with the 406 BR-induced genes (Fig. 2C). These genome-wide analyses revealed that AtIWS1 plays important roles in BL-induced gene expression.
Fig. 2.
BR induced gene expression is decreased in seb1 mutant. (A) Quantitative RT-PCR was performed with 2-week-old seedlings treated with or without 1,000 nM BL. The gene expression levels were significantly reduced in seb1 and increased in bes1-D as tested by two-way ANOVA F test (P < 0.01). The average and standard deviations were from three biological repeats. (B) Venn diagram shows the overlap genes among different groups. The lists of BL-induced genes and those reduced in seb1 with or without BL treatments are presented in Table S1, Table S2, and Table S3 (C) Clustering analysis of 406 BL-induced genes by GeneSpring program. Yellow rectangles indicated genes reduced in seb1 in the presence of BL.
To further understand the reduced growth phenotype of seb1, we examined genes implicated in cell elongation (Table 1). BRs promote cell elongation by inducing the expression of the genes involved in cell wall–loosening or -modifying enzymes, including xyloglucan endotransglycosylase/hydrolase (XTH), xyloglucan endotransglycosylase–related (XTR), pectin lyase, pectinesterase, and expansins (ATEXP) (22 –24). There are 27 cell wall–related genes that are induced by BL (Table S1); 11 of these genes have reduced expression in seb1 and up-regulated expression in bes1-D (Table 1). In most of the cases, the induction by BL is also reduced in seb1 compared with WT. Taken together, the global gene expression studies indicated that the expression of many BL-induced genes, including those involved in cell elongation, is reduced in seb1 mutant, which explains the impaired growth and BR response phenotypes of the mutants.
Table 1.
Expression levels of cell elongation related genes in seb1 and bes1-D mutants
| Gene | En-2 | seb1 | bes1-D | Gene | |||||||||
| no. | Mock | SD | BL | SD | Mock | SD | BL | SD | Mock | SD | BL | SD | name |
| AT1G10550 | 356 | 46 | 854 | 58 | 207 | 5 | 396 | 13 | 1,444 | 100 | 1,490 | 39 | XTH33 |
| AT4G30290 | 468 | 127 | 3,210 | 150 | 537 | 22 | 2,107 | 249 | 4,389 | 22 | 9,575 | 322 | XTH19 |
| AT1G65310 | 365 | 13 | 1,298 | 102 | 431 | 73 | 912 | 67 | 1,671 | 73 | 2,335 | 535 | XTH17 |
| AT3G07010 | 423 | 43 | 975 | 40 | 340 | 40 | 647 | 77 | 800 | 40 | 1,100 | 66 | Pectate lyase |
| AT3G23730 | 1,365 | 205 | 3,087 | 170 | 996 | 36 | 1,670 | 93 | 1,570 | 36 | 3,138 | 24 | XTR7 |
| AT4G25810 | 896 | 132 | 1,836 | 425 | 497 | 87 | 889 | 153 | 4,951 | 87 | 7,486 | 893 | XTR6 |
| AT5G57560 | 166 | 25 | 273 | 26 | 142 | 22 | 196 | 4 | 305 | 22 | 333 | 62 | XTH22/TCH4 |
| AT1G04680 | 1,012 | 131 | 1,657 | 90 | 676 | 65 | 938 | 89 | 813 | 65 | 1,164 | 199 | Pectate lyase |
| AT3G29030 | 381 | 32 | 551 | 63 | 225 | 34 | 302 | 26 | 549 | 34 | 420 | 59 | ATEXPA5 |
| AT2G37640 | 1,630 | 226 | 2,242 | 194 | 759 | 101 | 1,124 | 60 | 1,839 | 101 | 1,681 | 25 | ATEXPA3 |
| AT3G10720 | 2,237 | 201 | 3,030 | 288 | 2,828 | 115 | 2,127 | 53 | 5,244 | 115 | 4,917 | 134 | Pectinesterase |
Gene expression levels were determined by microarray analysis with GeneChip Affymetrix Arabidopsis ATH1 Genome Arrays. Averages and SDs are derived from three biological repeats.
AtIWS1 Interacts with BES1 Both in Vitro and in Vivo.
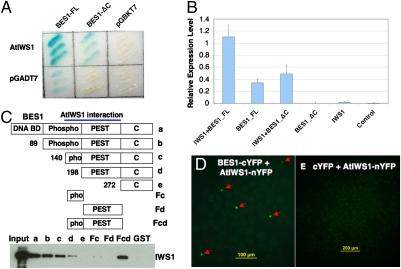
The suppression of bes1-D phenotype by seb1 and the reduction of BL-induced gene expression in the mutant prompted us to test the hypothesis that AtIWS1 directly interacted with BES1. We first tested the interaction using a yeast–two-hybrid system. Although BES1 by itself moderately increased the reporter gene expression, coexpression of AtIWS1 and BES1 significantly activated the reporter gene expression by both qualitative (Fig. 3A) and quantitative (Fig. 3B) assays. We also made a BES1-C-terminal deletion mutant that did not show activation by itself; however, coexpression with AtIWS1 still led to strong activation of the reporter gene, although AtIWS1 itself did not activate the reporter gene expression (Fig. 3 A and B). This observation indicated that BES1 and AtIWS1 interacted with each other in yeast. We next mapped the interaction site on BES1 by GST pull-down assays and found that the central part of BES1 (aa 140–271) was sufficient for interaction with AtIWS1 (Fig. 3C). We also tested whether BES1 interacts with AtIWS1 in vivo by Bimolecular Fluorescence Complementation (BiFC) experiments (25) using an Arabidopsis protoplast expression system (26). BES1 and AtIWS1 were fused to C-terminal YFP and N-terminal YFP, respectively. Strong fluorescence can be observed in the nucleus when BES1-cYFP and AtIWS1-nYFP were cotransformed into the protoplasts, indicating the in vivo interaction of BES1 with AtIWS1 proteins (Fig. 3D). No positive signals were observed when AtIWS1-nYFP was coexpressed with cYFP (Fig. 3E). Consistent with its interaction with BES1, AtIWS1-GFP is localized in the nucleus (Fig. S6).
Fig. 3.
AtIWS1 interacts with BES1 both in vitro and in vivo. In yeast two-hybrid experiments to detect direct protein–protein interactions, β-galactosidase (LacZ) activity was detected with X-gal (A) or with ONPG in a quantitative liquid-culture assay (B). (C) GST pull-down experiments using MBP-AtIWS1 and GST tagged full-length or truncated BES1. Full-length BES1 includes a DNA binding domain (DNA BD), BIN2 phosphorylation domain (Phospho), PEST motif, and the C-terminal domain. AtIWS1 was detected by a Western blot with anti-MBP antibody. (D) Cotransformation of Arabidopsis protoplasts with BES1-cYFP and AtIWS1-nYFP lead to the reconstitution of YFP activity in the nuclei, as indicated by arrows. (E) Cotransformation of cYFP and AtIWS1-nYFP did not produce any positive signals.
AtIWS1 Is Likely Involved in Transcription Elongation.
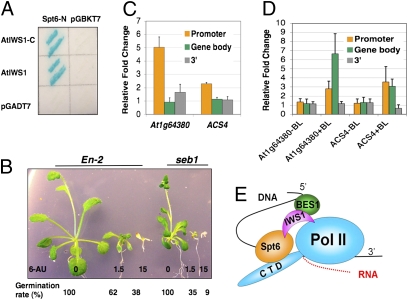
In yeast and human systems, IWS1/SPN1 is an important transcriptional regulator with multiple critical roles that are not fully understood. It is implicated in transcription elongation by interacting directly with elongation factor Spt6. We reasoned that if AtIWS1 were functionally similar to IWS1 from yeast and animal systems, it would interact with the conserved Spt6 (At1g65440) in Arabidopsis as well. We then tested the hypothesis and found that the conserved N terminus of AtSpt6 (1–482aa) interacts with either full-length or C terminus (203–502aa) of AtIWS1 in the yeast–two-hybrid assay (Fig. 4A).
Fig. 4.
AtIWS1 is likely involved in transcription elongation. (A) Yeast-two-hybrid assays to detect interactions between AtIWS1 or AtIWS1 C-terminal (IWS1 domain) and Arabidopsis Spt6 (At1g65440) N-terminal domain. (B) Four-week-old plants in the media with indicated concentrations (mg/L) of 6-AU. The germination rates were determined after 3-week of culture under light. The assay was repeated three times, and representative results are shown. (C) ChIP using bes1-D seedlings with anti-BES1 antibody and control IgG. (D) ChIP using AtIWS1-TAP transgenic and WT seedlings. The 5srRNA was used as internal control for C and D. The average and standard deviations were derived from three to four biological replicates. (E) A model for AtIWS1 function in BES1 mediated gene expression.
The association of AtIWS1 with AtSpt6 implies that AtIWS1 is involved in transcription elongation, like IWS1/SPN1 in other systems. To test the hypothesis, we examined the germination and growth of seb1 in medium containing 6-azauracil (6-AU), which is commonly used as transcription elongation inhibitor in yeast since it can cause depletion of free nucleotide levels therefore decrease transcription elongation efficiency (27). As the concentration of 6-AU was increased, both the germination rate and plant growth were inhibited. More significantly, seb1 was more sensitive to the inhibitor (Fig. 4B), supporting a role for AtIWS1 in transcription elongation.
If AtIWS1 is indeed involved in RNAPII postrecruitment and transcription elongation steps in Arabidopsis, the protein should be associated with the transcribed regions of the target genes in vivo, which should differ from the BES1’s binding patterns. We used Chromatin immunoprecipitation (ChIP)-qPCR to test the hypothesis. Two BES1 target genes (At1g64380 and At2g22810/ACS4) were chosen because of their long promoters and ORFs that would facilitate the ChIP assays (Fig. S7). As expected, BES1 was shown to interact mostly with the promoters but not the transcribed regions or the 3′ regions in both genes (Fig. 4C). In contrast, AtIWS1 showed strong association with the transcribed regions and moderate association with the promoters, only in the presence of BL (Fig. 4D). This demonstrated a correlation between the activation of BR signaling/BES1 accumulation and the association of AtIWS1 to the target genes. These results are consistent with the notion that IWS1 functions at postrecruitment and transcription elongation steps.
Discussion
In this study, we found that loss of function in SEB1/AtIWS1 gene significantly reduced BR response as measured by hypocotyl elongation assay and caused genome-wide decrease in BR target gene expression. AtIWS1 was found to directly interact with BES1 by yeast–two-hybrid and bimolecular fluorescence complementation assays as well as GST pull-down experiment. AtIWS1 interacted with transcription elongation factor Spt6 through conserved domains in each of the proteins. Moreover, loss-of-function mutant of IWS1 gene seb1 showed hypersensitive response to transcription elongation inhibitor 6-AU. Finally, the ChIP experiments demonstrated that AtIWS1 binding was enriched at both transcribed and promoter regions in the target genes, which was clearly different from that of BES1, which is enriched only in promoter regions. Based on these data, we propose a model in which BES1 binds directly to AtIWS1 and recruits it to BR-responsive gene promoters under BR-induced conditions (Fig. 4E). This model predicts that the seb1 phenotype would be more evident when the BR pathway was activated, which was proved by both the BR response and the genome-wide gene expression analyses of the seb1 mutant.
All of the available evidence suggested that IWS1 family proteins function after RNAPII recruitment steps to regulate gene expression, although the molecular mechanisms of IWS1 action remain to be fully defined. Yeast IWS1/SPN1 was originally identified in a genetic screen with a mutation that suppresses a postrecruitment-defective TBP (TATA binding protein) allele (14). Careful genetic studies indicated that IWS1/SPN1 functions at the postrecruitment step (14). IWS1 was also implicated in transcription elongation, as it interacts with transcription elongation factor and histone chaperone Spt6 and was copurified with several transcription elongation complexes (13, 15, 16). In this study, we showed several lines of evidence that, like its counterparts in yeast and animal systems, AtIWS1 is also involved in RNAPII postrecruitment/transcription elongation processes.
Our study provides compelling evidence that regulation of gene expression can occur after RNAPII recruitment. Transcription is a multiphase, highly concerted process, involving numerous factors at many stages. Although previous studies of transcription regulation were primarily focused on the initiation step, the importance of the regulation at transcription elongation has started to be recognized more recently (28 –32). Recent genomic studies suggest that a large portion of genes in a mammalian system can be regulated at postinitiation steps (29, 31). For example, it has been shown that hundreds of human genes had no detectable transcripts while their promoters were occupied by transcription preinitiation complex (31). Similarly, it was also observed in human cells that more than 30% of genes had active histone modification markers as well as initiating form of RNAPII at their promoters without detectable transcripts (29). The widespread phenomena may be explained by various mechanisms including abortive initiation, low processivity, and promoter-proximal pausing of RNAP II (29). Yet the molecular mechanisms by which the large number of genes is regulated after RNAPII recruitment by cellular signaling remain to be fully understood. Some transcriptional activators can influence subsequent steps including promoter clearance (33), promoter-proximal pausing through P-TEFb (34), and elongation rate of RNAPII possibly by interacting with TFIIH (35). For example, the heat shock activator HSF can indirectly recruit P-TEFb complex, which phosphorylates RNAP II and stimulates promoter-paused RNAPII to enter into productive elongation in response to heat shock (36). In addition, certain types of transcription activators were able to promote transcription elongation in vitro, correlating with their abilities to interact with TFIIH, the kinase activity of which can convert RNA polymerase from a nonprocessive to a processive form (32, 35, 37). The genetic, genomic, and molecular evidence presented in this study clearly demonstrates that the transcriptional factor BES1 can recruit AtIWS1 to promote plant steroid hormone–regulated gene expression. Our study provides genetic evidence that a transcription regulator involved in postrecruitment/transcription elongation can be directly targeted by a pathway-specific transcriptional factor, which reveals a unique regulatory mechanism for the control of gene expression.
Our results extended our understanding about the mechanisms of BES1-regulated gene expression. We previously showed that BES1 could interact with ELF6 (Early Flowering 6) and its homolog REF6 (Relative of Early Flowering 6), which are Jumonji N/C (JmjN/C) domain-containing histone demethylases, to modulate BR responses and other developmental processes such as flowering time control. The study of AtIWS1, on the other hand, shows that BES1 not only participates in histone modifications during transcription initiation but can also regulate postrecruitment/transcription elongation by interacting with factors such as AtIWS1. It has been reported that TRIP-1 (TGF-beta Receptor Interacting Protein-1), an essential subunit of eIF3 protein translation initiation complex, is phosphorylated by BR receptor BRI1, which may modulate its activity and influence protein translation (38). BR signaling, therefore, can affect the expression of target genes at multiple levels, including chromatin modification and transcription initiation/elongation, as well as translation.
Our results provide important insights into the biological function of IWS1 family proteins. Despite its apparent importance in the regulation of gene expression, the biological functions of IWS1 family proteins remain elusive. Yeast IWS1 is essential as complete loss-of-function mutant is lethal (14). Similarly, reduction of human IWS1 expression by RNAi leads to the arrest of cell growth (20). However, a nonlethal allele of yeast IWS1/SPN1 reveals that IWS1/SPN1 is required for the inducible expression of several genes, including PHO5 induced by phosphate starvation as well as both HIS3 and HIS4 by histidine deprivation (14). Similarly, we have observed that AtIWS1 was required for BL induction of many BR target genes, including those required for cell elongation (Table 1). Consistent with the reduced expression of the BR-induced genes, atiws1/seb1 mutant displayed defect in responding to BRs and overall stunted growth phenotype. Therefore, IWS1 may function to respond to various environmental conditions to regulate gene expression for optimal growth and fitness. In addition to affecting approximately one third of BR-regulated genes, AtIWS1 appears to influence up to 14% of all genes genome-wide (Fig. 3B and Fig. S5). Although some of the differential gene expression in seb1 mutant may be due to the pleiotropic effect, it is very likely that IWS1 also regulates gene expression in other processes, in addition to the established role in BR-regulated gene expression.
Unlike in yeast and human systems (14, 20), AtIWS1 is not essential for Arabidopsis survival. Although Arabidopsis seems to be the only species with two copies of IWS1 genes among the model organisms, the two genes do not seem to function redundantly (Fig. S4). The difference in phenotypic severity between Arabidopsis and other systems may be due to the possibility that the target genes of IWS1 are essential in yeast and human beings but not in plants, or may be due to the existence of possible functional redundant counterparts in Arabidopsis. Nevertheless, the viable seb1/atiws1 mutant provides a unique tool to dissect the functions of IWS1 family proteins by genetic and genomic approaches. Further studies should help to reveal the molecular functions of this important and conserved protein family in gene expression and their biological roles in growth and development in responding to environmental cues.
Materials and Methods
Plant Materials, Growth Conditions, and Hypocotyl Elongation Assays.
Arabidopsis thaliana ecotype Columbia (Col-0) and Enkheim-2 (En-2) were used as wild-type controls. Plant transformation, growth conditions, and hypocotyl elongation assays were carried out as previously described (11). The AtIWS1 and AtIWS2 T-DNA knockout lines are SALK_056238 and SALK_006346, respectively (39).
bes1-D Suppressor Screen.
Approximately 40,000 bes1-D seeds were treated with 0.2% EMS (ehyl methanesulfonate) for 14 h and grown in 40 pools (1,000/pool). M2 seeds from each pool were grown on 1/2MS medium that contains 1 μM Brassinazole (a gift from Prof. Tadao Asami) under light. Although bes1-D seedlings were resistant to the inhibitory effect of BRZ, suppressors that showed sensitivity to the inhibitor were selected. The putative suppressors were confirmed in M3 generation and backcrossed to bes1-D to determine the dominant/recessive nature of the mutations.
Map-Based Cloning.
The bes1-D seb1 double mutant in En-2 background was crossed to bes1-D single mutant in polymorphic Wassilewskija (Ws) background. All of the F1 offspring showed bes1-D phenotype, indicating that seb1 is a recessive mutation. In the F2 generation, approximately three fourths of plants showed bes1-D phenotype, whereas the remaining one fourth of plants showed bes1-D seb1 phenotype, suggesting that seb1 is a single gene mutation. Those bes1-D seb1 plants in F2 generation were used as mapping population. The mutation was narrowed down to 60 kb on chromosome 1 by using ∼800 bes1-D seb1 plants and 10 CAPS (40), dCAPS (41), or SSLP (42) markers (M1–M10; Table S4). Five candidate genes in this interval were then sequenced, and a single base pair change was detected.
Gene Expression and Microarray Analysis.
Total RNA was extracted and purified from seedlings of different genotypes and treatments using RNeasy Mini Kit (Qiagen) with on-column DNase digestion, following the manufacturers’ instructions. Microarray experiments were performed using Affymetrix ATH1 Genome Arrays with 2-week-old light grown En2, seb1 and bes1-D seedlings with long-day (16 h) conditions. The plants were treated with either 1 μM BL or DMSO as control in liquid 1/2 MS medium for 3 h. Triplicate samples were collected and processed for RNA extraction. Probe labeling, hybridization, and scanning were performed in GeneChip facility at Iowa State University according to the manufacturer’s instructions. Microarray data were normalized by R using MAS 5.0 method in the affy package, and the log2 transformed data set was then analyzed using the limma package. The false discovery rate (FDR) was controlled at 5% using the Benjamini–Hochberg method, and genes with adjusted P value <0.05 were considered to be significantly differentially expressed (43). The clustering analysis was done with GeneSpring program from Agilent Technologies. Quantitative RT-PCR was performed as previously described (11). The 5srRNA was used as reference gene. Three biological replicates and 2–3 technical replicates for each biological replicate were used.
Plasmid Construction.
All primers used in this study are listed in Table S5 For recombinant protein and GST pull-down assay, AtIWS1 coding region was amplified from Col-0 cDNA and incorporated into the pETMALc-H vector (44). BES1 and various deletion constructs were cloned into pET42a(+) (Novagen). For transgenic GFP plants, AtIWS1 ORF was fused with GFP tag flanked by BRI1 promoter (45) and RBCS terminator in pPZP211 (46). For transgenic AtIWS1-TAP(Tandem Affinity Purification)-tag plants, AtIWS1 ORF was fused into vector p35SCTAP with 35S-promoter and the tag containing a protein A module, a six His repeat (6× His) and nine copies of a myc repeat (9xmyc) (47). For bimolecular fluorescence complementation assay, the constructs of N or C -terminus of EYFP were as described previously (12). The coding region of AtIWS1 and BES1 were inserted upstream of YFP-N or YFP-C constructs, respectively.
Phylogenetic Analysis.
Protein sequences were obtained from NCBI, and aligned by ClustalX 2.0 (http://bips.u-strasbg.fr/fr/Documentation/ClustalX). Mrbayes program (http://mrbayes.csit.fsu.edu/index.php) was used to reconstruct the evolutionary tree. After 500,000 iterations in Mrbayes, the split frequency reached as low as 0.005, which suggested high confidence of convergence. The tree was viewed by TreeViewX (http://darwin.zoology.gla.ac.uk/_rpage/treeviewx).
ChIP Assays.
Chromatin immunoprecipitation (ChIP) was performed as described by Craig Pikaard lab based on published methods (48, 49) (http://www.biology.wustl.edu/pikaard/) with 2-week-old bes1-D seedlings using antibodies against BES1 or IgG (Millipore) control for immunoprecipitation. The method for ChIP with 2-week-old WT and AtIWS1-TAP seedlings was slightly modified, using IgG Sepharose beads for immunoprecipitation instead of antibody and protein A beads. The ChIP experiments were performed 3–4 independent times.
GST Pull-Down Assay.
The assay was performed as described previously (7). BES1 and its fragments fused with GST (GST) were purified with glutathione beads (Sigma). AtIWS1 fused with Maltose Binding Protein (MBP) was purified with Amylose resin (NEB). Approximately 5 μg proteins were used in each assay. The products then were detected by Western Blotting using antibody against MBP tag (NEB). Pull-down assays were repeated three times with similar results.
Bimolecular Fluorescence Complementation.
Arabidopsis mesophyll protoplasts were prepared and transformed by PEG-mediated transfection as described previously (26). Protoplasts were observed under an Olympus IX71 fluorescence microscope with an YFP filter, 16–24 h after transfection. The assay was repeated two times with different positive rates (5–10%), depending on transfection efficiency.
Yeast Two-Hybrid and lacZ Assays.
Yeast two-hybrid assay was done as described by Yeast Protocols Handbook (Clontech). Yeast (Y187) transformed with indicated constructs was grown in media lacking Trp and Leu. Positive clones were used for LacZ assay using X-gal (5-bromo-4-chloro-3-indolyl-b-D-galactopyranoside; Sigma) or quantitative liquid assay using ONPG (ortho-nitrophenyl-b-D-galactopyranoside; Sigma).
Supplementary Material
Acknowledgments
We thank ABRC for Arabidopsis T-DNA mutants, Tadao Asami (University of Tokyo) for providing BRZ, Bing Yang for vector p35SCTAP, and Jo Anne Powell-Coffman for critical comments on the manuscripts. This work was supported by National Science Foundation Grant IOS-0546503 and a faculty start-up fund from Iowa State University (to Y.Y.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0909198107/DCSupplemental.
References
- 1.Li J, Chory J. Brassinosteroid actions in plants. J Exp Bot. 1999;50:332–340. [Google Scholar]
- 2.Clouse SD. Molecular genetic studies confirm the role of brassinosteroids in plant growth and development. Plant J. 1996;10:1–8. doi: 10.1046/j.1365-313x.1996.10010001.x. [DOI] [PubMed] [Google Scholar]
- 3.Wang X, et al. Sequential transphosphorylation of the BRI1/BAK1 receptor kinase complex impacts early events in brassinosteroid signaling. Dev Cell. 2008;15:220–235. doi: 10.1016/j.devcel.2008.06.011. [DOI] [PubMed] [Google Scholar]
- 4.Tang W, et al. BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science. 2008;321:557–560. doi: 10.1126/science.1156973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li J, Jin H. Regulation of brassinosteroid signaling. Trends Plant Sci. 2007;12:37–41. doi: 10.1016/j.tplants.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 6.Zhao J, et al. Two putative BIN2 substrates are nuclear components of brassinosteroid signaling. Plant Physiol. 2002;130:1221–1229. doi: 10.1104/pp.102.010918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yin Y, et al. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell. 2002;109:181–191. doi: 10.1016/s0092-8674(02)00721-3. [DOI] [PubMed] [Google Scholar]
- 8.Wang ZY, et al. Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev Cell. 2002;2:505–513. doi: 10.1016/s1534-5807(02)00153-3. [DOI] [PubMed] [Google Scholar]
- 9.Yin Y, et al. A new class of transcription factors mediate brassinosteroid-regulated gene expression in Arabidopsis . Cell. 2005;120:249–259. doi: 10.1016/j.cell.2004.11.044. [DOI] [PubMed] [Google Scholar]
- 10.He JX, et al. BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science. 2005;307:1634–1638. doi: 10.1126/science.1107580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li L, et al. Arabidopsis MYB30 is a direct target of BES1 and cooperates with BES1 to regulate brassinosteroid target gene expression. Plant J. 2009;58:275–286. doi: 10.1111/j.1365-313X.2008.03778.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu X, Li L, Guo M, Chory J, Yin Y. Modulation of brassinosteroid-regulated gene expression by Jumonji domain-containing proteins ELF6 and REF6 in Arabidopsis. Proc Natl Acad Sci USA. 2008;105:7618–7623. doi: 10.1073/pnas.0802254105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krogan NJ, et al. RNA polymerase II elongation factors of Saccharomyces cerevisiae: A targeted proteomics approach. Mol Cell Biol. 2002;22:6979–6992. doi: 10.1128/MCB.22.20.6979-6992.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fischbeck JA, Kraemer SM, Stargell LA. SPN1, a conserved gene identified by suppression of a postrecruitment-defective yeast TATA-binding protein mutant. Genetics. 2002;162:1605–1616. doi: 10.1093/genetics/162.4.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ling Y, Smith AJ, Morgan GT. A sequence motif conserved in diverse nuclear proteins identifies a protein interaction domain utilised for nuclear targeting by human TFIIS. Nucleic Acids Res. 2006;34:2219–2229. doi: 10.1093/nar/gkl239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lindstrom DL, et al. Dual roles for Spt5 in pre-mRNA processing and transcription elongation revealed by identification of Spt5-associated proteins. Mol Cell Biol. 2003;23:1368–1378. doi: 10.1128/MCB.23.4.1368-1378.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang L, Fletcher AG, Cheung V, Winston F, Stargell LA. Spn1 regulates the recruitment of Spt6 and the Swi/Snf complex during transcriptional activation by RNA polymerase II. Mol Cell Biol. 2008;28:1393–1403. doi: 10.1128/MCB.01733-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoh SM, Lucas JS, Jones KA. The Iws1:Spt6:CTD complex controls cotranscrip-tional mRNA biosynthesis and HYPB/Setd2-mediated histone H3K36 methylation. Genes Dev. 2008;22:3422–3434. doi: 10.1101/gad.1720008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoh SM, Cho H, Pickle L, Evans RM, Jones KA. The Spt6 SH2 domain binds Ser2-P RNAPII to direct Iws1-dependent mRNA splicing and export. Genes Dev. 2007;21:160–174. doi: 10.1101/gad.1503107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Z, Zhou Z, Chen G, Bao S. A putative transcriptional elongation factor hIws1 is essential for mammalian cell proliferation. Biochem Biophys Res Commun. 2007;353:47–53. doi: 10.1016/j.bbrc.2006.11.133. [DOI] [PubMed] [Google Scholar]
- 21.Bres V, Yoh SM, Jones KA. The multi-tasking P-TEFb complex. Curr Opin Cell Biol. 2008;20:334–340. doi: 10.1016/j.ceb.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palusa SG, Golovkin M, Shin SB, Richardson DN, Reddy AS. Organ-specific, developmental, hormonal and stress regulation of expression of putative pectate lyase genes in Arabidopsis. New Phytol. 2007;174:537–550. doi: 10.1111/j.1469-8137.2007.02033.x. [DOI] [PubMed] [Google Scholar]
- 23.Becnel J, Natarajan M, Kipp A, Braam J. Developmental expression patterns of Arabidopsis XTH genes reported by transgenes and Genevestigator. Plant Mol Biol. 2006;61:451–467. doi: 10.1007/s11103-006-0021-z. [DOI] [PubMed] [Google Scholar]
- 24.Darley CP, Forrester AM, McQueen-Mason SJ. The molecular basis of plant cell wall extension. Plant Mol Biol. 2001;47:179–195. [PubMed] [Google Scholar]
- 25.Citovsky V, et al. Subcellular localization of interacting proteins by bimolecular fluorescence complementation in planta. J Mol Biol. 2006;362:1120–1131. doi: 10.1016/j.jmb.2006.08.017. [DOI] [PubMed] [Google Scholar]
- 26.Yoo SD, Cho YH, Sheen J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat Protoc. 2007;2:1565–1572. doi: 10.1038/nprot.2007.199. [DOI] [PubMed] [Google Scholar]
- 27.Exinger F, Lacroute F. 6-Azauracil inhibition of GTP biosynthesis in Saccharomyces cerevisiae. Curr Genet. 1992;22:9–11. doi: 10.1007/BF00351735. [DOI] [PubMed] [Google Scholar]
- 28.Core LJ, Lis JT. Transcription regulation through promoter-proximal pausing of RNA polymerase II. Science. 2008;319:1791–1792. doi: 10.1126/science.1150843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saunders A, Core LJ, Lis JT. Breaking barriers to transcription elongation. Nat Rev Mol Cell Biol. 2006;7:557–567. doi: 10.1038/nrm1981. [DOI] [PubMed] [Google Scholar]
- 31.Kim TH, et al. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kadonaga JT. Regulation of RNA polymerase II transcription by sequence-specific DNA binding factors. Cell. 2004;116:247–257. doi: 10.1016/s0092-8674(03)01078-x. [DOI] [PubMed] [Google Scholar]
- 33.Kumar KP, Akoulitchev S, Reinberg D. Promoter-proximal stalling results from the inability to recruit transcription factor IIH to the transcription complex and is a regulated event. Proc Natl Acad Sci USA. 1998;95:9767–9772. doi: 10.1073/pnas.95.17.9767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol Cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 35.Blau J, et al. Three functional classes of transcriptional activation domain. Mol Cell Biol. 1996;16:2044–2055. doi: 10.1128/mcb.16.5.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lis JT, Mason P, Peng J, Price DH, Werner J. P-TEFb kinase recruitment and function at heat shock loci. Genes Dev. 2000;14:792–803. [PMC free article] [PubMed] [Google Scholar]
- 37.Yankulov K, Blau J, Purton T, Roberts S, Bentley DL. Transcriptional elongation by RNA polymerase II is stimulated by transactivators. Cell. 1994;77:749–759. doi: 10.1016/0092-8674(94)90058-2. [DOI] [PubMed] [Google Scholar]
- 38.Ehsan H, et al. Interaction of Arabidopsis BRASSINOSTEROID-INSENSITIVE 1 receptor kinase with a homolog of mammalian TGF-beta receptor interacting protein. Plant J. 2005;43:251–261. doi: 10.1111/j.1365-313X.2005.02448.x. [DOI] [PubMed] [Google Scholar]
- 39.Alonso JM, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 40.Konieczny A, Ausubel FM. A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 1993;4:403–410. doi: 10.1046/j.1365-313x.1993.04020403.x. [DOI] [PubMed] [Google Scholar]
- 41.Neff MM, Neff JD, Chory J, Pepper AE. dCAPS, a simple technique for the genetic analysis of single nucleotide polymorphisms: Experimental applications in Arabidopsis thaliana genetics. Plant J. 1998;14:387–392. doi: 10.1046/j.1365-313x.1998.00124.x. [DOI] [PubMed] [Google Scholar]
- 42.Bell C, Ecker J. Assignment of thirty microsatellite loci to the linkage map of Arabidopsis. Genomics. 1994;19:137–144. doi: 10.1006/geno.1994.1023. [DOI] [PubMed] [Google Scholar]
- 43.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc, B. 1995;57:289–300. [Google Scholar]
- 44.Pryor KD, Leiting B. High-level expression of soluble protein in Escherichia coli using a His6-tag and maltose-binding-protein double-affinity fusion system. Protein Expr Purif. 1997;10:309–319. doi: 10.1006/prep.1997.0759. [DOI] [PubMed] [Google Scholar]
- 45.Li J, Chory J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell. 1997;90:929–938. doi: 10.1016/s0092-8674(00)80357-8. [DOI] [PubMed] [Google Scholar]
- 46.Hajdukiewicz P, Svab Z, Maliga P. The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol. 1994;25:989–994. doi: 10.1007/BF00014672. [DOI] [PubMed] [Google Scholar]
- 47.Rubio V, et al. An alternative tandem affinity purification strategy applied to Arabidopsis protein complex isolation. Plant J. 2005;41:767–778. doi: 10.1111/j.1365-313X.2004.02328.x. [DOI] [PubMed] [Google Scholar]
- 48.Nelson JD, Denisenko O, Sova P, Bomsztyk K. Fast chromatin immunopre-cipitation assay. Nucleic Acids Res. 2006;34:e2. doi: 10.1093/nar/gnj004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gendrel AV, Lippman Z, Yordan C, Colot V, Martienssen RA. Dependence of heterochromatic histone H3 methylation patterns on the Arabidopsis gene DDM1. Science. 2002;297:1871–1873. doi: 10.1126/science.1074950. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.