Abstract
The hepatitis C virus (HCV) life cycle involves multiple steps, but most current drug candidates target only viral replication. The inability to systematically discover inhibitors targeting multiple steps of the HCV life cycle has hampered antiviral development. We present a simple screen for HCV antivirals based on the alleviation of HCV-mediated cytopathic effect in an engineered cell line—n4mBid. This approach obviates the need for a secondary screen to avoid cytotoxic false-positive hits. Application of our screen to 1280 compounds, many in clinical trials or approved for therapeutic use, yielded >200 hits. Of the 55 leading hits, 47 inhibited one or more aspects of the HCV life cycle by >40%. Six compounds blocked HCV entry to levels similar to an antibody (JS-81) targeting the HCV entry receptor CD81. Seven hits inhibited HCV replication and/or infectious virus production by >100-fold, with one (quinidine) inhibiting infectious virus production by 450-fold relative to HCV replication levels. This approach is simple and inexpensive and should enable the rapid discovery of new classes of HCV life cycle inhibitors.
Keywords: cell death, drug, high throughput, rescue, virus assembly
Hepatitis C virus (HCV) infection is a major public health problem, putting ~180 million infected individuals worldwide (3% of the world’s population) at risk for developing cirrhosis, hepatocellular carcinoma, and liver failure. Chronic hepatitis C is a leading cause for liver transplantation in the Western world. The current standard IFN-based therapy is costly, time-consuming, riddled with serious side effects, and cures only ∼50% of patients infected with the most common genotype. Although it is anticipated that small molecules that effectively inhibit the HCV life cycle can complement or even replace IFN therapy, no anti-HCV drugs have yet been approved for human use. Efforts to develop HCV antivirals mostly target only viral replication due to widespread dependence on the HCV replicon model (1 –4), which effectively reproduces the RNA replication aspects of the HCV life cycle but does not allow screens to identify inhibitors of entry or infectious virus assembly and release.
The identification of a genotype 2a isolate of HCV that reproduces the entire viral life cycle in cell culture (5 –7) promised to accelerate the discovery of antivirals targeting all aspects of HCV growth in a more true-to-life model system. Several recent reports describe systems that can in principle screen for inhibitors targeting different steps of the HCV life cycle via the prevention of the spread of cell culture-derived hepatitis C virus (HCVcc) infection in a cell monolayer (8 –11). Very recently, one such screen based on a colorimetric readout following immunostaining with an anti-E2 antibody identified inhibitors of multiple stages of the HCV life cycle (12). Although effective, this approach is labor-intensive and not readily amenable to high-throughput application, requiring antibodies and 12 washing steps in addition to a secondary screen to assess drug cytotoxicity.
We previously created a cell line—n4mBid—that sensitively reports HCV infection via a cell death phenotype by introducing a HCV NS3-4A protease-cleavable derivative of the proapoptotic factor Bid, mBid (13), into the highly HCV-permissive hepatoma cell line Huh-7.5. In the present study, we demonstrate the utility of the n4mBid cell line as a medium for the discovery of inhibitors against different aspects of the HCV life cycle. In particular, we reasoned that n4mBid cells should be able to report the presence of anti-HCV molecules via increased cell viability relative to untreated cells in the presence of HCV infection. Such an assay should be simple, quick, cost-effective, and, importantly, should obviate the need for a separate assay to assess drug hits for cytotoxic effects because toxic drugs should not produce a positive cell viability readout.
Results and Discussion
Viral Inhibitors Effectively Protect n4mBid Cells from HCV Infection.
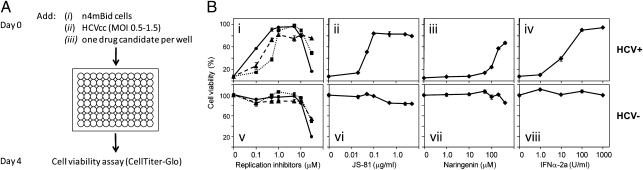
To verify that inhibition of different aspects of the HCV life cycle can alleviate the HCV-induced cytopathic effect experienced by n4mBid cells, we tested the effect of known HCV replication, entry, and assembly/release inhibitors on n4mBid cell viability in the presence of HCV infection. N4mBid cells were seeded in 96-well plates in the presence of both Jc1 HCVcc (14) and small molecules, and cell viability was quantified 4 days later (Fig. 1A). For small-molecule cytotoxicity profiles, regular growth media was used in place of HCVcc. The HCV replication inhibitors VX-950 (15), 2′-C-methyladenosine (2′CMA) (16), and cyclosporin A (17), targeting the NS3-4A protease, NS5B RNA-dependent RNA polymerase, and a required cellular factor [cyclophilin A (18)], respectively, each produced a strong dose-dependent increase in n4mBid cell viability following HCV infection. A recombinant monoclonal antibody (mAb JS-81) against the essential HCV entry factor CD81 (19) also exhibited a dose-dependent protection of n4mBid cells from the HCV cytopathic effect, as did the HCV assembly/release-specific inhibitor naringenin (20) and the cytokine IFNα-2a (Fig. 1B).
Fig. 1.
A n4mBid cell protection anti-HCV drug screen reports the presence of inhibitors targeting multiple stages of the HCV life cycle. (A) Schematic of screen. (B) Effect of known anti-HCV molecules on rescuing n4mBid cells from the HCV-induced cytopathic effect. (Upper) Drug dose–responses reporting n4mBid cell viability in the presence of HCV infection. (Lower) Drug cytotoxicity in the absence of HCV infection. (i and v) HCV replication inhibitors 2′CMA (•), VX-950 (▲), and cyclosporin A (■). (ii and vi) HCV entry-blocking anti-human CD81 mAb, JS-81. (iii and vii) The HCV assembly/release inhibitor naringenin. (iv and viii) IFNα-2a. Cell viability is presented as a percentage of mock-treated cells in the absence of both HCV infection and inhibitors. Values are the mean ± SD of two independent experiments carried out in duplicate.
Library Screening for Antivirals Using n4mBid Cells.
We next screened the LOPAC1280 library consisting of 1280 compounds known to be pharmacologically active in a variety of cellular processes for molecules able to protect n4mBid cells from the HCV cytopathic effect. Many of the compounds in this library are approved for human therapeutic use in clinical trials or are structurally related to clinically approved molecules. Fig. S1 shows a set of representative cell viability results obtained from this screen. In total, >200 molecules mediated a statistically significant increase in cell viability relative to untreated cells in response to HCV infection. Among the 56 hits that most strongly protected n4mBid cells from the HCV cytopathic effect, we identified the known HCV inhibitors cyclosporin A (17), MK-886 (12, 21), and TTNPB (22), as well as five additional compounds recently found in an anti-HCV drug screen targeting multiple aspects of the HCV life cycle (12). Numerous other molecules with reported anti-HCV activity appeared as weaker hits in our screen. A handful of known HCV inhibitors present within the library did not protect n4mBid cells from HCV-mediated cell death. This phenomenon is likely due to a combination of two factors: (i) an extent of HCV life cycle inhibition insufficient to alleviate the HCV cytopathic effect and (ii) drug cytotoxicity. For example, the estrogen receptor antagonist tamoxifen, a HCV replication inhibitor present in the library, did not appear as a hit, likely due to its significant cytotoxicity even while inhibiting HCV replication at 1 μM (23).
We commenced characterization of the leading 55 drug hits from our screen (excluding cyclosporin A) by measuring their dose-dependent effects on the protection of n4mBid cells from the HCV cytopathic effect, as well as on HCV-independent cellular cytotoxicity, confirming that all of the leading 55 drug hits mediated a dose-dependent protection phenotype from HCV-triggered cell death (Fig. S2). To probe the inhibitory effects on HCV replication, infectious virus assembly/release, and entry, the same 55 drug hits were further characterized at concentrations that mediated a significant protection of n4mBid cells from the HCV cytopathic effect while showing minimum cytotoxicity. For HCV replication and assembly/release studies, a HCV reporter virus encoding Gaussia luciferase (Gluc) was used (24), whereas Gluc reporter lentiviral particles pseudotyped with HCV envelope glycoproteins were used to assess small-molecule effects on HCV entry. Forty-seven of the 55 drug hits showed a >40% inhibitory effect on one or more of HCV replication, infectious virus production, and entry (Table 1), demonstrating an exceptionally high representation of true positive HCV life cycle inhibitors among hits produced by the n4mBid cell protection screening approach.
Table 1.
HCV inhibition characteristics of leading 55 hits from n4mBid cell protection small-molecule screen.
| Effect on HCV | ||||||||||
| Compound* | CC50 (μM) | EC50 (μM) | EC90 (μM) | CC50/EC50 | SDC (μM) | Replication | IVP | Entry | % Prt1 | % Prt10 |
| A5HT | >30 | ND | ND | ND | 30 | — | — | — | 12 | 74 |
| Amiodarone (12) | 14 | ND | ND | ND | 5 | — | ▿▿▿ | ▿ | 14 | 81 |
| Amitriptyline | 30 | ND | ND | ND | 10 | — | ▿▿▿ | ▿ | 19 | 75 |
| Amoxapine | 17 | ND | ND | ND | 7 | — | ▿▿▿ | — | 11 | 78 |
| Amperozide | 30 | ND | ND | ND | 5 | — | — | — | 46 | 69 |
| Apigenin | >30 | ND | ND | ND | 10 | — | — | — | 14 | 87 |
| Aprindine | 18 | ND | ND | ND | 10 | ▿ | ▿▿▿ | ▿ | 24 | 66 |
| Benztropine | 40 | ND | ND | ND | 10 | — | ▿▿ | ▿ | 51 | 93 |
| BIO | 8 | 0.70 | 1.3 | 11.4 | 5 | ▿▿▿▿ | ▿▿▿▿ | — | 72 | 0 |
| Biperiden | 30 | ND | ND | ND | 10 | — | ▿▿ | ▿ | 19 | 74 |
| Bromocriptine | >30 | 2.0 | 5.4 | >15 | 10 | ▿▿▿▿ | ▿▿▿ | ▿ | 12 | 70 |
| (±)-Butaclamol | 35 | ND | ND | ND | 10 | — | — | ▿ | 7 | 48 |
| CGS-12066A | 17 | 6.8 | 10.5 | 2.5 | 10 | ▿▿▿▿ | ▿▿▿▿ | ▿▿ | 12 | 75 |
| Cilnidipine | 20 | 4.9 | 10.0 | 4.1 | 10 | ▿▿▿▿ | ▿▿▿▿ | ▿▿ | 17 | 97 |
| Cinnarizine | >30 | ND | ND | ND | 10 | — | — | — | 16 | 68 |
| CPTA | >30 | ND | ND | ND | 10 | — | — | — | 15 | 86 |
| Cyproheptadine (12) | 17 | ND | ND | ND | 5 | — | ▿ | — | 24 | 74 |
| DKI | 18 | ND | ND | ND | 10 | — | — | — | 30 | 86 |
| Ebselen | >30 | ND | ND | ND | 30 | — | — | ▿ | 11 | 91 |
| Etazolate | >60 | 8.0 | 18.0 | >7.5 | 30 | ▿▿▿▿ | ▿▿▿▿ | ▿ | 15 | 80 |
| 5EIA | 17 | ND | ND | ND | 5 | — | ▿▿▿ | — | 8 | 58 |
| Felodipine | >30 | 9.5 | 23.0 | >3.2 | 30 | ▿▿▿▿ | ▿▿▿▿ | ▿▿ | 13 | 59 |
| Flunarizine | 18 | 2.1 | 5.2 | 8.6 | 10 | ▿ | ▿▿ | — | 73 | 85 |
| Fluphenazine (12) | 17 | 4.1 | 8.9 | 4.1 | 10 | ▿ | — | ▿▿▿▿ | 65 | 0 |
| Forskolin | >30 | ND | ND | ND | 10 | — | + | — | 17 | 71 |
| GR 127935 | 7 | 1.6 | 4.0 | 4.4 | 3 | ▿ | ▿ | ▿▿▿▿ | 84 | 0 |
| GR 89696 | 45 | ND | ND | ND | 10 | — | ▿ | ▿ | 11 | 58 |
| GW2974 | 25 | 3.1 | 9.0 | 8.1 | 10 | ▿▿▿▿ | ▿▿▿▿ | ▿▿ | 17 | 68 |
| Ifenprodil | 16 | ND | ND | ND | 10 | ▿ | — | ▿ | 14 | 58 |
| IPFME | 17 | ND | ND | ND | 10 | — | ▿▿▿ | ▿ | 12 | 59 |
| MK-886 (12, 21) | >30 | 7.5 | 11.0 | >4.0 | 10 | — | ▿ | ▿ | 12 | 96 |
| Nemadipine-A | >30 | 10.5 | 31.0 | >2.9 | 30 | ▿▿▿▿ | ▿▿▿▿ | ▿ | 16 | 69 |
| PCperazine (12) | 16 | 4.2 | 9.5 | 3.8 | 10 | ▿ | ▿ | ▿▿▿▿ | 20 | 62 |
| PD 404,182 | >60 | 12.0 | 41.0 | >5.0 | 30 | — | ▿▿ | ▿▿▿▿ | 18 | 70 |
| Perphenazine | 22 | ND | ND | ND | 10 | — | — | ▿▿ | 18 | 81 |
| Pimozide | 7 | ND | ND | ND | 5 | — | ▿ | ▿ | 49 | 79 |
| PPM | 16 | ND | ND | ND | 10 | — | ▿▿ | ▿▿ | 16 | 68 |
| Protriptyline | 16 | ND | ND | ND | 10 | — | ▿ | ▿ | 32 | 74 |
| Psora-4 | >30 | 5.7 | 10.0 | >5.3 | 10 | ▿▿▿ | ▿▿▿ | — | 14 | 66 |
| 6PTN | >60 | ND | ND | ND | 30 | ▿▿ | ▿▿ | — | 16 | 70 |
| Quinidine | 23 | 4.2 | 9.0 | 5.5 | 15 | ▿ | ▿▿▿▿ | ▿ | 44 | 60 |
| Quipazine-6N | 28 | ND | ND | ND | 10 | — | — | ▿▿ | 13 | 74 |
| Raloxifene | 7 | ND | ND | ND | 5 | ▿▿ | ++ | ▿▿ | 79 | 0 |
| Ritanserin | 17 | ND | ND | ND | 10 | — | — | ▿ | 57 | 72 |
| Ro 25–6981 | >30 | ND | ND | ND | 30 | — | — | ▿ | 18 | 65 |
| SB 222200 | >30 | 6.6 | 30.0 | >4.5 | 30 | ▿▿▿▿ | ▿▿▿▿ | ▿▿▿ | 16 | 73 |
| SB 224289 | 15 | ND | ND | ND | 5 | ▿ | ▿▿ | ▿▿ | 20 | 66 |
| SKF-38393AH | >30 | ND | ND | ND | 30 | ▿ | ▿▿ | — | 13 | 56 |
| SKF-95282 | 17 | ND | ND | ND | 10 | ▿ | ▿▿ | ▿ | 16 | 63 |
| TFP | 16 | ND | ND | ND | 8 | ▿ | ▿▿▿ | ▿ | 19 | 57 |
| (R,R)-THC | 14 | 4.5 | 9.7 | 3.1 | 10 | ▿▿▿▿ | ▿▿▿▿ | ▿▿ | 16 | 62 |
| TMB | 14 | ND | ND | ND | 10 | — | — | ▿ | 15 | 78 |
| Trifluoperazine (12) | 18 | 6.5 | 11.0 | 2.8 | 10 | ▿ | ▿▿ | ▿▿▿ | 15 | 78 |
| TTNPB (22) | 42 | 9.1 | 22.0 | 4.6 | 30 | ▿▿▿▿ | ▿▿▿▿ | — | 35 | 102 |
| U-74389G | >30 | ND | ND | ND | 5 | — | ++ | ▿ | 59 | 47 |
| 2′CMA | 18 | 0.09 | 0.24 | 200 | 1 | ▿▿▿▿ | ▿▿▿▿ | — | 94 | 71 |
| VX-950 | 30 | 0.20 | 0.60 | 150 | 1 | ▿▿▿▿ | ▿▿▿▿ | — | ND | ND |
| Mock | NA | NA | NA | NA | NA | — | — | — | 8 | 8 |
CC50 and EC50 values are from n4mBid dose–response profiles in the absence and presence of HCV infection, respectively (Fig. S2). SDC, selected drug concentration for HCV life cycle studies; IVP, infectious virus production; % Prt1 and % Prt10, n4mBid cell death protection in screen as a percentage of mock-infected cells at 1 and 10 μM, respectively (mock, 0.1% DMSO); ND, not determined; NA, not applicable; boldface entries are controls. ▿, 40–60% inhibition; ▿▿, 61–80% inhibition; ▿▿▿, 81–90% inhibition; ▿▿▿▿, >90% inhibition. +, 1.5- to 2-fold enhancement; ++, >2-fold enhancement; —, <40% inhibition or <50% enhancement. Replication and IVP levels are based on Fig. S4. Entry levels are based on Fig. S3. All inhibition and enhancement levels are relative to cells mock-treated with 0.1% DMSO in growth media.
*Complete names of compounds can be found in Table S1. References to compounds with known anti-HCV activity are indicated in parentheses.
A handful of drug hits failed to show significant inhibition of any of the probed stages of the HCV life cycle, whereas some drug hits even enhanced infectious HCV production despite clearly alleviating cellular cytopathic effects (Table 1 and Fig. S2). One possible explanation for screen hits that do not appear to inhibit any stage of the HCV life cycle might be that the molecules do in fact inhibit HCVcc entry, but in a genotype-specific manner. The HCVcc used in the initial screen is of genotype 2a origin, whereas our entry assays were carried out with lentiviral particles pseudotyped with the genotype 1a H77 E1/E2 envelope proteins. Although we also probed HCV entry using the genotype 2a HCJ6 E1/E2 glycoproteins, conclusive entry inhibition data could not be obtained due to a much-reduced efficiency of pseudoparticle production. Another possibility for screen hits that appear to be HCV-neutral is that they may inhibit a stage of the HCV life cycle not probed by any of our assays. For example, an inhibitor of capsid disassembly may act downstream of HCV entry and upstream of RNA replication (12). Finally, we cannot rule out the possibility that some molecules may interfere with mBid-mediated apoptosis, thus protecting cells from HCV-mediated cell death without specifically influencing the HCV life cycle. Screening hits that enhance infectious HCV production (Table 1) may act, at least in part, through alleviation of a cytopathic effect downstream of HCV infection rather than directly targeting the HCV life cycle, thus increasing HCV productivity by making the producer cells less susceptible to HCV-induced stress.
Effect of Selected Inhibitors on Viral Entry.
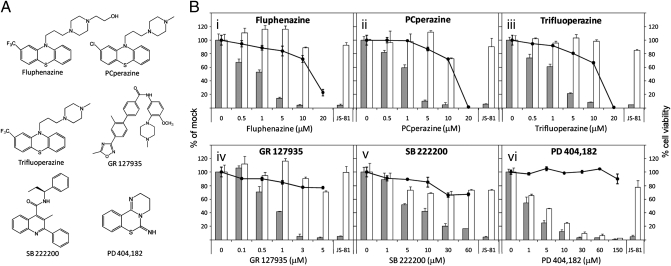
Several of the HCV inhibitor hits from our screen strongly inhibited entry of HCVpp into Huh-7.5 cells (Table 1 and Fig. S3). Six of these compounds exhibited a strong dose-dependent inhibition of HCVpp entry without significant cytotoxicity (Fig. 2). Five of these six compounds inhibited HCVpp entry without significantly affecting entry of VSV-G-pseudotyped lentivirus (VSV-Gpp), whereas PD 404,182 inhibited entry of both HCVpp and VSV-Gpp in a dose-dependent manner. Three compounds—fluphenazine, PCperazine, and trifluoperazine—are structurally similar to each other (Fig. 2A) and to chlorpromazine, an inhibitor of clathrin-coated pit formation (25), and were very recently identified as HCV entry inhibitors (12). The traditional function of these three molecules, however, is to antagonize D2 and/or D1 dopamine receptors (26, 27) and 5-HT2 serotonin receptors (28) in neural signaling networks. Interestingly, the structurally unrelated molecules GR 127935 and SB 222200 (Fig. 2A), reported antagonists of the 5-HT1B/1D serotonin receptor (29) and NK3 tachykinin receptor (30), respectively, also inhibited HCVpp entry without significantly affecting VSV-Gpp entry (Fig. 2B, iv and v). The NK3 receptor has been implicated in serotonergic pathways (31). Furthermore, it is known that human hepatocytes express 5-HT serotonin receptors (32, 33). Taken together, these observations point to a potential role of neurotransmission modulatory receptors in HCV entry into liver cells. In fact, it has been shown that the 5-HT2A receptor mediates infection of human glial cells by the polyomavirus, JCV (34). It is noteworthy that SB 222200 inhibited HCVcc replication by ∼15-fold (Table 1, Fig. 3A) in addition to its inhibition of HCVpp entry (Fig. 2B). PD 404,182, an inhibitor of bacterial KDO 8-P synthase with no known role in modulating human proteins (35), strongly inhibited entry of both HCVpp and VSV-Gpp (Fig. 2B, vi), suggesting interference with a process required for the entry of both pseudoparticles.
Fig. 2.
Leading six HCV entry inhibitors from small-molecule library screen. (A) Chemical structures. (B) Dose-dependent effects on entry of H77 HCVpp (shaded bars) and VSV-Gpp (open bars) into Huh-7.5 cells. The anti-CD81 antibody JS-81 (1 μg/mL) was included as a positive control for each dose–response. Drug cytotoxicity in the absence of pseudoparticle transduction (solid circles) was determined by CellTiter-Glo assay. Values are expressed as a percentage of 0.1% DMSO-treated cells.
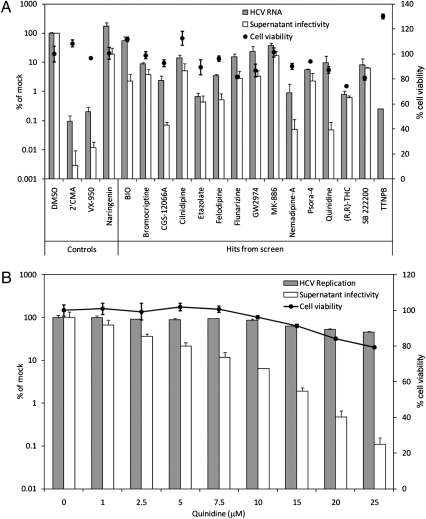
Fig. 3.
Leading inhibitors of HCV replication and infectious virus production from small-molecule library screen. (A) Intracellular HCV RNA levels and supernatant infectivity 96 h after electroporation of Huh-7.5 cells with the genomic RNA of Jc1 HCV and treatment with 15 leading replication/infectious virus production inhibitor hits at the concentrations (SDC) indicated in Table 1. Intracellular HCV RNA levels and supernatant infectivity were determined by quantitative RT–PCR and a cell-viability–adjusted median tissue culture infectious dose (TCID50)/mL assay, respectively. Note that a value is not shown for the supernatant infectivity of TTNPB-treated cells because no cells were infected at any of the serial dilutions of this supernatant. For inhibitor cytotoxicity determination, cell viabilities of Huh-7.5 cells electroporated with carrier (water) before inhibitor treatment were measured by CellTiter-Glo assay. Each point is the mean ± SD of at least three independent experiments. (B) Quinidine dose-dependently inhibits infectious HCV production in Huh-7.5 cells. Virus replication and supernatant infectivity were determined by a Gluc reporter HCV-based assay. Cytotoxicity to Huh-7.5 cells electroporated with only carrier was determined as in A. Each point is the mean ± SD of two independent experiments carried out in duplicate. Values in A and B are expressed as a percentage of 0.1% DMSO-treated cells.
Effect of Selected Inhibitors on HCV RNA Levels and Infectious Virus Production.
Fifteen of the HCV replication and/or infectious virus production inhibitors from our screen were subjected to quantitative RT–PCR and median infectious tissue culture dose (TCID50) analyses to verify the results of the reporter virus studies (Table 1 and Fig. S4). These studies were based on electroporation of Huh-7.5 cells with genomic RNA from the highly infectious Jc1 HCVcc (14). Overall, these experiments were in agreement with our Gluc reporter HCV assays. Fourteen of the 15 tested compounds inhibited HCV RNA replication and/or infectious virus production by >10-fold, and 7 compounds inhibited one or both aspects of the HCV life cycle by >100-fold, all at concentrations that are not significantly cytotoxic to Huh-7.5 cells (Fig. 3A). Among these, three compounds—bromocriptine (Parlodel), felodipine (Plendil), and quinidine—are Food and Drug Administration approved for use in treating various human disorders. Notably, quinidine shows strong anti-HCV activity at concentrations ranging from 10 to 25 μM (Fig. 3B), overlapping with the drug’s therapeutic range of 6–10 μM in human plasma (36). Etazolate, which showed little-to-no cytotoxicity to Huh-7.5 cells and effected an ∼150-fold inhibition of HCV RNA levels at 30 μM (Fig. 3A), is currently in phase II clinical trials for the treatment of Alzheimer’s disease (37). It should be noted that, although (2′Z,3′E)-6-bromoindirubin-3′-oxime (BIO) reproducibly appeared nontoxic to cells in Cell Titer-Glo assays at 5 μM (Fig. 3A and Fig. S2), visual inspection revealed clumped cellular formations 48 h after 5 μM BIO treatment of both n4mBid and Huh-7.5 cells in both the presence and the absence of HCV infection. The clumped formations were not observed at BIO concentrations of ≤1 μM.
A few drug hits inhibited the production of infectious HCVcc to a significantly greater degree than their inhibition of RNA replication. For example, BIO, CGS-12066A, felodipine, nemadipine-A, quinidine, and TTNPB, in addition to the two HCV replication inhibitory control molecules 2′CMA and VX-950, all inhibited infectious virus secretion over sixfold more than they inhibited HCV RNA replication (Fig. 3A). However, of these molecules, only quinidine inhibited infectious HCV production in a dose-dependent manner when assayed using a Gluc reporter virus (Fig. 3B). The other molecules inhibited infectious virus production at increasing concentrations, but only with an accompanying inhibition of HCV replication (Fig. S5). We suspect that the observed increased inhibition of virus production relative to replication for these inhibitors at a given concentration could be due to differential abilities of inhibitor-treated cells to achieve a minimum threshold level of virus replication and protein expression for successful infectious virus assembly and release.
It is worth noting that quinidine, a class I antiarrhythmic drug, inhibited HCV production by 450-fold relative to replication levels in the reporter virus assay at 25 μM (Fig. 3B). This potency is in contrast to the known HCV secretion inhibitors, naringenin (20) (Fig. 3A) and NN-deoxynojirimycin (38), which were able to effect only a ∼10-fold suppression of infectious virus titers. In addition, although quinidine exhibited a CC50 value of 23 μM in n4mBid cells (Table 1), it was only ∼20% cytotoxic to Huh-7.5 cells at 25 μM (Fig. 3B), suggesting a less pronounced cytotoxic action on Huh-7.5 relative to the derivative n4mBid cells.
The strongest replication/infectious virus production inhibitor identified by our screen was the synthetic retinoic acid analog TTNPB (Fig. 3A), which has been previously observed to play apparently conflicting roles in HCV replication. In one study, TTNPB was found to have a strong proreplication effect on HCV in a genotype 1b replicon model (39) whereas another study, using a different genotype 1b replicon, found that TTNPB down-regulated the replication of HCV (22). The latter study demonstrated only a moderate level of HCV inhibition with 10 μM TTNPB in the presence of sodium selenite and reported a stronger HCV inhibition by the natural compound all trans retinoic acid (ATRA), also in the presence of sodium selenite. ATRA appeared only as a relatively weak hit in our screen and so was not considered for further analysis. Despite the apparently conflicting reports on TTNPB in HCV replicon models, our results show that TTNPB strongly inhibits both HCV replication and virus production at 30 μM in the HCVcc model without sodium selenite supplementation. This inhibitory effect becomes much weaker at ≤10 μM (Fig. S5).
Conclusion.
In summary, we have developed an integrated cell protection screen for inhibitors targeting multiple aspects of the hepatitis C virus life cycle based on an engineered hepatoma cell line—n4mBid—that undergoes a significant cytopathic effect in response to HCV infection. We have successfully applied this screen to identify many previously undescribed HCV antivirals targeting virus replication, assembly/release, and/or cellular entry. Several potent HCV inhibitors from our screen are approved for human use as therapeutics and others are in clinical trials, warranting further studies of the potential utility of these molecules as HCV antivirals. The n4mBid cell protection screen is simple and inexpensive, and importantly, requires no separate assay to quantify drug cytotoxicity. Application of this screen to more complex small-molecule libraries, or to focused libraries based on molecular scaffolds with known anti-HCV potential, should lead to the discovery of many more classes of HCV antivirals.
Materials and Methods
Screening.
N4mBid cell viability assays were carried out in 96-well flat-bottom tissue culture plates at a seeding density of 2.4 × 104 cells/well. An equal volume of n4mBid cells (4.8 × 105 cells/mL) and Jc1 HCVcc (3 × 105–1 × 106 TCID50/mL) was premixed, and 100 μL of the mixture was added to individual wells containing 20 μL of 6× concentrated small molecules. For small-molecule cytotoxicity dose–response profiles, HCVcc was replaced with growth media. All small molecules were applied at two concentrations: 1 and 10 μM. For mock infections (providing an upper limit on cell viability in the HCV cytopathic effect protection assay), HCVcc was first inactivated by exposure to UV radiation for 2 min. Plates were incubated at 37 °C in a 5% CO2 atmosphere for 96 h. Cell viability was quantified with CellTiter-Glo reagent (Promega) diluted fivefold in water. Spent growth media was aspirated and replaced with 50 μL diluted CellTiter-Glo reagent. Microplates were then gently vortexed for 2 min and incubated at room temperature for an additional 8 min. Ten microliters of sample from each well was transferred to a white 96-well plate (Corning), and luminescence was measured in a Berthold Centro LB 960 luminometer for 0.1 s.
Additional methods are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge Taylor Sugg for assisting with analysis of data from the n4mBid HCV inhibitor screen and Catherine Murray for editing the manuscript. Financial support for this work came from new faculty start-up funds from the Artie McFerrin Department of Chemical Engineering, Texas A&M University, and from the Texas Engineering Experiment Station and National Institutes of Health Grant 1R21AI083965-01 (to Z.C., K.C., and R.S.), as well as from the National Institutes of Health (Grants CA057973 and AI072613, to C.M.R.) and generous gifts from the Greenberg Medical Research Institute, the Starr Foundation, and the Ellison Medical Foundation (C.M.R.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0915117107/DCSupplemental.
References
- 1.Lohmann V, et al. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science. 1999;285:110–113. doi: 10.1126/science.285.5424.110. [DOI] [PubMed] [Google Scholar]
- 2.Dansako H, et al. A new living cell-based assay system for monitoring genome-length hepatitis C virus RNA replication. Virus Res. 2008;137:72–79. doi: 10.1016/j.virusres.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 3.Mondal R, et al. Development of a cell-based assay for high-throughput screening of inhibitors against HCV genotypes 1a and 1b in a single well. Antiviral Res. 2009;82:82–88. doi: 10.1016/j.antiviral.2008.12.012. [DOI] [PubMed] [Google Scholar]
- 4.Huang P, et al. Discovery and characterization of substituted diphenyl heterocyclic compounds as potent and selective inhibitors of hepatitis C virus replication. Antimicrob Agents Chemother. 2008;52:1419–1429. doi: 10.1128/AAC.00525-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lindenbach BD, et al. Complete replication of hepatitis C virus in cell culture. Science. 2005;309:623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 6.Wakita T, et al. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med. 2005;11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhong J, et al. Robust hepatitis C virus infection in vitro. Proc Natl Acad Sci USA. 2005;102:9294–9299. doi: 10.1073/pnas.0503596102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iro M, et al. A reporter cell line for rapid and sensitive evaluation of hepatitis C virus infectivity and replication. Antiviral Res. 2009;83:148–155. doi: 10.1016/j.antiviral.2009.04.007. [DOI] [PubMed] [Google Scholar]
- 9.Murakami Y, et al. Identification of bisindolylmaleimides and indolocarbazoles as inhibitors of HCV replication by tube-capture-RT-PCR. Antiviral Res. 2009;83:112–117. doi: 10.1016/j.antiviral.2009.03.008. [DOI] [PubMed] [Google Scholar]
- 10.Pan KL, Lee JC, Sung HW, Chang TY, Hsu JT. Development of NS3/4A protease-based reporter assay suitable for efficiently assessing hepatitis C virus infection. Antimicrob Agents Chemother. 2009;53:4825–4834. doi: 10.1128/AAC.00601-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu X, Sainz B, Jr, Uprichard SL. Development of a cell-based hepatitis C virus infection fluorescent resonance energy transfer assay for high-throughput antiviral compound screening. Antimicrob Agents Chemother. 2009;53:4311–4319. doi: 10.1128/AAC.00495-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gastaminza P, Whitten-Bauer C, Chisari FV. Unbiased probing of the entire hepatitis C virus life cycle identifies clinical compounds that target multiple aspects of the infection. Proc Natl Acad Sci USA. 2009 doi: 10.1073/pnas.0912966107. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsu EC, et al. Modified apoptotic molecule (BID) reduces hepatitis C virus infection in mice with chimeric human livers. Nat Biotechnol. 2003;21:519–525. doi: 10.1038/nbt817. [DOI] [PubMed] [Google Scholar]
- 14.Pietschmann T, et al. Construction and characterization of infectious intragenotypic and intergenotypic hepatitis C virus chimeras. Proc Natl Acad Sci USA. 2006;103:7408–7413. doi: 10.1073/pnas.0504877103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen SH, Tan SL. Discovery of small-molecule inhibitors of HCV NS3-4A protease as potential therapeutic agents against HCV infection. Curr Med Chem. 2005;12:2317–2342. doi: 10.2174/0929867054864769. [DOI] [PubMed] [Google Scholar]
- 16.Carroll SS, et al. Inhibition of hepatitis C virus RNA replication by 2′-modified nucleoside analogs. J Biol Chem. 2003;278:11979–11984. doi: 10.1074/jbc.M210914200. [DOI] [PubMed] [Google Scholar]
- 17.Watashi K, Hijikata M, Hosaka M, Yamaji M, Shimotohno K. Cyclosporin A suppresses replication of hepatitis C virus genome in cultured hepatocytes. Hepatology. 2003;38:1282–1288. doi: 10.1053/jhep.2003.50449. [DOI] [PubMed] [Google Scholar]
- 18.Yang F, et al. Cyclophilin A is an essential cofactor for hepatitis C virus infection and the principal mediator of cyclosporine resistance in vitro. J Virol. 2008;82:5269–5278. doi: 10.1128/JVI.02614-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsu M, et al. Hepatitis C virus glycoproteins mediate pH-dependent cell entry of pseudotyped retroviral particles. Proc Natl Acad Sci USA. 2003;100:7271–7276. doi: 10.1073/pnas.0832180100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nahmias Y, et al. Apolipoprotein B-dependent hepatitis C virus secretion is inhibited by the grapefruit flavonoid naringenin. Hepatology. 2008;47:1437–1445. doi: 10.1002/hep.22197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aly HH, Shimotohno K, Hijikata M. 3D cultured immortalized human hepatocytes useful to develop drugs for blood-borne HCV. Biochem Biophys Res Commun. 2009;379:330–334. doi: 10.1016/j.bbrc.2008.12.054. [DOI] [PubMed] [Google Scholar]
- 22.Morbitzer M, Herget T. Expression of gastrointestinal glutathione peroxidase is inversely correlated to the presence of hepatitis C virus subgenomic RNA in human liver cells. J Biol Chem. 2005;280:8831–8841. doi: 10.1074/jbc.M413730200. [DOI] [PubMed] [Google Scholar]
- 23.Watashi K, et al. Anti-hepatitis C virus activity of tamoxifen reveals the functional association of estrogen receptor with viral RNA polymerase NS5B. J Biol Chem. 2007;282:32765–32772. doi: 10.1074/jbc.M704418200. [DOI] [PubMed] [Google Scholar]
- 24.Marukian S, et al. Cell culture-produced hepatitis C virus does not infect peripheral blood mononuclear cells. Hepatology. 2008;48:1843–1850. doi: 10.1002/hep.22550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang LH, Rothberg KG, Anderson RG. Mis-assembly of clathrin lattices on endosomes reveals a regulatory switch for coated pit formation. J Cell Biol. 1993;123:1107–1117. doi: 10.1083/jcb.123.5.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cai G, Gurdal H, Smith C, Wang HY, Friedman E. Inverse agonist properties of dopaminergic antagonists at the D(1A) dopamine receptor: Uncoupling of the D(1A) dopamine receptor from G(s) protein. Mol Pharmacol. 1999;56:989–996. doi: 10.1124/mol.56.5.989. [DOI] [PubMed] [Google Scholar]
- 27.Lummis SC, Baker J. Radioligand binding and photoaffinity labelling studies show a direct interaction of phenothiazines at 5-HT3 receptors. Neuropharmacology. 1997;36:665–670. doi: 10.1016/s0028-3908(97)00054-3. [DOI] [PubMed] [Google Scholar]
- 28.Herrick-Davis K, Grinde E, Teitler M. Inverse agonist activity of atypical antipsychotic drugs at human 5-hydroxytryptamine2C receptors. J Pharmacol Exp Ther. 2000;295:226–232. [PubMed] [Google Scholar]
- 29.De Vries P, Heiligers JPC, Villalón CM, Saxena PR. Blockade of porcine carotid vascular response to sumatriptan by GR 127935, a selective 5-HT1D receptor antagonist. Br J Pharmacol. 1996;118:85–92. doi: 10.1111/j.1476-5381.1996.tb15370.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sarau HM, et al. Nonpeptide tachykinin receptor antagonists. II. Pharmacological and pharmacokinetic profile of SB-222200, a central nervous system penetrant, potent and selective NK-3 receptor antagonist. J Pharmacol Exp Ther. 2000;295:373–381. [PubMed] [Google Scholar]
- 31.Spooren W, Riemer C, Meltzer H. Opinion: NK3 receptor antagonists: The next generation of antipsychotics? Nat Rev Drug Discov. 2005;4:967–975. doi: 10.1038/nrd1905. [DOI] [PubMed] [Google Scholar]
- 32.Lesurtel M, et al. Platelet-derived serotonin mediates liver regeneration. Science. 2006;312:104–107. doi: 10.1126/science.1123842. [DOI] [PubMed] [Google Scholar]
- 33.Ruddell RG, Mann DA, Ramm GA. The function of serotonin within the liver. J Hepatol. 2008;48:666–675. doi: 10.1016/j.jhep.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 34.Elphick GF, et al. The human polyomavirus, JCV, uses serotonin receptors to infect cells. Science. 2004;306:1380–1383. doi: 10.1126/science.1103492. [DOI] [PubMed] [Google Scholar]
- 35.Birck MR, Holler TP, Woodard RW. Identification of a slow tight-binding inhibitor of 3-deoxy-D-manno-octulosonic acid 8-phosphate synthase. J Am Chem Soc. 2000;122:9334–9335. [Google Scholar]
- 36.Lin C, Ke X, Ranade V, Somberg J. The additive effects of the active component of grapefruit juice (naringenin) and antiarrhythmic drugs on HERG inhibition. Cardiology. 2008;110:145–152. doi: 10.1159/000111923. [DOI] [PubMed] [Google Scholar]
- 37.Marcade M, et al. Etazolate, a neuroprotective drug linking GABA(A) receptor pharmacology to amyloid precursor protein processing. J Neurochem. 2008;106:392–404. doi: 10.1111/j.1471-4159.2008.05396.x. [DOI] [PubMed] [Google Scholar]
- 38.Steinmann E, et al. Antiviral effects of amantadine and iminosugar derivatives against hepatitis C virus. Hepatology. 2007;46:330–338. doi: 10.1002/hep.21686. [DOI] [PubMed] [Google Scholar]
- 39.Scholtes C, et al. Enhancement of genotype 1 hepatitis C virus replication by bile acids through FXR. J Hepatol. 2008;48:192–199. doi: 10.1016/j.jhep.2007.09.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.