Abstract
The livers of insulin-resistant, diabetic mice manifest selective insulin resistance, suggesting a bifurcation in the insulin signaling pathway: Insulin loses its ability to block glucose production (i.e., it fails to suppress PEPCK and other genes of gluconeogenesis), yet it retains its ability to stimulate fatty acid synthesis (i.e., continued enhancement of genes of lipogenesis). Enhanced lipogenesis is accompanied by an insulin-stimulated increase in the mRNA encoding SREBP-1c, a transcription factor that activates the entire lipogenic program. Here, we report a branch point in the insulin signaling pathway that may account for selective insulin resistance. Exposure of rat hepatocytes to insulin produced a 25-fold increase in SREBP-1c mRNA and a 95% decrease in PEPCK mRNA. Insulin-mediated changes in both mRNAs were blocked by inhibitors of PI3K and Akt, indicating that these kinases are required for both pathways. In contrast, subnanomolar concentrations of rapamycin, an inhibitor of the mTORC1 kinase, blocked insulin induction of SREBP-1c, but had no effect on insulin suppression of PEPCK. We observed a similar selective effect of rapamycin in livers of rats and mice that experienced an insulin surge in response to a fasting-refeeding protocol. A specific inhibitor of S6 kinase, a downstream target of mTORC1, did not block insulin induction of SREBP-1c, suggesting a downstream pathway distinct from S6 kinase. These results establish mTORC1 as an essential component in the insulin-regulated pathway for hepatic lipogenesis but not gluconeogenesis, and may help to resolve the paradox of selective insulin resistance in livers of diabetic rodents.
Keywords: Akt kinase, mTORC1 kinase, selective insulin resistance, SREBP-1c
Obesity frequently leads to insulin-resistance that, in turn, produces Type 2 diabetes (1, 2). In the prediabetic obese state, the ß-cells of the pancreas secrete excess insulin to compensate for the insulin-resistance, thereby maintaining normal blood glucose levels. Eventually, the pancreas can no longer produce sufficient insulin, the blood sugar rises and the full diabetic syndrome ensues. Insulin-resistance is manifest in three target organs: liver, adipose tissue, and muscle. Studies in rodent models revealed a peculiar feature of hepatic insulin-resistance, namely its selectivity (3). Insulin exerts two predominant actions in liver: it reduces glucose production (gluconeogenesis) and it increases the synthesis of fatty acids and triglycerides (lipogenesis). In the insulin-resistant state, only one of these actions is blocked in liver. The hormone loses its ability to reduce gluconeogenesis but it retains its ability to enhance lipogenesis (4, 5). These dual actions contribute to the lethal combination of hyperglycemia and hypertriglyceridemia that characterizes the diabetic state (3).
Both of the hepatic actions of insulin are mediated largely at the transcriptional level. In blocking gluconeogenesis, insulin reduces transcription of several crucial genes in glucose production, including phosphoenolpyruvate carboxykinase (PEPCK) and glucose-6-phosphatase (6). These actions are attributable, at least in part, to insulin-induced phosphorylation of the transcription factor FoxO1, an event that leads to its exclusion from the nucleus (7). In activating hepatic lipogenesis, insulin increases transcription of genes encoding acetyl-CoA carboxylase, fatty acid synthase, glycerol-3-phosphate acyltransferase, and others. These actions are caused by an insulin-induced increase in the active nuclear fragment of sterol regulatory element-binding protein-1c (SREBP-1c) (8). This increase occurs largely because insulin enhances the transcription of SREBP-1c (9, 10), and it also enhances the proteolytic processing of the membrane-bound SREBP-1c precursor, allowing it to enter the nucleus (11). The mechanism by which insulin enhances transcription of SREBP-1c is unknown, but the stimulation is known to require the participation of liver X receptors (LXR) and one of the nuclear SREBP isoforms, producing a feed-forward stimulation (12).
The simultaneous development of insulin-resistance in one transcriptional program (FoxO1-mediated gluconeogenesis) and sensitivity in another program (SREBP-1c-mediated lipogenesis) suggests that the insulin signaling pathway must bifurcate at some point. One branch must become resistant whereas the other remains responsive. The insulin signaling pathway is generally thought to proceed through receptor-mediated tyrosine phosphorylation of insulin receptor substrate-1 (IRS-1) and/or IRS-2. This leads to activation of phosphoinositide 3-kinase (PI3K), which phosphorylates and activates Akt (also known as protein kinase B) (Fig. 1A). Inhibitor studies in cultured cells, and gene knockout experiments in mice, have shown that this pathway is required both for inhibition of FoxO1 activity (13, 14) and for induction of SREBP-1c expression (15–17). Therefore, it is likely that the bifurcation must occur distal to Akt. One of the downstream targets of Akt is a kinase complex designated mammalian target of rapamycin complex 1 (mTORC1) (18). Recent evidence suggests that mTORC1 is required for activation of lipid synthesis in nonhepatic cells in response to growth factors (19, 20). In liver, lipids are synthesized not for cell growth, but for storage or export. The direct relationship between mTORC1, lipogenesis, and gluconeogenesis has not yet been studied either in insulin-responsive hepatocytes or in animals.
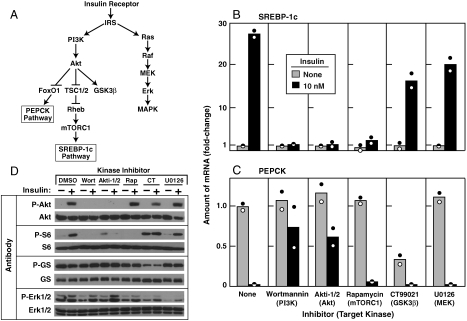
Fig. 1.
Insulin-mediated stimulation of SREBP-1c expression and inhibition of PEPCK expression in primary rat hepatocytes: effect of protein kinase inhibitors. (A) Insulin-activated protein kinase cascades as they relate to the SREBP-1c pathway (lipogenesis) and the PEPCK pathway (gluconeogenesis). (B–D) Hepatocytes were prepared and plated on day zero as described in Methods. On day one, cells were pretreated for 30 min with the following concentration of the indicated inhibitor: 0.1 μM wortmannin, 10 μM Akti-1/2, 0.1 μM rapamycin, 2 μM CT99021, or 10 μM U0126. Each well of cells then received insulin (final concentration, 10 nM), after which the cells were incubated for 6 h at 37 °C and then harvested. Duplicate wells of cells were pooled for measurement of mRNA by quantitative RT-PCR and phosphorylated proteins by immunoblot analysis. (B, C) Relative amounts of mRNAs for SREBP-1c (B) and PEPCK (C). Each value represents the amount of mRNA relative to that in cells receiving no inhibitor and no insulin. Each bar shows the average of two independent experiments (denoted by Circles) performed on different days. (D) Immunoblot analysis of phosphorylated Akt, S6 ribosomal protein (S6), glycogen synthase (GS), and Erk1/2. Filters were exposed to film for 2 sec (P-Akt, P-S6, and P-GS), 15 sec (Akt, S6, GS, and Erk1/2), or 30 sec (P-Erk1/2) at room temperature. Experiments in B–D were done three times with similar results.
In the current studies, we have utilized a robust system to search for a bifurcation point in insulin action in freshly isolated rat hepatocytes. In these cells, insulin addition increases SREBP-1c mRNA by more than 25-fold and decreases PEPCK mRNA by more than 95%. We use this system to show that subnanomolar concentrations of rapamycin, a specific inhibitor of the mTORC1 protein kinase complex (18), selectively block the insulin-mediated induction of SREBP-1c mRNA, while leaving PEPCK repression unaffected. A similar divergence was seen in livers of refed rats and mice that received rapamycin intraperitoneally. These data indicate that the two insulin signaling pathways in the liver diverge after Akt and prior to mTORC1, the latter being essential for activation of SREBP-1c, but not for inhibition of PEPCK. In the insulin-resistant state, insulin may continue to activate mTORC1 while losing its ability to inhibit FoxO1 and PEPCK.
Results
Fig. 1A shows a simplified representation of the kinase cascades activated by insulin in mammalian liver, highlighting the pathways that are felt to activate transcription of SREBP-1c (lipogenesis) or inhibit transcription of PEPCK (gluconeogenesis). Previous reports suggest that insulin-mediated stimulation of SREBP-1c expression is dependent on PI3K (15–17, 20). The downstream pathway by which PI3K regulates SREBP-1c transcription in liver remains unclear. To dissect this signaling pathway, we conducted a protein kinase inhibitor survey using freshly isolated primary rat hepatocytes as a model system.
Fig. 1B shows the relative mRNA levels of SREBP-1c in hepatocytes incubated with and without 10 nM insulin for 6 h in the absence or presence of various kinase inhibitors. SREBP-1c mRNA increased 28-fold after addition of insulin. This dramatic increase was blocked by wortmannin (PI3K inhibitor), Akti-1/2 (Akt inhibitor), and rapamycin (mTORC1 inhibitor), but not by CT99021 (GSK3ß inhibitor) or U0126 (MEK inhibitor). As a positive control in the same experiment, CT99021 and U0126 were shown by immunoblot analysis to inhibit the phosphorylation of glycogen synthase (GS) and Erk1/2, their respective substrates (Fig. 1D, Bottom Two).
PEPCK expression was examined in the same mRNA preparations used in Fig. 1B. In the absence of any kinase inhibitor, insulin decreased PEPCK mRNA by > 95% (Fig. 1C). This inhibition was largely overcome by wortmannin and Akti-1/2, but not by rapamycin, CT99021, or U0126. Taken together, the data in Fig. 1B and C indicate that PI3K and Akt are common mediators of insulin action on lipogenesis and gluconeogenesis. On the other hand, mTORC1 is required only for SREBP-1c activation and not for PEPCK suppression.
To verify the specificities of the 5 kinase inhibitors, we immunoblotted aliquots of whole-cell lysates from the experiments in Fig. 1B and C with antibodies to the phosphorylated forms of Akt and ribosomal S6 protein (Fig. 1D, Top Two). Consistent with the insulin kinase cascade shown in Fig. 1A, insulin-stimulated phosphorylation of Akt was blocked by the inhibitor of Akt itself (Akti-1/2) and that of its upstream activating kinase, PI3K (wortmannin), but not by the inhibitors of mTORC1, GSK3ß, or MEK. The inhibition of Akt phosphorylation by Akti-1/2 results from its prevention of auto-phosphorylation, which self activates the enzyme (21). Phosphorylation of S6 ribosomal protein, a downstream target of mTORC1 (18), was blocked by the inhibitors of mTORC1 and its two upstream activating kinases, Akt and PI3K, but not by the inhibitors of GSK3ß and MEK.
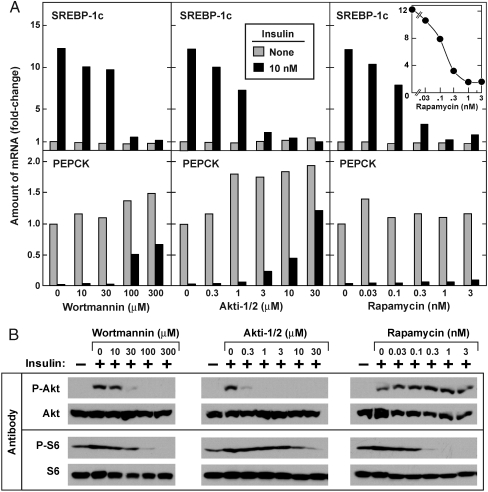
We next examined the dose response of the three inhibitors that blocked insulin-stimulated SREBP-1c expression. As shown in Fig. 2A, wortmannin and Akti-1/2 blocked the insulin-mediated SREBP-1c mRNA increase and the reciprocal PEPCK mRNA decrease at similar concentrations. In contrast, rapamycin inhibited the insulin-induced increase in SREBP-1c expression, but had no effect on the insulin-mediated decrease in PEPCK expression. The effect of rapamycin on insulin-induced SREBP-1c expression was extremely potent, a half-maximal effect occurring at ∼0.2 nM (Fig. 2A, Inset). We probed the same cell extracts with antibodies to the phosphorylated forms of Akt and ribosomal S6 protein (Fig. 2B). For each inhibitor, the expected effect was observed.
Fig. 2.
Dose-dependent effects of wortmannin, Akti-1/2, and rapamycin on SREBP-1c and PEPCK mRNA levels in primary rat hepatocytes. (A) Relative amounts of mRNAs for SREBP-1c (Top) and PEPCK (Bottom) in hepatocytes treated with the indicated concentration of the indicated inhibitor in the absence or presence of insulin as described in Fig. 1. (B) Immunoblot analysis of phosphorylated Akt and S6 ribosomal proteins. Whole-cell lysates were subjected to immunoblot analysis with the indicated antibody as described in Fig. 1D. Filters were exposed to film for 2 sec (P-Akt and P-S6) or 15 sec (Akt and S6). Experiments in A and B were done two times with similar results.
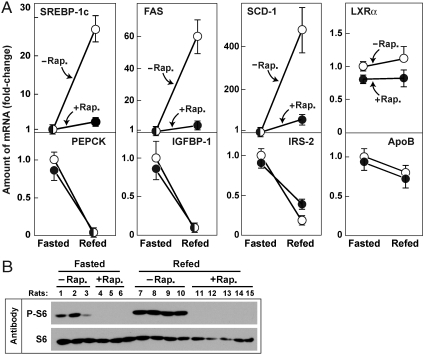
To determine whether mTORC1 is required for insulin-stimulated SREBP-1c expression in livers of living animals, we administered rapamycin to rats by intraperitoneal injection (Fig. 3). The rats were subjected to a fasting-refeeding protocol that has been shown previously to increase hepatic expression of SREBP-1c and its target genes as a result of the increase in blood insulin levels upon refeeding with a high carbohydrate diet (22, 23). In rats receiving vehicle alone, the hepatic mRNAs for SREBP-1c increased by 27-fold. The mRNAs for two SREBP target genes, fatty acid synthase (FAS), and stearoyl CoA desaturase-1 (SCD-1) were also increased dramatically (60- and 478-fold, respectively). All of these increases were reduced dramatically by rapamycin. The mRNAs for three genes that are negatively regulated by insulin (PEPCK, IGFBP-1, and IRS-2) were markedly decreased by refeeding, and none of these decreases was significantly affected by rapamycin. As controls, we measured mRNAs for two genes whose mRNAs are not significantly regulated by insulin, LXRα and apolipoprotein B. Neither was affected by rapamycin (Fig. 3A). Immunoblot analysis of whole-cell lysates from livers of the individual rats showed that phosphorylation of S6 protein was increased in all of the refed animals, and this increase was blocked in all of the rats treated with rapamycin (Fig. 3B). An experiment similar to that in Fig. 3 was repeated once in male Sprague–Dawley rats and once in male C57BL6 mice with similar results.
Fig. 3.
Effect of rapamycin on levels of mRNA encoding SREBP-1c and its target genes in livers of rats subjected to fasting and refeeding. Two groups of male Sprague–Dawley rats were fasted for 48 h. Six h prior to sacrifice, one of the groups received an intraperitoneal injection of 20 mg/kg rapamycin (•) and the other group received vehicle (○). One group continued fasting and the other group was refed with a high carbohydrate diet as described in Methods. After 6 h, all animals were anesthesized, and the livers were removed for measurement of mRNAs and phosphorylated proteins. (A) mRNA levels as determined by quantitative RT PCR. Each value represents the amount of mRNA relative to that of the vehicle-treated fasted group that is denoted arbitrarily as one. Values represent mean ± SEM of 3–5 rats in each group. (B) Immunoblot analysis of phosphorylated and total S6 ribosomal protein in livers of the rats used in (A). Filters were exposed to film for 2 sec (P-S6) or 15 sec (S6). Rap., rapamycin. A similar experiment was carried out in mice with similar results, and the experiment in rats was repeated once with similar results.
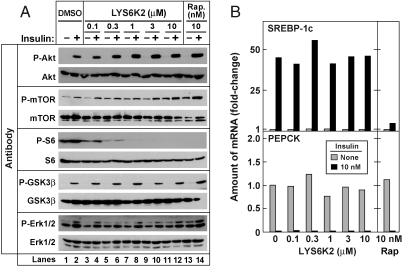
The only kinase known to be activated directly by mTORC1 is p70 ribosomal S6 kinase (S6K) (18, 24). To determine whether S6K activity is required for the insulin-mediated stimulation of SREBP-1c expression, we treated primary rat hepatocytes with a specific S6K inhibitor, LYS6K2, obtained from Eli Lilly and Company (Methods). As shown in Fig. 4A, phosphorylated S6 ribosomal protein was detected in the absence of insulin as well as in its presence. LYS6K2 at concentrations as low as 0.1–0.3 μM, blocked the phosphorylation of S6. LYS6K2 at concentrations as high as 10 μM did not block phosphorylation of other signaling kinases, including GSK3ß and Erk1/2. We noted that increasing concentrations of LYS6K2 increased the basal and insulin-induced phosphorylation of the upstream kinases, Akt, and mTOR. This phenomenon most likely results from a relief of the negative feedback effect of S6K that phosphorylates and inactivates IRS1, thereby inhibiting insulin signaling (25). Despite its potent inhibitory role on S6K activity, LYS6K2 did not block the insulin-induced increase in SREBP-1c expression (Fig. 4B, Upper). LYS6K2 also failed to block the insulin-mediated decrease in PEPCK expression (Lower). These results suggest that the action of mTORC1 in SREBP-1c expression is not mediated by activation of S6 kinase.
Fig. 4.
S6K inhibitor fails to block insulin-induced SREBP-1c mRNA expression in primary rat hepatocytes. (A) Immunoblot analysis of insulin-stimulated phosphorylation of the indicated protein in the presence of increasing concentrations of LYS6K2 (Lanes 1–12) or 10 nM rapamycin (Rap.) (Lanes 13, 14). Whole-cell lysates were subjected to immunoblot analysis with the indicated antibody. Filters were exposed to film for 2 sec (P-Akt and P-S6), 15 sec (Akt, mTOR, S6, GSK3ß, and Erk1/2), 30 sec (P-Erk1/2), or 60 sec (P-mTOR and P-GSK3ß). (B) Relative amounts of mRNAs of SREBP-1c (Top) and PEPCK (Bottom) in hepatocytes treated with the indicated concentration of LYS6K2 or 10 nM rapamycin in the absence or presence of insulin as described in Fig. 1. Data are expressed as described in Fig. 1B and C. This experiment was carried out in parallel with that in (A). Experiments in A and B were done three times with similar results.
Discussion
The current experiments were designed to search for the bifurcation point in the insulin signaling pathway in liver—one pathway leading to an increase in lipogenesis through SREBP-1c, and the other pathway leading to repression of gluconeogenesis through FoxO1. The existence of such a bifurcation point was postulated on the basis of the finding of specificity in the insulin-resistant state in rodent models of Type 2 diabetes (3). In this state, the lipogenic pathway remains sensitive to insulin, whereas the gluconeogenic pathway becomes resistant. The data reported here indicate that the lipogenic and gluconeogenic pathways bifurcate after Akt and prior to the mTORC1 complex (Fig. 1A). The activity of the mTORC1 complex is required for the insulin-mediated increase in SREBP-1c mRNA, but it is not necessary for insulin-mediated suppression of the gluconeogenic gene PEPCK (Fig. 3).
The current studies were facilitated by the use of freshly isolated rat hepatocytes that show a much greater SREBP-1c response to insulin as compared with long-term cultured hepatoma cells. The conclusion that mTORC1 is required for insulin-mediated SREBP-1c induction in liver is consistent with previous findings in retinal pigment epithelial cells by Porstmann, et al. (19), who showed that rapamycin blocks the increase in SREBP-1c protein that is induced by activation of Akt in these nonhepatic cells. The results are also consistent with the findings of Mauvoisin, et al. (26), who showed that rapamycin blocks the induction of SCD-1, an SREBP-1c target gene, in chicken embryo hepatocytes. All of these results differ from the finding of Azzout-Marniche, et al. (27), who failed to find an inhibitory effect of rapamycin on insulin-mediated increases in SREBP-1c mRNA and membrane-bound SREBP-1c protein in freshly isolated rat hepatocytes. In contrast with the current observation that rapamycin blocks SREBP-1c induction, we found that rapamycin failed to block the insulin-mediated repression of PEPCK in the same hepatocytes. This latter finding is consistent with the original observations of Sutherland, et al. (28).
Teleologically, the role of mTORC1 in mediating SREBP-1c activation is consistent with current models of the anabolic role of mTORC1. As pointed out by Sabatini and coworkers, the mTORC1 complex is activated under conditions of nutrient abundance (29). A major action of mTORC1 is to increase protein translation through phosphorylation and activation of S6 kinase and 4E-BP1. In addition, in growing cells mTORC1 activates lipid synthesis through induction of SREBP-1c (19). In liver, mTORC1 is activated by insulin, a hormone that signals nutrient abundance. Activation of mTORC1 leads to increased production of SREBP-1c, and this facilitates storage of excess nutrients as triglycerides. By inhibiting mTORC1, rapamycin acts as a mimic of starvation. The regulatory response to starvation is initiated by a fall in insulin and a rise in glucagon. Glucagon reduces the activity of the mTORC1 complex in rat liver (30). In starvation, the fall in insulin-mediated stimulation of mTORC1, coupled with the glucagon-mediated inactivation of this kinase complex, would further lead to a decrease in SREBP-1c mRNA and lipogenesis.
The pathway leading from the insulin receptor to mTORC1 is lengthy. The initial steps are shared with the anti-gluconeogenic pathway. Thus, the insulin-mediated increase in SREBP-1c mRNA and the decrease in PEPCK mRNA are both blocked by inhibitors of PI3K and Akt (Fig. 1B and C). Activated Akt phosphorylates Tuberous Sclerosis Complex 2 (TSC2), thereby initiating a chain of events that inactivates inhibitors of mTORC1 complex, leading to activation of the mTOR kinase (20). Rapamycin inhibits mTORC1 by binding to FKBP12 and the rapamycin/FKBP12 complex binds to mTORC1, inactivating it (18).
The current finding of an mTORC1 requirement for insulin-stimulated SREBP-1c expression is consistent with the recent findings of Leavens, et al. (17), who showed that genetic ablation of Akt2, the major hepatic Akt isoform, reduces hepatic SREBP-1c mRNA levels and prevents steatosis in insulin-resistant ob/ob mice. It is likely that Akt2 is required because it phosphorylates TSC2 and relieves the inhibition on mTORC1. While our results and those of Leavens, et al. appear clear, there are many conflicting observations in the literature relating Akt to lipogenesis and gluconeogenesis in liver (see the extensive discussion by Leavens, et al.) (17). Part of these discrepancies may relate to the fact that the liver must integrate other signals in addition to insulin. Of particular importance is glucagon, whose stimulation of adenylyl cyclase produces actions that oppose the actions of insulin, including the insulin-mediated increase in SREBP-1c mRNA (4).
In addition to mTORC1, mTOR kinase is also present in another complex designated mTORC2 (18). In our experiments with hepatocytes, the insulin-mediated increase in SREBP-1c mRNA was blocked at subnanomolar concentrations of rapamycin, a characteristic of reactions catalyzed by mTORC1 that is much more sensitive to rapamycin than is mTORC2 (18). Rapamycin also blocked the increase in hepatic SREBP-1c that is associated with refeeding in vivo, a finding that indicates that mTORC1 activation is required for the SREBP-1c increase in a setting in which insulin is increasing while glucagon is declining.
The mechanism by which activated mTORC1 induces SREBP-1c mRNA remains to be elucidated. Our study with the Lilly S6 kinase inhibitor indicates that S6 kinase is not required (Fig. 4). Insulin-mediated SREBP-1c induction is known to require the action of at least two transcription factors that bind to well-defined sequences in the SREBP-1c enhancer. These transcription factors are LXR and SREBP itself (12, 31). mTORC1 could mediate the insulin induction by activating either of these transcription factors or the coactivators that interact with them.
Previous reports raised the possibility that PKCλ, an atypical protein kinase C, may play a role in mediating the insulin-mediated increase in SREBP-1c transcription (32–34), although these results have been controversial (17). Inasmuch as a specific inhibitor for PKCλ is not available, we were not able to directly examine its role in mediating the insulin stimulation of SREBP-1c expression in rat hepatocytes. If PKCλ does play a role, it clearly cannot function in the absence of mTORC1 activity as indicated by the nearly complete inhibition achieved by rapamycin.
A growing body of evidence supports the notion that insulin action in liver is required for the steatosis and hypertriglyceridemia characteristic of the insulin-resistant state. When Biddinger, et al. (35) examined mice with a liver-specific knockout of the insulin receptor, they showed that the animals had marked hyperglycemia and hyperinsulinemia as in other insulin-resistant states, yet the total ablation of insulin action in the liver prevented the steatosis and hypertriglyceridemia that accompany insulin resistance when the insulin receptor is intact. In a similar vein, Semple, et al. (36) studied humans with hyperglycemia and hyperinsulinemia secondary to inactivating mutations in the insulin receptor. These individuals also lacked the steatosis and hypertriglyceridemia that is invariant in the usual forms of insulin resistance in which the insulin receptor is intact.
The above studies indicate that active insulin receptors are required in the liver for hyperinsulinemia to produce steatosis and hypertriglyceridemia in humans as well as mice. Less clear is the requirement for Akt. Early work from our laboratory reported a decrease in insulin-stimulated phosphorylation of Akt in livers of mice with insulin-resistant diabetes, owing to lipodystrophy (4). A decreased phosphorylation of Akt should lead to decreased mTORC1 activity and, therefore, decreased SREBP-1c mRNA. Yet, the livers of these animals manifest elevated SREBP-1c mRNA levels. The regulation of Akt is extremely complex, involving multiple phosphorylation sites and multiple interacting proteins (37). The antibody used in studies of Shimomura, et al., (4) measured phosphorylation of serine 473 in Akt. It is possible that a serine other than serine 473 becomes phosphorylated in livers of insulin-resistant mice, or that another pathway becomes activated that bypasses the need for active Akt in stimulating mTORC1. In this regard, Semple, et al. (36) studied two humans who are heterozygous for an inactivating mutation in Akt2. These individuals had hyperglycemia and hyperinsulinemia. Excess triglycerides were found in liver and plasma. The authors provided evidence that the mutant Akt2 might exert a dominant-negative effect on the normal Akt2 produced by the normal allele, but it is fair to state that one cannot be certain about the level of Akt2 activity in the livers of these heterozygous subjects. Now that the role of mTORC1 in hepatic lipogenesis has been established, it will be necessary to perform detailed studies of the relation between Akt2 and mTORC1 activity in rodent models of insulin-resistance and diabetes.
Materials and Methods
Protein Kinase Inhibitors.
We obtained wortmannin (catalog number W1628), Akti-1/2 (number A6730), rapamycin (number R0395), and U0126 (number U-120) from Sigma; CT99021 (number 1386) from Axon Medchem (Groningen, Netherlands); and LYS6K2 from Eli Lilly and Company. For cell culture studies, all inhibitors were prepared in dimethyl sulfoxide and stored at -20 °C. For whole animal studies, a stock solution of rapamycin (50 mg/ml) was prepared in 100% ethanol and stored at -20 °C.
Diet Studies.
Male Sprague–Dawley rats were used at 4 months of age. All animal experiments were performed with the approval of the Institutional Animal Care and Research Advisory Committee at UT Southwestern Medical Center. For the fasting and refeeding experiments, rats were divided into two groups: fasted and refed. The fasted group was fasted for 48 h, and the refed group was fasted for 48 h and then refed with a high carbohydrate/low fat diet (catalog number TD 88122; Harlan Teklad Diets) for 6 h prior to study. The starting times for the experiments were staggered so that all rats were sacrificed at the same time, which was at the end of the dark cycle. 6 h prior to sacrifice, all animals were injected intraperitoneally with 1.2–1.4 ml of either vehicle alone (14% ethanol, 5% (v/v) Tween 80, and 5% (v/v) polyethylene glycol 400) or vehicle containing rapamycin at a dose of 20 mg/kg.
Other Methods.
Additional Materials and Methods and Table S1 are described in SI Text.
Supplementary Material
Acknowledgments.
We thank our colleagues Guosheng Liang, Guoxun Chen, and Jay Horton for helpful discussions; Jeff Cormier for excellent technical assistance; Richard Gibson for invaluable help with animals; and Eli Lilly and company for the gift of LYS6K2. This work was supported by grants from the National Institute of Health (Grant HL20948) and the Perot Family Foundation. S.L. is a recipient of a postdoctoral fellowship from the American Diabetes Association.
Footnotes
The authors declare no conflict of interest.
See Commentary on page 3281.
This article contains supporting information online at www.pnas.org/cgi/content/full/0914798107/DCSupplemental.
References
- 1.Reaven GM. Why syndrome X? From Harold Himsworth to the insulin resistance syndrome. Cell Metab. 2005;1:9–14. doi: 10.1016/j.cmet.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 2.Kahn BB, Flier JS. Obesity and insulin resistance. J Clin Invest. 2000;106:473–481. doi: 10.1172/JCI10842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown MS, Goldstein JL. Selective vs total insulin resistance: A pathogenic paradox. Cell Metab. 2008;7:95–96. doi: 10.1016/j.cmet.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 4.Shimomura I, et al. Decreased IRS-2 and increased SREBP-1c lead to mixed insulin resistance and sensitivity in livers of lipodystrophic and ob/ob mice. Mol Cell. 2000;6:77–86. [PubMed] [Google Scholar]
- 5.Matsumoto M, Han S, Kitamura T, Accili D. Dual role of transcription factor FoxO1 in controlling hepatic insulin sensitivity and lipid metabolism. J Clin Invest. 2006;116:2464–2472. doi: 10.1172/JCI27047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Granner D, Andreone T, Sasaki K, Beale E. Inhibition of transcription of the phosphoenolpyruvate carboxykinase gene by insulin. Nature. 1983;305:549–551. doi: 10.1038/305549a0. [DOI] [PubMed] [Google Scholar]
- 7.Nakae J, et al. Regulation of insulin action and pancreatic β-cell function by mutated alleles of the gene encoding forkhead transcription factor Foxo1. Nat Genet. 2002;32:245–253. doi: 10.1038/ng890. [DOI] [PubMed] [Google Scholar]
- 8.Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shimomura I, et al. Insulin selectively increases SREBP-1c mRNA in livers of rats with streptozotocin-induced diabetes. P Natl Acad Sci USA. 1999;96:13656–13661. doi: 10.1073/pnas.96.24.13656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferre P, et al. Sterol-regulatory-element-binding protein 1c mediates insulin action on hepatic gene expression. Biochem Soc Trans. 2001;29:547–552. doi: 10.1042/bst0290547. [DOI] [PubMed] [Google Scholar]
- 11.Hegarty BD, et al. Distinct roles of insulin and liver X receptor in the induction and cleavage of sterol regulatory element-binding protein-1c. P Natl Acad Sci USA. 2005;102:791–796. doi: 10.1073/pnas.0405067102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen G, et al. Central role for liver X receptor in insulin-mediated activation of Srebp-1c transcription and stimulation of fatty acid synthesis in liver. P Natl Acad Sci USA. 2004;101:11245–11250. doi: 10.1073/pnas.0404297101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell. 2007;129:1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gross DN, Wan M, Birnbaum MJ. The role of FOXO in the regulation of metabolism. Curr Diabetes Rep. 2009;9:208–214. doi: 10.1007/s11892-009-0034-5. [DOI] [PubMed] [Google Scholar]
- 15.Fleischmann M, Iynedjian PB. Regulation of sterol regulatory-element binding protein 1 gene expression in liver: Role of insulin and protein kinase B/cAkt. Biochem J. 2000;349:13–17. doi: 10.1042/0264-6021:3490013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Porstmann T, et al. PKB/Akt induces transcription of enzymes involved in cholesterol and fatty acid biosynthesis via activation of SREBP. Oncogene. 2005;24:6465–6481. doi: 10.1038/sj.onc.1208802. [DOI] [PubMed] [Google Scholar]
- 17.Leavens KF, et al. Akt2 is required for hepatic lipid accumulation in models of insulin resistance. Cell Metab. 2009;10:405–418. doi: 10.1016/j.cmet.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sabatini DM. mTOR and cancer: insights into a complex relationship. Nature Rev Cancer. 2006;6:729–734. doi: 10.1038/nrc1974. [DOI] [PubMed] [Google Scholar]
- 19.Porstmann T, et al. SREBP activity is regulated by mTORC1 and contributes to Akt-dependent cell growth. Cell Metab. 2008;8:224–236. doi: 10.1016/j.cmet.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laplante M, Sabatini DM. An emerging role of mTOR in lipid biosynthesis. Curr Biol. 2009;19:R1046–R1052. doi: 10.1016/j.cub.2009.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bain J, et al. The selectivity of protein kinase inhibitors: a further update. Biochem J. 2007;408:297–315. doi: 10.1042/BJ20070797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsuda M, et al. SREBP cleavage-activating protein (SCAP) is required for increased lipid synthesis in liver induced by cholesterol deprivation and insulin elevation. Genes Dev. 2001;15:1206–1216. doi: 10.1101/gad.891301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liang G, et al. Diminished hepatic response to fasting/refeeding and liver X receptor agonists in mice with selective deficiency of sterol regulatory element-binding protein-1c. J Biol Chem. 2002;277:9520–9528. doi: 10.1074/jbc.M111421200. [DOI] [PubMed] [Google Scholar]
- 24.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 25.Tremblay F, et al. Identification of IRS-1 SER-1101 as a target of S6K1 in nutrient- and obesity-induced insulin resistance. P Natl Acad Sci USA. 2007;104:14056–14061. doi: 10.1073/pnas.0706517104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mauvoisin D, et al. Role of the PI3-kinase/mTor pathway in the regulation of the stearoyl CoA desaturase (SCD1) gene expression by insulin in liver. J Cell Commun Signal. 2007;1:113–125. doi: 10.1007/s12079-007-0011-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Azzout-Marniche D, et al. Insulin effects on sterol regulatory-element-binding protein-1c (SREBP-1c) transcriptional activity in rat hepatocytes. Biochem J. 2000;350:389–393. [PMC free article] [PubMed] [Google Scholar]
- 28.Sutherland C, O'Brien RM, Granner DK. Phosphatidylinositol 3-kinase, but not p70/p85 ribosomal S6 protein kinase, is required for the regulation of phosphoenolpyruvate carboxykinase (PEPCK) gene expression by insulin. J Biol Chem. 1995;270:15501–15506. doi: 10.1074/jbc.270.26.15501. [DOI] [PubMed] [Google Scholar]
- 29.Peng T, Golub TR, Sabatini DM. The immunosuppressant rapamycin mimics a starvation-like signal distinct from amino acid and glucose deprivation. Mol Cell Biol. 2002;22:5575–5584. doi: 10.1128/MCB.22.15.5575-5584.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baum JI, Kimball SR, Jefferson LS. Glucagon acts in a dominant manner to repress insulin-induced mammalian target of rapamycin complex 1 signaling in perfused rat liver. Am J Physiol-Endoc M. 2009;297:E410–E415. doi: 10.1152/ajpendo.00042.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Repa JJ, et al. Regulation of mouse sterol regulatory element-binding protein-1c gene (SREBP-1c) by oxysterol receptors, LXRα and LXRβ. Genes Dev. 2000;14:2819–2830. doi: 10.1101/gad.844900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matsumoto M, et al. PKCλ in liver mediates insulin-induced SREBP-1c expression and determines both hepatic lipid content and overall insulin sensitivity. J Clin Invest. 2003;112:935–944. doi: 10.1172/JCI18816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taniguchi CM, et al. Divergent regulation of hepatic glucose and lipid metabolism by phosphoinositide 3-kinase via Akt and PKCλ/ζ. Cell Metab. 2006;3:343–353. doi: 10.1016/j.cmet.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 34.Sajan MP, et al. Role of atypical protein kinase C in activation of sterol regulatory element binding protein-1c and nuclear factor kappa B (NFκB) in liver of rodents used as a model of diabetes, and relationships to hyperlipidaemia and insulin resistance. Diabetologia. 2009;52:1197–1207. doi: 10.1007/s00125-009-1336-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Biddinger SB, et al. Hepatic insulin resistance is sufficient to produce dyslipidemia and susceptibility to atherosclerosis. Cell Metab. 2008;7:125–134. doi: 10.1016/j.cmet.2007.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Semple RK, et al. Postreceptor insulin resistance contributes to human dyslipidemia and hepatic steatosis. J Clin Invest. 2009;119:315–322. doi: 10.1172/JCI37432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brazil DP, Yang Z-Z, Hemmings BA. Advances in protein kinase B signalling: AKTion on multiple fronts. Trends Biochem Sci. 2004;29:233–242. doi: 10.1016/j.tibs.2004.03.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.