Abstract
Transcription of the eukaryotic genomes is carried out by three distinct RNA polymerases I, II, and III, whereby each polymerase is thought to independently transcribe a distinct set of genes. To investigate a possible relationship of RNA polymerases II and III, we mapped their in vivo binding sites throughout the human genome by using ChIP-Seq in two different cell lines, GM12878 and K562 cells. Pol III was found to bind near many known genes as well as several previously unidentified target genes. RNA-Seq studies indicate that a majority of the bound genes are expressed, although a subset are not suggestive of stalling by RNA polymerase III. Pol II was found to bind near many known Pol III genes, including tRNA, U6, HVG, hY, 7SK and previously unidentified Pol III target genes. Similarly, in vivo binding studies also reveal that a number of transcription factors normally associated with Pol II transcription, including c-Fos, c-Jun and c-Myc, also tightly associate with most Pol III-transcribed genes. Inhibition of Pol II activity using α-amanitin reduced expression of a number of Pol III genes (e.g., U6, hY, HVG), suggesting that Pol II plays an important role in regulating their transcription. These results indicate that, contrary to previous expectations, polymerases can often work with one another to globally coordinate gene expression.
Keywords: ChIP-Seq, RNA-Seq, transcription, gene regulation
All nuclear genes in eukaryotes are transcribed by three RNA polymerases, Pol I, II, and III. Although polymerases are known to share subunits (1) and even a small number of targets (2, 3), they transcribe distinct classes of genes: Pol I transcribes 18S, 28S, and 5.8S rDNA genes. Pol II transcribes protein coding genes and many noncoding RNA genes, and Pol III transcribes three different classes of genes including 5S (Class I), tRNA (Class II), and many small noncoding genes (Class III). The latter include U6, hY, 7SK, and vault (HVG) genes and others. For all three classes of Pol III genes, sequences upstream of the genes have been found to be required for their optimal expression, although the factors that bind upstream of Pol III genes are largely not known (4).In general, transcription by the different polymerases is believed to be largely independent of one another. However, components normally associated with Pol II have been found to be associated with Pol III subunits and Pol III-transcribed genes. c-Myc, which is normally associated with Pol II-transcribed genes, has been found associated with TFIIIB, a Pol III component (5) and TFIIS, a transcription elongation factor for Pol II, was recently found to bind near Pol III genes in yeast (6). In addition, Pol II has been found to bind upstream and enhance the transcription of several U6 genes (3). However, the extent to which Pol II and its partner proteins coassociate with Pol III and coordinate expression has not been studied on genome scale. This information is valuable for both understanding the functional organization of the human genome and whether basic cellular processes are coordinated with one another.
Here, we describe a study to examine the genome-wide binding of Pol III throughout the human genome, which had not been performed in metazoans. Pol III binding was found to be associated with many known Pol III-transcribed genes as well as several previously unidentified Pol III target genes. We examined the association of Pol II and three other factors, c-myc, c-Jun, and c-Fos, with Pol III genes and found that Pol II and all three factors bind near Class II and III Pol III genes. The influence of Pol II transcription on the expression of Pol III-transcribed genes was examined by α-amanitin treatment, which revealed that transcription by Pol II is required for the optimal expression of a number of Class III Pol III genes. Thus, our results indicate that rather than functioning independently, Pol II binds near, and can influence the expression of Pol III genes.
Results
Genome-Wide in Vivo Mapping of Pol III Binding Sites.
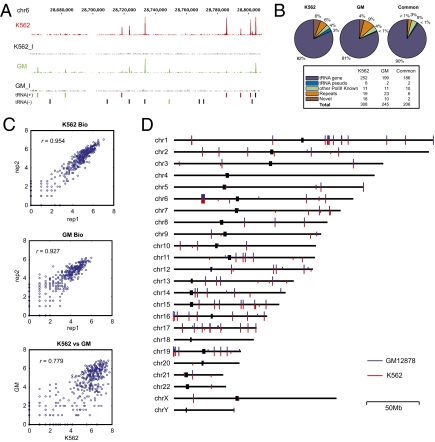
To systematically explore the coordination between Pol II and Pol III, we first mapped the binding sites of Pol III throughout the human genome in two cell lines K562 and GM12878 using chromatin immunoprecipitation (ChIP) followed by deep sequencing (ChIP-Seq). For these experiments, the specificity of the antibodies was first examined by immunoblot analysis and mass spectrometry. Immunoprecipitation (IP) followed by immunoblot analysis (IP/Western blot) showed a single band of expected molecular mass (Fig. S1), whereas mass spectrometric (MS) analysis detected Pol III subunits (Table S1). Importantly, subunits specific to other polymerases (Pol I and Pol II) were not detected in either of these analyses. ChIP was performed on sonicated nuclear extracts prepared from cross-linked K562 and GM12878 cells, and the resulting DNA was sequenced by using the Illumina Genome Analyzer II (Materials and Methods). Two biological replicates were prepared from each cell type. As controls, DNA isolated from the same sonicated extracts without IP was analyzed in parallel (i.e., “Input DNA”). For each cell line a total of 7.5–10 million (M) and 11–16 M uniquely mapped reads were generated for the Pol III ChIP and Input DNAs, respectively (see Table S5). In contrast to Input DNA, Pol III ChIP DNA peaks were observed over many known Pol III genes, including tRNAs, HVGs, hYs, and U6 in both cell types (Fig. 1A shows an example; Fig. 1D shows the genome-wide distribution). In addition, a high concordance was observed for the biological replicates (signal tracks Fig. S2; Fig. 1C, see below).
Fig. 1.
Signal tracks of Pol III binding in K562 and GM12878 cells. (A) Signal tracks show enrichment of Pol III ChIP-Seq signals near tRNA genes in K562 and GM12878 (GM) cells. The area under a peak represents the number of sequence reads mapped in that chromosomal region. The y axis (the number of reads) was normalized for total number of uniquely mapped reads for each sample. K562_I and GM_I are the signal tracks of control Input DNAs of K562 and GM12878 cells. Green and red bars underneath the signal tracks indicate genomic location of differentially bound Pol III peaks in GM12878 and K562, respectively. Black bars indicate tRNA genes that are not differentially bound by Pol III. (B) A summary of the different classes of Pol III targets in K562 and GM12878 cells and their overlap in the two cell lines. (C) Correlation of the signal intensities (normalized number of mapped reads) of Pol III peaks between two biological replicates of K562 (K562 Bio) and GM12878 (GM Bio) cells and between K562 and GM12878 peaks. (D) Genome-wide map of Pol III binding sites. Each bar represents a Pol III binding site, in red for K562 and in blue for GM. The presence of Pol II binding sites near Pol III genes are indicated by the longer bar.
To find potential Pol III binding peaks, the ChIP-Seq raw data were first scored by using the PeakSeq algorithm (7). To further reduce false discoveries, we used a stringent criteria to filter the resulting peaks, while maintaining a 5% false-negative rate based on known Pol III targets (Materials and Methods and Fig. S3). As a result, we detected 308 Pol III peaks in K562 cells and 245 peaks in GM12878 cells (Fig. 1B). As expected, the majority (≈80%) of Pol III binding peaks reside in tRNA genes (252/308 in K562 and 199/245 in GM12878 cells); other targets include pseudotRNA genes, U6, Vault (HVG), hY RNA genes, and a number (18 in K562 and 10 in GM12878 cells) of previously unidentified Pol III target genes (discussed below). A complete list of targets for the two cell types is in Table S2. In K562 cells we detected Pol III binding at ≈40% of all known tRNA genes (a detailed list of all human tRNA genes can be found in http://lowelab.ucsc.edu/GtRNAdb/Hsapi/). This percentage is likely a slight underestimate, because we may have failed to detect binding at the tRNA genes for which no unique sequence tags are available within the gene or flanking regions. It is also possible that increased depth of sequencing beyond the 7.5–10 M mapped reads used in this study could have helped to detect additional Pol III binding sites that are weak.
The target genes identified in K562 and GM12878 cells overlap extensively for known Pol III targets (Figs. 1B and 2); in fact, ≈93% of the GM12878 Pol III binding sites are common with K562 cells. In contrast, the previously unidentified Pol III targets are largely specific for each cell line and only two overlap. Likewise the peak ChIP signals exhibit a reasonable correlation between the two cell types (r = 0.78; Fig. 1C). Nonetheless, there are significant differences in binding at some regions as the correlation coefficient of peak signals between cell types is significantly lower than that between the two biological replicates (0.93–0.95; Fig. 1C). Examples of differentially bound genes are shown in Fig. 1A and a complete list is shown in Table S3. A >2-fold difference in Pol III binding was observed for 114 Pol III peaks. These results indicate that differential Pol III binding or accessibility occurs on a number of genes.
Fig. 2.
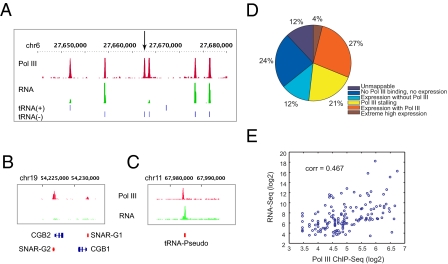
Expression analysis of Pol III-bound genes by RNA-Seq. Nonprotein coding RNA (ncRNA) purified from K562 cells (Materials and Methods) was converted to double-stranded cDNA and sequenced using an Illumina Genome Analyzer II. The reads were mapped to the genome. Signal tracks show enrichment of Pol III ChIP-Seq, and RNA-Seq mapped reads near tRNA genes (the arrow points to a presumably stalled tRNA gene) (A), SNAR-G2 (B), and a tRNA pseudogene (C). The positions of these genes on different chromosomes are indicated on x axis, and the signal intensities of each peak, a function of number of mapped sequence tags, are plotted on y axis. (D) Different classes of mappable Pol III genes based on RNA expression. (E) Moderate correlation of Pol III binding with expression of ncRNAs (RNA after removing polyA+ RNAs and ribosomal RNAs from total RNA).
tRNA Pseudogenes and Previously Unidentified Genomic Regions Are Bound by Pol III.
In addition to known genes, we also found Pol III binding at genes not known to be transcribed by Pol III as well as a number of loci that had not been annotated. Pol III occupancy was detected near tRNA pseudogenes; eight were bound in K562 cells and two in GM12878 cells; the latter are common for both cell types. All of the pseudogenes are nuclear encoded, but one bound in K562 cells is similar to a mitochondrial tRNA gene.
In addition to tRNA pseudogenes, binding was observed near a U6 gene (U6atac) that had not been annotated. Another previously unidentified Pol III target is SNAR-G2 (small ILF3/NF90 associated RNA G2), which encodes a ncRNA of 117 nt. Pol III binding was also detected near the homologous SNAR-G1 gene but was just below our stringent threshold. SNAR-G1 and G2 genes lie ≈100 nt upstream of chorionic gonadotrophin hormone genes (CGB1 and CGB2) and are hypothesized (8) to regulate CGB gene expression (Fig. 2B). Although neither SNAR-G2 nor SNAR-G1 were previously identified as Pol III targets, another gene with similar characteristics, SNAR-A1, is known to be transcribed by Pol III (9). Another previously unidentified target of Pol III detected in both K562 and GM12878 is hsa-mir-886, which encodes a miRNA, suggesting that this gene may be regulated by Pol III. A study (10) indicated regulation of a number of miRNA genes by Pol III. However, hsa-mir-886 is the only known pri-miRNA gene that overlapped with a Pol III binding target, indicating that transcription of miRNAs by Pol III is not a general mechanism. In summary, these results revealed several previously unidentified Pol III gene targets.
Many Pol III Bound Genes Are Expressed.
To examine the expression of the different Pol III bound regions, we used RNA Sequencing (RNA-Seq). Total RNA was isolated from K562 cells and ribosomal RNAs and polyA RNAs were removed. Double-stranded cDNA was prepared and subjected to deep sequencing using the Illumina Genome Analyzer II (Materials and Methods). A total of ≈25 M uniquely mapped reads were obtained. Fig. 2A shows a signal map of Pol III binding and expression of tRNA genes in K562 cells. Overall, a very high proportion of Pol III-bound genes [224/308 (72.7%)] were found to be expressed (10 or more sequence tags) in K562 cells, including tRNA, hY, U6, HVG, and other genes (Table S4). The highest level of expression was found for the RNase MRP gene, which is involved in processing of precursor rRNA. Among the hY RNA genes, hY5 is expressed at high level, hY1 is expressed at much lower level, and little or no expression of the hY4 and hY3 genes was evident. All three HVG genes are expressed in K562 cells, and the HVG2 gene is the most abundantly expressed. Most of the Pol III bound tRNA genes, 204/252 (≈81%), are expressed (>10 tag sequences). Based on unique reads, the level of expression of tRNA genes varied widely, even for genes encoding tRNAs with the same codon specificity. Because these tRNA genes contain nearly identical sequences, it is likely that flanking sequences influence the expression level of these tRNA genes. Interestingly, the U6atac gene, the has-mir-886 gene and 7 of 8 tRNA pseudogenes (Fig. 2C) were also expressed. The level of expression of nuclear tRNA pseudogenes is comparable to tRNA genes and the mitochondrial tRNA pseudogene expression level is highest among all tRNA pseudogenes. The expression data of ≈500 known Pol III genes (tRNA, hY, HVG, 7SK, and others) are summarized in Fig. 2D. In general, for the genes that are expressed, the level of expression correlated reasonably well with the level of Pol III ChIP-Seq signal (Fig. 2E) although many exceptions are observed.
Interestingly, we detected the presence of many genes (21%) that were bound by Pol III but not expressed. An example is shown in Fig. 2A. This lack of detectable expression cannot be due to a mapping artifact because all of these genes contain unique sequences that are mappable by short reads. Perhaps Pol III is stalled on these transcriptionally inactive genes, analogous to what has been observed for a subset of Pol II genes (11, 12). Alternatively, Poll II may not have initiated transcription, or perform very rapid initiation and termination cycles. Regardless, Pol III is located at many genes whose expression is below the level of detection.
Pol II Is Often Associated with Pol III Genes.
To examine the association of Pol II with Pol III-bound genes, we mapped the in vivo binding locations of Pol II by using ChIP-Seq and an antibody (8WG16) to the hypophosphorylated form of the Pol II carboxy terminal domain. This isoform primarily resides at the site of transcription initiation and the antibody was validated by using immunoblot analysis and mass spectrometry. Importantly the antibody recognized Pol II and did not react with subunits specific to Pol I and III (Table S1). ChIP-Seq was performed by using two biological replicates for each cell type; a total of 18.1 M and 13.2 M uniquely mapped reads were generated from K562 and GM12878 cells, respectively. Peaks were scored at P < 0.001 as described in the Materials and Methods.
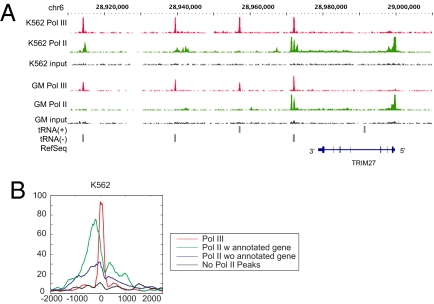
Correlation of Pol II bound regions with those occupied by Pol III revealed a striking coassociation near all types of Pol III genes throughout the human genome. Fig. 1D shows a genome-wide view of Pol II association with Pol III and Fig. 3A shows an example of Pol II binding near tRNA genes for K562 and GM12878 cells. Pol II binding was detected near most class III Pol III genes, including U6, hY, and HVG, for which 2 of 2, 2 of 4, and 3 of 3 genes were bound, respectively. In all of these cases, binding of Pol II correlated with expression, As an example, the two hY genes that lack Pol II binding were not expressed or expressed at very low level. The majority of Pol III-bound tRNA genes (57% for K562 cells) also bound Pol II, but unlike Class III genes, a strong correlation of expression and the presence of a Pol II peak was not observed. Pol II occupancy was detected near four tRNA pseudogenes and the mitochondrial tRNA pseudogene in K562 cells, and the two tRNA pseudogenes specifically bound by Pol III in GM12878 cells. The previously unidentified genes, including U6atac, has-mir-886, and SNAR genes also contain a nearby Pol II binding site. In total, 182 (59%) and 60 (25%) of the Pol III bound genes contain a nearby Pol II peak in K562 and GM12878 cells, respectively. The binding of Pol II near fewer Pol III genes in GM12878 cells may reflect the reproducibly lower overall signal intensity of Pol II peaks in these cells and therefore reduced sensitivity. The Pol II binding signal is not an artifact of open regions as analysis of input signals revealed few peaks in the bound regions and those that were present were of very low magnitude. In general, the Pol II peaks lay close, but usually immediately upstream of the Pol III peaks, which are typically directly on the Pol III genes (Fig. 4B).
Fig. 3.
Pol II binds near Pol III genes. (A) Signal tracks show enrichment of K562 and GM12878 Pol II (in green), and Pol III ChIP-Seq reads (in red) near tRNA genes. The position of tRNA genes on chromosome 6 are shown by gray bars on the x axis, and the signal intensities are plotted on y axis. (B) An aggregate plot of Pol II signal and Pol III signals relative to the transcription start site (indicated by 0) of Pol III-transcribed genes in K562 cells.
Fig. 4.
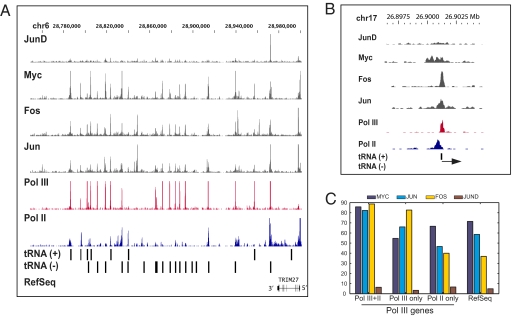
Transcription factors bind near Pol III genes. (A) Signal tracks showing enrichment of Pol III, Pol II, c-Myc, c-Fos, c-Jun, and JunD ChIP-Seq reads near tRNA genes. The position of tRNA genes on chromosome 6 are shown by black bars on x axis, and the signal intensities are plotted on y axis, (B) Close-up view of transcription factors, Pol III and Pol II binding near a tRNA gene on chromosome 17. (C) TF binding data for all mappable Pol III and RefSeq genes. The y axis indicates the percentage of mappable Pol III or RefSeq genes that contain a particular TF binding sites within ±500 bases from the TSS.
To examine the location of Pol II binding regions more systematically we determined the aggregated binding signal relative to the Pol III peaks. As shown in Fig. 3B, a strong Pol II signal is located ≈172 bp upstream of the transcription start site (TSS) of the Pol III genes. Interestingly, although a stronger peak is present at the 5′ end of the gene, Pol II binding is also evident at the 3′ end of the Pol III genes. An example of Pol II binding 3′ to a tRNA-Arg gene is shown in Fig. S4A, and the distribution of all Pol II peaks located within and 500 bases of Pol III genes is shown in Fig. S4B. These results indicate that a large fraction of Pol III genes contain associated Pol II binding.
It is possible that the Pol II peaks that lay near the Pol III genes are transcribing protein coding or genes encoding other polyA+ RNAs in these regions. We found that only a small subset of Pol II peaks near the Pol III genes also mapped near annotated RefSeq genes (16% and 22% in K562 and GM12878, respectively), indicating that most Pol II peaks are not near known genes. To determine whether the remainder might also lay near previously unidentified mRNA-encoded genes we performed RNA-Seq on polyA+ RNA from K562 cells. ≈23 M uniquely mapped reads were generated and mapped to the genome, and transcribed regions identified using criteria described in Materials and Methods. PolyA transcription (>10 tags) was detected within <500 bp of only 25 of 182 (14%) of Pol II-bound regions located at Pol III bound genes; all 25 are annotated RefSeq genes and no previously unidentified genes encoding polyA+ RNAs were found. Thus, Pol II binding is primarily associated with Pol III genes and not other genes encoding polyA messages.
Transcription Factors Associated with Pol II Are Often Associated with Pol III Promoters.
The close association of Pol II with Pol III genes prompted us to examine the relationship of other Pol II-associated factors with Pol III genes. We therefore used ChIP-Seq to map the location of three other factors associated with Pol II including the proto-oncogenes c-Myc, c-Fos and c-Jun, as well as another factor, Jun-D. The distribution of these proteins was determined in K562 cells by using antibodies characterized by immunoblot analysis, and for c-Myc, also by mass spectrometry. A total of 12–17 M uniquely mapped reads were generated from two to three highly correlated biological replicate experiments and 23,369, 35,746, 22,728, and 2,325 significant binding sites were detected for c-Fos, c-Jun, c-Myc, and Jun-D, respectively (Table S5). A large fraction of the binding sites map to the promoter regions of annotated genes as shown previously for c-myc (13, 14), although many binding sites in intronic and intergenic regions are also detected. For the purposes of this study we determined the location of these factors relative to the Pol III genes. Overall, c-Myc, c-Fos, and c-Jun binding was detected near 74%, 86% and 76% of Pol III targets and all three factors resided near 60% of the Pol III targets (Fig. 4 A and B). Binding of these three factors detected for both Class II and III Pol III genes. Nearly all (>80%) Pol III genes with Pol II contained a c-Myc, Fos and c-Jun binding site; however, many of these sites also lay near Pol III-bound genes lacking Pol II (Fig. 4C). In contrast, JunD binding was detected near only 15% of Pol III targets. Interestingly, inspection of signal tracks and analysis of aggregate signals revealed that, unlike Pol II peaks which lay upstream of the Pol III peaks, the binding sites of c-Myc, c-Fos, and c-Jun resided directly at or extremely close to the Pol III peaks (Fig. S5). A similar analysis for RefSeq genes indicated that the binding sites of these transcription factors map at or extremely close to Pol II binding sites. We searched for the presence of Myc (E-box sequence) and Fos and Jun binding motifs (AP-1 site) within 100 bp upstream of Pol III and RefSeq genes. These recognition sequences were found within 100 bp upstream of Pol II and Pol III-transcribed genes including tRNA, U6, HVG, and hY and their pseudogenes. Thus, the vast majority of Pol III-bound genes often contain nearby binding sites for many transcription factors known to be involved in transcribing Pol II-transcribed genes, indicating a significant level of sharing of transcriptional machinery in eukaryotes by the two different RNA polymerases.
α-Amanitin Treatment Affects Transcription of a Subset of Pol III Genes.
To understand the significance of the Pol II binding near Pol III-transcribed genes, Pol II transcription elongation was inhibited with α-amanitin and the effect on Pol III transcript levels was examined by using RNA-Seq (15, 16). Among the three RNA polymerases, Pol II is the most sensitive to α-amanitin; however, higher doses (250 μg/mL) (17) of α-amanitin can affect transcription by Pol III and Pol I is largely resistant. We first determined the minimum concentration of α-amanitin which dramatically reduced the steady-state level of c-myc RNA by treating K562 cells with three different concentrations (25 μg/mL, 50 μug/mL, and 100 μg/mL) of α-amanitin. At a concentration of 50 μg/mL α-amanitin decreased c-Myc RNA level ≈70 fold; no further reduction of c-Myc RNA level was observed at higher α-amanitin concentrations. This concentration (50 μg/mL) is consistent with the published value of α-amanitin that selectively inhibits Pol II relative to Pol III in HeLa cells (18). ChIP-Seq experiments performed on cross-linked K562 cells treated with α-amanitin revealed that Pol II remained bound to the genome, consistent with a role for α-amanitin in blocking Pol II elongation (19).
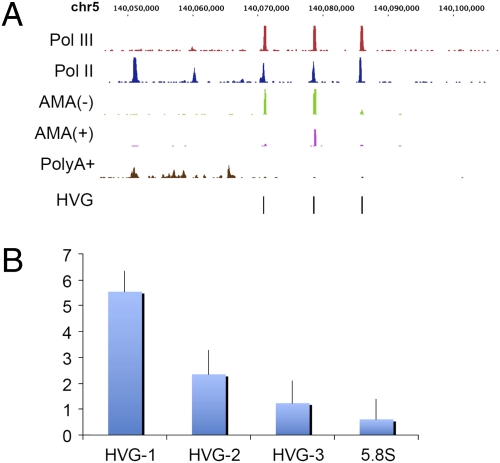
Total RNA was next prepared from untreated (AMA−) K562 cells and those treated with α-amanitin (AMA+); three biological replicate experiments were performed. RT-qPCR analysis revealed the depletion (30- to 60-fold) of c-myc transcript in the treated cells relative to untreated cells. No significant change in 18S and 28S rRNA level was evident after α-amanitin treatment when measured by using an Agilent Bioanalyzer. For one replicate a spike-in control RNA prepared by in vitro transcription of pTRI mouse beta actin DNA was added in equal amounts to both AMA− and AMA+ total RNA. Ribosomal and polyA+ RNAs were removed from the samples and the remaining nonprotein coding RNAs were converted to cDNAs and subjected to RNA-Seq. For each replicate experiment, ≈10–27 M and 10–24 M uniquely mapped sequence reads were generated from untreated and α-amanitin-treated cells, respectively. The effect of α-amanitin treatment on expression of Pol III-transcribed genes was measured by comparing the number of sequence reads normalized to the spike-in control and/or 5.8S RNA (similar results were observed for both reference RNA). A decrease of greater than 1.5-fold in steady-state RNA levels was observed in at least two replicate experiments for 20 different genes including U6 (2 genes), HVG (3 genes), 7SK and the hY1 Class III genes, one previously unidentified Pol III gene, 2 tRNA pseudogenes and 10 tRNA genes in α-amanitin treated cells relative to untreated cells (Fig. 5 and Table S6). RT-qPCR performed on purified (ribo−, polyA−) ncRNA with primers specific to HVG genes validated the reduced expression after α-amanitin treatment (Fig. 5B). Thus, our results indicate that several members of both Class III and Class II genes depend on Pol II for optimal expression.
Fig. 5.
Effect of α-amanitin on Pol III gene expression. (A) The top two signal tracks show enrichment of sequence reads of Pol III and Pol II ChIP DNA near HVG genes. AMA− and AMA+ tracks show expression of HVG genes in untreated and α-amanitin-treated cells, respectively. The polyA track shows expression of RefSeq genes in K562 cells. Unique sequence reads from ncRNA samples were mapped to Pol III-transcribed genes. The position of HVG genes on chromosome 5 are indicated by black bars on x axis, and the signal intensities are plotted on y axis. The decrease in RNA level is evident for all three HVG genes in α-amanitin-treated cells. The polyA+ RNA-Seq track shows mapping of polyA RNA sequence reads to HARS2 and ZMAT2 genes. (B) qPCR validation of RNA-Seq Data. Single-stranded cDNA was prepared from purified ncRNA (ribo minus/polyA minus) by using a primer sequence conserved among all three HVG genes. qPCR was performed by using primers specific to each HVG gene. The Y-axis indicates fold reduction in AMA- cells relative to AMA+ cells.
Discussion
These studies analyzing two different cell lines by ChIP-Seq demonstrate a surprisingly close association of Pol II and Pol II-associated transcription factors, c-Jun, c-Fos, and c-Myc with most Pol III-transcribed genes throughout the human genome. It is possible that either binding, transcription, or the act of transcription initiation by Pol II helps open chromatin, thereby allowing accessibility of Pol III or its associated factors to their nearby targets. Because transcripts are not normally evident at these regions, presumably transcription by Pol II in these regions must either not occur or produce very short or unstable transcripts. Mechanistically the activation of expression of Pol III genes by Pol II might occur through Pol II stalling, which produces short RNAs (20) and has been hypothesized to require TFIIS (21), a factor recently found to be also associated with Pol III genes (6). For a small number of Class III genes, transcription by Pol II rather than binding per se is required for efficient gene expression because α-amanitin, which blocks elongation and still retains Pol II on the DNA, inhibits expression.
Recently, TSS-associated RNAs (TSSa RNA) have been described for Pol II genes (22, 23). These RNAs, 20–90 nt long, are transcribed from both sense and antisense strands and mapped close to 50 nt downstream and 250 nt upstream of TSS, respectively. Our discovery of Pol II binding located ≈172 nt upstream of TSS of Pol III-transcribed genes, suggests that Pol II may be binding upstream of both Pol III and Pol II TSSes.
Interestingly, Class III Pol III genes contain upstream elements that resemble elements found in many Pol II-transcribed genes, including TATA boxes, PSE (proximal sequence elements), and “octamer elements” (24), which are required for their expression. Our results indicate that many transcription factors normally associated with Pol II transcription bind very close to transcription start sites of Pol III genes and may be the activators that bind these upstream elements.
For many Pol III genes with nearby Pol II binding, particularly the tRNA genes, α-amanitin had no effect or the effect was modest. This result may be due to the long half life of these RNAs, although similar results were observed from the analysis of tags of the tRNA precursors. tRNA genes, unlike many of the Class III Pol III genes, are either less reliant on Pol II for their expression or alternatively Pol II initiation, rather than elongation facilitates expression of the Pol III genes; because α-amanitin primarily affects elongation, its effect would be modest.
Overall, these results demonstrate a close association of Pol II and Pol III at Pol III genes throughout the human genome. Not only are the subunits shared by these enzymes, but we demonstrate that Pol II can facilitate the expression of a number of Class III Pol III gene transcription. These results indicate that the extensive mechanisms that regulate Pol II transcription likely have very broad effects upon gene regulation.
The fact that c-myc, c-Jun, and c-Fos bind near Pol III genes as well as Pol II genes indicates that these factors may have a greater general role in eukaryotic gene expression than previously appreciated. c-Myc has been shown to bind TFIIIB, a Pol III-associated transcription factor in W138 human lung fibroblast cells (5). Furthermore, Drosophila S2 cells treated with Myc siRNA exhibited a modest decrease (27 ± 2% and 34 ± 6%) in tRNA Leu and snoRNA U3 levels, respectively (25). Our results indicate that c-Myc, Jun, and Fos may play a role in transcription of most Pol III genes. Increased expression of Pol III-transcribed genes has been documented in many types of cancer including breast, ovarian, skin, and cervical carcinoma (26). It is plausible that the activation of c-Myc, Jun, Fos, and other factors play a global role in activating Pol II and Pol III gene expression during cell proliferation and oncogenesis when rapid cell growth and division occurs.
Finally, our study suggests evidence for Pol III stalling because we find Pol III binding at genes that have no evidence of transcription. This observation raises the possibility that the expression of many Pol III genes may be regulated through mechanisms downstream of binding or initiation as shown previously for Pol II (11, 12). It will be interesting to determine whether common factors and mechanisms are involved in the postinitiation steps of the regulation gene expression of both Pol II and Pol III genes.
Materials and Methods
Cell Culture.
K562 and GM12878 cells were obtained from American Type Culture Collection (lot no. 4607240 for K562 and Expansion A for GM12878) and cultured by using standard conditions.
Antibody Validation.
RNA Pol II (8WG16) and Pol III antibodies were validated by both immunoprecipitation followed by Western blot (IP/Western) and mass spectrometry. c-Myc, c-Fos, JunD, and c-Jun were validated by IP/Western. See SI Materials and Methods for details.
ChIP-Seq.
ChIP DNA for each biological replicate was prepared from 5 × 107 formaldehyde cross-linked GM12878 and K562 cells (27), except after RNase and Proteinase-K digestion, ChIP DNA was purified by using QIAquick PCR Purification Kit (Qiagen). The adapters (Illumina) were ligated to ChIP DNA and sequenced (7). Peaks from the unique reads with two mismatches or less were scored using PeakSeq at a threshold of P < 0.001 and a minimum signal of 2-fold. In addition, nine Pol III peaks that mapped to two regions, one on chromosome 22 and the other on chromosome 9, were excluded because of high background that was observed in all ChIP samples (Pol III, Pol II, Myc, Fos, JunD, and Jun and may represent either mapping problems or amplified regions in these cell lines).
RNA-Seq.
K562 cells were incubated in the presence and absence of 50 μg/mL α-amanitin for 9 h. Total RNA was extracted using TRIzol reagents (Invitrogen), following the manufacturer’s protocol. A spike-in control RNA was synthesized by in vitro transcription of pTRI mouse β-actin DNA using T7 RNA polymerase. The RNA was purified after DNase I digestion. Ribosomal RNAs were removed from total RNA by using RiboMinus Eukaryote Kit (Invitrogen) and polyA RNA was purified from Ribo(-) RNA fraction using the Applied Biosystem (Ambion) MicroPoly(A) Purist kit. PolA RNA was fragmented by using Fragmentation Reagent (Ambion). Double-stranded cDNA synthesis was performed using the SuperScript double-stranded cDNA synthesis kit (Invitrogen). First-strand cDNA was synthesized from polyA RNA by using a mixture of oligo dT and random primer (Invitrogen) and for ncRNA using random primer only. RNA-Seq libraries were prepared for sequencing using the Illumina protocol (15).
Mapping Short Reads to Reference Genome.
ChIP-Seq short reads were mapped to reference human genome (UCSC hg18, excluding haplotype sequences) by using ELAND from the Illumina data analysis software, allowing up to 2 mismatches. RNA-Seq short reads were mapped by SRMapper (15).
Defining Pol III/II Peaks.
ChIP-Seq peaks for Pol II and Pol III were first determined by Peak-Seq. (7). To further reduce false-positive Pol III peaks, we did further filtering by using the most stringent criteria, allowing a maximum of 5% false-negative rate estimated from a set of known Pol III targets. The criteria include peak width, peak height, and signal/noise ratio (peak height/input height). This process is visualized in Fig. S2. Finally, we manually excluded peaks in two regions, chr22 17–22 M and chr9 132–133 M, from K562 Pol III ChIP-Seq experiments because they show unusually high background.
Pol III target regions were annotated based on UCSC annotation for repetitiveness (percent covered by repetitive elements), RNA and RefSeq genes (whose TSSs are within 50 bp). Targets that have greater than 30% repetitive elements were considered repeats, and those have no annotations were considered previously unidentified targets. The ChIP Seq data are available at http://genome.ucsc.edu/ENCODE/ and GEO.
Supplementary Material
Acknowledgments
We thank Youhan Xu, Minyi Shi, and Hannah Monahan Giovanelli for technical assistance. This work was supported by National Institutes of Health grants.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE 19551).
This article contains supporting information online at www.pnas.org/cgi/content/full/0911315106/DCSupplemental.
References
- 1.Sentenac A. Eukaryotic RNA polymerases. CRC Crit Rev Biochem. 1985;18:31–90. doi: 10.3109/10409238509082539. [DOI] [PubMed] [Google Scholar]
- 2.Sussman DJ, Chung J, Leder P. In vitro and in vivo analysis of the c-myc RNA polymerase III promoter. Nucleic Acids Res. 1991;19:5045–5052. doi: 10.1093/nar/19.18.5045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Listerman I, Bledau AS, Grishina I, Neugebauer KM. Extragenic accumulation of RNA polymerase II enhances transcription by RNA polymerase III. PLoS Genet. 2007;3:e212. doi: 10.1371/journal.pgen.0030212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dieci G, Fiorino G, Castelnuovo M, Teichmann M, Pagano A. The expanding RNA polymerase III transcriptome. Trends Genet. 2007;23:614–622. doi: 10.1016/j.tig.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 5.Gomez-Roman N, Grandori C, Eisenman RN, White RJ. Direct activation of RNA polymerase III transcription by c-Myc. Nature. 2003;16:290–294. doi: 10.1038/nature01327. [DOI] [PubMed] [Google Scholar]
- 6.Ghavi-Helm Y, et al. Genome-wide location analysis reveals a role of TFIIS in RNA polymerase III transcription. Genes Dev. 2008;15:1934–1947. doi: 10.1101/gad.471908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rozowsky J, et al. PeakSeq enables systematic scoring of ChIP-seq experiments relative to controls. Nat Biotechnol. 2009;1:66–75. doi: 10.1038/nbt.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mathews MB, Parrott AM. NF90 binds novel human non-coding RNAs. FASEB J. 2008;22:995.1. (meeting abstract) [Google Scholar]
- 9.Parrott AM, Mathews MB. Novel rapidly evolving hominid RNAs bind nuclear factor 90 and display tissue-restricted distribution. Nucleic Acids Res. 2007;35:6249–6258. doi: 10.1093/nar/gkm668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat Struct Mol Biol. 2006;13:1097–1101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- 11.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;13:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu JQ, Snyder M. RNA polymerase II stalling: Loading at the start prepares genes for a sprint. Genome Biol. 2008;9:220. doi: 10.1186/gb-2008-9-5-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim J, Lee JH, Iyer VR. Global identification of Myc target genes reveals its direct role in mitochondrial biogenesis and its E-box usage in vivo. PloS ONE. 2008;12:e1798. doi: 10.1371/journal.pone.0001798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Z, et al. A global transcriptional regulatory role for c-Myc in Burkitt’s lymphoma cells. Proc Natl Acad Sci USA. 2003;100:8164–8169. doi: 10.1073/pnas.1332764100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagalakshmi U, et al. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 17.Haeusler RA, Engelke DR. Spatial organization of transcription by RNA polymerase III. Nucleic Acids Res. 2006;34:4826–4836. doi: 10.1093/nar/gkl656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee Y, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chafin DR, Guo H, Price DH. Action of alpha-amanitin during pyrophosphorolysis and elongation by RNA polymerase II. J Biol Chem. 1995;270:19114–19119. doi: 10.1074/jbc.270.32.19114. [DOI] [PubMed] [Google Scholar]
- 20.Rasmussen EB, Lis JT. Short transcripts of the ternary complex provide insight into RNA polymerase II elongational pausing. J Mol Biol. 1995;252:522–535. doi: 10.1006/jmbi.1995.0517. [DOI] [PubMed] [Google Scholar]
- 21.Pokholok DK, Hannett NM, Young RA. Exchange of RNA polymerase II initiation and elongation factors during gene expression in vivo. Mol Cell. 2002;9:799–809. doi: 10.1016/s1097-2765(02)00502-6. [DOI] [PubMed] [Google Scholar]
- 22.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seila AC, et al. Divergent transcription from active promoters. Science. 2008;322:1849–1851. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murphy S, Moorefield B, Pieler T. Common mechanisms of promoter recognition by RNA polymerases II and III. Trends Genet. 1989;5:122–126. doi: 10.1016/0168-9525(89)90043-7. [DOI] [PubMed] [Google Scholar]
- 25.Steiger D, Furrer M, Schwinkendorf D, Gallant P. Max-independent functions of Myc in Drosophila melanogaster. Nat Genet. 2008;40:1084–1091. doi: 10.1038/ng.178. [DOI] [PubMed] [Google Scholar]
- 26.Marshall L, White RJ. Non-coding RNA production by RNA polymerase III is implicated in cancer. Nat Rev Cancer. 2008;8:911–914. doi: 10.1038/nrc2539. [DOI] [PubMed] [Google Scholar]
- 27.Euskirchen GM, et al. Mapping of transcription factor binding regions in mammalian cells by ChIP: Comparison of array- and sequencing-based technologies. Genome Res. 2007;17:898–909. doi: 10.1101/gr.5583007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.