Abstract
Ectopic Myc expression plays a key role in human tumorigenesis, and Myc dose-dependent tumorigenesis has been well established in transgenic mice, but the Myc target genes that are dependent on Myc levels have not been well characterized. In this regard, we used the human P493-6 B cells, which have a preneoplastic state dependent on the Epstein–Barr viral EBNA2 protein and a neoplastic state with ectopic inducible Myc, to identify putative ectopic Myc target genes. Among the ectopic targets, JAG2 that encodes a Notch receptor ligand Jagged2, was directly induced by Myc. Inhibition of Notch signaling through RNAi targeting JAG2 or the γ-secretase Notch inhibitor N-[N-(3,5-difluorophenacetyl)-L-alanyl]-(S)-phenylglycine t-butyl ester (DAPT) preferentially inhibited the neoplastic state in vitro. Furthermore, P493-6 tumorigenesis was inhibited by DAPT in vivo. Ectopic expression of JAG2 did not enhance aerobic cell proliferation, but increased proliferation of hypoxic cells in vitro and significantly increased in vivo tumorigenesis. Furthermore, the expression of Jagged2 in P493-6 tumors often overlapped with regions of hypoxia. These observations suggest that Notch signaling downstream of Myc enables cells to adapt in the tumor hypoxic microenvironment.
Keywords: neoplasia, Notch, target genes, transcription, hypoxia
The Myc proto-oncogene encodes a transcription factor, c-Myc (herein termed Myc), that is implicated in genesis of many human malignancies (1, 2). Deregulated Myc expression induces tumorigenesis, presumably via its downstream targets comprising about 10–15% of the human genome (2). It is therefore essential to determine whether the Myc target gene network is both quantitatively and qualitatively different between nontumorigenic and tumorigenic states.
The chromosomal translocations of Burkitt lymphoma deregulate the expression of the Myc transcription factor. Ectopic Myc dosage effects on tumorigenesis are best exemplified by many transgenic mouse models, in which the targeted tissues succumb to tumor formation (3). The spectrum of hematologic neoplasms depends on the dosage of Myc in bone marrow cells (4). In fact, Myc-induced hematologic transgenic tumors regressed when Myc levels are diminished (5); however, the Myc dose-dependent target genes have not been elucidated. Though these animal models show the importance of Myc dosage on tumorigenesis, they are not easily amenable to molecular analysis to identify the putative Myc target genes that are induced in the tumorigenic state.
The critical Myc target genes that are necessary for cellular transformation and tumorigenesis are beginning to emerge, but the effects of cell type and cellular context on target gene response to Myc remain poorly understood. The technical limitation has been the dearth of models of Myc-mediated tumorigenesis that provide both nontumorigenic and tumorigenic states in the same system. In this regard, the human B-cell line P-493-6 provides an experimentally tractable system to identify putative endogenous and ectopic Myc target genes (6).
The P493-6 cells were derived from human peripheral blood B cells immortalized by an Epstein–Barr viral (EBV) genome that is complemented with an EBV nuclear antigen-estrogen receptor (EBNA2-ER) fusion protein and a tetracycline-repressible Myc transgene (6). We selected P493-6 cells because they exist in at least three states. With tetracycline, the P-493-6 cells withdraw from the cell cycle in a state with very low Myc levels, arbitrarily termed the “NO Myc” state. In the presence of tetracycline and estradiol, which activates EBNA2-ER, the cells proliferate with induction of endogenous Myc by EBNA2, achieving a “LOW Myc” nontumorigenic state that is equivalent to EBV-immortalized peripheral B lymphocytes (7). In the absence of tetracycline and estradiol, ectopic Myc is induced in a “HIGH Myc” tumorigenic state that resembles human Burkitt lymphoma. This system has allowed us to delineate a subset of genes that specifically responded to ectopic Myc expression, potentially representing target genes that contribute to lymphomagenesis.
Here, we report JAG2, which encodes the Notch ligand Jagged2, as one of the genes most highly up-regulated upon ectopic Myc expression in the P493-6 cells. Although the role of Notch signaling has not been thoroughly dissected in B-cell lymphomas, Jagged2 overexpression is prevalent in the B-cell malignancy multiple myeloma (8). Recent evidence suggests that Notch may synergize with the B-cell receptor to enhance B-cell activation (9). In this report, we document that JAG2 is a direct Myc target and that Jagged2 and Notch signaling participate in P493-6 lymphomagenesis.
Results and Discussion
Human P493-6 B-Cell Neoplasm Model.
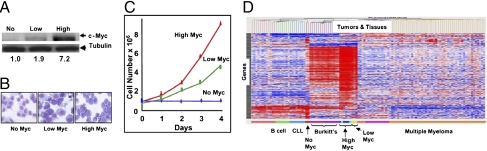
The P493-6 cells were engineered with a tetracycline-regulated Myc and an EBNA2-ER fusion protein to achieve NO, LOW, and HIGH Myc states as described previously (10). As shown in Fig. 1 A–C, P493-6 cells in the NO Myc state grew minimally with more compact chromatin and pale cytoplasm, whereas those with LOW Myc grew with the appearance of lymphoblastoid cells. Untreated cells expressing HIGH Myc grew even better, with the appearance of Burkitt cells characterized by perinuclear lipid vacuoles and frequent mitotic figures. Because P-493-6 cells were derived from mature circulating B cells, they do not express Bcl-6, which is usually found in germinal center-derived Burkitt lymphoma cells. In this regard, we sought to molecularly characterize the P493-6 cell line by arbitrarily selecting 445 gene probes (Table S1) whose expression is >5-fold different between the NO and HIGH Myc states for clustering analysis using expression profiles of 169 primary human mature B-cell malignancies and 21 normal B-cell samples (11). The P493-6 cells with LOW and HIGH Myc clustered together and are closest to Burkitt lymphoma (Fig. 1D), thus validating P493-6 cells as a model of Burkitt lymphoma. The LOW Myc state, however, also resembles multiple myeloma. The NO Myc state is closest to chronic lymphocytic leukemia, which has a low proliferative status. Though many genes up-regulated in P493-6 cells are shared with Burkitt lymphoma, there are subsets of genes unique to P493-6 cells. Moreover, a small subset of genes (at the top of the clustering diagram; Fig. 1D) is shared with multiple myeloma. As such, the P493-6 cells most closely resemble Burkitt lymphoma; however, their expression profiles indicate that they also share gene expression patterns found in other B-cell malignancies
Fig. 1.
c-Myc levels influence cell morphology, proliferation, and gene expression in P493-6 human B cells. (A) Immunoblot of Myc in NO (0.1 μg/mL tetracycline), LOW (0.1 μg/mL tetracycline and 1 μM estradiol), or HIGH (untreated) Myc states of P493-6 cells. Tubulin serves as loading control. Relative expression levels to LOW Myc state are shown (numbers below the panel) as determined by densitometry of Western blot bands. (B) Histological appearance of P493-6 cells in NO, LOW, or HIGH Myc states. Note several mitotic cells, as well as deep basophilic cytoplasm with vacuoles in the ectopic Myc-expressing cells. (C) Proliferation of P493-6 cells with NO Myc, LOW Myc, or HIGH Myc. Experiment was repeated thrice, and a representative graph is shown (D). Unsupervised hierarchical cluster analysis of P493-6 and human B-cell malignancies using 445 gene probes with >5-fold expression changes that discriminate HIGH Myc from LOW Myc states. Analysis was performed with normal and malignant B-cell samples together with P493-6 in NO, LOW, or HIGH Myc states. Expression of the genes in the different samples is represented by the heat map where every row is a gene and every column is a sample. Red represents genes that are overexpressed, blue underexpressed, and white median expression. The diagnoses for the samples are indicated below the heat map.
Immunophenotyping of HIGH Myc P493-6 cells by flow cytometry reveals high CD19 (surface kappa chain) and low CD10 expression, which is characteristic of activated mature B cells. All of these markers are consistent with a Burkitt phenotype. However, these cells also express high CD38 and CD9, markers indicative of plasma cell differentiation—perhaps reflecting the peripheral blood origin of these cells. These markers indicate that the P493-6 cell system does not fully recapitulate the classic Burkitt phenotype, because they have a partial plasma cell phenotype. However, LOW or NO Myc P493-6 cells have significantly diminished CD38 as well as CD9, suggesting that the P493-6 cells in the HIGH Myc state acquire a partial plasma cell phenotype.
Gene Expression Profiling of NO, LOW, and HIGH Myc States.
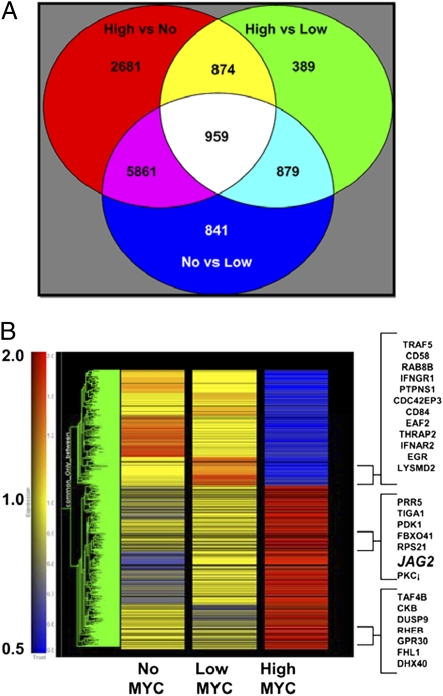
To determine differences in gene expression profiles between the P493-6 cells expressing ectopic (HIGH Myc), EBNA-2 driven (LOW Myc), or very low Myc (NO Myc) levels, we analyzed multiple biological samples using oligonucleotide microarrays. As shown in Fig. 2A, gene expression profiles under the three conditions revealed that signals of >10,000 probe sets are different between the HIGH and NO Myc states, and >8,000 are different between the LOW and NO Myc states, whereas just over 3,100 are different between EBNA-2 driven LOW Myc and ectopic HIGH Myc states. The intersection of all three sets yields 959 probe sets whose signals are altered in common. We focused on genes whose profiles are different between the HIGH and LOW Myc states and different between the HIGH and NO Myc states, but not between the LOW and NO Myc states. We surmise that this set would contain genes that could be pathologic, because they are significantly induced in the HIGH Myc but not the LOW Myc state. The 874 probe sets, as shown in the diagram, represent 603 putative Myc targets that are differentially regulated only upon ectopic Myc expression (Table S2 with list of 874 probe sets identified).
Fig. 2.
Microarray gene expression analysis of P493-6 cells reveals an ectopic Myc target gene set. (A) Venn diagram of differentially expressed genes reveals 874 putative pathologic Myc target gene probe sets. Top left circle represents probe sets differentially expressed between NO and HIGH Myc states, upper right circle encompasses probe sets differentially expressed between LOW Myc and HIGH Myc, and lower middle circle represents the sets differentially expressed for NO and LOW Myc. Replicate biological samples under each condition were analyzed. (B) Heat map of 874 probe sets from A comparing expression of genes in NO, LOW, and HIGH Myc states.
The heat map in Fig. 2B displays the relative expression intensities of this set of 603 ectopic Myc-driven genes under NO, LOW, and HIGH Myc states. The map reveals a set of genes that are down-regulated in a stepwise manner from NO to LOW to HIGH Myc (Table S3). This set is distinct from the set that is down-regulated by HIGH Myc but is apparently induced by EBNA2 (yellow to red to blue, going from NO to LOW to HIGH Myc). A number of known EBNA2 targets, such as IL4I1, CD84, and MFN1 (Table S4) are found in this group (12). Likewise, there is a set of genes (toward the bottom of the heat map) that appears to be repressed by EBNA2 in the LOW Myc state, but is increased between the NO and HIGH Myc states (Table S5). Thus, under the condition of β-estradiol and tetracycline treatment, the genes are not differentially expressed solely based upon Myc expression levels but are likely modulated by EBNA2. Notwithstanding this caveat, we sought among the 603 genes a set that is only expressed highly in the HIGH Myc state.
We have identified a set of putative Myc-dependent genes that display a distinct stepwise increase between LOW and HIGH Myc states (yellow – yellow – red in the heat map from NO to LOW to HIGH Myc states; Table S6). Among these genes, several are previously reported Myc targets (Table S7). Furthermore, gene set enrichment analysis (GSEA) of 167 annotated up-regulated genes unique to the HIGH Myc state reveal an overlap with the ZHAN_MMPC_LATEVS set, which contains genes expressed in tonsilar plasma cells and the STEMCELL_HEMATOPOIETIC-UP set, which are genes increased in hematopoietic stem cells as compared with differentiate brain and bone marrow cells (Table S8). Seventy-eight annotated down-regulated Myc targets overlap with the BASSO_GERMINAL_CENTER_CD40_UP set (Table S8). In addition, we have been able to further validate, with quantitative real-time PCR, the differential expression of several of these genes up-regulated only upon ectopic Myc expression (Fig. S1). Among these genes is JAG2, which encodes the Notch ligand Jagged2 and is highly up-regulated in the HIGH Myc state.
HIGH Myc State Increases NOTCH Signaling Components with JAG2 as a Direct c-Myc Target.
Given our observation that JAG2 is induced more than 5-fold only upon ectopic Myc expression, we focused on the Notch signaling cascade. We found from replicated biological experiments that several members of the Notch pathway are also up-regulated upon ectopic HIGH Myc expression, including MFNG, MIB1, Jagged-1, and Notch1.
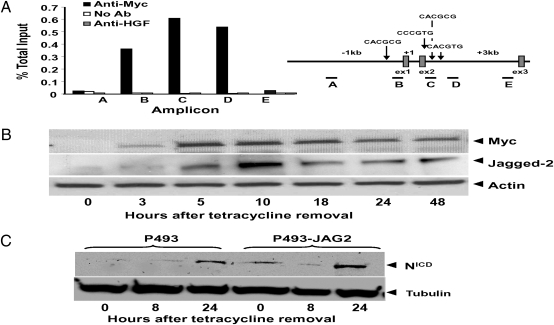
We sought to determine whether JAG2 is a direct Myc target. Because JAG2 is not expressed in the Myc-ER fibroblast system, we are unable to determine whether JAG2 induction by 4-hydroxytamoxifen would still occur with cycloheximide treatment—a hallmark of some direct Myc target genes. Thus, we evaluated JAG2 regulatory regions for potential Myc binding sites and found E boxes within the promoter region and intron 2 of the JAG2 genomic sequence. Chromatin immunoprecipitation (ChIP) documented that Myc bound both regions containing the noncanonical and canonical binding sites (Fig. 3A).
Fig. 3.
Myc binds to the JAG2 gene, induces Jagged2, and activates Notch. (A) Chromatin immunoprecipitation (ChIP) assay for Myc binding to JAG2 in P493-6 B cells (Left). Anti-human growth factor (HGF) antibody serves as a nonspecific antibody control. Schematic diagram of JAG2 gene highlighting consensus Myc binding sites (Right). Canonical E-box was noted in intron 2 (CACGTG) and predicted Myc-Max binding site within promoter region (CACGCG). (B) Immunoblot of Jagged2 and Myc upon tetracycline release of P493-6 cells. P493-6 cells were treated with 0.1 μg/mL of tetracycline for 72 h, washed twice in PBS, and then placed in RPMI with 10% serum for the noted times. Twenty-five micrograms of whole-cell lysate protein were run on a 10% SDS/PAGE gel. Actin serves as a sample loading control. (C) Immunoblot of Notch intracellular domain (NICD) on removal of tetracycline from P493-6 cells or P493-6 cells overexpressing Jagged2 (P493-JAG2). Tubulin serves as a sample loading control.
To determine the response of JAG2 to Myc induction, we measured protein levels of Jagged2 following removal of tetracycline from cells in the NO Myc state (Fig. 3B). Jagged2 protein briskly increased upon reinduction of Myc (Fig. 3B). The correlation between Myc and Jagged2 expression in B cells was further documented in human CB33-Myc B cells, EBV-negative human CA46, and Ramos Burkitt lymphoma cells, which have deregulated Myc expression (Fig. S2). The human Raji EBV-positive Burkitt lymphoma cell line, however, had virtually undetectable Jagged2 expression. Intriguingly, EBNA2 protein behaves like an activated intracellular Notch receptor, thereby bypassing the need for canonical Notch signaling or Jagged2 expression to sustain EBV-positive cell proliferation (13).
The rapid induction of JAG2 mRNA within 3–5 h posttetracycline release in P493-6 cells is compatible with the response of a direct target gene (Fig. S2C). We have also found from array-based nuclear run-on studies that JAG2 is among genes that are robustly increased at the transcriptional level in response to Myc in the P493-6 cell system (Fig. S2D). These observations suggest that JAG2 mRNA expression could correlate with Myc status in primary human B lymphomas bearing Myc chromosomal translocations to Ig gene loci. In this regard, we used the Oncomine database (https://www.oncomine.org/resource/login.html) to examine the relationship between Myc chromosomal translocation, Myc expression, and JAG2 expression in the study of Burkitt lymphomas by Hummel et al. (14). Among 200 lymphomas that were karyotyped, 59 samples (including 37 Burkitt specimens) had Myc chromosomal translocations. Comparison between lymphomas with rearranged Myc versus negative controls reveals a highly statistically significant increase in the expression of both Myc and JAG2 in the lymphomas with Myc translocations (Fig. S3A). The link between genetic aberrations of Myc and increased JAG2 expression suggests that Myc activation of JAG2 may also occur in primary human lymphomas.
In addition to the evidence that Myc directly transactivates JAG2, we found the functional activation of the Notch pathway via detection of the intracellular NICD, following Myc activation. Jagged2 is a ligand for the Notch family of receptors (15). Upon ligand-receptor binding, the Notch receptors undergo a series of γ-secretase-mediated proteolytic cleavages that releases an intracellular portion of the Notch receptor, termed NICD (16, 17), which subsequently translocates to the nucleus and regulates gene expression (18), including the up-regulation of target genes such as Hes-1, Hey-1, Nrarp, and Deltex (19). We document that upon tetracycline release and subsequent Myc induction, there is increased formation of the Notch1 intracellular domain NICD. Ectopic overexpression of JAG2 in P493-6 cells (P493-JAG2) further increased the amount of NICD with Myc induction (Fig. 3C).
Jagged2 Involvement in Cell Proliferation and Tumorigenesis.
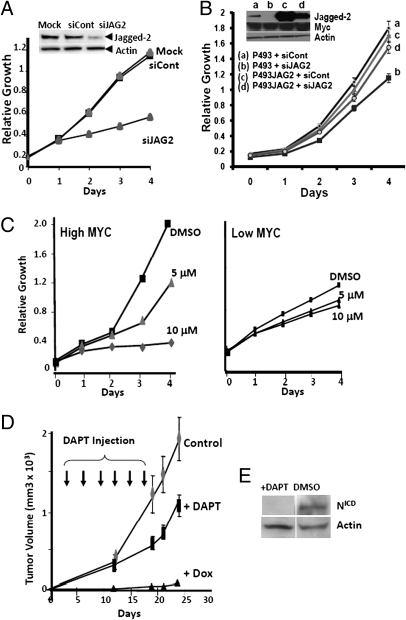
Having established that JAG2 behaves as a direct Myc target gene, we sought to determine the role of JAG2 in P493-6 cell proliferation and tumorigenesis. We performed both gain- and loss-of-function studies by overexpressing JAG2 or using interference RNA, respectively. Fig. 4A documents that siRNA knockdown of JAG2 decreased proliferation rate when compared with scrambled oligonucleotide controls (siCont). Overexpression of Jagged2 rescued the diminished growth rate of P493-6 cells caused by siRNA (Fig. 4B), indicating that the knockdown effect was not due to off-target siRNA effects. These findings suggest that JAG2 is necessary for maximal P493-6 cell proliferation in vitro. Previous studies have shown that inhibition of the γ-secretase Presenillin-1 reduces the production of the Notch intracellular domain, and subsequent downstream signaling, often resulting in apoptosis (20). To further study JAG2 loss-of-function in vitro and in vivo, we used the γ-secretase inhibitor (GSI) DAPT, which blocks Notch signaling by preventing the processing of the Notch1 receptor to form NICD, but not the levels of Myc expression. There was a dose-dependent inhibition of cell proliferation of the HIGH Myc cells by DAPT as compared with DMSO vehicle-treated controls (Fig. 4C). It is remarkable that the slower growth of the LOW Myc cells was proportionally much less inhibited by DAPT, suggesting that the augmented expression of Jagged2 in the HIGH Myc state sensitizes the HIGH Myc cells to DAPT inhibition. Similar findings were obtained using a second γ-secretase inhibitor, L685458. These observations further underscore the potential role of JAG2 as a pathological Myc target gene, which stimulates the Notch signaling pathway in the HIGH Myc state.
Fig. 4.
JAG2 participates in the proliferation of P493-6 B-cell line. (A) P493-6 proliferation upon knockdown of JAG2 (siJAG2) versus control mock electroporated cells or those treated with control oligonucleotides (siCont). Experiments were performed three times with similar results. A representative experiment is shown. Immunoblot for Jagged2 after transient knockdown with JAG2 siRNA (siJAG2) smartpool or control oligonucleotides (siCont). We loaded 25 μg of cellular proteins obtained at 72 h postelectroporation with siRNA (final concentration 100 nm) per well. Actin serves as a sample loading control. (B) P493-6 and P493-6 JAG2 cell proliferation upon knockdown of JAG2 (siJAG2) versus siCont. Relative growth is shown based on absorbance values as mean ± SD (n = 3). Day 4 P value using the two-tailed Student’s t test was 0.02 for siJAG2-treated P493-6 JAG2 versus P493-6 WT (control) cells. No significance difference was found between these cells when treated with siCont. (Inset) Immunoblots for Jagged2 and c-Myc, with actin serving as loading control. A total of 30 μg of cellular proteins obtained at 96 h postelectroporation with siRNA were loaded per well. (C) In vitro growth of ectopic Myc-expressing (HIGH Myc) and EBNA2-driven Myc (LOW Myc)-expressing P493-6 cells treated with vehicle (DMSO; dimethyl sulfoxide) control or the γ-secretase inhibitor DAPT at 5 μM and 10 μM. (D) In vivo volume of tumor xenografts in SCID mice treated with 3×/week s.c. DAPT injections over 2 weeks (designated by arrows). DAPT SC treatments started on day +3 posttumor injections of 25 × 106 cells/mouse. Five mice per condition were used, and experiment was repeated with similar results. Tumor volumes determined by caliper measurements are shown as mean ± SEM. (E) Immunoblot of Notch intracellular domain (NICD) to evaluate Notch inhibition via use of the gamma-secretase inhibitor DAPT. P493-6 cells were treated with 0.1 μg/mL of tetracycline for 72 h, washed twice in PBS, and then placed in RPMI with 10% serum with DAPT or control DMSO (solvent) for 24 h. Actin serves as a sample loading control.
Because anaplastic medulloblastoma is a Myc-driven cancer distinguishable from classic medulloblastoma by high Myc expression (21), we sought to determine the level of Jagged2 expression in human primary medulloblastoma tissue microarrays (Fig. S3B). We document that with high Myc expression in anaplastic medulloblastoma, there is a higher level of Jagged2 expression as compared with classic medulloblastoma (Fig. S3B). The increased Jagged2 expression in high Myc-expressing B cells, Burkitt lymphoma cell lines, and anaplastic medulloblastoma (Fig. S2B) suggests that high Myc expression in different tumor types can be associated with elevated Jagged2 expression.
We next determined whether inhibition of the Notch signaling cascade would inhibit P493-6 tumorigenesis in vivo. We have previously shown that P493-6 cells form tumors in SCID mice (22). These tumors are dependent on Myc, such that doxycycline (a tetracycline analog) oral administration diminishes Myc levels and inhibit tumor formation (Fig. 4D). Treatment of P493-6 cells with thrice weekly s.c. injections of DAPT for 2 weeks led to a significant decrease in tumor volume when compared with control vehicle-treated mice (Fig. 4D). Furthermore, we document that upon induction of Myc expression, DAPT inhibits Notch activation, resulting in decreased activated NICD levels (Fig. 4E). These observations support the hypothesis that JAG2 is an ectopic Myc target and that inhibition of the Notch signaling pathway diminishes P493-6 tumorigenesis.
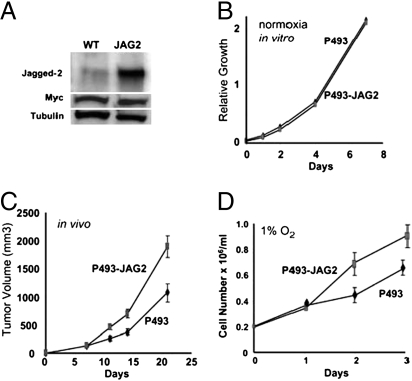
Because our studies demonstrate the necessity of JAG2 expression and Notch signaling pathway for full tumorigenic potential, we sought to determine whether JAG2 gain of function would increase the growth and tumorigenic potential of P493-6 cells. Although JAG2 overexpression (Fig. 5A) in P493-6 cells did not affect in vitro cell proliferation (Fig. 5B), the in vivo xenograft growth of P493-6 JAG2 cells is significantly increased with an average P493-6 JAG2 tumor size nearly 50% larger than the control P493-6 cells (Fig. 5C). From these observations, we surmise that JAG2 overexpression provides an in vivo advantage through the Notch signaling pathway in the tumor microenvironment.
Fig. 5.
Overexpression of JAG2 in P493-6 cells enhances tumorigenicity and growth under hypoxic conditions. (A) Immunoblot of Jagged2 for parental P493-6-expressing empty vector (WT) and polyclonal P493-6 JAG2-overexpressing cells grown in the absence of tetracycline. Ten micrograms of whole-cell lysate proteins were used for each lane. Cells were grown in normal media conditions without tetracycline. Film was exposed for only ≈5 seconds. Also shown are immunoblots for Myc and tubulin that serves as a loading control. (B) P493-6 parental and JAG2 overexpressing cells in vitro proliferation curve. Experiment was performed three times with similar results. (C) P493-6 parental and JAG2 overexpressing cells tumor volume comparison. 20 × 106 cells were injected s.c. Five SCID mice per condition were used, and experiment was repeated with similar results. Tumor volumes shown as mean ± SEM. (D) Growth assay for P493-6 parental and JAG2 overexpressing cells under hypoxic conditions (1% oxygen) for the indicated times. Cell numbers, mean ± SD, were counted in triplicates and averaged. Experiment was repeated and produced similar results.
It has been reported that the hypoxia inducible factor 1 (HIF-1) could augment Notch signaling during development (23, 24). However, the role of Notch in the hypoxic tumor microenvironment has not been addressed. In this regard, we determined whether JAG2 gain-of-function activity could be unmasked under hypoxic conditions in vitro. P493-6 cells have diminished proliferation rate in hypoxia (1% oxygen) as compared with nonhypoxic (20% oxygen) conditions. However, ectopic expression of JAG2 could increase the proliferation of hypoxic P493-6 cells (Fig. 5D), suggesting that JAG2 expression provides a growth advantage in hypoxia, particularly for in vivo tumorigenesis. By fluorescent immunohistochemistry, we further document the expression of Jagged2 in regions of hypoxia in P493-6 tumors, suggesting that JAG2 may be an important component of tumorigenesis under in vivo hypoxic conditions (Fig. S3C).
Our studies to identify the downstream Notch and HIF1 targets under hypoxic conditions alone have not revealed dramatic alterations of Notch target genes in JAG2-overexpressing cells, except for the known Notch target gene, HEY1. HEY1 is induced >3-fold by hypoxia in JAG2-overexpressing P493-6 cells (Fig. S4A). However, upon comparison of in vitro versus in vivo expression profiles for P493-6 cells, we have observed that mRNA levels for Notch signaling components or targets, such as Notch 2 and MAML2, increased significantly in the tumor xenograft samples. Furthermore, P493-6 JAG2 tumors have increased expression of Notch downstream targets genes NRARP and Deltex, when compared with parental P493-6 tumors (Fig. S4B). These findings are supported by the recent findings that heterozygosity for hypoxia inducible factor 1α decreases the incidence of thymic lymphomas, which are associated with decreased Notch signaling and diminished expression of downstream target genes NRARP and Deltex (25). The detailed events downstream of JAG2 in the Notch signaling pathway are clearly important to delineate for a richer understanding of the Myc, JAG2, and Notch connection in additional studies, which are beyond the scope of the current report. The Notch signaling cascade is involved in the development of several hematological malignancies, particularly T-cell leukemias (26) and multiple myeloma (8). In addition to previous studies showing that endogenous Myc is a downstream target of Notch and that Myc and Notch could collaborate in T-cell leukemogenesis (27, 28), our findings provide evidence that ectopic Myc transactivation of JAG2 contributes to P493-6 cell tumorigenesis. Our observation, via the Oncomine database, of a statistically significant correlation between Myc chromosomal translocations and mRNA levels of Myc and JAG2 among 200 primary human lymphomas (14) suggests that Myc could also induce JAG2 expression in human Burkitt or Burkitt-like lymphomas.
EBV immortalization of lymphocytes involves the activation of the Notch pathway by EBNA2, which interacts with the Notch cofactor CBF1 to stimulate target genes (13). When EBNA2-ER is activated in P493-6 cells by estradiol, EBNA2 direct activation of Notch target genes would not require ligand-activated Notch, and hence EBNA2-ER-driven P493-6 cells would be resistant to γ-secretase inhibitors, as we have documented. A recent study reports that selected B-cell lines are sensitive to the GSIs, suggesting a therapeutic role for GSIs in B lymphoid malignancies (20). Interestingly, Jagged2 levels are increased in samples from multiple myeloma patients (8), which often have Myc overexpression (8, 11). Our demonstration of an increased expression of JAG2 in the higher grade, HIGH-Myc-expressing anaplastic medulloblastoma versus classic medulloblastoma further suggests the potential importance of the direct Myc target gene JAG2 in tumorigenesis.
Because P493-6 cells were derived from mature B lymphocytes that express cell surface markers indicative of Burkitt lymphoma as well as CD9, a marker of plasma cell differentiation, the response of JAG2 to HIGH Myc could have resulted from a more mature B cellular state that partially mimics multiple myeloma cells. Intriguingly, a large study showed that Burkitt lymphomas have frequent expression of multiple myeloma 1/IRF4 (MUM1), suggesting a partial plasma cell phenotype in a subset of Burkitt lymphomas (29). Thus, in addition to defining Jagged2 as a direct Myc target gene in P493-6 cells, we also provide the proof of concept through these functional studies that JAG2 is candidate pathological target gene that deserves further study in other systems.
Our JAG2 loss-of-function studies suggest that Jagged2 is necessary for the full growth and tumorigenic potential of P493-6 cells. However, Jagged2 overexpression is insufficient to increase P493-6 cell proliferation under nonhypoxic conditions. Intriguingly, a hypoxic in vitro growth advantage of Jagged2 overexpressing P493-6 cells correlated with an increase in the tumorigenicity of the P493-6 JAG2 cells in vivo. Immunohistochemistry of P493-6 tumors further showed abundant expression of JAG2 in hypoxic regions. These observations agree with a previous study suggesting that Notch signaling is increased in hypoxia through the direct interaction between the intracellular Notch receptor domain NICD and HIF-1 (24). We speculate that increased NICD levels through Jagged2 expression could reciprocally inhibit factor-inhibiting HIF (FIH) and augment HIF function to provide a hypoxic growth advantage independent of Notch target genes (30). Further evidence suggests that NICD enhances HIF1 recruitment to its target gene promoters, resulting in increased HIF1 activity in vitro (23). HIF function would further increase the tumorigenicity of P493-6 as we have previously observed with P493-6 cells overexpressing a stabilized HIF-1 mutant (22).
Materials and Methods
Chemicals and reagents were purchased from Sigma-Aldrich. Smartpool siRNAs were purchased from Dharmacon, Inc. and used according to manufacturer’s instructions. Burkitt lymphoma cell lines were obtained from ATCC. Lymphoid cell lines and P493-6 human B-cell line was cultured as described (31). Chromatin immunoprecipitation, immunoblotting, and RNA isolation from P493-6 cells was as described (31). Immunohistochemistry for hypoxia was performed using pimonidazole hydrochloride (Hypoxyprobe) from Natural Pharmacia International and according to instructions. Goat anti-Jagged2 antibody (Santa Cruz) was used according to instructions. Tissue microarrays were stained with the anti-Jagged2 antibody (1:400) using Dako Biotin Blocking System. Microarray analyses were performed with Affymetrix GeneChip Human 133 2.0 Arrays. Gene expression analysis for gene clustering was performed as described (11). The Oncomine database is accessible at https://www.oncomine.org/resource/login.html. Full materials and methods are found in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank D. Eick (Munich, Germany) for the P493-6 cells and R. Dalla-Favera (New York, NY) for the CB33 and CB33-Myc cell lines. This work was supported by a Maryland TEDCO Grant 2008-MSCRFE-0156-00 and National Institutes of Health Grants R01 CA57341 and R01 CA51497 (to C.V.D.) and the Leukemia Lymphoma Society (C.V.D). J.T.Y is supported by National Institutes of Health Institutional Training Grant T32CA60441.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo/ (accession no. GSE19703).
This article contains supporting information online at www.pnas.org/cgi/content/full/0901230107/DCSupplemental.
References
- 1.Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat Rev Cancer. 2008;8:976–990. doi: 10.1038/nrc2231. [DOI] [PubMed] [Google Scholar]
- 2.Eilers M, Eisenman RN. Myc’s broad reach. Genes Dev. 2008;22:2755–2766. doi: 10.1101/gad.1712408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murphy DJ, et al. Distinct thresholds govern Myc’s biological output in vivo. Cancer Cell. 2008;14:447–457. doi: 10.1016/j.ccr.2008.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith DP, Bath ML, Metcalf D, Harris AW, Cory S. MYC levels govern hematopoietic tumor type and latency in transgenic mice. Blood. 2006;108:653–661. doi: 10.1182/blood-2006-01-0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Felsher DW, Bishop JM. Reversible tumorigenesis by MYC in hematopoietic lineages. Mol Cell. 1999;4:199–207. doi: 10.1016/s1097-2765(00)80367-6. [DOI] [PubMed] [Google Scholar]
- 6.Pajic A, et al. Cell cycle activation by c-myc in a Burkitt lymphoma model cell line. Int J Cancer. 2000;87:787–793. doi: 10.1002/1097-0215(20000915)87:6<787::aid-ijc4>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 7.Kaiser C, et al. The proto-oncogene c-myc is a direct target gene of Epstein-Barr virus nuclear antigen 2. J Virol. 1999;73:4481–4484. doi: 10.1128/jvi.73.5.4481-4484.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Houde C, et al. Overexpression of the NOTCH ligand JAG2 in malignant plasma cells from multiple myeloma patients and cell lines. Blood. 2004;104:3697–3704. doi: 10.1182/blood-2003-12-4114. [DOI] [PubMed] [Google Scholar]
- 9.Thomas M, et al. Notch activity synergizes with B-cell-receptor and CD40 signaling to enhance B-cell activation. Blood. 2007;109:3342–3350. doi: 10.1182/blood-2006-09-046698. [DOI] [PubMed] [Google Scholar]
- 10.Lombardi L, Newcomb EW, Dalla-Favera R. Pathogenesis of Burkitt lymphoma: Expression of an activated c-myc oncogene causes the tumorigenic conversion of EBV-infected human B lymphoblasts. Cell. 1987;49:161–170. doi: 10.1016/0092-8674(87)90556-3. [DOI] [PubMed] [Google Scholar]
- 11.Chesi M, et al. AID-dependent activation of a MYC transgene induces multiple myeloma in a conditional mouse model of post-germinal center malignancies. Cancer Cell. 2008;13:167–180. doi: 10.1016/j.ccr.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spender LC, et al. Cell target genes of Epstein-Barr virus transcription factor EBNA-2: Induction of the p55alpha regulatory subunit of PI3-kinase and its role in survival of EREB2.5 cells. J Gen Virol. 2006;87:2859–2867. doi: 10.1099/vir.0.82128-0. [DOI] [PubMed] [Google Scholar]
- 13.Hayward SD. Viral interactions with the Notch pathway. Semin Cancer Biol. 2004;14:387–396. doi: 10.1016/j.semcancer.2004.04.018. [DOI] [PubMed] [Google Scholar]
- 14.Hummel M, et al. Molecular Mechanisms in Malignant Lymphomas Network Project of the Deutsche Krebshilfe. A biologic definition of Burkitt’s lymphoma from transcriptional and genomic profiling. N Engl J Med. 354:2419–2430. doi: 10.1056/NEJMoa055351. [DOI] [PubMed] [Google Scholar]
- 15.Luo B, Aster JC, Hasserjian RP, Kuo F, Sklar J. Isolation and functional analysis of a cDNA for human Jagged2, a gene encoding a ligand for the Notch1 receptor. Mol Cell Biol. 1997;17:6057–6067. doi: 10.1128/mcb.17.10.6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fortini ME. Gamma-secretase-mediated proteolysis in cell-surface-receptor signalling. Nat Rev Mol Cell Biol. 2002;3:673–684. doi: 10.1038/nrm910. [DOI] [PubMed] [Google Scholar]
- 17.Shimizu K, et al. Integrity of intracellular domain of Notch ligand is indispensable for cleavage required for release of the Notch2 intracellular domain. EMBO J. 2002;21:294–302. doi: 10.1093/emboj/21.3.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ehebauer M, Hayward P, Martinez-Arias A. Notch signaling pathway. Sci STKE. 2006;2006:cm7. doi: 10.1126/stke.3642006cm7. [DOI] [PubMed] [Google Scholar]
- 19.Iso T, Kedes L, Hamamori Y. HES and HERP families: Multiple effectors of the Notch signaling pathway. J Cell Physiol. 2003;194:237–255. doi: 10.1002/jcp.10208. [DOI] [PubMed] [Google Scholar]
- 20.Kogoshi H, Sato T, Koyama T, Nara N, Tohda S. Gamma-secretase inhibitors suppress the growth of leukemia and lymphoma cells. Oncol Rep. 2007;18:77–80. [PubMed] [Google Scholar]
- 21.Stearns D, et al. c-myc overexpression causes anaplasia in medulloblastoma. Cancer Res. 2006;66:673–681. doi: 10.1158/0008-5472.CAN-05-1580. [DOI] [PubMed] [Google Scholar]
- 22.Gao P, et al. HIF-dependent antitumorigenic effect of antioxidants in vivo. Cancer Cell. 2007;12:230–238. doi: 10.1016/j.ccr.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng X, et al. Interaction with factor inhibiting HIF-1 defines an additional mode of cross-coupling between the Notch and hypoxia signaling pathways. Proc Natl Acad Sci USA. 2008;105:3368–3373. doi: 10.1073/pnas.0711591105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gustafsson MV, et al. Hypoxia requires notch signaling to maintain the undifferentiated cell state. Dev Cell. 2005;9:617–628. doi: 10.1016/j.devcel.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 25.Bertout JA, et al. Heterozygosity for hypoxia inducible factor 1alpha decreases the incidence of thymic lymphomas in a p53 mutant mouse model. Cancer Res. 2009;69:3213–3220. doi: 10.1158/0008-5472.CAN-08-4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chiang MY, et al. Leukemia-associated NOTCH1 alleles are weak tumor initiators but accelerate K-ras-initiated leukemia. J Clin Invest. 2008;118:3181–3194. doi: 10.1172/JCI35090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weng AP, et al. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev. 2006;20:2096–2109. doi: 10.1101/gad.1450406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palomero T, et al. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc Natl Acad Sci USA. 2006;103:18261–18266. doi: 10.1073/pnas.0606108103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gualco G, et al. Frequent expression of multiple myeloma 1/interferon regulatory factor 4 in Burkitt lymphoma. Hum Pathol. 2009;40:565–571. doi: 10.1016/j.humpath.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coleman ML, et al. Asparaginyl hydroxylation of the Notch ankyrin repeat domain by factor inhibiting hypoxia-inducible factor. J Biol Chem. 2007;282:24027–24038. doi: 10.1074/jbc.M704102200. [DOI] [PubMed] [Google Scholar]
- 31.Kim JW, Gao P, Liu YC, Semenza GL, Dang CV. Hypoxia-inducible factor 1 and dysregulated c-Myc cooperatively induce vascular endothelial growth factor and metabolic switches hexokinase 2 and pyruvate dehydrogenase kinase 1. Mol Cell Biol. 2007;27:7381–7393. doi: 10.1128/MCB.00440-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.