Abstract
In vertebrates, Evx homeodomain transcription factor-encoding genes are expressed in the posterior region during embryonic development, and overexpression experiments have revealed roles in tail development in fish and frogs. We analyzed the molecular mechanisms of posterior neural development and axis formation regulated by eve1. We show that eve1 is involved in establishing trunk and tail neural ectoderm by two independent mechanisms: First, eve1 posteriorizes neural ectoderm via induction of aldh1a2, which encodes an enzyme that synthesizes retinoic acid; second, eve1 is involved in neural induction in the posterior ectoderm by attenuating BMP expression. Further, eve1 can restore trunk neural tube formation in the organizer-deficient ichabod −/− mutant. We conclude that eve1 is crucial for the organization of the antero-posterior and dorso-ventral axis in the gastrula ectoderm and also has trunk- and tail-promoting activity.
Keywords: bone morphogenic protein, neural induction, organizer, posterior development, retinoic acid
The molecular mechanisms of neural induction and patterning in chordate embryos have been extensively studied in animals such as ascidians, amphibians, fish, chick, and mouse (1, 2). Initial analyses in amphibians revealed that the dorsal organizer (Spemann’s organizer) induces neural (CNS) fates in dorsal ectoderm, and subsequently vegetal marginal signals posteriorize proximal neural ectoderm to induce trunk and tail neural cell fates such as spinal cord and caudal hindbrain, whereas distant animal pole cells give rise to rostral neural tissues including the forebrain, midbrain, and part of the hindbrain (3). Molecular analyses of organizer activity have uncovered multiple molecules crucial for neural induction, including the secreted bone morphogenic protein (BMP) antagonists Chordin, Noggin, and Follistatin, leading to the conclusion that BMP inhibition is crucial for neural induction (1, 2). In addition to the BMP antagonists, FGF has an important role in neural induction in many species (4 –8).
Concomitant with and subsequent to neural induction, neural ectoderm is posteriorized by the activity of several factors, among them FGF (5, 7, 9 –11), Wnt (11 –14). and retinoic acid (RA) (11, 15 –17). RA is essential for posterior neural development in vertebrates, being required for the specification of the future hindbrain and anterior spinal cord (18, 19). In zebrafish FGF and Wnt signaling posteriorize neural ectoderm by suppressing anterior-specific gene expression independently of RA and inducing posterior genes in an RA-dependent process (11).
Some of the transcription factors acting downstream of posteriorizing signals are known and include Homeodomain proteins of the Hox cluster (6, 20), Cdx (21, 22), and Evx (7, 23) families. In the zebrafish gastrula, posterior neural ectoderm and mesoderm are marked by the expression of eve1, a member of the eve/evx family of homeobox genes that encode transcriptional repressors. Evx genes have been implicated in a conserved role in posterior body patterning in a variety of species, including fly, mouse, worm, frog, and zebrafish (24 –26). In zebrafish, overexpression of eve1 disrupts antero-posterior (A/P) patterning in a concentration-dependent manner, leading to loss of head structures and tail duplications and to mispatterning of posterior tissue (25). At the gastrula stage, eve1 expression is restricted to the ventral side and is maintained by BMP signaling, a key ventralizing molecule. Eve1 has been regarded as a ventral marker gene with a presumed role in tail development. However, eve1 expression begins at blastula stage at around 30% epiboly, when it covers most of the margin with the exception of the presumptive organizer (27), suggesting a potentially wider role for eve1 in posterior development. Thus it is not clear if eve1 is involved in trunk development in addition to its accepted role in tail development. In addition, little is known about the mechanism of eve1 function, and no loss-of-function data in fish have been reported so far. Using loss- and gain-of-function strategies, we show here that eve1 regulates trunk and tail development. We find that eve1 affects the formation of trunk and tail neural ectoderm via two molecular mechanisms: induction of the neural ectoderm in both trunk and tail regions at the gastrula stage, at least partly by titration of BMP levels; and posteriorization of neural ectoderm via an RA signal. Furthermore our data provide evidence that eve1 exerts its organizing activity as a transcriptional repressor.
Results
Overexpression of Eve1 Causes Anterior Truncation, Induces Posterior Neural Markers, and Suppresses Markers for Anterior Neural and Nonneural Tissues.
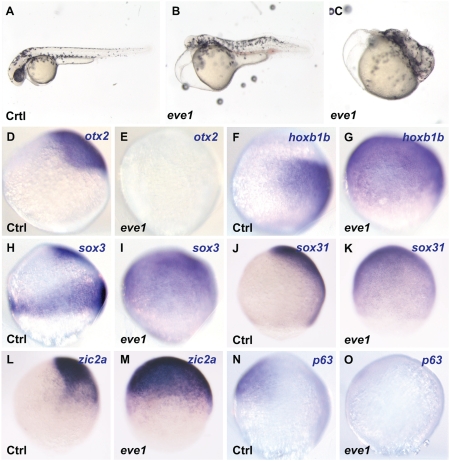
To determine the role of eve1 in A/P patterning, we overexpressed eve1 in vivo and analyzed the expression of otx2, an anterior neural marker, and hoxb1b, a marker for prospective posterior (trunk and tail) neural tissue (7, 20, 28). Phenotypic analysis confirmed previous results (25, 29) such as truncation of head structures (75%, n = 32) (Fig. 1B), and some embryos showed more severe effects with loss of head and trunk (13%, n = 32) (Fig. 1C). The only remaining anteriorly positioned structure was the heart, which continued to beat. Consistent with the lack of anterior structures, otx2 is suppressed in embryos injected with eve1 mRNA (94%, n = 17) (Fig. 1E), whereas hoxb1b expression is partially (15%) or circumferentially (85%) expanded (n = 20) (Fig. 1G).
Fig. 1.
Eve1 overexpression causes anterior truncation, suppression of anterior markers, and induction of posterior markers. Zebrafish embryos were injected with eve1 mRNA (as indicated at the bottom left corner of each panel; Ctrl, uninjected controls). (A–C) Eve1 mRNA injected embryos at 48 hpf showing anterior truncation and progressive loss of posterior patterning. (D–O) In situ hybridization of control and eve1 mRNA-injected embryos at 80% epiboly (lateral views, dorsal to the right, where discernible). Genes analyzed are indicated in the top right corner. Expression of the anterior gene otx2 and epidermal gene p63 was suppressed (D, E; N, O), whereas the expression of hoxb1b was expanded by eve1 overexpression (F, G). Expansion also was observed for sox3, zic2a, and sox31 (H–M).
Surprisingly, in eve1-injected embryos hoxb1b expression expanded to include the prospective epidermal domain, raising the possibility that eve1 may have a role in neural induction in addition to its role in A/P patterning. To test this notion, we analyzed the expression of three additional neural markers, sox3, zic2a, and sox31, and the epidermal marker, p63. In embryos injected with eve1 mRNA, the sox3- (95%, n = 21), zic2a- (95%, n = 20), and sox31- (83%, n = 18) positive domain covers most of the embryo, including the prospective epidermal domain (Fig. 1 I, K, and M), with the concomitant suppression of p63 expression (100%, n = 24) (Fig. 1O). Together, these results suggest that eve1 acts both as a posteriorizing and a posterior neural-promoting factor.
Zebrafish Eve1 Functions as a Repressor in Posterior Neural Development.
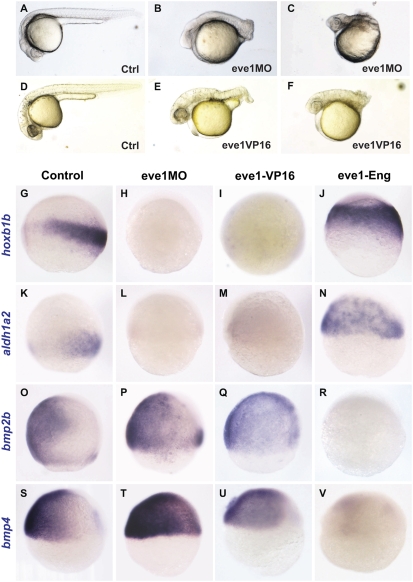
To carry out loss-of-function analyses, we first used an eve1 antisense morpholino (MO) directed at the intron1/exon2 acceptor splice site (eve1MO) (Materials and Methods). This MO led to the reduction of mature mRNA and the appearance of an alternatively spliced form of mRNA in the embryo (Fig. S1A). The phenotypes in embryos injected with eve1MO complement those of eve1 overexpression, namely a loss of posterior structures with largely unaffected head structures (Fig. 2 B and C). In the most severe phenotype, most of the trunk and tail tissue is absent (Fig. 2C and Fig. S1B). Because eve1 is thought to function as a transcriptional repressor (30 –32), we reasoned that fusion of the eve1 homeodomain (DNA binding) to the activator domain of the viral protein VP16 would generate an antimorphic construct (Materials and Methods). Similar to the effect of eve1MO, injection of eve-VP16 led to a variable reduction of the posterior axis (71%, n = 28) (Fig. 2 E and F).
Fig. 2.
Eve1 depletion suppresses trunk and tail development, and eve1 acts as a repressor. Zebrafish embryos were injected with eve1MO (B, C, H, L, P, T) or with eve1-VP16 mRNA (E, F, I, M, Q, U) as shown at bottom right of panels B, C, E, and F and at the top of the columns for the remainder. Embryos at 24 hpf (A–C) and at 28 hpf (D–F), show variable loss of trunk and tail tissue. (G–V) In situ staining of embryos at 70–80% epiboly (G–R) and 60% epiboly (S–V): lateral views, dorsal to the right (where discernible), with probes shown at the left of the rows. Hoxb1b and aldh1a2 are suppressed by eve1MO (H and L) and eve1-VP16 (I and M, compare with G and K), and expression of both genes is expanded in embryos injected with eve1-Eng (J and N). Conversely, bmp2b and bmp4 expression domains are expanded in embryos injected with eve1MO (P and T) and eve1-VP16 (Q and U, compare with O and S), whereas eve1-Eng suppresses expression of both BMPs (R and V).
We next examined the expression of otx2 and hoxb1b in eve1MO-injected embryos. Consistent with the gain-of-function analysis, hoxb1b expression was strongly suppressed (96%, n = 28) (Fig. 2H), but we found no noticeable difference in otx2 expression. We further examined the expression of aldh1a2 (formerly raldh2), which codes for an RA synthesizing enzyme expressed in posterior paraxial mesoderm (33), as well as meis3, another posterior-specific neural gene (34). The expression of aldh1a2 was much reduced (71%) or absent (29%) in eve1MO-injected embryos (n = 24) (Fig. 2L), as was the expression of meis3 (Fig. S2E). To test the specificity of the eve1MO, we coinjected eve1 mRNA and found that the expression of both aldh1a2 (95%, n = 21) and hoxb1b (96%, n = 28) was restored and slightly expanded as compared with uninjected embryos (Fig. S1C 1 –6). Furthermore, we found that epiboly defects caused by eve1MO were rescued by coinjection of eve1 mRNA (Fig. S1D). These results indicate that the MO is specific for eve1. In further analysis of eve1 loss of function, we found that the expression of hoxb1b (75%, n = 16) and aldh1a2 (88%, n = 16) was suppressed in embryos injected with eve1-VP16 (Fig. 2 I and M).
Eve1-VP16 and eve1MO show a similar phenotype that is complementary to that of eve1 overexpression, suggesting that eve1 exerts its posteriorizing influence as a repressor. To explore this possibility, we fused the eve1 homeodomain to the repressor domain of the Drosophila Engrailed (Eng) protein (Materials and Methods) (32, 35). Injection of eve1-Eng led to expansion of hoxb1b (70%, n = 20) and aldh1a2 (56%, n = 16) expression (Fig. 2 J and N), phenotypes complementary to those elicited by eve1MO and eve1-VP16. Because the results of overexpression suggested that eve1 might have a role in neural induction, we also looked at bmp2b and bmp4 expression under conditions of eve1 gain and loss of function. Injection of eve1MO expanded both bmp2b (83%, n = 18) and bmp4 (63%, n = 16) expression (Fig. 2 P and T), as did injection of eve1-VP16 (76%, n = 17, and 70%, n = 23, respectively) (Fig. 2 Q and U), whereas injection of eve1-Eng suppressed both bmp2b (74%, n = 19) (Fig. 2R) and bmp4 (90%, n = 20) (Fig. 2V) expression. Together these data suggest that eve1 acts as a transcriptional repressor in promoting posterior neural development.
Eve1 Induces Hoxb1b Expression via RA Signaling.
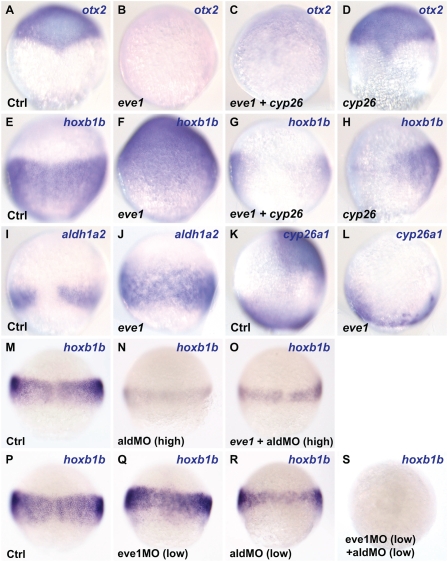
Because eve1 induces hoxb1b expression (Fig. 1G), and eve1MO injection led to loss of aldh1a2 expression (Fig. 2L), we investigated whether this effect is mediated by the RA pathway by injecting eve1 and cyp26a1 mRNAs in different combinations and examining hoxb1b as well as otx2 expression. Cyp26a1 is an RA-degrading enzyme, and overexpression of cyp26a1 allows examination of eve1 function when RA signaling is suppressed (11). Otx2 expression was suppressed by injection of eve1 mRNA alone, whereas hoxb1b expression was expanded (Fig. 3 B and F). However, when eve1 and cyp26a1 mRNAs were coinjected, both otx2 and hoxb1b were suppressed (Fig. 3 C and G), suggesting that suppression of otx2 by eve1 is RA-independent, but expansion of hoxb1b is dependent on RA. As previously reported (11), cyp26a1 injection alone had no significant effect on otx2 expression (Fig. 3D) but suppressed hoxb1b (Fig. 3H). To examine further eve1 function upstream of RA in hoxb1b induction, we injected eve1 mRNA and analyzed the expression of aldh1a2 and cyp26a1. Eve1 causes the expansion of the aldh1a2 expression domain and suppression of cyp26a1 in anterior neural ectoderm (Fig. 3 J and L). Given that RA is a long-range signaling molecule (36), the induction of aldh1a2 and suppression of cyp26a1 may provide the mechanism of hoxb1b induction in the animal pole by overexpression of eve1.
Fig. 3.
Eve1 induces hoxb1b expression via an RA signal. (A–J and M–S) Dorsal and (K and L) lateral views (where discernible, dorsal to the right) of zebrafish embryos fixed for in situ staining at 80% epiboly (A–L) and 60% epiboly (M–S) Injections are indicated at the bottom left of each panel, and genes analyzed are given at the top right. (A–D) Suppression of otx2 by eve1 does not depend on RA because it resists overexpression of the RA-metabolizing enzyme Cyp26a1. (E–H) Eve1-mediated induction of hoxb1b does not occur when eve1 and cyp26a1 mRNAs are coinjected (G), and cyp26a1 injection alone suppresses hoxb1b expression (H; only one of two cells was injected in this embryo). (I and J) Eve1 induces aldh1a2 expression. (K and L) Anterior expression of cyp26a1 is suppressed by eve1 but remains unaffected at the margin. (M–O) Injection of high concentrations of aldMO and eve1 mRNA (Materials and Methods). Eve1 cannot rescue hoxb1b expression in aldMO-injected embryos (O). (P–S) Injection of low concentrations of eve1MO (2 ng/nL) and aldMO showed synergism in the suppression of hoxb1b.
To complement the cyp26a1 and eve1MO data, we sought further confirmation that eve1 functions upstream of aldh1a2 and, presumably, of RA in inducing hoxb1b. Injection of an antisense MO directed against the aldh1a2 gene (aldMO; Materials and Methods) alone resulted in a marked reduction of hoxb1b expression (95%, n = 22) (Fig. 3N), and this inhibition could not be rescued by coinjection of eve1 mRNA (100%, n = 26) (Fig. 3O). A similar result was obtained with another RA-responsive gene, meis3 (11) (Fig. S2 A–D). Further, there is synergism between eve1MO (2 ng/nL) and aldMO (low) action in the regulation of hoxb1b expression. Low concentrations of either MO alone led to a partial and variable reduction of the hoxb1b signal, whereas coinjection of both MOs at the same low doses led to the complete abolition of hoxb1b expression in most injected embryos (72%, n = 25); in the remainder, hoxb1b expression was variably reduced (Fig. 3 P–S). Again, we obtained similar results with meis3 (Fig. S2 E–H). These data provide strong evidence that eve1 functions upstream of RA in positively regulating the expression of hoxb1b and meis3 and possibly of other RA-responsive genes as well.
Eve1 Promotes Neural Ectoderm by Antagonizing BMP Expression.
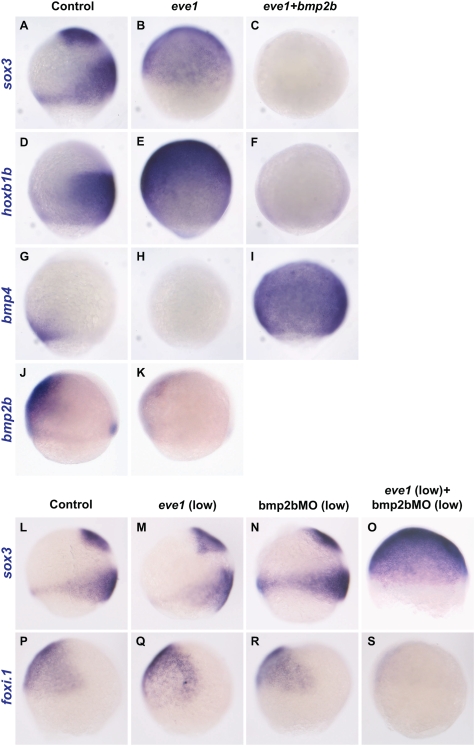
Overexpression of eve1 can suppress epidermal and induce neural marker genes (Fig. 1), suggesting that eve1 and BMP signaling have antagonistic roles in neural versus epidermal specification in the ectoderm. We used eve1 and bmp2b mRNA injection to determine whether BMP signaling can suppress eve1-mediated induction of the neural markers hoxb1b and sox3. When eve1 and bmp2b mRNAs are coinjected, both sox3 (100%, n = 24) and hoxb1b (96%, n = 25) expression is suppressed compared with control and eve1-injected embryos (Fig. 4 A–F). Because eve1MO injection leads to the variable expansion of both bmp2b and bmp4 (Fig. 2 P and T), we tested for possible inhibition of bmp4 and bmp2b expression in embryos overexpressing eve1 and found that eve1 mRNA injection inhibited bmp4 (88%, n = 17) and bmp2b (89%, n = 19) expression (Fig. 4 H and K). Thus eve1 can regulate expression levels of BMP in gastrula embryos, suggesting that BMP signaling occurs downstream of eve1. This suggestion is supported by the fact that injection of bmp2b mRNA strongly induced bmp4 expression in the presence of exogenous eve1 (100%, n = 20) (Fig. 4I). Likewise, injection of a low concentration of bmp2b RNA (7 pg/nL) together with eve1 abolished the ability of eve1 to induce neural markers or suppress bmp4 (Fig. S3).
Fig. 4.
Interactions between eve1 and BMP. Lateral views (where discernible, dorsal to the right) of zebrafish embryos at 70–80% epiboly (A–S). Embryos were injected at the one-cell stage (A–K, M, and Q) or at the one– to four-cell stage (N and R). For coinjection of eve1 mRNA and bmp2bMO, embryos first were injected with eve1 at the one-cell stage and then were injected with bmp2bMO at the four- to eight-cell stage (O and S). Genes analyzed are indicated at the left of the rows; injections are indicated at the top of columns. Bmp2b suppresses neural markers sox3 and hoxb1b even in the presence of eve1 (A–F). Bmp4 expression is suppressed by eve1 (H) but is ubiquitously induced by coinjection of bmp2b mRNA (I, compare to G). Bmp2b expression also is suppressed by eve1 (J, K). Eve1 and bmp2bMO synergize in ectodermal fate specification (L–S). Low levels of eve1 mRNA (10 pg/nL) or bmp2bMO (100 pg/nL) injected individually do not affect sox3 or foxi.1 expression (M, N, Q, and R), but coinjection at the same concentrations induced sox3 (O) and suppressed foxi.1 (S).
To explore further the antagonistic nature of eve1 and BMP signaling in ectodermal fate specification, we examined their combined effects on the expression of the neural marker sox3 and the epidermal marker foxi.1. At low (10 pg/nL) concentrations, neither eve1 mRNA (100%, n = 25) nor bmp2bMO (100%, n = 26) can induce sox3 (Fig. 4 M and N) or suppress foxi.1 (Fig. 4 Q and R). However, when eve1 and bmp2bMO were coinjected at low concentrations, sox3 was expanded (95%, n = 19) (Fig. 4O), whereas foxi.1 was suppressed (94%, n = 17) (Fig. 4S). These results support the view that eve1 has a role in posterior neural induction via antagonism of BMP signaling.
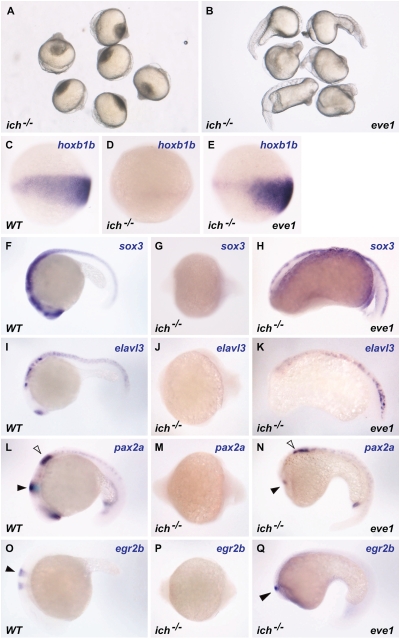
Eve1 Rescues Posterior Dorsal Axis and Expression of Hoxb1b in Ichabod−/− Mutants.
Ichabod−/− (ich−/−) mutants have reduced expression of beta-catenin 2 that leads to loss of the organizer, ventralization, and loss of head and trunk structures (37 –39) (Fig. 5A). In such embryos, BMP expression is expanded dorsally, and this expansion is thought to account for the observed ventralization. Because eve1 antagonizes BMP signaling and has a role in posteriorization, we injected ich−/− embryos with eve1 mRNA to test whether eve1 could rescue trunk and tail development. Eve1-injected ich−/− embryos at 24 h postfertilization (hpf) showed a partial rescue of the posterior dorsal axis in the trunk and tail (100%, n = 38) (Fig. 5B) when compared with uninjected embryos (100%, n = 28) (Fig. 5A). Expression of hoxb1b that is absent in ich−/− mutants at gastrula (Fig. 5D) (39) is restored in eve1-injected embryos (Fig. 5E). Expression of the neural marker sox3 initially occurs in the trunk/tail domain of gastrula-stage ich−/− embryos and is reduced gradually and becomes faint by 24 hpf (Fig. 5G), but sox3 expression is retained in the rescued trunk and tail neural tube in eve1-injected ich−/− embryos (Fig. 5H). Similarly, the neural expression domains of elavl3 (formerly huC), pax2a, and egr2b (formerly krox20), which are variably reduced or lost in ich−/− embryos 24 hpf (Fig. 5 J, M, and P), are partially restored in the trunk and tail after eve1 injection (penetrance = 100%) (Fig. 5 K, N, and Q). These data indicate that eve1 can induce and maintain some posterior dorsal structures as well as neural gene expression in the trunk and tail of organizer-defective ich−/− embryos.
Fig. 5.
Eve1 rescues posterior neural development in ich−/− mutants. Homozygous ich−/− embryos were injected with eve1 mRNA. Genotype is indicated at the bottom left of each panel, injections at the bottom right, and in situ probes at the top right. Uninjected ich−/− embryos at 24 hpf (A). Eve1 mRNA injection leads to varying levels of rescue of posterior dorsal axis (C–E). Embryos stained for hoxb1b at 80% epiboly, presumed lateral view, dorsal to the right. Expression of hoxb1b is absent in uninjected ich−/− embryos (C, D) but is rescued by injection of eve1 mRNA (E). In situ hybridization of wild-type and ich−/− embryos at 24 hpf (anterior to the left) (F–Q). Neural gene expression and posterior dorsal axis formation was partially rescued by the injection of eve1 mRNA. Rescue of pax2a appears to extend to the midbrain–hindbrain boundary (L–N, arrowheads), whereas egr2b expression appears to extend to rhombomere 5 (O–Q, arrowheads).
Discussion
Eve1 Promotes Posterior Development as a Transcriptional Repressor.
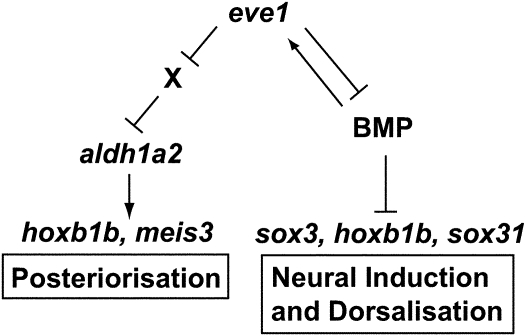
The Evx proteins have been shown previously to function as transcriptional repressors in Drosophila development (31, 32, 40, 41). Our data suggest that Eve1 also functions as a transcriptional repressor in vertebrates in promoting posterior development and that eve1-VP16 acts as a dominant-negative form. Overexpression of wild-type eve1 and eve1-VP16 results in opposite phenotypes: Eve1 suppressed head formation, whereas eve1-VP16 suppressed trunk and tail formation, with consistent effects on the expression of marker genes. Further, inhibition of eve1 expression by an MO phenocopied the eve1-VP16 phenotype, whereas eve1-Eng phenocopied the effects of eve1 overexpression on posterior neural markers and BMP expression. As a result, it is likely that the up-regulation of marker genes by eve1 is indirect. In neural induction and dorsalization, suppression of BMP by eve1 could explain the induction of neural-specific genes, whereas in posteriorization eve1 may well repress an as yet unidentified repressor of aldh1a2 (Fig. 6)
Fig. 6.
Role of eve1 in posteriorization and neural induction. See text for discussion.
Eve1 Induces Posterior Cell Fates via Retinoic Acid.
Through gain- and loss-of-function analyses of eve1, we explored the mechanisms of eve1 function in zebrafish trunk and tail development. Overexpression of eve1 suppressed head structures and in the trunk and tail expanded neural and suppressed epidermal cell fates. These data indicate a role for eve1 in both posteriorization and neural induction (Fig. 6). The regulation of RA levels via induction of aldh1a2 and suppression of cyp26a1 is necessary and sufficient for the induction of the posterior gene hoxb1b by eve1. This conclusion is supported by the observations that eve1MO inhibits aldh1a2 expression and that eve1 induction of hoxb1b and meis3 is mediated by aldh1a2, because neither gene could be induced by eve1 in aldMO-injected embryos. Eve1 suppresses the anterior gene otx2 via an RA-independent route, suggesting that there are two separate mechanisms for eve1-mediated posteriorization: RA-dependent posterior induction and RA-independent anterior suppression. This distinction may assist in creating a border between anterior (RA-negative) and posterior (RA-positive) gene-expression domains. Analogous separable mechanisms already have been observed for two other posteriorizing factors, FGF (9 –11) and Wnt (13, 14, 42). Similar to the situation after reduction of RA signaling (11), but unlike the effect of FGF and Wnt (11), no posterior expansion of anterior gene expression was seen in eve1 morphants, suggesting that suppression of anterior genes may not be a primary role of eve1. Because suppression of RA alone does not expand otx2, these results further support the idea that eve1 posteriorizes embryos via the RA pathway. We have shown that otherwise eve1 functions in a similar manner to FGF, RA, and Wnt posterior signaling, and, because eve1 is induced by FGF (11, 23), it is tempting to suggest that eve1 acts downstream of FGF in mediating posteriorization signals.
A Role for eve1 in Neural Induction.
A surprising finding was a role for eve1 in the induction of posterior neural markers. In embryos where eve1 is overexpressed, the expression of sox3, sox31, and other neural markers (Fig. 1) is expanded through the entire ectoderm, including the animal pole and presumptive epidermis. In these embryos the epidermal marker p63 is suppressed, suggesting that prospective epidermal tissue has been respecified as neural. This conclusion is supported by the finding that eve1 is necessary for the expression of hoxb1b (Fig. 2) and meis3 (Fig. S2). In addition, eve1 suppresses BMP expression in the gastrula embryo, but eve1 cannot induce the expression of either sox3 or hoxb1b in the presence of BMP. Thus it appears that Eve1 does not antagonize BMP signaling but rather suppresses BMP expression. As a consequence, a synergistic relationship exists between eve1 and Bmp2bMO in the induction of sox3 and suppression of foxi.1, a marker for epidermal tissue (Fig. 4). Together these data suggest that eve1 enhances neural induction by reducing the expression of BMP in the gastrula ectoderm (Fig. 6).
Further evidence for eve1-mediated neural induction and maintenance comes from the experiment using ventralized ich−/− embryos. In ich−/− embryos, the expression of hoxb1b, elavl3, pax2a, and egr2b is low or absent, sox3 is expressed only weakly, and neural tissue is greatly reduced at 24 hpf. Injection of eve1 mRNA into these mutant embryos rescued hoxb1b expression and partially restored the expression of elavl3, pax2, and egr2b with a penetrance of 100%. Likewise, overexpression of eve1 restored a posterior dorsal axis in ich−/− embryos. We suggest that eve1 elicits these effects at least partially by a reduction of BMP expression, thereby substituting in the posterior domain for the absence of organizer-derived BMP antagonists.
Taken together the data suggest that eve1 has dorsalizing activity (including neural induction) via regulation of BMP expression in the gastrula ectoderm. Many genes that regulate BMP expression and signaling along the dorso-ventral axis have been reported (for review, see ref. 43). For example, positive regulators of BMP, such as Bmp2b and Bmp4 in zebrafish (44) and ADMP (45) in Xenopus, are expressed in the organizer and may contribute to fine tuning of BMP expression and signaling. Besides secreted molecules, many transcription factors also have been shown to suppress BMP expression and to dorsalize the embryo [e.g., hex (46), iro3 (47)]. Here, we propose to add eve1 as another regulator of BMP activity that is unique in the sense that eve1 expression is maintained by the BMP signal in the ventral side and in turn limits BMP expression (negative feedback). The variety of mechanisms regulating BMP expression levels indicates that precise control of the timing and level of BMP signaling is crucial in regulating neural versus nonneural patterning, A/P patterning, cell migration, and some aspects of gastrulation.
Eve1 as an Effector of the Posterior Organizer.
Eve1 has been thought to play an important role in tail development, because overexpression of eve1 induces ectopic tail structures (25), and induction of ectopic tails by Wnt, BMP, and Nodal induces eve1 expression (48). Although eve1 is expressed in the prospective tail region only in the late gastrula, eve1 expression is much wider in the blastula and early gastrula, being expressed in prospective trunk mesoderm and neural ectoderm at that stage (7, 29). Eve1 is positively regulated by FGF (7, 23) and Wnt (49, 50), two signaling pathways that are critical for induction of both trunk and tail structures. Furthermore, it has been proposed that in both Xenopus and zebrafish tail formation is a continuation of trunk formation (7, 51) and that both occur as interactions between dorsal and ventral cells. Considering these ideas and our current data, we propose that eve1 acts as a posterior organizer in regulating posterior specificity as well as dorso-ventral specificity for trunk and tail tissue. Eve1 may function as a posterior dorsal gene in the sense that it induces caudal neural tissue. The contrasting function of eve1 might be understood in the light of the observation that it represses BMP but enhances RA (through aldh1a2). Thus eve1 would be required for posterior development (RA, and possibly other functions) but would limit the ventralizing action of BMP to facilitate formation of caudal neural tissue.
Materials and Methods
RNA Probe Synthesis and in Situ Hybridization.
Probes used (except aldh1a2), antisense RNA probe synthesis, and in situ hybridization procedures have been previously described (34). RZPD clone IMAGp998B2417171Q1 in pExpress1 was used for synthesis of the aldh1a2 probe.
Constructs, mRNA Synthesis, and mRNA Injection.
Capped mRNAs were synthesized using the mMessage mMachine SP6 kit (Ambion) according to the manufacturer’s instructions. Unless otherwise stated, the mRNA concentrations used for injections were bmp2b, 50 pg/nL; eve1, 20 pg/nL; eve1-VP16, 300 pg/nL; eve1-Eng, 300 pg/nL; and cyp26a1, 500 pg/nL. mRNAs were injected through the intact chorion into all blastomeres at the one- to two-cell stage. To make the eve1-VP16 and the eve1-Eng fusion constructs, the eve1 homeodomain was amplified by PCR (forward primer: GCCCTCGAGCAAGAATACTGCAAAGAAAGT; reverse primer: GCCTCTAGAGTGGATTTGGCCAGTGTAGAC) and subcloned into a pCS2_VP16 and pCS2_Eng vector (52).
Morpholino Analysis and Injection.
Eve1 mRNA (mildly) and eve1MO (more severely) affected epiboly movements, making the analysis of gene expression difficult in later stages in eve1MO-injected embryos; therefore in these embryos we concentrated on earlier marker analyses. The eve1MO (GeneTools LLC) corresponds to the intron1/exon2 acceptor splice site: 5′-CTGTCCTCTGCTACTGAAAAGAATA-3′. The eve1MO was injected at 5 ng/nL unless otherwise indicated. The bmp2bMO (GeneTools LLC) 5′-GCGGACCACGGCGACCATGATC-3′ targets the transcription start site; it was used at 0.1 ng/nL. The aldh1a2 MO (Open Biosystems) (53) has the sequence 5′-GTTCAACTTCACTGGAGGTCATCGC-3′and was used at 1:2 (high) and 1:4 (low) dilutions from a 1-mM stock. In all cases, 1–2 nL of solution was injected into the yolk as close as possible to the cells of embryos at the one- to four-cell stage.
Supplementary Material
Acknowledgments
We thank Dr. Aya Takesono for help and advice provided throughout the research period and writing of the manuscript. We thank the fish facility personnel at the University of Exeter and University College London. This work was supported in part by the Intramural Program of the National Institute of Child Health and Human Development, National Institutes of Health (I.B.D.) and by the Welcome Trust (S.W.W.). C.C. is a PhD student at the University of Exeter and his PhD studentship is provided by the Biotechnology and Biological Sciences Research Council in the United Kingdom.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/1000389107/DCSupplemental.
References
- 1.Stern CD. Neural induction: Old problem, new findings, yet more questions. Development. 2005;132:2007–2021. doi: 10.1242/dev.01794. [DOI] [PubMed] [Google Scholar]
- 2.Muñoz-Sanjuán I, Brivanlou AH. Neural induction, the default model and embryonic stem cells. Nat Rev Neurosci. 2002;3:271–280. doi: 10.1038/nrn786. [DOI] [PubMed] [Google Scholar]
- 3.Nieuwkoop PD, Nigtevecht GV. Neural activation and transformation in explants of competent ectoderm under the influence of fragments of anterior notochord in Urodeles. J Embryol Exp Morphol. 1954;2:175–193. [Google Scholar]
- 4.Delaune E, Lemaire P, Kodjabachian L. Neural induction in Xenopus requires early FGF signalling in addition to BMP inhibition. Development. 2005;132:299–310. doi: 10.1242/dev.01582. [DOI] [PubMed] [Google Scholar]
- 5.Kengaku M, Okamoto H. bFGF as a possible morphogen for the anteroposterior axis of the central nervous system in Xenopus. Development. 1995;121:3121–3130. doi: 10.1242/dev.121.9.3121. [DOI] [PubMed] [Google Scholar]
- 6.Koshida S, et al. Inhibition of BMP activity by the FGF signal promotes posterior neural development in zebrafish. Dev Biol. 2002;244:9–20. doi: 10.1006/dbio.2002.0581. [DOI] [PubMed] [Google Scholar]
- 7.Kudoh T, Concha ML, Houart C, Dawid IB, Wilson SW. Combinatorial Fgf and Bmp signalling patterns the gastrula ectoderm into prospective neural and epidermal domains. Development. 2004;131:3581–3592. doi: 10.1242/dev.01227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Streit A, Berliner AJ, Papanayotou C, Sirulnik A, Stern CD. Initiation of neural induction by FGF signalling before gastrulation. Nature. 2000;406:74–78. doi: 10.1038/35017617. [DOI] [PubMed] [Google Scholar]
- 9.Lamb TM, Harland RM. Fibroblast growth factor is a direct neural inducer, which combined with noggin generates anterior-posterior neural pattern. Development. 1995;121:3627–3636. doi: 10.1242/dev.121.11.3627. [DOI] [PubMed] [Google Scholar]
- 10.Cox WG, Hemmati-Brivanlou A. Caudalization of neural fate by tissue recombination and bFGF. Development. 1995;121:4349–4358. doi: 10.1242/dev.121.12.4349. [DOI] [PubMed] [Google Scholar]
- 11.Kudoh T, Wilson SW, Dawid IB. Distinct roles for Fgf, Wnt and retinoic acid in posteriorizing the neural ectoderm. Development. 2002;129:4335–4346. doi: 10.1242/dev.129.18.4335. [DOI] [PubMed] [Google Scholar]
- 12.Fekany-Lee K, Gonzalez E, Miller-Bertoglio V, Solnica-Krezel L. The homeobox gene bozozok promotes anterior neuroectoderm formation in zebrafish through negative regulation of BMP2/4 and Wnt pathways. Development. 2000;127:2333–2345. doi: 10.1242/dev.127.11.2333. [DOI] [PubMed] [Google Scholar]
- 13.Kiecker C, Niehrs C. A morphogen gradient of Wnt/beta-catenin signalling regulates anteroposterior neural patterning in Xenopus. Development. 2001;128:4189–4201. doi: 10.1242/dev.128.21.4189. [DOI] [PubMed] [Google Scholar]
- 14.Yamaguchi TP. Heads or tails: Wnts and anterior-posterior patterning. Curr Biol. 2001;11:R713–R724. doi: 10.1016/s0960-9822(01)00417-1. [DOI] [PubMed] [Google Scholar]
- 15.Blumberg B, et al. An essential role for retinoid signaling in anteroposterior neural patterning. Development. 1997;124:373–379. doi: 10.1242/dev.124.2.373. [DOI] [PubMed] [Google Scholar]
- 16.Conlon RA. Retinoic acid and pattern formation in vertebrates. Trends Genet. 1995;11:314–319. doi: 10.1016/s0168-9525(00)89089-7. [DOI] [PubMed] [Google Scholar]
- 17.Durston AJ, et al. Retinoic acid causes an anteroposterior transformation in the developing central nervous system. Nature. 1989;340:140–144. doi: 10.1038/340140a0. [DOI] [PubMed] [Google Scholar]
- 18.Gavalas A, Krumlauf R. Retinoid signalling and hindbrain patterning. Curr Opin Genet Dev. 2000;10:380–386. doi: 10.1016/s0959-437x(00)00100-3. [DOI] [PubMed] [Google Scholar]
- 19.Maden M. Retinoid signalling in the development of the central nervous system. Nat Rev Neurosci. 2002;3:843–853. doi: 10.1038/nrn963. [DOI] [PubMed] [Google Scholar]
- 20.Koshida S, Shinya M, Mizuno T, Kuroiwa A, Takeda H. Initial anteroposterior pattern of the zebrafish central nervous system is determined by differential competence of the epiblast. Development. 1998;125:1957–1966. doi: 10.1242/dev.125.10.1957. [DOI] [PubMed] [Google Scholar]
- 21.Skromne I, Thorsen D, Hale M, Prince VE, Ho RK. Repression of the hindbrain developmental program by Cdx factors is required for the specification of the vertebrate spinal cord. Development. 2007;134:2147–2158. doi: 10.1242/dev.002980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shimizu T, Bae YK, Hibi M. Cdx-Hox code controls competence for responding to Fgfs and retinoic acid in zebrafish neural tissue. Development. 2006;133:4709–4719. doi: 10.1242/dev.02660. [DOI] [PubMed] [Google Scholar]
- 23.Griffin K, Patient R, Holder N. Analysis of FGF function in normal and no tail zebrafish embryos reveals separate mechanisms for formation of the trunk and the tail. Development. 1995;121:2983–2994. doi: 10.1242/dev.121.9.2983. [DOI] [PubMed] [Google Scholar]
- 24.Ahringer J. Posterior patterning by the Caenorhabditis elegans even-skipped homolog vab-7. Genes Dev. 1996;10:1120–1130. doi: 10.1101/gad.10.9.1120. [DOI] [PubMed] [Google Scholar]
- 25.Barro O, et al. Widespread expression of the eve1 gene in zebrafish embryos affects the anterior-posterior axis pattern. Dev Genet. 1995;17:117–128. doi: 10.1002/dvg.1020170204. [DOI] [PubMed] [Google Scholar]
- 26.Ruiz i Altaba A, Melton DA. Involvement of the Xenopus homeobox gene Xhox3 in pattern formation along the anterior-posterior axis. Cell. 1989;57:317–326. doi: 10.1016/0092-8674(89)90969-0. [DOI] [PubMed] [Google Scholar]
- 27.Joly JS, Maury M, Joly C, Boulekbache H, Condamine H. Ventral and posterior expression of the homeo box gene eve1 in zebrafish (Brachydanio rerio) is repressed in dorsalized embryos. C R Seances Soc Biol Fil. 1993;187:356–363. [PubMed] [Google Scholar]
- 28.Alexandre D, et al. Ectopic expression of Hoxa-1 in the zebrafish alters the fate of the mandibular arch neural crest and phenocopies a retinoic acid-induced phenotype. Development. 1996;122:735–746. doi: 10.1242/dev.122.3.735. [DOI] [PubMed] [Google Scholar]
- 29.Joly JS, Joly C, Schulte-Merker S, Boulekbache H, Condamine H. The ventral and posterior expression of the zebrafish homeobox gene eve1 is perturbed in dorsalized and mutant embryos. Development. 1993;119:1261–1275. doi: 10.1242/dev.119.4.1261. [DOI] [PubMed] [Google Scholar]
- 30.Li C, Manley JL. Even-skipped represses transcription by binding TATA binding protein and blocking the TFIID-TATA box interaction. Mol Cell Biol. 1998;18:3771–3781. doi: 10.1128/mcb.18.7.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Briata P, et al. Transcriptional repression by the human homeobox protein EVX1 in transfected mammalian cells. J Biol Chem. 1995;270:27695–27701. doi: 10.1074/jbc.270.46.27695. [DOI] [PubMed] [Google Scholar]
- 32.Fujioka M, Yusibova GL, Patel NH, Brown SJ, Jaynes JB. The repressor activity of Even-skipped is highly conserved, and is sufficient to activate engrailed and to regulate both the spacing and stability of parasegment boundaries. Development. 2002;129:4411–4421. doi: 10.1242/dev.129.19.4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grandel H, et al. Retinoic acid signalling in the zebrafish embryo is necessary during pre-segmentation stages to pattern the anterior-posterior axis of the CNS and to induce a pectoral fin bud. Development. 2002;129:2851–2865. doi: 10.1242/dev.129.12.2851. [DOI] [PubMed] [Google Scholar]
- 34.Kudoh T, et al. A gene expression screen in zebrafish embryogenesis. Genome Res. 2001;11:1979–1987. doi: 10.1101/gr.209601. [DOI] [PubMed] [Google Scholar]
- 35.Vickers ER, Sharrocks AD. The use of inducible engrailed fusion proteins to study the cellular functions of eukaryotic transcription factors. Methods. 2002;26:270–280. doi: 10.1016/S1046-2023(02)00031-2. [DOI] [PubMed] [Google Scholar]
- 36.White RJ, Nie Q, Lander AD, Schilling TF. Complex regulation of cyp26a1 creates a robust retinoic acid gradient in the zebrafish embryo. PLoS Biol. 2007;5(11):e304. doi: 10.1371/journal.pbio.0050304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kelly C, Chin AJ, Leatherman JL, Kozlowski DJ, Weinberg ES. Maternally controlled (beta)-catenin-mediated signaling is required for organizer formation in the zebrafish. Development. 2000;127:3899–3911. doi: 10.1242/dev.127.18.3899. [DOI] [PubMed] [Google Scholar]
- 38.Bellipanni G, et al. Essential and opposing roles of zebrafish beta-catenins in the formation of dorsal axial structures and neurectoderm. Development. 2006;133:1299–1309. doi: 10.1242/dev.02295. [DOI] [PubMed] [Google Scholar]
- 39.Maegawa S, Varga M, Weinberg ES. FGF signaling is required for beta-catenin-mediated induction of the zebrafish organizer. Development. 2006;133:3265–3276. doi: 10.1242/dev.02483. [DOI] [PubMed] [Google Scholar]
- 40.Biggin MD, Tjian R. A purified Drosophila homeodomain protein represses transcription in vitro. Cell. 1989;58:433–440. doi: 10.1016/0092-8674(89)90424-8. [DOI] [PubMed] [Google Scholar]
- 41.Han KY, Manley JL. Transcriptional repression by the Drosophila even-skipped protein: Definition of a minimal repression domain. Genes Dev. 1993;7:491–503. doi: 10.1101/gad.7.3.491. [DOI] [PubMed] [Google Scholar]
- 42.Kazanskaya O, Glinka A, Niehrs C. The role of Xenopus dickkopf1 in prechordal plate specification and neural patterning. Development. 2000;127:4981–4992. doi: 10.1242/dev.127.22.4981. [DOI] [PubMed] [Google Scholar]
- 43.De Robertis EM, Kuroda H. Dorsal-ventral patterning and neural induction in Xenopus embryos. Annu Rev Cell Dev Biol. 2004;20:285–308. doi: 10.1146/annurev.cellbio.20.011403.154124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nikaido M, Tada M, Saji T, Ueno N. Conservation of BMP signaling in zebrafish mesoderm patterning. Mech Dev. 1997;61:75–88. doi: 10.1016/s0925-4773(96)00625-9. [DOI] [PubMed] [Google Scholar]
- 45.Reversade B, De Robertis EM. Regulation of ADMP and BMP2/4/7 at opposite embryonic poles generates a self-regulating morphogenetic field. Cell. 2005;123:1147–1160. doi: 10.1016/j.cell.2005.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ho CY, Houart C, Wilson SW, Stainier DYR. A role for the extraembryonic yolk syncytial layer in patterning the zebrafish embryo suggested by properties of the hex gene. Curr Biol. 1999;9:1131–1134. doi: 10.1016/s0960-9822(99)80485-0. [DOI] [PubMed] [Google Scholar]
- 47.Kudoh T, Dawid IB. Role of the iroquois3 homeobox gene in organizer formation. Proc Natl Acad Sci USA. 2001;98:7852–7857. doi: 10.1073/pnas.141224098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Agathon A, Thisse C, Thisse B. The molecular nature of the zebrafish tail organizer. Nature. 2003;424:448–452. doi: 10.1038/nature01822. [DOI] [PubMed] [Google Scholar]
- 49.Ramel M-C, Lekven AC. Repression of the vertebrate organizer by Wnt8 is mediated by Vent and Vox. Development. 2004;131:3991–4000. doi: 10.1242/dev.01277. [DOI] [PubMed] [Google Scholar]
- 50.Ueno S, et al. Biphasic role for Wnt/beta-catenin signaling in cardiac specification in zebrafish and embryonic stem cells. Proc Natl Acad Sci USA. 2007;104:9685–9690. doi: 10.1073/pnas.0702859104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gont LK, Steinbeisser H, Blumberg B, de Robertis EM. Tail formation as a continuation of gastrulation: The multiple cell populations of the Xenopus tailbud derive from the late blastopore lip. Development. 1993;119:991–1004. doi: 10.1242/dev.119.4.991. [DOI] [PubMed] [Google Scholar]
- 52.Kessler DS. Siamois is required for formation of Spemann’s organizer. Proc Natl Acad Sci USA. 1997;94:13017–13022. doi: 10.1073/pnas.94.24.13017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dobbs-McAuliffe B, Zhao Q, Linney E. Feedback mechanisms regulate retinoic acid production and degradation in the zebrafish embryo. Mech Dev. 2004;121:339–350. doi: 10.1016/j.mod.2004.02.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.