Abstract
When introduced or cultivated plants or animals hybridize with their native relatives, the spread of invasive genes into native populations might have biological, aesthetic, and legal implications. Models suggest that the rate of displacement of native by invasive alleles can be rapid and inevitable if they are favored by natural selection. We document the spread of a few introduced genes 90 km into a threatened native species (the California Tiger Salamander) in 60 years. Meanwhile, a majority of genetic markers (65 of 68) show little evidence of spread beyond the region where introductions occurred. Using computer simulations, we found that such a pattern is unlikely to emerge by chance among selectively neutral markers. Therefore, our results imply that natural selection has favored both the movement and fixation of these exceptional invasive alleles. The legal status of introgressed populations (native populations that are slightly genetically modified) is unresolved by the US Endangered Species Act. Our results illustrate that genetic and ecological factors need to be carefully weighed when considering different criteria for protection, because different rules could result in dramatically different geographic areas and numbers of individuals being protected.
Keywords: conservation, California Tiger Salamander, genetics, hybridization, Ambystoma
Biological invasions typically involve introduced species that increase in number and negatively impact native species or crops through ecological interactions (1, 2). Some invasions also involve genetic interactions, primarily through hybridization with native species (3, 4). Hybrid invasions can be difficult to detect without molecular data, but their impacts can be severe, including both conventional ecological effects and effects that depend specifically on genetic mixture (5, 6). These uniquely genetic issues include (i) the question of whether introgression of introduced alleles should be considered a minor evolutionary change (7 –9) or a “genomic extinction” (4, 10), (ii) the problem of how to define the legal status of introgressed populations (11 –13), and (iii) the possibility that recombinant genotypes might express novel phenotypes with novel ecological consequences (14, 15).
Similar concerns arise over hybridization between native plants and genetically modified crops (16, 17). Mathematical analyses have repeatedly shown that natural selection has a great impact on the probability and rate of introgression (16, 18–20), and the joint effects of selection and dispersal must be understood to predict the movement of nonnative genes. The unsettling conclusions from this theory are that (i) advantageous alleles can overcome almost any barrier to gene flow, eventually becoming fixed throughout all available gene pools (19, 21), and (ii) little can be predicted without specific information on the ecological and genetic determinants of fitness in a given system (22). Thus, a critical question for invasion biology is whether rapid introgression (and consequent genetic transformation of native species) is likely to be common on spatial and temporal scales relevant to contemporary conservation planning.
When a hybrid invasion starts from a relatively small, localized introduction, most native alleles are expected to remain at high relative frequency when genetic differences are selectively neutral or native alleles are locally adapted (23, 24). The limited existing data appear consistent with these predictions (23). However, after the F1 generation, recombination allows different loci to respond to different selection pressures and advantageous introduced alleles might spread rapidly into populations that continue to be recognized phenotypically as native (19). Such patterns are unlikely to be detected with the small number of molecular markers typically available for nonmodel systems. Here, we survey a large number of molecular markers to assess the potential for heterogeneous introgression of invasive genes into a threatened native species and address some of the challenges raised for conservation when a protected taxon becomes genetically modified through hybridization.
In California, introduced Barred Tiger Salamanders (Ambystoma tigrinum mavortium, sometimes recognized as A. mavortium) hybridize with threatened native California Tiger Salamanders (A. californiense). Divergence of mtDNA sequences and allozymes are consistent with 3–10 my of isolation before the introductions, with geological evidence favoring a 3–5 my divergence (25, 26) Barred Tiger Salamanders were deliberately introduced into central California in 1940s and ’50s because their large aquatic larvae (“waterdogs”) are valued as live bait by bass fishermen (27, 28). Barred Tiger Salamander larvae were imported primarily from Texas and New Mexico (9, 28). Almost all of the introductions occurred in the northern Salinas Valley, where hundreds of larvae were released into multiple ponds (9). Barred Tiger Salamanders were introduced because they have a longer larval period and reach a much larger laval size than native California Tiger Salamanders. The extended larval period of Barred Tiger Salamanders is plastic, with greatest expression in perennial ponds where some individuals forgo metamorphosis entirely to become sexually mature paedomorphs (29).
Previous research has used putatively diagnostic molecular markers to document hybridization between the introduced and native lineages (28). Drastic allele frequency differences imply greater invasion success in perennial ponds (artificially constructed for livestock and irrigation) relative to more natural seasonal ponds and vernal pools (9, 30). Some hybrid genotypes have high fitness in the wild (31), and laboratory experiments demonstrated that F1, backcross, and wild hybrids have higher growth rates than natives and higher rates of predation on native amphibians (6).
Populations near the known introduction sites in the Salinas Valley remain variable for native and introduced alleles (8, 9). However, a few markers have become fixed for introduced alleles at five ecologically varible breeding sites that were examined in detail (8). Here, we examine the spatial spread of introduced alleles by estimating allele frequencies in 44 breeding ponds along a geographic gradient extending over 200 km from the known introduction sites into the range of pure California Tiger Salamanders, and an additional 18 ponds on a small scale gradient within an intact patch of habitat (Fig. 1). We used 68 single nucleotide markers diagnostic for native vs. introduced ancestry to quantify introgression (Methods).
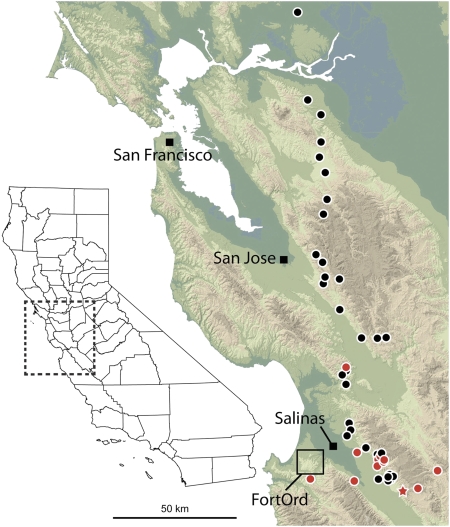
Fig. 1.
Transect sampling locations (Table S2). Red circles denote known or suspected introduction sites (9). Fort Ord area is shown as a box, but individual localities are not shown (see Fig. 2). Red star indicates reference population for cline position distances.
Results and Discussion
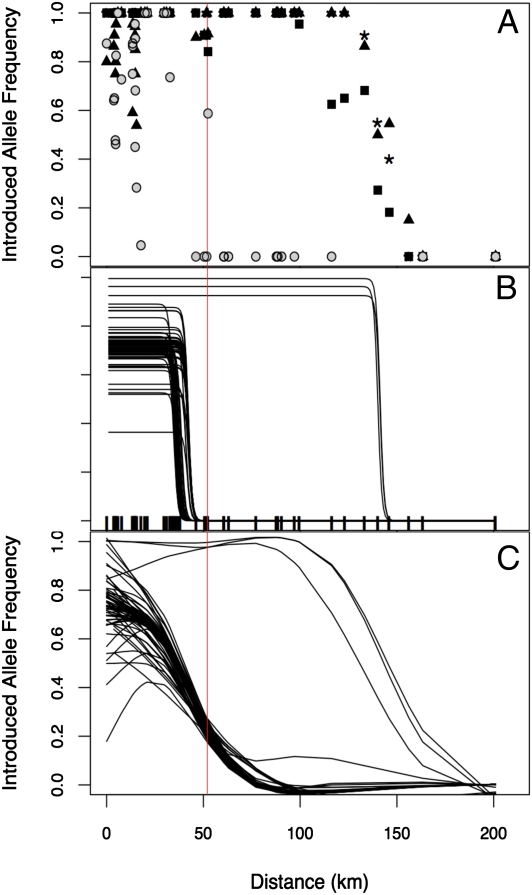
Most markers (65 of 68) show an abrupt transition from a mixture of native and introduced alleles to pure native alleles in the vicinity of the most northerly known introduction site (Fig. 2). The remaining three markers show very high frequencies of introduced alleles in ponds up to 94 km farther north, at which point they appear to shift to pure native (Fig. 2). These three markers (E6E11, E12C11, and E23C6) are unlinked and map to linkage groups 11, 14, and 4, respectively, in the Ambystoma mexicanum x tigrinum linkage map (32). It is possible that these three exceptional markers are not diagnostic and we are mistakenly attributing a natural pattern of variation to introgression. We think this is unlikely for two reasons. First, in vetting our putatively diagnostic markers, we scored California Tiger Salamanders from throughout the native range (8); it would be a remarkable coincidence that native alleles identical in state to introduced alleles were fixed only in or adjacent to the region where introductions have been documented. Second, the geographical concordance and steepness of the clines exhibited by the three markers imply a shared, nonequilibrium history (see below).
Fig. 2.
Introduced allele frequencies vs. distance from the southernmost introduction site. (A) Gray circles show the median introduced allele frequency for each breeding pond included in the transect. Introduced allele frequencies of three exceptional markers are illustrated separately: E6E11 (squares), E12C11 (triangles), and E23C6 (asterisks). (B) Sigmoid clines fitted to each of the 68 markers. Marks along the bottom illustrate the position of each sampled pond. The red vertical line at 52 km indicates the northernmost introduction site. (C) Cubic splines fitted to each of the 68 markers.
Although any large sample of markers is expected to show variation in allele frequencies and distances of geographic spread, the pattern seen in these three markers is remarkable. Two lines of inference support the conclusion that these three exceptional markers have experienced a significantly faster rate of introgression than expected by chance alone. First, prior analysis of five breeding sites shows that the same three markers cause significant deviations from neutral expectations for the variance in allele frequencies within admixed populations, implying selection for fixation of introduced alleles at the local scale (8). Second, the geographic distances in this study allowed us to evaluate whether the disparate spatial patterns illustrated in Fig. 2 can be explained as variants of a common stochastic process (described by dispersal and drift) or whether the additional influence of natural selection is likely.
To test whether the patterns for these three markers could be explained by sampling error, we used Akaike’s information criterion (AIC) to compare the likelihoods of cline models where (i) all markers were constrained to have the same parameters, (ii) all were allowed to have independent parameters, and (iii) the most deviant markers were allowed to differ in average allele frequency and cline location by using partially constrained models (33). The best fit was obtained by allowing four markers to have deviant parameters (Table S1, Model 2). Marker E12G5 had the lowest average frequency of introduced alleles in the hybrid region; however, this result does not appear extreme relative to the range of outcomes observed in neutral simulations (Fig. 3). The other three markers (E6E11, E12C11, and E23C6) had the highest frequencies of introduced alleles, and introduced alleles of these markers were distinctly more widespread than introduced alleles of any other markers in the dataset (Fig. 2).
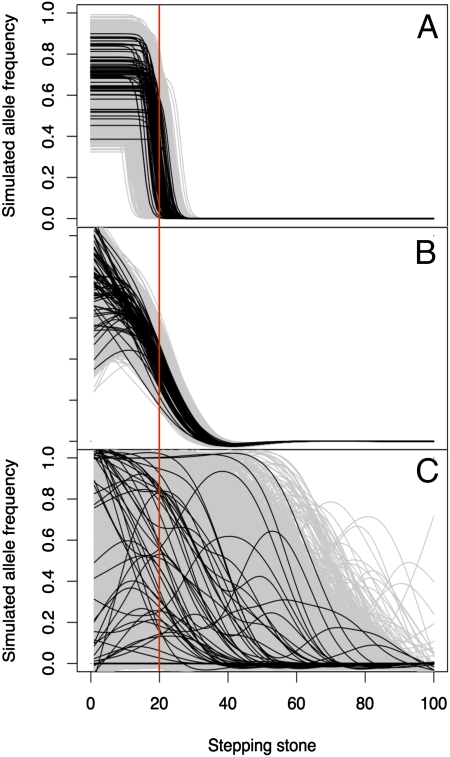
Fig. 3.
Clines fitted to simulated data from a neutral stepping stone model. Initial allele frequency was 0.71 in the first 20 demes (up to the vertical red line) and zero elsewhere. Ten thousand replicates are shown in gray and a random 68 in black. (A) Sigmoid clines after 60 time steps with n = 50 per deme and m = 0.25 per time step. (B) Cubic splines after 60 time steps with n = 50 and m = 0.25. (C) Cubic splines after 1,000 time steps with m = 0.50 and n = 10.
This analysis accounts for sampling variance, but not “evolutionary variance,” owing to the real, stochastic nature of drift and dispersal (34). To investigate the expected variation among single locus clines without selection, we performed stochastic simulations of a stepping stone model (35) of 100 demes in a linear array. We initialized the system by introducing a new allele to the first 20 demes at a frequency of 0.71 (based on field data; Table S1) and simulating dispersal and drift in discrete time. These simulations demonstrate that independent neutral markers exhibit an abrupt and consistent drop to almost zero introduced alleles within a few steps of the introduction sites (Fig. 3 A and B). This concordant front persists for hundreds of generations and aberrant clines like those in Fig. 2 were never observed in tens of thousands of simulations (Figs. S1 and S2). For a wide range of population sizes, dispersal rates, and initial allele frequencies, it takes thousands of generations before the spatial distribution of alleles flattens out and different markers begin to show widely different patterns. Even then, the variance among markers is recognizable as a continuum (Fig. 3C). In contrast, our data exhibit a clear discontinuity between the spatial patterns observed for most markers (which conform well to the drift/dispersal simulations—also see refs. 23 and 36) and the three rapidly introgressing markers.
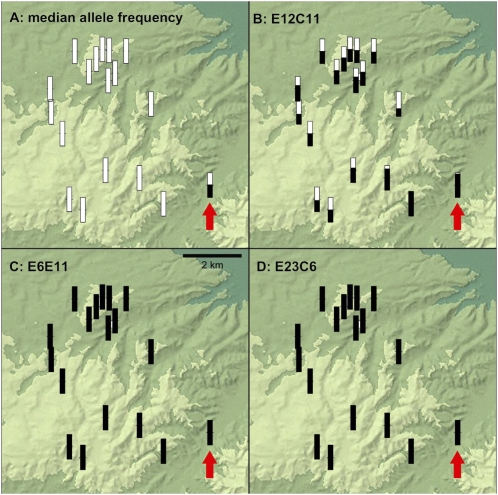
To provide a smaller scale view of introgression between introduced and native tiger salamanders, we studied an additional 18 breeding ponds on Old Fort Ord Public Lands, a semi-isolated ≈40 km2 patch of habitat just south of the Salinas River on Monterey Bay (Fig. 1). Toro Creek Pond is a suspected introduction site on Old Fort Ord where admixture was detected by using three markers (9). Our analysis revealed patterns similar to those seen on the 200-km transect scale. For the same set of 65 markers, we found no evidence of introduced alleles outside of Toro Creek Pond (Fig. 4A). Once again, markers E6E11 and E23C6 were fixed for introduced alleles throughout the 18 breeding ponds, whereas E12C11 was fixed at ponds adjacent to the introduction site and showed intermediate introduced allele frequencies in all other sites (Fig. 4). These observations reinforce our inference that most introduced alleles have remained within the immediate vicinity of the introductions while a consistent set of three have introgressed into otherwise native populations.
Fig. 4.
Frequencies of introduced alleles in ponds at Fort Ord Public Lands (see Fig. 1). (A) For 65 of 68 markers, introduced alleles were detected only in Toro Creek Pond, a suspected introduction site (indicated by the arrow in each image). The black portion of each thermometer illustrates the introduced allele frequency. (B–D) Introduced alleles of E6E11, E12C11, and E23C6, the three rapidly introgressing markers.
The determinants of hybrid fitness in this system are complex, and we still lack a complete understanding of the fitness consequences of differential hybridization in the field. F1 hybrids suffer high rates of embryonic mortality (8, 31), but F1 and later generation hybrids have high survival as larvae, high growth rates, and early onset of reproductive maturity relative to native California Tiger Salamanders (6, 8, 31). These factors likely interact with habitat to affect relative fitness of various recombinant genotypes (6, 30). The pattern documented here is not obviously tied to pond hydroperiod (the three introgressive alleles are fixed in both seasonal and perennial breeding ponds), and potential correlations with ecologically important phenotypes await future association studies.
The specific genetic and ecological causes of these extraordinary rates of introgression remain unknown. But the observation that three of 68 randomly selected genomic segments have been almost entirely replaced by introduced alleles over such broad geographic and short temporal scales has tremendous implications for management. First, our results provide critical empirical support for a prediction from population genetics theory (16, 18–20): Natural selection can cause rapid changes in the distribution and abundance of introduced alleles not accounted for by rates of dispersal and outcrossing alone. Second, the pace of evolutionary change after introduction (under 60 years, in this case), and the fact that it might not be observed without extensive genetic analysis, implies that management of certain introduced genes will often be impossible once they are “out of the bottle” (16).
Other studies of hybridization between native and introduced species have documented replacement of native gene pools by admixed or even predominantly introduced genotypes (4, 12, 37–40). Demographic imbalances between domestic and wild populations might create asymmetrical gene flow and deterministic spread of selectively neutral or even deleterious alleles (41). Spread can also be enhanced by differences in dispersal behavior, as in the case of Rainbow Trout and Westslope Cutthroat Trout (42), or hybrid vigor as in the case of the crayfish Orconectes rusticus and O. propinquus (39). However, most studies have relied on a relatively small number of molecular markers that severely limits the statistical probability of detecting heterogeneous patterns of introgression among markers. Our observation of the spread of a few invasive alleles far beyond a more generally recognizable hybrid zone illustrates how the genetic influence of hybridization might be underestimated in many cases. Although rapid, selection-driven introgression might affect relatively few markers (and, therefore, be hard to detect), it could be common given the large genomes of most sexually reproducing organisms (43).
With genomic heterogeneity of introgression, strict genetic criteria for protection might have unintended consequences. If all tiger salamanders containing the three introgressive alleles were classified as nonnative, then those animals and their habitat (hundreds of km2 of prime California range land) would not qualify for protection under the US Endangered Species Act. By the same token, if we consider California Tiger Salamanders genetically extinct throughout this area, then the geographic range of the protected species would be reduced and the consequent increase in extinction risk might warrant a change from threatened to endangered status, carrying with it more restrictive land use practices (44). We think these considerations make genetic purity an impractical conservation goal (not to mention the statistical challenge of evaluating “purity”; ref. 9). The genetic influence of introduced Barred Tiger Salamanders beyond the Salinas Valley is negligible for most markers, such that there remains a sharp distinction between mostly pure native populations and the admixed populations of the Salinas Valley. We feel that this demarcation should be one guide for assigning legal protection. Further, assessment of the conservation value of introgressed California Tiger Salamanders should be based on the phenotypic and ecological consequences of introgression. An appropriate conservation goal, in this and other cases of introduced × native introgression, might be to maximize ecological authenticity. That is, it would be better to protect individuals and populations that function like the native species, even if they are not genetically authentic, rather than to have no tiger salamanders present (7). This idea is appropriate only if introgressed individuals do not have negative impacts on other community members (which would make them ecologically inauthentic). Future research should investigate potential associations between introgressing alleles and traits of ecological significance, particularly when a small number of highly selected alleles challenge the view of absolute genetic purity.
Methods
We captured tiger salamander larvae with two-person seines, clipped ≈1 cm of tissue from the end of each tail, took field Global Positioning System locality information, and released the animals immediately. We extracted DNA by using phenol-chloroform, amplified previously described sequences by using PCR, and scored single nucleotide markers on a Victor3 reader by using the FP-TDI system (8). Markers were developed (8) by sequencing mapped EST markers sampled at ≈50 cM intervals based on ref. 32. Candidate single nucleotide differences were identified in a panel of four California Tiger Salamanders (from Jepson Prairie, well outside the introgression zone) and four Barred Tiger Salamanders. Candidate markers were declared diagnostic if the single nucleotide differences were fixed in a larger sample of 20 California Tiger Salamanders from 10 ponds from throughout the native range and 12 Barred Tiger Salamanders from a known site of pure introduced animals (Lake County, CA) outside the native range of California Tiger Salamanders (8). Detailed analysis of Hardy-Weinberg and linkage disequilbria in five ponds with large sample sizes is presented elsewhere (8). Here, we focus entirely on estimates of allele frequencies based on 10–56 individuals per pond (Table S2).
To quantify the relationship between spatial position and introduced allele frequency, we used nonlinear regression in R (www.r-project.org) to fit a sigmoid cline PX = L / [1 + exp(X − M)], where PX is the estimated frequency of introduced alleles at distance X, L is the average frequency of introduced alleles within the region of the introductions, and M is the inflection point of the cline. For X we used the straight-line distance in kilometers between each pond and the southernmost pond in the transect. This formulation assumes that the allele frequency approaches a zero asymptote when X >M and that the cline width is 1 km. When we attempted to estimate cline width, the algorithm did not converge because the likelihood plateaus as width approaches zero (i.e., all cline widths smaller than a few kilometers are equally likely, given the data). This likelhood plateau reflects the fact that the transition from admixed to essentially pure native populations is abrupt relative to the scale of interpond distances and it is impossible to sample at a fine enough grain to differentiate a step-cline from a very steep cline (9). To illustrate that our results are not artifacts of the parametric model, we also fitted cubic splines to the data for each marker (Fig. 2C).
To evaluate discordance among markers (33), we compared the AIC of a constrained cline model where all markers were modeled to have the same L and the same M, an unconstrained model where L and M were estimated separately for each marker, and a set of partially constrained models where the three or four most deviant markers (4.4 and 5.9 percentiles, respectively) were allowed to have different parameters (Table S1).
We also explored the expected nonequilibrium spatial dynamics of selectively neutral alleles introduced into a previously occupied range using a stochastic stepping stone model (SI Text). Analytical theory (18, 20) and simulations (36) show that the expected change in position of a neutral cline is zero. The important question is whether a few deviant clines like those in Fig. 2A are likely to arise in the absence of selection. We simulated 100 demes in a stepping-stone pattern with dispersal between adjacent demes (each deme exchanges alleles with its two neighbors with probability m/2 per time step). The first 20 demes were initialized with an allele frequency of 0.71 (the average introduced allele frequency in the admixture zone) and the last 80 had zero introduced alleles. We simulated 10,000 independent loci for 60 time steps (the number of years since the initial introduction). We think years is the appropriate timescale for dispersal, but using generations (≈2–4 years, implying 15–30 time steps) does not change the results. Simulations were repeated for each of nine parameter combinations: diploid effective population sizes of 10, 25, and 50 per deme and dispersal probabilities of m = 0.10, 0.25, and 0.50 (interpond dispersal of California Tiger Salamanders averages less than m = 0.20; refs. 45 and 46). For each of these 90,000 simulated datasets, we fitted the sigmoid model above. We then replicated the simulations and fitted cubic splines to the simulated data. We inspected fitted curves to evaluate the range of patterns when admixture is followed by dispersal and drift alone. For the parameter combination with the widest variance in cline position (m = 0.50 and n = 10), we also simulated 10,000 loci for 1,000 time steps to gain a broader perspective on the variance of outcomes when there has been greater opportunity for spread. Simulations were intended to bracket dispersal estimates for California Tiger Salamanders and provide a general assessment of how often neutral markers are fixed for introduced alleles at various distances (relative to the dispersal distance of the organism) at timescales relevant to biological invasions.
Supplementary Material
Acknowledgments
We thank A .M. Picco, A. T. Chang, O. J. Abramyan, W. K. Savage, T. P. Heah, S. Micheletti, L. N. Gray for assistance in the field and laboratory. Our work was funded by the Environmental Protection Agency (U 91572401 and R 828896), US Department of Agriculture (04XN022, EPDRP), National Science Foundation (DEB 0516475, DEB 0213155), CALFED (01-N43), the University of Tennessee, and the University of California, Davis Agricultural Experiment Station.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0911802107/DCSupplemental.
References
- 1.Pimentel D, et al. Economic and environmental threats of alien plant, animal, and microbe invasions. Agric Ecosyst Environ. 2001;84:1–20. [Google Scholar]
- 2.Simberloff D. Confronting introduced species: A form of xenophobia? Biol Invasions. 2003;5:179–192. [Google Scholar]
- 3.Allendorf FW, Leary RF, Spruell P, Wenburg JK. The problems with hybrids: Setting conservation guidelines. Trends Ecol Evol. 2001;16:613–622. [Google Scholar]
- 4.Rhymer JM, Simberloff D. Extinction by hybridization and introgression. Annu Rev Ecol Syst. 1996;27:83–109. [Google Scholar]
- 5.Brusati ED, Grosholz ED. Native and introduced ecosystem engineers produce contrasting effects on estuarine infaunal communities. Biol Invasions. 2006;8:683–695. [Google Scholar]
- 6.Ryan ME, Johnson JR, Fitzpatrick BM. Invasive hybrid tiger salamander genotypes impact native amphibians. Proc Natl Acad Sci USA. 2009;106:11166–11171. doi: 10.1073/pnas.0902252106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daniels MJ, Corbett L. Redefining introgressed protected mammals: When is a wildcat a wild cat and a dingo a wild dog? Wildl Res. 2003;30:213–218. [Google Scholar]
- 8.Fitzpatrick BM, et al. Rapid fixation of non-native alleles revealed by genome-wide SNP analysis of hybrid tiger salamanders. BMC Evol Biol. 2009;9:176. doi: 10.1186/1471-2148-9-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fitzpatrick BM, Shaffer HB. Introduction history and habitat variation explain the landscape genetics of hybrid tiger salamanders. Ecol Appl. 2007;17:598–608. doi: 10.1890/06-0369. [DOI] [PubMed] [Google Scholar]
- 10.Allendorf FW, Luikart G. Conservation and the genetics of populations. Malden, MA: Blackwell; 2007. [Google Scholar]
- 11.Allendorf FW, et al. Cutthroat trout hybridization and the the U. S. Endangered Species Act: One species, two policies. Conserv Biol. 2005;19:1326–1328. [Google Scholar]
- 12.Allendorf FW, et al. Intercrosses and the U.S. Endangered Species Act: Should hybridized populations be included as Westslope cutthroat trout? Conserv Biol. 2004;18:1203–1213. [Google Scholar]
- 13.Campton DE, Kaeding LR. Westslope cutthroat trout, hybridization, and the U. S. Endangered Species Act. Conserv Biol. 2005;19:1323–1325. [Google Scholar]
- 14.Arnold ML. Evolution Through Genetic Exchange. Oxford: Oxford Univ Press; 2006. [Google Scholar]
- 15.Ellstrand NC, Schierenbeck KA. Hybridization as a stimulus for the evolution of invasiveness in plants? Proc Natl Acad Sci USA. 2000;97:7043–7050. doi: 10.1073/pnas.97.13.7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chapman MA, Burke JM. Letting the gene out of the bottle: The population genetics of genetically modified crops. New Phytol. 2006;170:429–443. doi: 10.1111/j.1469-8137.2006.01710.x. [DOI] [PubMed] [Google Scholar]
- 17.Hails RS, Morley K. Genes invading new populations: A risk assessment perspective. Trends Ecol Evol. 2005;20:245–252. doi: 10.1016/j.tree.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 18.Fisher RA. The wave of advance of advantageous genes. Ann Eugen. 1937;7:355–369. [Google Scholar]
- 19.Pialek J, Barton NH. The spread of an advantageous allele across a barrier: The effects of random drift and selection against heterozygotes. Genetics. 1997;145:493–504. doi: 10.1093/genetics/145.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Slatkin M. In: Population genetics and ecology. Karlin S, Nevo E, editors. New York: Academic; 1976. pp. 767–780. [Google Scholar]
- 21.Morjan CL, Rieseberg LH. How species evolve collectively: Implications of gene flow and selection for the spread of advantageous alleles. Mol Ecol. 2004;13:1341–1356. doi: 10.1111/j.1365-294X.2004.02164.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hall RJ, Ayres DR. What can mathematical modelling tell us about hybrid invasions? Biol Invasions. 2009;11:1217–1224. [Google Scholar]
- 23.Currat M, Ruedi M, Petit RJ, Excoffier L. The hidden side of invasions: Massive introgression by local genes. Evolution. 2008;62:1908–1920. doi: 10.1111/j.1558-5646.2008.00413.x. [DOI] [PubMed] [Google Scholar]
- 24.Zayed A, Whitfield CW. A genome-wide signature of positive selection in ancient and recent invasive expansions of the honey bee Apis mellifera . Proc Natl Acad Sci USA. 2008;105:3421–3426. doi: 10.1073/pnas.0800107105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shaffer HB. Evolution in a paedomorphic lineage. I. An electrophoretic analysis of the Mexican ambystomatid salamanders. Evolution. 1984;38:1194–1206. doi: 10.1111/j.1558-5646.1984.tb05643.x. [DOI] [PubMed] [Google Scholar]
- 26.Shaffer HB, McKnight ML. The polytypic species revisited: Genetic differentiation and molecular phylogenetics of the tiger salamander Ambystoma tigrinum (Amphibia: Caudata) complex. Evolution. 1996;50:417–433. doi: 10.1111/j.1558-5646.1996.tb04503.x. [DOI] [PubMed] [Google Scholar]
- 27.Collins JP. Distribution, habitats, and life history variation in the tiger salamander, Ambystoma tigrinum, in east-central and southeast Arizona. Copeia. 1981;1981:666–675. [Google Scholar]
- 28.Riley SPD, Shaffer HB, Voss SR, Fitzpatrick BM. Hybridization between a rare, native tiger salamander (Ambystoma californiense) and its introduced congener. Ecol Appl. 2003;13:1263–1275. [Google Scholar]
- 29.Petranka JW. Salamanders of the United States and Canada. Washington, DC: Smithson Inst; 1998. [Google Scholar]
- 30.Fitzpatrick BM, Shaffer HB. Environment-dependent admixture dynamics in a tiger salamander hybrid zone. Evolution. 2004;58:1282–1293. doi: 10.1111/j.0014-3820.2004.tb01707.x. [DOI] [PubMed] [Google Scholar]
- 31.Fitzpatrick BM, Shaffer HB. Hybrid vigor between native and introduced salamanders raises new challenges for conservation. Proc Natl Acad Sci USA. 2007;104:15793–15798. doi: 10.1073/pnas.0704791104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith JJ, Kump DK, Walker JA, Parichy DM, Voss SR. A comprehensive expressed sequence tag linkage map for tiger salamander and Mexican axolotl: Enabling gene mapping and comparative genomics in Ambystoma. Genetics. 2005;171:1161–1171. doi: 10.1534/genetics.105.046433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gay L, Crochet P-A, Bell DA, Lenormand T. Phenotypic traits in hybrid zones: A window on tension zone models. Evolution. 2008;62:2789–2806. doi: 10.1111/j.1558-5646.2008.00491.x. [DOI] [PubMed] [Google Scholar]
- 34.Long JC. The genetic structure of admixed populations. Genetics. 1991;127:417–428. doi: 10.1093/genetics/127.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kimura M, Weiss GH. The stepping stone model of population structure and the decrease of genetic correlation with distance. Genetics. 1964;49:561–576. doi: 10.1093/genetics/49.4.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Griebeler EM, Müller JC, Seitz A. Spatial genetic patterns generated by two admixing genetic lineages: A simulation study. Conserv Genet. 2006;7:753–766. [Google Scholar]
- 37.Ayres DR, Smith DL, Zaremba K, Klohr S, Strong DR. Spread of exotic cordgrasses and hybrids (Spartina sp.) in the tidal marshes of San Francisco Bay, California, USA. Biol Invasions. 2004;6:221–231. [Google Scholar]
- 38.Rosenfield JA, Nolasco S, Lindauer S, Sandoval C, Kodric-Brown A. The role of hybrid vigor in the replacement of Pecos Pupfish by its hybrids with Sheepshead Minnow. Conserv Biol. 2004;18:1589–1598. [Google Scholar]
- 39.Perry WL, Feder JL, Dwyer G, Lodge DM. Hybrid zone dynamics and species replacement between Orconectes crayfishes in a northern Wisconsin lake. Evolution. 2001;55:1153–1166. doi: 10.1111/j.0014-3820.2001.tb00635.x. [DOI] [PubMed] [Google Scholar]
- 40.Oliveira R, Godinho R, Randi E, Alves PC. Hybridization versus conservation: Are domestic cats threatening the genetic integrity of wildcats (Felis silvestris silvestris) in Iberian Peninsula? Philos Trans R Soc Lond B Biol Sci. 2008;363:2953–2961. doi: 10.1098/rstb.2008.0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haygood R, Ives AR, Andow DA. Consequences of recurrent gene flow from crops to wild relatives. Proc R Soc Lond B Biol Sci. 2003;270:1879–1886. doi: 10.1098/rspb.2003.2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boyer MC, Muhlfeld CC, Allendorf FW. Rainbow trout (Onchorhynchus mykiss) invasion and the spread of hybridization with native westslope cutthroat trout (Oncorhynchus clarkii lewisi) Can J Fish Aquat Sci. 2008;65:658–669. [Google Scholar]
- 43.Gregory RT, et al. Eukaryotic genome size databases. Nucleic Acids Res. 2007;35:D332–D338. doi: 10.1093/nar/gkl828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fish and Wildlife Service (2004) Endangered and Threatened Wildlife and Plants; Determination of Threatened Status for the California Tiger Salamander; and Special Rule Exemption for Existing Routine Ranching Activities; Final Rule. 69. Federal Register. 2004;149:47211–47248. [Google Scholar]
- 45.Trenham PC, Koenig WD, Shaffer HB. Spatially autocorrelated demography and interpond dispersal in the salamander Ambystoma californiense . Ecology. 2001;82:3519–3530. [Google Scholar]
- 46.Wang IJ, Savage WK, Shaffer HB. Landscape genetics and least-cost path analysis reveal unexpected dispersal routes in the California tiger salamander (Ambystoma californiense) Mol Ecol. 2009;18:1365–1374. doi: 10.1111/j.1365-294X.2009.04122.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.