Abstract
The butterfly Heliconius erato can see from the UV to the red part of the light spectrum with color vision proven from 440 to 640 nm. Its eye is known to contain three visual pigments, rhodopsins, produced by an 11-cis-3-hydroxyretinal chromophore together with long wavelength (LWRh), blue (BRh) and UV (UVRh1) opsins. We now find that H. erato has a second UV opsin mRNA (UVRh2)—a previously undescribed duplication of this gene among Lepidoptera. To investigate its evolutionary origin, we screened eye cDNAs from 14 butterfly species in the subfamily Heliconiinae and found both copies only among Heliconius. Phylogeny-based tests of selection indicate positive selection of UVRh2 following duplication, and some of the positively selected sites correspond to vertebrate visual pigment spectral tuning residues. Epi-microspectrophotometry reveals two UV-absorbing rhodopsins in the H. erato eye with λmax = 355 nm and 398 nm. Along with the additional UV opsin, Heliconius have also evolved 3-hydroxy-DL-kynurenine (3-OHK)-based yellow wing pigments not found in close relatives. Visual models of how butterflies perceive wing color variation indicate this has resulted in an expansion of the number of distinguishable yellow colors on Heliconius wings. Functional diversification of the UV-sensitive visual pigments may help explain why the yellow wing pigments of Heliconius are so colorful in the UV range compared to the yellow pigments of close relatives lacking the UV opsin duplicate.
Keywords: adaptive evolution, color vision, opsin, rhodopsin
Evolutionary biologists have long framed our understanding of morphological variation between closely related species like the wing color patterns of butterflies or bright plumage of birds as products of both natural and sexual selection (1). Yet from the beginning of the field 150 years ago, Darwin and others wondered about the contribution of the sensory mechanisms of animals to the process of morphological differentiation between species. Whereas there is much evidence to suggest that the visual systems and signals of aquatic animals (especially fish) evolve in tandem, the extent to which such correlated evolution exists in terrestrial animals is largely an open question. Except for mammals (2), simply not enough has been known about either the physiological or genetic basis of vision in groups of closely related animals to probe this relationship. Correlated evolution between butterfly wing pigment color and visual pigments has never been demonstrated, so we sought to examine whether such a relationship might exist.
We focused our efforts on the visual pigments and wing pigments in Heliconius or “passion-vine” butterflies and their close relatives. Heliconius form numerous mimetic color-pattern races throughout Mexico and Central and South America and are an example of an adaptive radiation (3). Like other terrestrial animals, passion-vine butterflies rely on visual cues when searching for food (4) and potential mates (4 –8). Unlike most other butterflies, there is the added selection pressure that they must also recognize conspecific from heterospecific comimics to successfully reproduce (9).
Early work on Heliconius erato identified three major photoreceptor types in the compound eye with spectral sensitivity peaks at 370 nm (but see below), 470 nm, and 560–570 nm (10, 11). Electroretinograms (12, 13) and electrophysiological recordings in the brain (5) further indicated a second long wavelength receptor with peak sensitivity at 610–640 nm.
Opsins together with an 11-cis-3-hydroxyretinal chromophore comprise the visual pigments found in the photoreceptor cells of the adult compound eye of butterflies (14). Through cDNA screening we previously characterized three opsin-encoding mRNAs from H. erato that cluster with other known insect long wavelength- (LWRh), blue- (BRh), and UV-sensitive (UVRh) opsins (15). As in other butterflies, the ommatidial units of the H. erato eye contain nine photoreceptor cells, R1–R9. We found the R1 and R2 photoreceptor cells express either UVRh or BRh mRNAs and the R3–R8 photoreceptor cells express LWRh. In total, we found three subtypes of ommatidia in the main retina with respect to opsin mRNA expression: UVRh-BRh-LWRh, UVRh-UVRh-LWRh, and BRh-BRh-LWRh (15).
In addition to these three rhodopsins, the H. erato eye contains non-opsin red filter pigments in the photoreceptor cells of some ommatidia but not others (15). These pigments selectively absorb short wavelength light and produce the red-colored ommatidia observed in the butterflies’ eye glow (15, 16). Yellow facets correspond to ommatidia that lack the red filter pigment. Because all R3–R8 cells in the main retina contain one LWRh rhodopsin with a wavelength of peak absorbance λmax = 555 nm (17), two kinds of long wavelength receptor are present in the eye: the photoreceptor class expressing the LWRh opsin alone and the ∼620 nm photoreceptor class produced by colocalization of LWRh with the red filter pigment.
These studies bring the total number of known photoreceptor classes in the H. erato eye to four but there are hints in older work that there may be more. Spectral sensitivity of the compound eye of Heliconius numata measured by electroretinograms indicated three major peaks at 370 nm, 470 nm, and 570 nm (11), identical to those observed in H. erato. Intriguingly, intracellular recordings indicated an additional photoreceptor type at 390 nm (18). This suggests the presence of two UV receptors in the eye of H. numata but until now molecular evidence for two UV opsins has been lacking.
Because only one UV opsin has been reported in other butterflies (19 –21), we were surprised to find a second UV opsin transcript (UVRh2) in the course of screening H. erato eye cDNA (22). We investigate the evolutionary origins of UVRh2 by screening eye cDNAs from eight additional Heliconius species representing each of the major lineages in the genus and five basal heliconiines. We examine the selective forces shaping the evolution of this previously unreported duplicate gene and put the amino acid changes following duplication into a structural context by homology modeling. We then provide unique physiological evidence for the presence of two UV-absorbing rhodopsins in the eye of H. erato and reanalyze the older electrophysiological work in light of our findings. Lastly, we examine the hypothesis that wing pigment coloration in the UV is more likely to occur in species with two UV opsins than in species with only one.
Results and Discussion
Heliconius Have a Second UV Opsin Gene that Is Positively Selected.
The two H. erato UV opsin cDNAs, UVRh1 and UVRh2, encode proteins 378 amino acids in length with a sequence identity of 86%. Together with LWRh and BRh opsin mRNAs (see ref. 15), the H. erato eye contains four visual pigments. This finding contrasts with the visual system of all other butterflies studied (23) in which only one UV opsin mRNA is expressed in the retina.
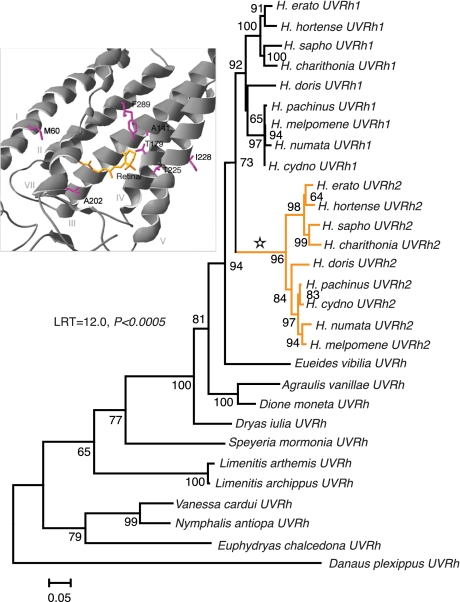
We screened cDNAs from five species belonging to different genera in the Heliconiinae and eight additional Heliconius species representing the major lineages within the genus (24) to pinpoint the origins of the novel opsin (Table S1). Only one UV opsin mRNA was detected in the eyes of the non-Heliconius butterflies, whereas copies of both UVRh1 and UVRh2 were found in all nine Heliconius species examined (Fig. 1). Phylogenetic reconstruction of the UV opsin family of nymphalid butterflies (Fig. 1) showed good (73%) bootstrap support for a single clade of Heliconius UV opsins that includes both UVRh1 and UVRh2. Support for individual UVRh1 and UVRh2 clades reached 92 and 96%, respectively. These results suggest that the evolutionary origin of the gene duplication occurred at the base of the genus Heliconius.
Fig. 1.
Gene tree of UV opsins from Heliconiinae and outgroup taxa. Based upon maximum likelihood analysis of 1,134 characters using GTR + Γ + I model with gamma-shape parameter = 1.932 and proportion of invariant sites = 0.485. All codon positions were used in the reconstruction and the reliability of the tree was tested using 500 bootstrap replicates. Bar corresponds to the number of substitutions/site. Star indicates positively selected branch leading to orange UVRh2 clade. (Inset) Homology model of Heliconius erato UVRh2 opsin. Positively selected sites in close proximity to the chromophore indicated by magenta, retinal chromophore indicated by orange. Residues 60, 141, 179, 202, 225, 228, and 289 correspond to human red cone pigment residues 59, 141, 180, 202, 227, 230, and 277.
A longer branch leads to the UVRh2 clade compared to the UVRh1 clade. To test whether this might represent adaptive change, we performed a phylogeny-based test of selection (25). We found that a branch-sites model of evolution, one that permitted along the branch leading to UVRh2 (indicated by a star in Fig. 1) a class of positively selected codon with nonsynonymous-to-synonymous substitution rate ratio (ω) > 1, was a significantly better fit to the data (likelihood score = −7375.33) than a branch-sites model (likelihood score = −7381.31) in which this class of codon was restricted to having ω = 1 (likelihood ratio test, LRT = 2∆ℓ = 12.0, df = 1, P = 0.0005). Since the LRT suggested the presence of positively selected sites, the method of Bayes empirical Bayes (26) was implemented to calculate posterior probabilities for site classes on the positively selected branch. A total of 28 codons were identified as having a ≥95% posterior probability of ω > 1 along the branch leading to the UVRh2 clade (Table S2).
Positively Selected Sites Map to the Chromophore Binding Pocket.
To examine the distribution of positively selected amino acids in relation to the chromophore, a homology model was made of both UV opsins against the bovine rhodopsin crystal structure (27). Of the 28 identified positively selected sites, 19 fall in transmembrane domains, 4 correspond to known spectral tuning sites in vertebrate visual pigments, and several contain the class of substitution that causes spectral tuning effects: amino acids 180, 227, 230, and 277 in the human red cone pigment numbering system (Fig. 1 Inset and Table S2). Most notably, the two Heliconius pigments differ in having amino acid changes A180T and F277Y, homologous to naturally occurring polymorphic sites in primate cone pigments. In site-directed mutagenesis experiments, amino acid changes A180S and F277Y increased λmax values of the human green pigment by ∼7 and 10 nm, respectively, and the effects of these amino acid substitutions on λmax were approximately additive (28, 29). The human red and green cone pigments are also polymorphic at minor tuning site 230 (29) whereas UVRh1 and UVRh2 differ by amino acid change F230I.
Positively selected amino acid 227 is in close proximity to the chromophore (27). Mutations at site 227 (equivalent to H211F and H211C in bovine rhodopsin) reduce λmax values by 3 and 5 nm, respectively (30). The UV opsins of H. melpomene, H. sapho, and H. numata differ by amino acid change A227S, which may also affect spectral tuning because it involves the substitution of a hydroxyl for a nonhydroxyl-bearing residue. Lastly, we note that the pigments differ by a S141A change, though this site was not identified as positively selected. Substitutions at this site are associated with autosomal dominant retinitis pigmentosa in humans and an L141A substitution caused a 3-nm spectral shift in in vitro studies (31).
H. erato Has Two UV-Absorbing Rhodopsins in the Eye.
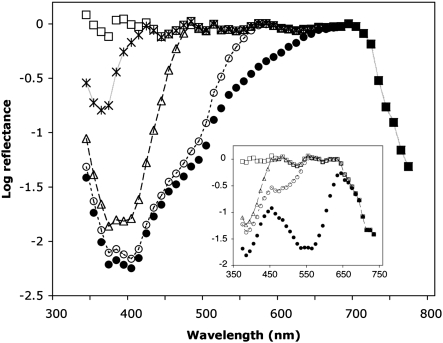
To investigate further the presence of two functionally distinct rhodopsins implied by our homology modeling data, we analyzed epi-microspectrophotometric data obtained from the eyes of H. erato compared to similar data obtained from the eyes of Dryas iulia. We used similar procedures to those used to obtain λmax for the UV rhodopsin of other butterflies (see refs. 20, 32) and found evidence for two UV-absorbing rhodopsins in H. erato, with λmax = 355 nm and 398 nm, and evidence for only one UV-absorbing rhodopsin in D. iulia, with λmax = 385 nm (Fig. 2). We also reanalyzed the older electrophysiological data for H. numata (11, 18) and found that these data are best fit using least-squares regression by a model in which four rhodopsins are present in the eye, rather than three, with λmax = 349 ± 6 nm, 399 ± 3 nm, 467 ± 2 nm, and 554 ± 3 nm, respectively (SI Text and Fig. S1).
Fig. 2.
Densitometric analysis of an epi-microspectrophotometric reflectance spectrum of Heliconius erato and Dryas iulia eyeshine. Solid black circles, H. erato experimental eyeshine data. Open circles, spectrum after having computationally stripped optical density (OD) 0.40 of rhodopsin with λmax = 555 (R555); open triangles, spectrum after then stripping OD 1.20 of R470; stars, spectrum after having also stripped OD 1.62 of R398; open squares, spectrum after having also stripped OD 0.73 of R355. (Inset) Solid black circles, D. iulia experimental eyeshine data. Open circles, spectrum after having computationally stripped OD 1.7 of R555; open triangles, spectrum after then stripping OD 0.6 of R470; open squares, spectrum after having also stripped OD 1.2 of R385. The squares represent the average reflectance spectrum of tracheolar tapeta that optically terminate each rhabdom.
Duplicate UV Opsins in the Eyes Co-Occur with UV-Yellow Pigments on the Wings.
All butterflies examined prior to this study have a single UV-absorbing visual pigment. Together with blue and green receptors, this implies that butterflies can see from 300 to 600 nm or greater. Adding one type of photoreceptor and appropriate neuronal wiring typically means increasing color discrimination. The fact that the UV opsin duplicates we describe here have been selectively maintained within Heliconius predicts that color signals for mating, foraging, and egg laying may be more variable in the spectral range of 310–390 nm, which may be better discriminated if there is more than one receptor type sampling this part of the spectrum. Following this prediction, we measured 506 wing reflectance spectra from the nine Heliconius species from which we cloned the opsins and compared them with 290 wing reflectance spectra from the five non-Heliconius species (Table S1).
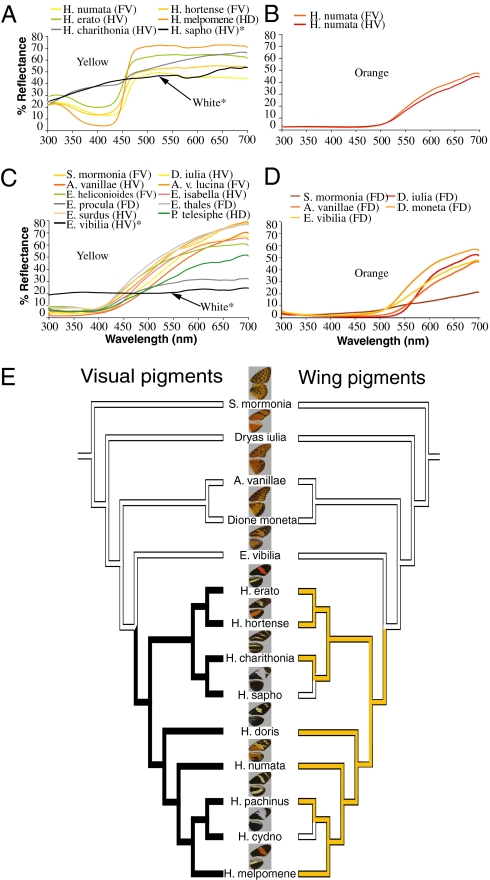
What is striking is that Heliconius do indeed have wing colors with more spectral variation in the UV compared to the other heliconiine species (Fig. 3 A–D and Fig. S2). To quantify this variation, we first averaged spectral reflectance data sampled from the same wing location from n = 3–14 individuals per species and then calculated both slope and maximum reflectance in the 310–390 nm (UV) range for each of these averaged spectra. For a reflectance spectrum to be considered variable in the UV, the maximum reflectance and slope needed to exceed a threshold, so that the strength of the UV signal is significantly high enough for visual perception (SI Text).
Fig. 3.
Reflectance spectra of wing colors in the UV and visible range and UV opsin(s) in the adult heliconiine eye. Representative white, yellow, and orange reflectance spectra of (A and B) Heliconius and (C and D) non-Heliconius wing pigment colors. (A) Heliconius yellow pigments contain a UV component, unlike the yellow pigments found in other heliconiines shown in B. (E) Species tree derived from a maximum likelihood analysis of mitochondrial (COI) and nuclear (EF-1α) genes. Although yellow wing pigments occur in both Heliconius and non-Heliconius species, yellow pigments with a UV component (yellow, Right) were found to be more likely to occur in species with two UV opsins (black, Left) in the eyes than in species with only one UV opsin (white, Left) in a test of correlated trait evolution (LRT, P = 0.01). F, forewing; H, hindwing; D, dorsal; V, ventral.
We found unexpectedly that the yellow wing pigments of Heliconius contributed the most to their spectral variability in the UV. The reflectance spectra of these yellow pigments, apparently composed primarily of 3-hydroxy-DL-kynurenine (3-OHK) (33), are fundamentally different from those of other heliconiine butterflies in the UV (analogous to the difference in human vision between purple, a mixture of red and blue, and red) (Fig. 3 A and C). To examine whether this difference in fact extends to other closely related heliconiines not sampled for opsins, we measured 222 wing reflectance spectra from five additional species of Eueides, the sister genus to Heliconius (Fig. 3C), with prominent yellow on the wings (a yellow which is purportedly not 3-OHK) (34), as well as the subspecies Agraulis vanillae lucina and Podotricha telesiphe, a close relative to D. iulia (Table S1). In all instances, except for the forewing dorsal patches of Eueides heliconioides (Fig. S2) where there is minor variation in the UV, the yellow of non-Heliconius are flat below 450 nm (Fig. 3C).
To further explore the biochemical basis of the observed differences in color we performed UV-visible spectrophotometry and mass spectrometry on the yellow pigments extracted from the wings of Heliconius erato, H. melpomene, Eueides thales, E. surdus, and E. heliconioides (SI Text) and found that whereas the yellow wing pigments of the two Heliconius do indeed contain 3-OHK, this pigment is completely absent from the wings of the three Eueides species (Figs. S3 and S4).
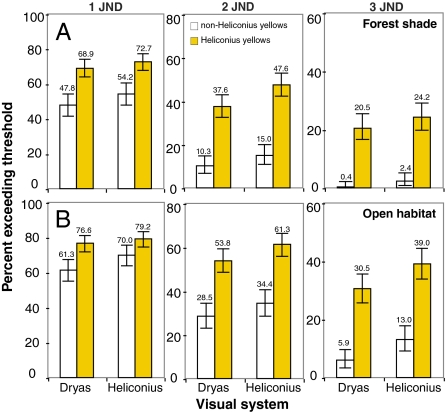
To understand the possible functional advantage of deploying 3-OHK yellow on the wings compared to the other yellow pigments, we next modeled the color space of all possible classes of trichromatic ommatidia of H. erato and D. iulia using a receptor noise model (35) that has been used to model the visual system of other butterflies (36). We divided all yellow reflectance spectra into two categories, Heliconius yellow and non-Heliconius yellow and calculated color differences (ΔS) in units of just noticeable differences (JNDs) for all pairwise comparisons within each of these categories under field measurements of the forest shade (Fig. 4A) and open habitat (Fig. 4B) illuminating conditions where we observed heliconiines to fly (Fig. S5 and SI Text). The ΔS values represent the Euclidean distance separating two colors in Heliconius or Dryas color space and numbers exceeding a threshold represent colors that are more likely to be discriminated. To our surprise, we found for both visual systems a higher percentage of pairs of Heliconius yellows differed by one, two, and three JNDs compared to non-Heliconius yellows under both dim (Fig. 4) and bright illumination (Table S3) making it very likely that more of these colors can be discriminated. This indicates by evolving a new mechanism for producing yellow, Heliconius has likely increased the number of distinct yellow colors on the wing compared to non-Heliconius species. We also found that under all conditions studied except one, Heliconius may have a slight advantage over Dryas if they compare signals from photoreceptor cells expressing UVRh1, UVRh2, and LWRh, especially when discriminating Heliconius from non-Heliconius yellows (Fig. S6 and Table S3). It is also plausible that other combinations of photoreceptors are used for species recognition, in which case the Dryas visual system may be superior (Table S3). In either case, it will be intriguing to see whether these modeling results can be validated using behavioral tests.
Fig. 4.
Percentage of pairs of Heliconius yellows (n = 351 pairs) (yellow) and non-Heliconius yellows (n = 253) (white) that differ as modeled through the eyes of H. erato and D. iulia under dim illumination. (A) Forest shade and (B) open habitat irradiance spectra measurements used in the calculations were obtained at field sites in Oaxaca, Mexico. Results for the Heliconius UVRh1, UVRh2, and LWRh receptor combination are compared to the Dryas UVRh, BRh, and LWRh receptor combination. Other possible receptor combinations are shown in Table S3. Threshold (ΔS) units of just noticeable differences (JNDs) of one, two, and three were chosen to account for the difficulty in estimating true noise values in the butterfly (44). For both visual systems, more Heliconius yellows differ by one, two, or three JNDs making it very likely that more of these colors can be discriminated. Bars indicate 95% confidence intervals.
These results strongly suggest that Heliconius have in their molecular evolutionary toolkit the ability to deploy distinct yellow colors on the wings, and assuming they are used for species recognition, novel UV opsins in their eyes that may help them distinguish these UV yellows from other yellows, conferring on them a selective advantage. As Fig. S6 indicates, this color difference may be especially significant for identifying conspecific from heterospecific co-mimics, such as Eueides isabella, which has a yellow that is completely flat in the UV range compared to its co-mimic H. numata (Fig. 3 A and C) with a yellow that contains a UV component. Even within the genus Heliconius, where both UV receptors are present, co-mimics that to our eyes look similar may in fact be distinguishable through the eyes of Heliconius.
Because two UV photoreceptors in the eye might enhance discrimination of yellow wing pigments with a UV component from yellow pigments without, we were interested in asking whether UV-yellow wing pigments are more likely to evolve in the presence of two UV opsins than in the presence of only one. Using a species phylogeny derived from independent molecular characters (24), we classified species into discrete categories according to whether they had one UV opsin or two, and according to whether they had yellow pigments with a UV component or yellow pigments without. We reconstructed ancestral states using maximum parsimony and found that the UV opsin duplication likely occurred in the ancestor to extant Heliconius (Fig. 3E, Left), coincident with the appearance of the UV-yellow wing pigments (Fig. 3 A and E, Right, and Fig. S2). A likelihood-based test of correlated trait evolution (37) indicated that the UV-yellow pigment was more likely to occur on the wings of species whose eyes contain two UV visual pigments than in species whose eyes contained only one (P = 0.01) (Fig. 3E). We found the correlation to hold as well when extended to all UV colors (P = 0.004), although we note that while intriguing, evidence for correlated trait evolution does not establish causality.
Heliconius are known to use wing color as a cue in mate recognition (4, 7, 38) and between two closely related species, recognizing the mimetic color pattern of one another is important in reproductively isolating the species in hybrid zones (7). Even between H. erato races with differing wing color patterns, butterfly paper models lacking UV reflecting components were less attractive to males, whose own wings contained UV reflecting components, than were models made with hexane-washed odorless real wings that contained UV components (9). This suggests that spectral properties of H. erato wings in the UV may play an important role in mate recognition, in the context of males choosing among different wing color races of their own species. In Heliconius pachinus and its sister species the polymorphic H. cydno, yellow males prefer to mate with yellow females, facilitated by a strong genetic association between these traits (8). In these and other Heliconius, mate preferences have been shown to co-evolve with wing color (39). Our discovery of spectrally distinct UV rhodopsins in the eyes of Heliconius suggests a possible sensory mechanism for discriminating the striking Heliconius yellows and other UV colors from similar colors lacking a UV component, further facilitating the link between cue and preference, and contributing to adaptive radiation of the genus.
Conclusion
In conclusion, we provide molecular and physiological evidence for two distinct UV-absorbing visual pigments in the adult compound eye of H. erato and evidence that one of these genes, UVRh2, evolved via positive selection following recent duplication. We also provide statistical and biochemical evidence that a molecular evolutionary innovation in the eyes of Heliconius has evolved coincidentally with wing pigment coloration in the UV. Importantly, we show using environmental light measurements and computational modeling that by deploying the novel 3-OHK-based pigment, Heliconius has significantly expanded the palette of discriminable colors used on their wings compared to close relatives. This is in contrast to other examples of adaptive evolution from visual ecology where a spectral shift in the λmax of a single visual pigment has been linked to an increase in sensitivity to a conspecific color signal, as in the case of cichlid fish (40). Our discovery of a positively selected UV opsin gene together with the expanded number of UV colors on the wing suggests a new sensory mechanism of adaptive evolution between closely related species.
Methods
PCR, Cloning, and Sequencing of Opsins.
H. erato cDNA was synthesized from total RNA extracted from a single adult head (TRIzol; Gibco-BRL) using a Marathon cDNA amplification kit (BD Biosciences, Clontech). Rapid amplification of cDNA end (5′ and 3′ RACE) products were amplified using BD Advantage Polymerase (BD Biosciences) and degenerate primers. PCR products were cloned into pGEM-T Easy vector (Promega) and sequenced with Big Dye 3.1 sequencing kit (Applied Biosystems) at the DNA core facility in the University of California, Irvine.
One-step RT-PCR was performed to amplify UVRh1 and UVRh2 from total RNAs from one to two specimens each of the other Heliconius species using AffinityScript multiple temperature cDNA synthesis kit (Stratagene) and gene-specific primers. Where direct sequencing of RT-PCR products was not possible, the PCR products were cloned into pGEM and individual clones were sequenced. Individual cDNA pools were synthesized from one specimen each for the non-Heliconius species and screened using 5′ and 3′ RACE as described above.
Phylogenetic Analysis.
The UV opsin sequences were aligned along with other lepidopteran UV opsins downloaded from GenBank (SI Text). A total of 1,134 nucleotide characters were used in the phylogenetic reconstruction. For maximum likelihood analysis, the best-fit DNA substitution model (GTR + Γ + I) was identified using the Akaike information criterion. Estimates of the gamma-shape parameter and proportion of invariant sites were then used in the program PHYML (41) to obtain the 500 bootstrap replicates.
Branch-Sites Test of Selection.
Branch-sites tests of selection are phylogeny-based tests that require the a priori identification of a foreground branch that is hypothesized to be evolving at a different rate than the background branches in the rest of the tree. We specified the branch leading to the Heliconius UVRh2 opsin gene as the foreground branch and then proceeded to test the hypothesis, using a likelihood ratio test that a model which included one class of sites with ω > 1, indicating positive selection, was superior to a model where ω = 1. We estimated codon frequencies on the basis of the actual nucleotide frequencies at each of the three codon positions, which has been suggested as a procedure that will reduce the level of false positives due to potential nucleotide biases. We also used the Bayes empirical Bayes method (26) as implemented in PAML 4.1 to identify the positively selected sites along this branch.
Homology Modeling.
Homology models of H. erato UVRh1 and UVRh2 were generated by uploading amino acid sequences to the protein homology/analogy recognition engine (PHYRE) website (42). Alignments generated by the server were visually inspected and then models for each protein that used the bovine rhodopsin crystal structure (PDB no. 1U19) as a template were downloaded. The modeled UVRh1 and UVRh2 structures were then aligned in Swiss-Pdb Viewer DeepView v. 4.0 (43) and sites that differed between the two proteins were mapped and compared with homologous sites in bovine rhodopsin and human red cone pigment (Fig. 1 Inset and Table S2).
Epi-Microspectrophotometric Densitometry.
Our experiment with H. erato began by mounting a butterfly in a slotted plastic tube in a goniometric stage, orienting the eye so a patch of ommatidia looked into the microscope objective. The focus was adjusted for optimal collection of eyeshine and discrimination against stray light. An 80% bleach of the LWRh rhodopsin R555 (17) was achieved with 11 hours of periodic flashes from a 45-watt halogen lamp covered by a Hoya R62 orange filter. After waiting for metarhodopsin photoproducts to decay from the rhabdoms the reflectance spectrum shown in Fig. 2 was measured with dim monochromatic flashes.
Densitometric analysis following a procedure described for other butterflies (20, 32) started by stripping from Fig. 2 a density 0.40 of R555 so that the residual spectrum (open circles) was flat for 570 nm and greater—wavelengths for which the other visual pigments have no significant absorbance. Stripping density 1.20 of the BRh rhodopsin R470 produced a residual spectrum (triangles) that was flat for 480 nm and greater and has a large dip in the UV that is poorly fit by a single UV visual pigment. Stripping density 1.62 of UVRh rhodopsin R398 leaves a residual (stars) that is well fit by density 0.73 of UVRh rhodopsin R355. The final residual (squares) is the average tapetal reflectance spectrum. A similar approach was used for determining the λmax values of the D. iulia rhodopsins except the experimental eyeshine data of D. iulia was best fit by a model in which only one UV opsin was present rather than two (SI Text).
Wing Pigment Extraction and Spectrophotometry.
Whole, intact yellow color pattern fragments were cut from dried adult wings. Each wing fragment was gently rocked in 800 μL of acidified methanol (0.5% HCl) until visual inspection revealed most or all of the yellow pigment had been extracted (∼1–5 minutes per wing fragment). The acid methanol extractions were dried down in a vacuum centrifuge, resuspended in 100 μL of methanol, dried down again, and then resuspended in 100 μL of methanol. These resuspended samples were used for UV-VIS spectrophotometer analysis (Hitachi U-3310) and further diluted 1:20 in methanol for analysis on a Micromass LCT mass spectrometer using positive ion electrospray ionization.
Measuring Irradiance Spectra.
We visited two sites in the Mixe region of Oaxaca, Mexico (Choapam, 760 m and Amaltepec, 1,600 m) in October 2009 where we first caught a number of heliconiines with nets and identified them to the species level. We then took irradiance measurements at these sites using an Ocean Optics USB2000 spectrometer and either a CC-3 cosine-corrected 100 μm or 400 μm fiber. Data were smoothed using a Gaussian function (3 nm bandwidth), transformed using LS-1-CAL calibrated lamp data, and then normalized for color space modeling (see below).
Color Space Modeling.
The color space models of Vorobyev and Osorio (1998) (35) as implemented in the program SPEC written by Jarrod Hadfield were used to estimate the discriminability 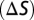 of pairs of wing reflectance spectra. Details of the equations used to derive
of pairs of wing reflectance spectra. Details of the equations used to derive 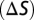 are given in the SI Text, but begin by first calculating von Kries’ transformed quantum catches,
are given in the SI Text, but begin by first calculating von Kries’ transformed quantum catches, 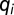 , for each photoreceptor type to account for color constancy, as follows:
, for each photoreceptor type to account for color constancy, as follows:
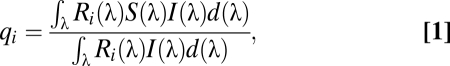 |
where 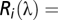 sensitivity of the receptor type
sensitivity of the receptor type 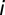 ,
, 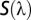 = the reflectance spectrum of the wing color,
= the reflectance spectrum of the wing color, 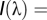 the irradiance spectrum, and
the irradiance spectrum, and 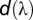 = 5 nm from the interval of 310–695 nm. Spectral sensitivity curves,
= 5 nm from the interval of 310–695 nm. Spectral sensitivity curves, 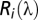 , for H. erato and D. iulia were generated using rhodopsin templates based on our experimentally determined λmax values shown in Fig. 2. Reflectance data used for color comparisons are shown in Fig. 3 and Fig. S2. Both open habitat and forest shade irradiance spectra shown in Fig. S5 were used in the calculations along with the parameters specified in the SI Text. Threshold values of
, for H. erato and D. iulia were generated using rhodopsin templates based on our experimentally determined λmax values shown in Fig. 2. Reflectance data used for color comparisons are shown in Fig. 3 and Fig. S2. Both open habitat and forest shade irradiance spectra shown in Fig. S5 were used in the calculations along with the parameters specified in the SI Text. Threshold values of 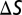 ≥ 1, 2, and 3 were chosen to account for the difficulty in estimating true photoreceptor noise values in the butterfly and their potential impact on modeling results (44).
≥ 1, 2, and 3 were chosen to account for the difficulty in estimating true photoreceptor noise values in the butterfly and their potential impact on modeling results (44).
Ancestral State Reconstruction and Tests of Correlated Evolution.
The question of whether UV wing color was more likely to appear in species with the duplicated UV opsins was tested using the correlated evolution test based on maximum likelihood and a species phylogeny (37) as implemented in Mesquite. Since the most recent molecular phylogeny of Heliconiinae (24) lacks our outgroup taxon, Speyeria mormonia, we downloaded COI and EF-1α data used in that paper and combined it with other sequences found in GenBank. We used the resulting maximum likelihood tree with branch lengths and compared the likelihoods when the traits were assumed to evolve independently (four parameter model) to the likelihoods when they were assumed to evolve dependently (eight parameter model). P-values were calculated by running Monte Carlo tests using 1,000 simulated data sets.
Supplementary Material
Acknowledgments
We dedicate this paper to the memory of Guillermo Zaccardi, whose anatomical work on the H. erato eye was essential to the discovery of the second UV opsin gene. We thank Almut Kelber and Misha Vorobyev for discussions; Larry Gilbert for providing butterflies; Monica Ramstetter, Fabio Macciardi, Matt McHenry, John Greaves, Robert Montgomerie, Emily Daniels, Omar Ávalos, Claudia Hernández, and Maita Kuvhenguhwa for research assistance; Brian Brown for access to Los Angeles Natural History Museum collections; and three anonymous reviewers for their comments. This work was supported by National Science Foundation IOS-0819936 (to A.D.B.), Programa de Apoyo a Proyectos de Investigación e Inovación Tecnológica IN-203509 (to J.L.B.), and a University of California Institute for Mexico and the United States/Consejo Nacional de Ciencia y Tecnología grant (to J.L.B. and A.D.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY918905 and GQ451890–GQ451908).
This article contains supporting information online at www.pnas.org/cgi/content/full/0910085107/DCSupplemental.
References
- 1.Ellers J, Boggs CL. The evolution of wing color: Male mate choice opposes adaptive wing color divergence in Colias butterflies. Evolution. 2003;57:1100–1106. doi: 10.1111/j.0014-3820.2003.tb00319.x. [DOI] [PubMed] [Google Scholar]
- 2.Jacobs GH. The distribution and nature of colour vision among the mammals. Biol Rev Camb Philos Soc. 1993;68:413–471. doi: 10.1111/j.1469-185x.1993.tb00738.x. [DOI] [PubMed] [Google Scholar]
- 3.Jiggins CD, McMillan WO. The genetic basis of an adaptive radiation: Warning colour in two Heliconius species. Proc R Soc Lond B Biol Sci. 1997;264:1167–1175. [Google Scholar]
- 4.Crane J. Imaginal behavior of a Trinidad butterfly, Heliconius erato hydara Hewitson, with special reference to the social use of color. Zoologica NY. 1955;40:167–196. [Google Scholar]
- 5.Swihart SL. The neural basis of color-vision in the butterfly, Heliconius erato . J Insect Physiol. 1972;18:1015–1025. [Google Scholar]
- 6.Sweeney A, Jiggins C, Johnsen S. Insect communication: Polarized light as a butterfly mating signal. Nature. 2003;423:31–32. doi: 10.1038/423031a. [DOI] [PubMed] [Google Scholar]
- 7.Jiggins CD, Naisbit RE, Coe RL, Mallet J. Reproductive isolation caused by colour pattern mimicry. Nature. 2001;411:302–305. doi: 10.1038/35077075. [DOI] [PubMed] [Google Scholar]
- 8.Kronforst MR, et al. Linkage of butterfly mate preference and wing color preference cue at the genomic location of wingless . Proc Natl Acad Sci USA. 2006;103:6575–6580. doi: 10.1073/pnas.0509685103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Estrada C, Jiggins CD. Interspecific sexual attraction because of convergence in warning colouration: Is there a conflict between natural and sexual selection in mimetic species? J Evol Biol. 2008;21:749–760. doi: 10.1111/j.1420-9101.2008.01517.x. [DOI] [PubMed] [Google Scholar]
- 10.Langer H, Struwe G. Spectral absorption by screening pigment granules in the compound eye of butterflies (Heliconius) J Comp Physiol. 1972;79:203–212. [Google Scholar]
- 11.Struwe G. Spectral sensitivity of the compound eye in butterflies (Heliconius) J Comp Physiol. 1972;79:191–196. [Google Scholar]
- 12.Bernhard CG, Boëthius J, Gemne G, Struwe G. Eye ultrastructure, colour reception and behavior. Nature. 1970;226:865–866. doi: 10.1038/226865a0. [DOI] [PubMed] [Google Scholar]
- 13.Swihart SL, Gordon WC. Red photoreceptor in butterflies. Nature. 1971;231:126–127. doi: 10.1038/231126a0. [DOI] [PubMed] [Google Scholar]
- 14.Smith CW, Goldsmith TH. Phyletic aspects of the distribution of 3-hydroxyretinal in the Class Insecta. J Mol Evol. 1990;30:72–84. doi: 10.1007/BF02102454. [DOI] [PubMed] [Google Scholar]
- 15.Zaccardi G, Kelber A, Sison-Mangus MP, Briscoe AD. Color discrimination in the red range with only one long-wavelength sensitive opsin. J Exp Biol. 2006;209:1944–1955. doi: 10.1242/jeb.02207. [DOI] [PubMed] [Google Scholar]
- 16.Stavenga DG. Reflections on colourful ommatidia of butterfly eyes. J Exp Biol. 2002;205:1077–1085. doi: 10.1242/jeb.205.8.1077. [DOI] [PubMed] [Google Scholar]
- 17.Frentiu FD, Bernard GD, Sison-Mangus MP, Brower AVZ, Briscoe AD. Gene duplication is an evolutionary mechanism for expanding spectral diversity in the long-wavelength photopigments of butterflies. Mol Biol Evol. 2007;24:2016–2028. doi: 10.1093/molbev/msm132. [DOI] [PubMed] [Google Scholar]
- 18.Struwe G. Spectral sensitivity of single photoreceptors in the compound eye of a tropical butterfly (Heliconius numata) J Comp Physiol. 1972;79:197–201. [Google Scholar]
- 19.Sauman I, et al. Connecting the navigational clock to sun compass input in monarch butterfly brain. Neuron. 2005;46:457–467. doi: 10.1016/j.neuron.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 20.Briscoe AD, Bernard GD, Szeto AS, Nagy LM, White RH. Not all butterfly eyes are created equal: Rhodopsin absorption spectra, molecular identification and localization of ultraviolet-, blue-, and green-sensitive rhodopsin-encoding mRNAs in the retina of Vanessa cardui . J Comp Neurol. 2003;458:334–349. doi: 10.1002/cne.10582. [DOI] [PubMed] [Google Scholar]
- 21.Vanhoutte KJA, Eggen BJL, Janssen JJM, Stavenga DG. Opsin cDNA sequences of a UV and green rhodopsin of the satyrine butterfly Bicyclus anynana . Insect Biochem Mol Biol. 2002;32:1383–1390. doi: 10.1016/s0965-1748(02)00058-9. [DOI] [PubMed] [Google Scholar]
- 22.Zaccardi G, Kelber A, Sison-Mangus MP, Briscoe AD. Opsin expression in the eyes of Heliconius erato . Perception. 2006;35(Suppl. S):142–143. [Google Scholar]
- 23.Briscoe AD. Reconstructing the ancestral butterfly eye: Focus on the opsins. J Exp Biol. 2008;211:1805–1813. doi: 10.1242/jeb.013045. [DOI] [PubMed] [Google Scholar]
- 24.Beltran M, Jiggins CD, Brower AVZ, Bermingham E, Mallet J. Do pollen feeding, pupal-mating and larval gregariousness have a single origin in Heliconius butterflies? Inferences from multilocus DNA sequence data. Biol J Linn Soc. 2007;92:221–239. [Google Scholar]
- 25.Zhang JZ, Nielsen R, Yang ZH. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol. 2005;22:2472–2479. doi: 10.1093/molbev/msi237. [DOI] [PubMed] [Google Scholar]
- 26.Yang ZH, Wong WSW, Nielsen R. Bayes empirical Bayes inference of amino acid sites under positive selection. Mol Biol Evol. 2005;22:1107–1118. doi: 10.1093/molbev/msi097. [DOI] [PubMed] [Google Scholar]
- 27.Palczewski K, et al. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 28.Neitz M, Neitz J, Jacobs GH. Spectral tuning of pigments underlying red-green color vision. Science. 1991;252:971–974. doi: 10.1126/science.1903559. [DOI] [PubMed] [Google Scholar]
- 29.Asenjo AB, Rim J, Oprian DD. Molecular determinants of human red/green color discrimination. Neuron. 1994;12:1131–1138. doi: 10.1016/0896-6273(94)90320-4. [DOI] [PubMed] [Google Scholar]
- 30.Nathans J. Determinants of visual pigment absorbance: Role of charged amino acids in the putative transmembrane segments. Biochemistry. 1990;29:937–942. doi: 10.1021/bi00456a013. [DOI] [PubMed] [Google Scholar]
- 31.Garriga P, Liu X, Khorana HG. Structure and function in rhodopsin: Correct folding and misfolding in point mutants at and in proximity to the site of the retinitis pigmentosa mutation Leu-125→Arg in the transmembrane helix C. Proc Natl Acad Sci USA. 1996;93:4560–4564. doi: 10.1073/pnas.93.10.4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bernard GD, Remington CL. Color vision in Lycaena butterflies: Spectral tuning of receptor arrays in relation to behavioral ecology. Proc Natl Acad Sci USA. 1991;88:2783–2787. doi: 10.1073/pnas.88.7.2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reed RD, McMillan WO, Nagy LM. Gene expression underlying adaptive variation in Heliconius wing patterns: Non-modular regulation of overlapping cinnabar and vermillion prepatterns. Proc R Soc Lond B Biol Sci. 2008;275:37–46. doi: 10.1098/rspb.2007.1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brown KS. Chemotaxonomy and chemomimicry: The case of 3-hydroxykynurenine. Syst Zool. 1967;16:213–216. [Google Scholar]
- 35.Vorobyev M, Osorio D. Receptor noise as a determinant of colour thresholds. Proc R Soc Lond B Biol Sci. 1998;265:351–358. doi: 10.1098/rspb.1998.0302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koshitaka H, Kinoshita M, Vorobyev M, Arikawa K. Tetrachromacy in a butterfly that has eight varieties of spectral receptors. Proc R Soc Lond B Biol Sci. 2008;275:947–954. doi: 10.1098/rspb.2007.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pagel M. Detecting correlated evolution on phylogenies: A general method for the comparative analysis of discrete characters. Proc R Soc Lond B Biol Sci. 1994;255:37–45. [Google Scholar]
- 38.Mavarez J, et al. Speciation by hybridization in Heliconius butterflies. Nature. 2006;441:868–871. doi: 10.1038/nature04738. [DOI] [PubMed] [Google Scholar]
- 39.Jiggins CD, Estrada C, Rodrigues A. Mimicry and the evolution of premating isolation in Heliconius melpomene . J Evol Biol. 2004;17:680–691. doi: 10.1111/j.1420-9101.2004.00675.x. [DOI] [PubMed] [Google Scholar]
- 40.Seehausen O, et al. Speciation through sensory drive in cichlid fish. Nature. 2008;455:620–626. doi: 10.1038/nature07285. [DOI] [PubMed] [Google Scholar]
- 41.Guindon S, Lethiec F, Duroux P, Gascuel O. PHYML Online–a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res. 2005;33:W557–W559. doi: 10.1093/nar/gki352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bennett-Lovsey RM, Herbert AD, Sternberg MJ, Kelley LA. Exploring the extremes of sequence/structure space with ensemble fold recognition in the program Phyre. Proteins. 2008;70:611–625. doi: 10.1002/prot.21688. [DOI] [PubMed] [Google Scholar]
- 43.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 44.Lind O, Kelber A. Avian colour vision: Effects of variation in receptor sensitivity and noise data on model predictions as compared to behavioural results. Vision Res. 2009;49:1939–1947. doi: 10.1016/j.visres.2009.05.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.