Abstract
Objective:
It has been reported that single-nucleotide polymorphism (SNP) in the proximal promoter region of apolipoprotein M (apoM) gene may confer the risk in the development of type 2 diabetes (T2D) and coronary artery disease (CAD) in the Han Chinese. However, in a recent study demonstrated that plasma apoM level did not correlated to the coronary heart disease. In the present studies, we investigated the SNP T-778C of apoM gene in CAD patients and controls in the Han Chinese population. Moreover we examined whether serum apoM levels could be influenced by this promoter mutation.
Material and methods:
One hundred twenty-six CAD patients and 118 non-CAD patients were subjected in the present study. All patients were confirmed by the angiography. The genotyping of polymorphisms T-778C in apoM promoter was determined by real-time polymerase chain reaction. Serum apoM levels were semi-quantitatively determined by the dot-blotting analysis.
Results:
Distribution of apoM T-778C genotype in non-CAD patients was as following: 84.7% were T/T, 15.3% were T/C and 0.0% was C/C. T allele frequencies were 92.4% and C allele, 7.6%. In the CAD patients, 99 patients (78.6%) had the T/T genotype, 25 patients (19.8%) with T/C genotype and 2 patients (1.6%) with C/C genotype. The allele frequency was 88.5% for the T allele and 11.5% for the C allele. There was no statistical significant difference of serum apoM levels found in these three genotypes.
Conclusions:
There was no significant difference in allele or genotype frequencies between CAD patients and non-CAD patients. Binary logistic regression analysis with adjustments for age, gender, triglycerides, total cholesterol, low-density lipoprotein, high-density lipoprotein, apoAI, apoB, and LP(a) indicated that the TC and CC genotypes in SNP T-778C were not significantly associated with the development of CAD (odds ratio = 1.510, 95% confidence interval: 0.756–3.017; p = 0.243).
Keywords: apolipoprotein M, single-nucleotide polymorphism, coronary artery disease, real-time PCR
Introduction
Association of difference genes in relation to the predisposition of cardiovascular diseases has been widely examined. Traditional candidate genes of cardiovascular disease, such as angiotensin-converting enzyme (ACE) and angiotensinogen (AGT) are well known to be correlated to coronary heart disease (CAD), and certain new loci that have been recently added to the growing list of the candidate genes, ie, myocyte enhancer factor 2 (MEF2A), apolipoprotein M (apoM), phosphodiesterase 4 (PDE4D), etc.1 It may conclude that CAD is both multifactorial and polygenic in nature. Atheroma formation, the pathological hallmark of CAD, is also an inflammatory process. The disorder of lipid profile, especially lower high-density lipoprotein (HDL) cholesterol and higher low-density lipoprotein (LDL) are important risk factors involved in this process.2
ApoM is one of the latest additions to the apolipoprotein family, first identified and characterized in 1999.3 The apoM gene is located in a highly conserved segment in the major histocompatibility complex (MHC) class III locus on chromosome 6, and codes for a 22-kDa protein that structurally belongs to the lipocalin superfamily. Human apoM cDNA (734 base pairs) contains six exons and encodes 188-amino acid residue-long protein.4 The 5′-untranslated region was 33 nucleotides and the 3′-untranslated region 120 nucleotides, not including the poly (A) tail. In plasma, apoM is mainly confined to HDL, with a small proportion present in triglyceride-rich lipoproteins (TGRLP) and LDL. In vivo, apoM is selectively expressed in hepatocytes and in the tubular epithelium of kidney.5 The anti-atherogenic function of HDL is well established. The ability of HDL to promote cholesterol efflux from foam cells in atherosclerotic lesions is generally regarded as one of the key mechanisms behind the anti-atherosclerotic function. Data from apoM-silencing RNA showed that apoM is critical for the formation of HDL, notably pre-β-HDL1. Overexpression of apoM had a protective effects against atherosclerosis.6
A variety of candidate genes have been investigated as predisposing factors of CAD, including those involved in lipid metabolisms.7 It has been reported that single-nucleotide polymorphism (SNP T-778C) in the proximal promoter region of apoM gene might be associated with the levels of plasma cholesterol and fasting plasma glucose, which may confer the risk in the development of T2D in Han Chinese.8 Jiao and collaeagues9 reported that apoM T-778C SNP could associate to the CAD and might be a risk factor of CAD, whereas Ahnström and colleagues10 recently demonstrated contradictorily that plasma apoM was not a predictor of CAD events. In order to elucidate whether apoM is a candidate gene involving in the pathogenesis of CAD and whether the promoter mutation could influence the expression of apoM, in the present study, we investigated the genetic polymorphisms of T-778C of apoM gene in the CAD patients and control subjects in Han Chinese. Moreover we examined the serum levels of apoM in the different genotypes.
Materials and methods
Subjects
Two hundred forty-four patients who had obtained coronary angiography between Jan 2006 and March 2007 were subjects of the present study. One hundred twenty-six patients were diagnosed as CAD according to the results of angiography (a lesion was classed as being significant when stenosis was >50%). And 118 patients with normal coronary arteries were excluded CAD. All patients were from the third affiliated hospital of Suzhou University and were informed by written consent.
Genotyping of apoM T-778C polymorphisms by using real-time PCR
A 250 μl-sample of peripheral blood was obtained from each subject, and DNA was extracted by DNA extract Kit (Shenerg Biocolor, Shanghai, China). The SNP ID number and detail sequence information of apoM T-778C is publicly available (http://www.ncbi.nlm.nih.gov/SNP/rs805296). According to the sequence we designed the primers and probes as: forward primer: ACTGACACATTCACTCAACATTTATTACTA; reverse primer: AGGGGTTGGTGGTGTTTTGTT; sensor probe: CTACAAAAGTACAAAAATT-FAM; and anchor probe: GTGGTGGCACACGC-TAMRA. All specific primers and probes were synthesized and fluorescence modified (Invitrogen, Shanghai, China). Taq DNA polymerase, 4 × dNTPs, 10 × polymerase chain reaction (PCR) buffer, and MgCl2 were purchased from Shenerg Biocolor (Shanghai, China). In brief, PCR was performed as below: 40 to 80 ng of genomic DNA template, 2.5 μl of 10 × PCR buffer, 1.5 mM MgCl2, 0.5 μ l of 4 × dNTPs, 1.25U Taq DNA polymerase, 10 pmol of each primer, and 2 pmol of each probe in a final reaction volume of 25 μl. Thermal cycling was performed in a LightCycler® (Roche, Swaziland). The cycling program consisted of 1 min of initial denaturation at 95 °C, followed by 40 cycles at 95 °C for 0 s (temperature transition rate 20 °C/s), 61 °C for 10 s (temperature transition rate 20 °C/s), and 72 °C for 10 s (temperature transition rate 20 °C/s). The analytical melting program involved melting the PCR products at 95 °C for 30 s, 30 °C for 4 min, and increased to 80 °C at temperature transition rate of 0.1 °C/s, with continuous acquisition of fluorescence data.
Semi-quantitative determination of serum apoM
2 μl of serum was applied to the Hybond-C membrane in duplicate (Amersham Biosciences, Piscataway, NJ, USA). All samples were applied to the same membrane in order to prevent experimental bias. The membrane was quenched in the Tris-HCl buffer in presence of 4% Tween and 3% BSA overnight at 4 °C, and sequentially incubated with primary antibody (dilution, 1:2000 in the Tris-HCl buffer) for one hour and then incubated with alkaline phosphatase (AP) conjugated secondary antibody for one hour at room temperature. The development of AP activity was performed with a commercial visualization system according to the manufacturer’s instructions (Huamei, Shanghai, China). The relative amount of apoM were analyzed with a Macintosh computer by using the software of Quantity One (version 4.2.1, Bio-Rad Laboratories, Hercules, CA, USA), and presented as arbitrary units, being equivalent to volume density (intensity/mm2).
Statistical analysis
Results are expressed as mean ± SD. Comparisons of the general characteristics in two groups and serum apoM levels between different genotypes were statistically evaluated by the unpaired Student’s t-test (Prism software, version 4; GraphPad Inc., La Jolla, CA, USA). Significance was established at a p value less than 0.05. Binary logistic regression analyses were performed by the SPSS statistical package (version 10.0, SPSS Inc., Chicago, IL, USA). Hardy–Weinberg equilibrium was tested for the T-778C polymorphism. Allele frequencies, genotype frequencies, odds ratios (ORs) and 95% confidence intervals (CIs) were all estimated by Chi-squared analyses.
Results
The general characteristics of non-CAD and CAD patients were shown in Table 1. Mean age of non-CAD patients and CAD patients were 63.0 ± 10.8 and 62.5 ± 9.4, respectively. The non-CAD patients and the CAD patients were matched in age and sex (Table 1). It is demonstrated that CAD patients had higher triglyceride (TG) levels (2.23 ± 1.57 mmol/L; p = 0.001), and lower HDL-C (1.12 ± 0.38 mmol/L; p = 0.000) than non-CAD patients (Table 1).
Table 1.
Characteristics of CAD and non-CAD patients
| CAD patients N = 126 | Non-CAD patients N = 118 | p-value | |
|---|---|---|---|
| Male/Female | 98/28 | 88/30 | 0.557 |
| Age (means ± SD) | 62.5 ± 9.4 | 63.0 ± 10.8 | 0.619 |
| TG (mmol/L) | 2.23 ± 1.57* | 1.71 ± 1.10 | 0.001 |
| TC (mmol/L) | 4.50 ± 1.14 | 4.48 ± 1.01 | 0.916 |
| LDL-C (mmol/L) | 2.22 ± 1.08 | 2.33 ± 0.91 | 0.244 |
| HDL-C (mmol/L) | 1.12 ± 0.38* | 1.22 ± 0.31 | 0.000 |
| ApoAI (g/L) | 1.19 ± 0.16 | 1.20 ± 0.14 | 0.284 |
| ApoB (g/L) | 0.93 ± 0.27 | 0.88 ± 0.20 | 0.121 |
| LP(a) (g/L) | 105 ± 73* | 104 ± 103 | 0.034 |
Notes: Data are means ± SD.
p < 0.05 vs control group.
Abbreviations: CAD, coronary artery disease; LDL-C, low-density lipoprotein cholesterol; LP(a), lipoprotein(a); HDL-C, high-density lipoprotein cholesterol; SD, standard deviation; TC, total cholesterol; TG, triglycerides.
The allele frequencies and genotype distributions of T-778C in non-CAD and CAD patients were summarized in Table 2. ApoM T-778C genotype distribution in non-CAD patients was as follows: 84.7% were homozygous for the T allele (T/T), 15.3% were heterozygous (T/C) and 0.0% was homozygous for the C allele (C/C). Allele frequencies were 92.4% for the T allele and 7.6% for the C allele, which was in compliance with Hardy–Weinberg equilibrium (p = 0.370). In CAD patients, 99 patients (78.6%) had the T/T genotype, 25 patients (19.8%) had the T/C genotype, and two patients (1.6%) had the C/C genotype. The allele frequency was 88.5% for the T allele and 11.5% for the C allele, which was also in the Hardy–Weinberg equilibrium (p = 0.772). There was no statistical significant difference in allele frequencies of T and C between CAD and non-CAD patients (OR = 1.575, 95% CI: 0.850–2.919; p = 0.146) (Table 3).
Table 2.
The allele frequencies and genotype distributions of T-778C in non-CAD and CAD patients
| Groups |
Genotypes (n) |
Allele frequencies (%) |
|||
|---|---|---|---|---|---|
| T/T | T/C | C/C | T | C | |
| Non-CAD patients | 100 (84.7%) | 18 (15.3%) | 0 (0.0%) | 92.4 | 7.6 |
| CAD patients | 99 (78.6%) | 25 (19.8%) | 2 (1.6%) | 88.5 | 11.5 |
Abbreviation: CAD, coronary artery disease.
Table 3.
Tests for association of CAD patients and non-CAD patients
| p value | X2 | OR | 95% CI | |
|---|---|---|---|---|
| T/C | 0.146 | 2.11 | 1.575 | 0.850–2.919 |
| TT/TC | 0.318 | 1.00 | 1.403 | 0.720–2.732 |
| TT/CC | 0.157 | 2.00 | 5.050 | 0.239–106.530 |
| TT/(TC + CC) | 0.214 | 1.54 | 1.515 | 0.785–2.926 |
Abbreviations: CAD, coronary artery disease; CI, confidence interval; OR, odds ratio.
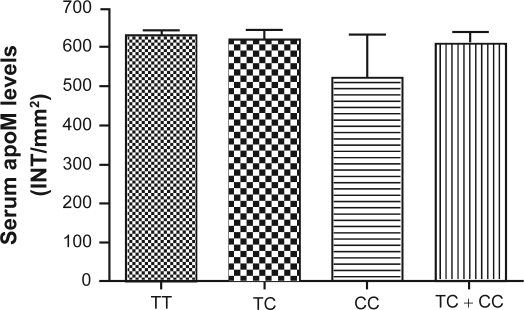
Furthermore, we compared the plasma lipid levels in the non-CAD patients and CAD patients according to the genotypes of SNP T-778C, respectively (Table 4). There were no statistical significant differences between the genotypes in terms of age, gender and plasma lipid levels, either in non-CAD patients or in CAD patients. However, Mean TG level was higher in CAD patients with TT genotype compared to the non-CAD patients (2.29 ± 1.62 vs 1.61 ± 0.86 mmol/L; p < 0.001). The HDL level was lower in CAD patients than in non-CAD control group with TT (1.12 ± 0.41 vs 1.20 ± 0.30 mmol/L; p < 0.001) and C allele carriers (1.11 ± 0.27 vs 1.32 ± 0.35 mmol/L; p = 0.030) (Table 5). Binary logistic regression analysis with adjustments for age, gender, TG, total cholesterol (TC), LDL, HDL, apoAI, apoB, and lipoprotein(a) (LP(a)) indicated that the TC and CC genotypes in SNP T-778C were not significantly associated with CAD (OR = 1.510, 95% CI: 0.756–3.017; p = 0.243). However, TG levels (OR = 1.525, 95% CI: 1.064–2.187; p = 0.022) and HDL levels (OR = 0.308, 95% CI: 0.098–0.969; p = 0.044) were significantly associated with CAD. As shown in Figure 1, there was no statistical significant difference of serum apoM levels found in these three genotypes.
Table 4.
Characteristics of non-CAD and CAD patients according to the genotypes of SNP T-778C (Data are means ± SD)
|
Non-CAD patients |
p value |
CAD patients |
p value | |||
|---|---|---|---|---|---|---|
| TT | TC + CC | TT | TC + CC | |||
| Age (years) | 63.0 ± 10.9 | 63.5 ± 10.3 | 0.920 | 62.6 ± 9.5 | 62.5 ± 9.4 | 0.739 |
| TG (mmol/L) | 1.61 ± 0.86 | 2.24 ± 1.90 | 0.252 | 2.29 ± 1.62 | 2.01 ± 1.39 | 0.150 |
| TC (mmol/L) | 4.45 ± 0.91 | 4.65 ± 1.47 | 0.428 | 4.49 ± 1.16 | 4.54 ± 1.08 | 0.842 |
| LDL-C (mmol/L) | 2.32 ± 0.90 | 2.44 ± 1.01 | 0.539 | 2.17 ± 1.09 | 2.41 ± 1.04 | 0.287 |
| HDL-C (mmol/L) | 1.20 ± 0.30 | 1.32 ± 0.35 | 0.265 | 1.12 ± 0.41 | 1.11 ± 0.27 | 0.658 |
| ApoAI (g/L) | 1.20 ± 0.13 | 1.23 ± 0.15 | 0.530 | 1.19 ± 0.15 | 1.18 ± 0.19 | 0.589 |
| ApoB (g/L) | 0.87 ± 0.18 | 0.90 ± 0.29 | 0.860 | 0.93 ± 0.28 | 0.94 ± 0.24 | 0.677 |
| LP(a) (g/L) | 105 ± 99 | 97 ± 123 | 0.479 | 108 ± 77 | 95 ± 53 | 0.327 |
Abbreviations: ApoAI, apolipoprotein AI; ApoB, apolipoprotein B; CAD, coronary artery disease; LDL-C, low-density lipoprotein cholesterol; LP(a), lipoprotein(a); HDL-C, high-density lipoprotein cholesterol; SD, standard deviation; SNP, single nucleotide polymorphism; TC, total cholesterol; TG, triglycerides.
Table 5.
Characteristics of non-CAD patients and CAD patients according to the common genotypes (Data are means ± SD)
|
TT |
p value |
TC + CC |
p value | |||
|---|---|---|---|---|---|---|
| Non-CAD patients | CAD patients | Non-CAD patients | CAD patients | |||
| Age (years) | 63.0 ± 10.9 | 62. 6 ± 9.5 | 0.713 | 63.5 ± 10.3 | 62.5 ± 9.4 | 0.685 |
| TG (mmol/L) | 1.61 ± 0.86 | 2.29 ± 1.62* | 0.000 | 2.24 ± 1.90 | 2.01 ± 1.39 | 0.790 |
| TC (mmol/L) | 4.45 ± 0.91 | 4.49 ± 1.16 | 0.615 | 4.65 ± 1.47 | 4.54 ± 1.08 | 0.372 |
| LDL-C (mmol/L) | 2.32 ± 0.90 | 2.17 ± 1.09 | 0.204 | 2.44 ± 1.01 | 2.41 ± 1.04 | 0.781 |
| HDL-C (mmol/L) | 1.20 ± 0.30 | 1.12 ± 0.41* | 0.001 | 1.32 ± 0.35 | 1.11 ± 0.27* | 0.030 |
| ApoAI (g/L) | 1.20 ± 0.13 | 1.19 ± 0.15 | 0.481 | 1.23 ± 0.15 | 1.18 ± 0.19 | 0.331 |
| ApoB (g/L) | 0.87 ± 0.18 | 0.93 ± 0.28 | 0.203 | 0.90 ± 0.29 | 0.94 ± 0.24 | 0.366 |
| LP(a) (g/L) | 105 ± 99 | 108 ± 77 | 0.052 | 97 ± 123 | 95 ± 53 | 0.281 |
Note:
p < 0.05 vs control group.
Abbreviations: ApoAI, apolipoprotein AI; ApoB, apolipoprotein B; CAD, coronary artery disease; LDL-C, low-density lipoprotein cholesterol; LP(a), lipoprotein(a);HDL-C, high-density lipoprotein cholesterol; SD, standard deviation; TC, total cholesterol; TG, triglycerides.
Figure 1.
Serum apoM levels in different three genotypes. ApoM concentrations were determined by the dot-blotting analyses as described in Materials and methods. Data are expressed as the intensity/mm2 (means ± SD) that was analyzed by Quantity One software. Data are means ± SE.
Abbreviations: apom, apolipoprotein M; TC, total cholesterol; INT, intensity; SD, standard deviation; SE, standard error.
Discussion
HDL is generally regarded as an antiatherogenic component in the circulation, mainly ascribed to its role in the “reverse cholesterol transport”. In genetically modified mice, apoM is critical for the formation of HDL particles, notably in the pre-β-HDL.6 Christoffersen and colleagues11 compared the properties of human HDL particles that contain apoM with those that did not. ApoM+ particles were significantly more efficient in promoting the efflux of cholesterol from prelabelled THP-1 cells, lending support to the notion that one mechanism behind the anti-atherogenic effect of apoM reflects a role in the reverse cholesterol transport. The ability of HDL to promote cholesterol efflux from foam cells in atherosclerotic lesions is generally regarded as one of the key mechanisms behind its protective function. However, as a new potential marker for atherosclerotic diseases in man, apoM was still little investigated.
More recently, a potential involvement of apoM polymorphisms with certain diseases was of interest. Kabbara and colleagues12 excluded that apoM was a candidate gene for Alzheimer’s disease in the French population. Study of the SNPs located in apoM proximal promoter region found that the SNP T-778C conferred the risk of development of type 2 diabetes and CAD.8,9 However, a population-based nested case-control study showed that apoM had no association with coronary heart disease.10
As it is difficult to obtain appropriate control groups for the association studies in CAD,13 in our initial study we chose participants without any symptoms suggestive of CAD as the control group. Data showed that there was no significant difference in the allele frequencies of T and C between CAD patients and healthy volunteers (OR = 1.019, 95% CI: 0.659–1.575; p = 0.934. Data not shown). The healthy group may have contained some volunteers who had potential stenoses of their coronary arteries, but were not showing symptoms. Compared to the CAD group, this control group was much younger. Perhaps these individuals may have certain genes to initiate and develop CAD in the future, therefore the younger subjects are not proper controls. Furthermore we chose age- and gender-matched patients with normal coronary arteries who were excluded from CAD as controls. The results confirmed our previous finding that there was no association in the allele frequencies of T and C between CAD patients and non-CAD patients (OR = 1.575, 95% CI: 0.850–2.919; p = 0.146). Binary logistic regression analyses with adjustments for age, gender, TG, TC, LDL, HDL, apoAI, apoB, and LP(a) indicated that the TC and CC genotypes in SNP T-778C were not significantly associated with CAD (OR = 1.510, 95% CI: 0.756–3.017, p = 0.243).
Given the evidence implicating apoM in the pathogenesis of CAD, there are several possible explanations for the lack of association observed in the present study. First, it was possible that the lack of any association with CAD could be the lack of power of the study to observe a difference in only 126 patients. Second, this polymorphism may be functionally silent. The results of the serum apoM levels between different genotype groups suggested the mutant promoter might not effect apoM gene expression. In the present study, we used semi-quantitative method to estimated serum apoM levels, more precise methods are guaranteed to be applied in further study. Third, Marenberg and colleagues suggested that at younger ages, death from coronary heart disease is influenced by genetic factors in both women and men, but the genetic effect decreases at older ages and a genetic effect may therefore be diluted.14 Thus, perhaps the effect of apoM polymorphism in our study was decreased in CAD patients because of age factors. Finally, initiation and development of CAD are quite complicated; several risk factors might influence its process, such as body mass index, smoking history, family history, and environmental effects.15 The complicated progression of CAD suggested that CAD is a multifactorial and multigenetic disease.
Although direct clinical studies of human apoM in relation to CAD are still scarce, more and more data indicate that there is a linkage between apoM and risk factors of cardiovascular disease such as diabetes and obesity. It is a challenge to define the roles of apoM in man, and to elucidate its physiopathological role on regulation of HDL metabolism and on anti-atherosclerotic processes.
Acknowledgments
This work was supported by the National Nature Science Foundation of China (Project No. 30570752) and Changzhou Health Bureau (047182, ZD200709). The authors report no conflicts of interest in this work.
References
- 1.Visvikis-Siest S, Marteau JB. Genetic variants predisposing to cardiovascular disease. Curr Opin Lipidol. 2006;17:139–151. doi: 10.1097/01.mol.0000217895.67444.de. [DOI] [PubMed] [Google Scholar]
- 2.Whitney EJ, Krasuski RA, Personius BE, et al. A randomized trial of a strategy for increasing high-density lipoprotein cholesterol levels: effects on progression of coronary heart disease and clinical events. Ann Intern Med. 2005;142:95–104. doi: 10.7326/0003-4819-142-2-200501180-00008. [DOI] [PubMed] [Google Scholar]
- 3.Xu N, Dahlback B. A novel human apolipoprotein (apoM) J Biol Chem. 1999;274:31286–31290. doi: 10.1074/jbc.274.44.31286. [DOI] [PubMed] [Google Scholar]
- 4.Xie T, Rowen L, Aguado B, et al. Analysis of the gene-dense major histocompatibility complex class III region and its comparison to mouse. Genome Res. 2003;13:2621–2636. doi: 10.1101/gr.1736803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang XY, Dong X, Zheng L, et al. Specific tissue expression and cellular localization of human apolipoprotein M as determined by in situ hybridization. Acta Histochem. 2003;105:67–72. doi: 10.1078/0065-1281-00687. [DOI] [PubMed] [Google Scholar]
- 6.Wolfrum C, Poy MN, Stoffel M. Apolipoprotein M is required for prebeta-HDL formation and cholesterol efflux to HDL and protects against atherosclerosis. Nat Med. 2005;11:418–422. doi: 10.1038/nm1211. [DOI] [PubMed] [Google Scholar]
- 7.Hamsten A. Molecular genetics as the route to understanding, prevention, and treatment. Lancet. 1996;348(Suppl 1):S17–S19. doi: 10.1016/s0140-6736(96)98006-x. [DOI] [PubMed] [Google Scholar]
- 8.Niu N, Zhu X, Liu Y, et al. Single nucleotide polymorphisms in the proximal promoter region of apolipoprotein M gene (apoM) confer the susceptibility to development of type 2 diabetes in Han Chinese. Diabetes Metab Res Rev. 2007;23:21–25. doi: 10.1002/dmrr.641. [DOI] [PubMed] [Google Scholar]
- 9.Jiao GQ, Yuan ZX, Xue YS, et al. A prospective evaluation of apolipoprotein M gene T-778C polymorphism in relation to coronary artery disease in Han Chinese. Clin Biochem. 2007;40:1108–1112. doi: 10.1016/j.clinbiochem.2007.04.023. [DOI] [PubMed] [Google Scholar]
- 10.Ahnstrom J, Axler O, Jauhiainen M, et al. Levels of apolipoprotein M are not associated with the risk of coronary heart disease in two independent case-control studies. J Lipid Res. 2008;49(9):1912–1917. doi: 10.1194/jlr.M700471-JLR200. [DOI] [PubMed] [Google Scholar]
- 11.Christoffersen C, Nielsen LB, Axler O, Andersson A, Johnsen AH, Dahlback B. Isolation and characterization of human apolipoprotein M-containing lipoproteins. J Lipid Res. 2006;47:1833–1843. doi: 10.1194/jlr.M600055-JLR200. [DOI] [PubMed] [Google Scholar]
- 12.Kabbara A, Payet N, Cottel D, Frigard B, Amouyel P, Lambert JC. Exclusion of CYP46 and APOM as candidate genes for Alzheimer’s disease in a French population. Neurosci Lett. 2004;363:139–143. doi: 10.1016/j.neulet.2004.03.066. [DOI] [PubMed] [Google Scholar]
- 13.Samani NJ. Molecular genetics of coronary artery disease: measuring the phenotype. Clin Sci (Lond) 1998;95:645–646. [PubMed] [Google Scholar]
- 14.Marenberg ME, Risch N, Berkman LF, Floderus B, de Faire U. Genetic susceptibility to death from coronary heart disease in a study of twins. N Engl J Med. 1994;330:1041–1046. doi: 10.1056/NEJM199404143301503. [DOI] [PubMed] [Google Scholar]
- 15.Allen RA, Lee EM, Roberts DH, Park BK, Pirmohamed M. Polymorphisms in the TNF-alpha and TNF-receptor genes in patients with coronary artery disease. Eur J Clin Invest. 2001;31:843–851. doi: 10.1046/j.1365-2362.2001.00907.x. [DOI] [PubMed] [Google Scholar]