Abstract
Gram-negative bacteria can enter the bloodstream and interact with serum cationic proteins. The character of interaction will depend on the surface characteristics of bacterial cells, which are determined by bacterial chemotype and density of lipopolysaccharide (LPS) packing in the cell wall. It was shown that the lysozyme treatment resulted in the increase sensitivity to hypotonic shock. Significant differences to this effect were found between Escherichia coli strain D21 and D21f2 under treatment with physiological protein concentration. On the basis of electrokinetic measurements and studies of the interaction of cells with lysozyme, the hypothesis was formed that the cell wall of the E. coli strain D21f2 contains more LPS and has a higher density of their packing than the cell wall of the E. coli D21 cells. The effect of lysozyme and lactoferrin on the viability of E. coli cells of two different strains was examined. Lysozyme was found to more effectively inhibit the growth of the E. coli D21 bacteria, and lactoferrin suppressed mainly the growth of the E. coli D21f2 bacteria. These results indicate that the differences in LPS core structure of bacterial R-chemotype, which determines surface charge and density of LPS packing, plays an essential role in the mechanisms of interaction of the cationic proteins with the cell wall.
Keywords: lipopolysaccharide, Escherichia coli, chemotype, lysozyme, lactoferrin, colony-forming units
Introduction
The cell wall of Gram-negative bacteria is a highly organized structure, which supports structural rigidity and viability of the cell. The outer leaflet of the outer membrane of the bacterial cell is primarily formed by lipopolysaccharide (LPS).1 Aside from LPS, the outer membrane contains proteins, in particular porins, which form channels for transport of low-molecular substances into the cell.2 The composition of LPS determines the chemotype and surface characteristics of bacteria, in particular their electrokinetic potential (EKP). The study of EKP of Escherichia coli and Salmonella bacteria of different chemotypes revealed the maximal EKP values to be observed in the deep Re-Rd-mutants.3 The lowest EKP values were registered in the S-chemotype cells; these cells are covered with full-length O-polysaccharide chains, which shield surface charge. Enterobacterial LPS composition determine significantly the cell surface properties that follows from coincidence of EKP values of LPS preparations and initial EKP values of cells from which these LPSs have been isolated. The value of cell EKP depends not only on the composition of LPS molecules; it is also affected by their number in the cell wall.4 Evidently, these factors should influence the interaction of bacteria with blood cationic proteins, such as lysozyme or lactoferrin. These proteins are major components of specific granules of human polymorphonuclear leucocytes. During inflammatory response, lactoferrin and lysozyme are actively secreted by neutrophils. Both proteins possess antimicrobial activity.5,6
As known from the literature, the effects of bactericidal proteins, the bactericidal/permeability-increasing protein (BPI) in particular, will depend on the chemotype of bacteria that these proteins interact with.7 The studies conducted in this area considered mainly the S- and R-bacterial chemotypes. There is, however, almost no data on how the interaction of Gram-negative bacteria with cationic proteins is influenced by the composition of the bacterial LPS core.
In this connection, the objective of our work was to study the effect of the structure of LPS core of Gram-negative E. coli bacteria, belonging to the R-chemotype, on the interaction of bacterial cells with the plasma cationic proteins, particularly, with lysozyme and lactoferrin.
Materials and methods
Chemicals
The following commercial protein preparations were used in our experiments: chicken egg white lysozyme and bovine milk lactoferrin (Sigma Aldrich, St. Louis, MO, USA).
Cell culture
Two strains E. coli K-12: D21 and D21f2 (obtained from DSMZ GmbH, Braunschweig, Germany), which differ in the structure of the LPS core, were used in the experiments. E. coli D21 cells, the wild type, were grown on the agar medium M9,8 while E. coli D21f2 cells in the medium 382 (DSMZ GmbH, Braunschweig, Germany) at 37 °C for 24 h. The contents of Ca2+ and Mg2+ in the growth media were adjusted specifically, because these ions affect the formation of cell wall during bacterial growth, as well as the release of LPS from the cell wall.9
Determination of electrokinetic properties of bacterial cells
For electrokinetic measurements bacterial cells were washed from the surface of the agar medium and rinsed twice in a phosphate-citrate buffer Mac-Ilvena with ionic strength 0.02, pH 7.0.10 Washed cells were stored at 20 °C as a thick suspension in the buffer (1010 cells/ml) and were used within 2 h. Right before measuring the pH-dependence of the cell electrophoretic mobility (EPM), the suspension of cells was diluted by phosphate-citrate buffer of required pH to a concentration of 5 × 106 cells/ml. EPM of 20–25 cells was measured with a “Parmoquant-2” microscope (Carl Zeiss, Jena, Germany) at 20 °C. The EKP of bacterial cells was calculated by the Smoluchowski formula, taking no account of surface polarization.10
Determination of turbidity changes of the E. coli cell suspension after treatment with lysozyme
E. coli cells were washed from the agar medium and their concentration was adjusted to 108 cells/ml. After centrifugation and rinsing with phosphate-buffered saline (PBS), cells were resuspended in the same buffer, containing lysozyme at a concentration of 150–1000 μg/ml. The control cells were resuspended in the pure PBS-buffer. Incubation of cells with lysozyme was carried out at 37 °C for 1 h under gentle stirring. Then control and lysozyme-treated cells were subjected to hypotonic shock by rinsing in bidistilled water, pH 6.0. The degree of cell lysis and aggregation after incubation with lysozyme was estimated by the turbidity change of the cell suspension. Turbidity was measured at 540 nm using a photoelectric colorimeter FEC-56M (OMP, Zagorsk, Russia), with cells orienting chaotically relative to the light beam; or use was made of an experimental photoelectric colorimeter with electrooptical cell,11 in which cells were oriented in a uniform variable (104 Hz) electrical field (25 V·cm−1), with their long axis directed perpendicular to the light beam.
Interaction of bacterial cells with serum proteins
Cells were washed from the agar medium with PBS pH 7.4, and their concentration was adjusted to 108 cells/ml. After centrifugation and rinsing with the buffer, cells were resuspended in PBS-buffer solution of either lysozyme or lactoferrin, each protein taken at the corresponding physiological concentration (150 μg/ml for lysozyme and 100 μg/ml for lactoferrin). Control cells were resuspended in the pure PBS-buffer. Incubation of cells with proteins was carried out at 37 °C for 1 h under gentle stirring.
Determination of the number of CFU
After incubation and the subsequent centrifugation of bacterial cells, the cell pellet was resuspended in the PBS, and the number of colony-forming units (CFU) was determined for control and protein-treated cells by inoculating the corresponding agar medium and counting the colonies formed.
The point in the graphs is the means ± standard deviation. Statistical significance of the results was estimated by Student–Fisher’s criterion. The values shown are average results of three independent experiments.
Results
Electrosurface properties of bacterial cells
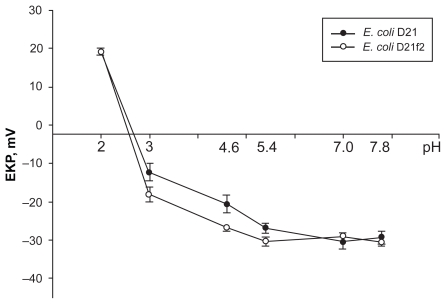
The pH-profiles of cell EKP of the two bacterial strains are given in Figure 1. The curve shapes are typical for the case when the surface of a bacterial cell is covered with both negatively and positively charged (acidic and basic) groups.10 As the figure shows, both strains have close values of negative cell EKP at pH 5.4–7.8. A statistically significant difference in the strain’s cell EKP values appeared only at a more acidic pH (5.4–4.6), when the value of EKP is mainly determined by the ionization of phosphate groups on the cell surface.12 In this pH region, cells of the E. coli D21f2 strain have a larger negative charge. Under further decrease of the suspension pH, the cells pass the isoelectric point and acquire positive EKP values, which may indicate the presence of basic groups on their surface, eg, amine groups of porins. In the acidic medium, the phosphate groups of lipid A and heptose residues as well as carboxyl groups of KDO (3-deoxy-D-manno-octulosonic acid) are completely protonated; therefore, it is only basic protein groups that remain charged at low pH values.
Figure 1.
pH-dependence of EKP of the E. coli D21 and D21f2 cells in the phosphate-citrate buffer with the ionic strength 0.02 (conductivity, 1.30 ± 0.12 mSm·cm−1). At pH 5.4 the E. coli D21 and D21f2 cells had EKP −26.8 ± 0.8 and −30.3 ± 0.6 respectively (P < 0.01); At pH 4.6 these values were −20.60 ± 1.40 and −26.8 ± 0.4 (P < 0.001); and at pH 3.0, they were −12.37 ± 1.35 and −18.18 ± 1.10 (P < 0.01). The values are means ± SD of three independent experiments.
Abbreviations: EKP, electrokinetic potential; SD, standard deviation.
Sensitivity of the E. coli cell suspension after its treatment with lysozyme to hypotonic shock
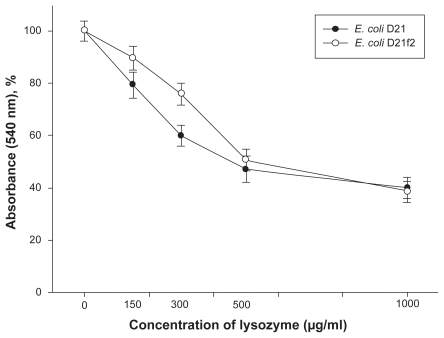
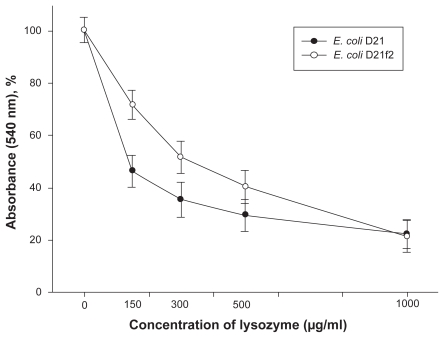
After incubation with lysozyme the bacterial cells were subjected to osmotic shock, that is, they were resuspended and washed in bidistilled water. Figure 2 shows turbidity changes of E. coli cell suspensions treated with lysozyme comparatively to the control. As seen from the figure, the decrease in turbidity at the lysozyme concentrations 150–300 μg/ml is more pronounced in the suspension of the D21-strain cells, as compared to that in the cell suspensions of the E. coli D21f2 strain. Using the technique of orienting cells in a uniform varialbe field with their long axis directed perpendicular to the light beam upon turbidity measurements enabled us to distinguish better between the two strains in their sensitivity to lysozyme (Figure 3).
Figure 2.
Changes in the turbidity of the lysozyme-treated E. coli D21 and D21f2 cell suspensions (%) comparatively to the control suspensions, the cells oriented chaotically relative to the light beam. At the concentration of lysozyme 150 μg/ml, changes in the turbidity of the E. coli D21 and D21f2 cell suspensions were 79.3 ± 3.1 and 89.6 ± 2.6, respectively (P < 0.1); at the concentration of lysozyme 300 μg/ml, 60.0 ± 2.4 and 76.0 ± 2.4 (P < 0.02). The values are means ± SD of three independent experiments.
Abbreviation: SD, standard deviation.
Figure 3.
Changes in the turbidity of the lysozyme-treated E. coli D21 and D21f2 cell suspensions (%) comparatively to the control suspensions, the cells oriented with their long axis being perpendicular to the light beam. At the concentration of lysozyme 150 μg/ml, changes in the turbidity of the E. coli D21 and D21f2 cell suspensions were 46.1 ± 3.5 and 71.5 ± 2.6 respectively (P < 0.01); at the concentration of lysozyme 300 μg/ml, 35.2 ± 3.8 and 51.4 ± 3.6 (P < 0.01). The values are means ± SD of three independent experiments.
Abbreviation: SD, standard deviation.
Generation of CFU by bacteria after incubation of bacterial cells with serum proteins
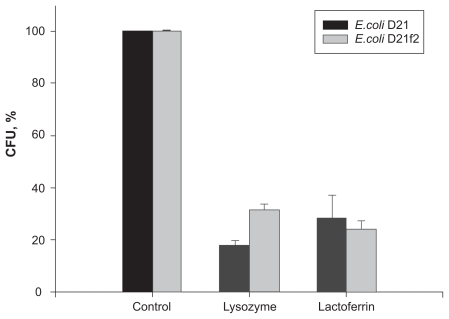
The experiments performed showed that the ability of cells of the two E. coli strains to generate CFU decreased remarkably after treatment of the cells with lysozyme or lactoferrin (Figure 4). The strain that turned out to be most liable to the lysozyme growth-suppressing effect was the E. coli D21 strain: the number of colonies formed by the lysozyme-treated cells was significant lower than that in the control samples. The effect of lysozyme on cells of the E. coli D21f2 strain was less pronounced. The cationic protein lactoferrin suppress mainly the growth of the E. coli D21f2 bacteria.
Figure 4.
Effect of cationic proteins on the CFU-generating ability of the E. coli D21 and D21f2 cells (% of control). A statistically significant difference (P < 0.01) in the CFU-generating ability was revealed only for the lysozyme-treated E. coli strains of two different chemotypes. The values are means ± SD of three independent experiments.
Abbreviations: CFU, colony-forming units; SD, standard deviation.
Discussion
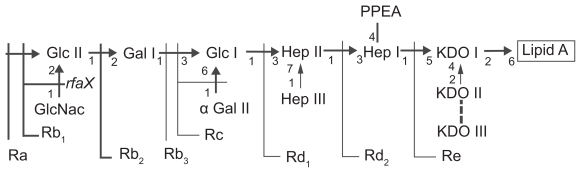
The studied E. coli strains are of the R-chemotype (their LPS lacks O-antigen) and differ in the LPS core structure. E. coli D21 contains LPS with a full-length Ra-structured core, whereas LPS from E. coli D21f2 contains a minimally structured Re-core.13 The structure of the core determines the size and charge of the LPS molecule, which could be important for the interaction of LPS with serum proteins. Owing to the internal core, LPS from E. coli D21 contain, in contrast to E. coli D21f2 LPS, two additional phosphate groups of phosphorylated glycosaccharides (Figure 5).14 Together with the carboxyl groups of KDO and phosphate groups of lipid A, these phosphate groups can ionize and contribute to the total negative charge of the cell surface. Our electrokinetic measurements showed that for cells of both strains, negative EKP values were practically the same at neutral pH, whereas at more acidic pH values, EKP of the E. coli D21f2 cells was more negative than that of the E. coli D21 cells (Figure 1). These results allow us to assume that more negative charge on the surface of the E. coli D21f2 cells may be due to a larger amount of LPS molecules and, correspondingly, a higher density of their packing in the outer membrane, as compared to the E. coli D21 cells.
Figure 5.
The structures of LPS cores from E. coli K-12 (from Ra to Re) Copyright © 1994. Adapted with permission from Reeves P. Biosynthesis and assembly of lipopolysaccharide. In: Ghuysen J-M, Hakenbeck R, editors. Bacterial Cell Wall. London, UK: Elsevier; 1994. p. 281–317.
Abbreviation: LPS, lipopolysaccharides.
To check our hypothesis that the number and packing density of LPS molecules on the cell surface depend on the length of the LPS core, we conducted experiments with the treatment of bacterial cells with lysozyme (Figures 2, 3). It was found that lysozyme treatment resulted in a decrease in the turbidity of cell suspensions comparatively to the control, the effect being observed for both strains examined, yet being more apparent in the cells of the E. coli D21 strain. Thus, on the basis of turbidimetric assay can conclude about significant differences of cell sensitivity of studied strains to hypotonic shock after treatment with lysozyme.
The antimicrobial activity of cationic proteins includes their ability to suppress the growth of bacteria. Here we examined how two cationic proteins taken at physiological concentrations affect the number of CFU generated by the E. coli cells. We found that lysozyme suppressed growth of the E. coli D21 colonies more effectively than that of the E. coli D21f2 colonies (Figure 4). To damage the peptidoglycan layer, lysozyme has to penetrate through the outer leaflet of the outer membrane. The tight LPS packing, which, according to our data, the E. coli D21f2 strain seems to possess, should make this task more difficult for lysozyme. There was a statisitcally significant difference (P < 0.01) between the lysozyme-treated E. coli cells of different chemotypes in regard to their CFU-generating ability.
The experiments in vitro showed that lactoferrin inhibits bacterial growth. It binds to porins, whose number is about 105 per E. coli cell,15 and decrease viability of bacteria.16 In the E. coli H10407 strain, the transition from the S- to R-chemotype enhances the interaction of lactoferrin with the cell surface.17 The LPS O-chains are assumed to shield porins, protecting them from interaction with lactoferrin.16 As our data show, the Ra-core oligosaccharides also can hinder the access of lactoferrin to porins, unlike the two KDO of the reduced Re-core (Figure 4). We observed that lactoferrin suppress mainly the growth of the Re-chemotype bacteria. This corresponds well to the recent data on the interaction of lactoferrin with bacteria of the S- and Ra-chemotypes.18
In total, the results obtained indicate an essential role of the structure of LPS core of E. coli bacteria of R-chemotype, which determines the cell surface charge and the density of LPS packing, in the mechanism of interaction of cationic serum proteins with the bacterial cell wall. Obtained results may to be of interest on the therapy with bactericidal cationic proteins.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Nikaido H, Vaara M. The gram-negative bacterial outer membrane. In: Neidhardt F, editor. Escherichia coli and Salmonella typhimurium. Washington, DC: ASM Publications; 1987. pp. 7–44. [Google Scholar]
- 2.Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Molecular Biol Rev. 2003;67(4):593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gremyakova TA, Fomchenkov VM, Fursova NK, et al. Comparative description of physico-chemical properties of lipopolisaccharides from Yersinia pestis and R-mutants of enterobacteria. Microbiology (Moscow) 1996;65(6):763–767. [PubMed] [Google Scholar]
- 4.Ivanov AYu, Fomchenkov VM. Dependence of the damaging effect of surface-active substances on Escherichia coli cells on the culture growth phase. Microbiology (Moscow) 1989;85(6):969–975. [PubMed] [Google Scholar]
- 5.Ellison RT, III, Giehl TJ. Killing of Gram-negative bacteria by lactoferrin and lysozyme. J Clin Invest. 1991;88:1080–1091. doi: 10.1172/JCI115407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Naidu AS, Arnold RR. Influence of lactoferrin on host-microbe interaction. In: Hutchens TW, Lonnerdal B, editors. Lactoferrin: Interactions and biological functions. New York, NY: Humana Press; 1997. pp. 259–275. [Google Scholar]
- 7.Capodici C, Chen S, Sidorczyk Z, et al. Effect of lipopolysaccharide (LPS) chain length on interactions of bactericidal/permeability-increasing protein and its bioactive 23-kilodalton NH2-terminal fragment with isolated LPS and intact Proteus mirabilis and . Escherichia coli Infect Immun. 1994;62(1):259–265. doi: 10.1128/iai.62.1.259-265.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clowes RC, Heyes W. Methods in Genetics of Microorganisms. Moscow, Russia: Meditsina (Russ); 1970. [Google Scholar]
- 9.Rossi P, Giansanti F, Boffi A, et al. Ca2+ binding to bovine lactoferrin enhances protein stability and influences the release of bacterial lipopolysaccharide. Biochem Cell Biol. 2002;80:41–48. doi: 10.1139/o01-209. [DOI] [PubMed] [Google Scholar]
- 10.James AM. The electrical properties and topochemistry of bacterial cells. Adv Colloid Interface Sci. 1982;15(3/4):171–221. [Google Scholar]
- 11.Ivanov AYu, Novikov VV, Fesenko EE. Effect of an aqueous salt solution exposed to weak magnetic fields on the bacterial plasma membrane sensitivity to reactive oxygen species. Biophysics (Moscow) 2002;47(2):297–302. [PubMed] [Google Scholar]
- 12.Beavan MJ, Belk DM, Stewart GG, Rose AH. Changes in electrophoretic mobility and lytic enzyme activity associated with development of flocculating ability in Saccharomyces cerevisiae. Can J Microbiol. 1979;25(8):888–895. doi: 10.1139/m79-132. [DOI] [PubMed] [Google Scholar]
- 13.Amor K, Heinrichs DE, Frirdich E, et al. Distribution of core oligosaccharide types in lipopolysaccharides from Escherichia coli. Infect Immun. 2000;68(3):1116–1124. doi: 10.1128/iai.68.3.1116-1124.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reeves P. Biosynthesis and assembly of lipopolysaccharide. In: Ghuysen J-M, Hakenbeck R, editors. Bacterial Cell Wall. London, UK: Elsevier; 1994. pp. 281–317. [Google Scholar]
- 15.Rosenbuch JP. Characterization of the major envelope protein from Escherichia coli. Regular arrangement on the peptidoglycan and unusual dodecyl sulfate binding. J Biol Chem. 1974;249:8019–8029. [PubMed] [Google Scholar]
- 16.Erdei L, Forsgren A, Naidu AS. Lactoferrin binds to porins OmpF and OmpC in Escherichia coli. Infect Immun. 1994;62:1236–1240. doi: 10.1128/iai.62.4.1236-1240.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gado I, Erdei J, Laszlo VG, et al. Correlation between human lactoferrin binding and colicin susceptibility in Escherichia coli. Antimicrob Agents Chemother. 1991;35:2538–2543. doi: 10.1128/aac.35.12.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andra J, Lohner K, Blondelle SE, et al. Enhancement of endotoxin neutralization by coupling of a C12-alkyl chain to a lactoferricin-derived peptide. Biochem J. 2005;385:135–143. doi: 10.1042/BJ20041270. [DOI] [PMC free article] [PubMed] [Google Scholar]