Abstract
Colorectal cancer is a leading cause of cancer death worldwide. To identify molecular targets for colorectal cancer therapy, we tested small interfering RNAs (siRNAs) against 97 genes whose expression was elevated in human colorectal cancer tissues for the ability to promote apoptosis of human colorectal cancer cells (HT-29 cells). The results indicate that the downregulation of PSMA7 (proteasome subunit, α-type, 7) and RAN (ras-related nuclear protein) most efficiently induced apoptosis of HT-29 cells. PSMA7 and RAN were highly expressed in colorectal cancer cell lines compared with normal colon tissues. Furthermore, PSMA7 and RAN were overexpressed in not only colon tumor tissues but also the other tumor tissues. Moreover, in vivo delivery of PSMA7 siRNA and RAN siRNA markedly induced apoptosis in HT-29 xenograft tumors in mice. Thus, silencing of PSMA7 and RAN induces cancer cells to undergo apoptosis, and PSMA7 and RAN might be promising new molecular targets for drug and RNA interference-based therapeutics against colorectal cancer.
Keywords: colorectal cancer, molecular target, RNAi, PSMA7, RAN
Introduction
Colorectal cancer is one of the most common cancers in women and men worldwide. Nearly 1.2 million cases of colorectal cancer were expected to occur in 2007.1 The highest incidence rates are found in Japan, North America, parts of Europe, New Zealand, and Australia.1 Worldwide, some 630,000 people die from colorectal cancer per year, accounting for 8% of all cancer deaths.1 The five-year survival for persons with colorectal cancer is about 65% in Japan; however, when this cancer is detected at advanced stages, the five-year survival rate decreases to 10%,2 necessitating effective therapeutic targets.
A tumor is characterized by uncontrolled growth and spread of abnormal cells, which invade adjacent normal tissue and spread to other organs, a process that causes death.3 Multiple molecular alterations are involved in a transformation from a normal cell into a cancerous cell and a progression from a pre-cancerous lesion to malignant tumors.
Angiogenesis is critical in tumor growth and survival. Its inhibition is a promising target for cancer therapy.4–6 Vascular endothelial growth factor (VEGF) plays a key role in angiogenesis in cancer. In many human tumors including colorectal cancer, VEGF amd VEGF receptors (VEGFR) are overexpressed. Regulating the VEGF/VEGFR pathway is an effective approach to treat cancer.6,7 A humanized anti-VEGF monoclonal antibody, Bevacizumab, is the first approved biological inhibitor against VEGF for the treatment of metastatic colorectal cancer.4–6,8 Small-molecule tyrosine kinase inhibitors against VEGFRs are also being developed for cancer therapy.4,6,9,10
Furthermore, the survival of abnormal cells is a characteristic feature of cancer. In colorectal tumors, some signal transduction pathways drive abnormal cell growth. The most important factor promoting cell survival is epidermal growth factor (EGF).11,12 Its signaling is a potential target for cancer therapy.4,5 In treating colorectal cancer, a monoclonal antibody against EGF receptor (EGFR) such as a cetuximab is active,4,5,12–14 and small-molecular tyrosine kinase inhibitors of EGFRs have been shown to be effective.4,12
Additionally, one of the hallmarks of human carcinogenesis is the breakdown of cell apoptotic machinery.15 Overexpression of anti-apoptotic Bcl-2 family members frequently relates to decreased sensitivity to anticancer drugs and radiotherapy in many types of cancer.16 The antisense oligonucleotide drug targeting Bcl-2 mRNA expression such as an oblimersen is being investgated in some cancers.17 The BH3-domain of anti-apoptotic Bcl-2 family proteins is required for the antiapoptotic function. BH-3 mimic peptides that interfere with Bcl-2 signaling are currently under development.16 Moreover, therapies based on tumor necrosis factor-related apoptosis-inducing ligand (TRAIL), which induces programmed cell death, have been studied.18 Monoclonal antibodies against TRAIL receptors with an agonistic effect on the TRAIL pathway have been generated.18,19 Thus, inducing apoptosis is a promising approach in the development of a molecular targeted therapy for cancer.
In this paper, we focused on apoptosis induction to identify molecular targets for colorectal cancer therapy. We tested siRNAs against 97 genes whose expression was elevated in human colorectal cancer tissues for the ability to promote apoptosis of human colon cancer cells (HT-29 cells). The results showed that the downregulation of proteasome subunit, α-type, 7 (PSMA7) and ras-related nuclear protein (RAN) strongly caused apoptosis of HT-29 cells. PSMA7 siRNA and RAN siRNA markedly induced apoptosis in HT-29 xenograft tumor tissues in mice. This silencing of PSMA7 and RAN that induces cancer cells to undergo apoptosis suggests that PSMA7 and RAN are potential key targets for future RNA interference (RNAi)-based therapeutics against colorectal cancer.
Materials and methods
Cell culture
Five colorectal cancer cell lines were obtained from the American Type Culture Collection (ATCC), and maintained at 37 °C under 5% CO2 in a humidified incubator. Caco-2 (human colorectal adenocarcinoma) cells were cultured in Eagle’s minimum essential medium (EMEM; Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS; Invitrogen) and nonessential amino acids (Invitrogen). Human colorectal carcinoma (HCT116) and human colorectal adenocarcinoma (HT-29) cells were cultured in McCoy’s 5A medium (Invitrogen) containing 10% FBS. LoVo (human colorectal adenocarcinoma, derived from supraclavicular lymph node metastatic site) cells were cultured in Ham’s F12 medium (Invitrogen) containing 10% FBS. T84 (human colorectal carcinoma, derived from lung metastatic site) cells were cultured in DMEM/Ham’s F-12 medium (Invitrogen) containing 10% FBS. To obtain total RNA from these cell lines we plated cells at 1 × 105 cells per well (6-well plate) and culture for three days.
Design and synthesis of siRNAs
We designed siRNAs and synthesized them with four siRNA duplexes for each gene target (Dharmacon, Chicago, IL, USA). The siRNA sequences were described in Table 1.
Table 1.
The sequences of siRNAs
| Sense strand | Antisense strand | |
|---|---|---|
| PSMA7 | ||
| #1 | 5′-GAAGUAUGUUGCUGAAAUUUU-3′ | 5′-AAUUUCAGCAACAUACUUCUU-3′ |
| #2 | 5′-GAAGAGACAUUGUUGUUCUUU-3′ | 5′-AGAACAACAAUGUCUCUUCUU-3′ |
| #3 | 5′-GAAGAUCUGUGCUUUGGAUUU-3′ | 5′-AUCCAAAGCACAGAUCUUCUU-3′ |
| #4 | 5′-CAUCGUGGGUUUCGACUUUUU-3′ | 5′-AAAGUCGAAACCCACGAUGUU-3′ |
| RAN | ||
| #1 | 5′-AGAAGAAUCUUCAGUACUAUU-3′ | 5′-UAGUACUGAAGAUUCUUCUUU-3′ |
| #2 | 5′-GUGAAUUUGAGAAGAAGUAUU-3′ | 5′-UACUUCUUCUCAAAUUCACUU-3′ |
| #3 | 5′-CCUAUUAAGUUCAAUGUAUUU-3′ | 5′-AUACAUUGAACUUAAUAGGUU-3′ |
| #4 | 5′-ACAGGAAAGUGAAGGCGAAUU-3′ | 5′-UUCGCCUUCACUUUCCUGUUU-3′ |
Abbreviation: siRNA, small interfering RNAs.
Cell transfection array
For RNAi-based functional screening of genes, we used a reverse transfection based-cell transfection array.20 HT-29 cells were plated into the cell transfection array in a 96-well format and transfected with siRNA. We evaluated the effects of the downregulation of genes on promotion of apoptosis, as mentioned below.
Measurement of cell proliferation
We plated HT-29 cells into a cell transfection array at a density of 5 × 103 cells per well and cultured. Three days after, we measured cell proliferation by resazurin reduction assay using CellTiter-Blue Reagent (Promega, Madison, WI, USA). Cells were incubated with CellTiter-Blue Reagent for one hour at 37 °C, and the fluorescence was then measured at 560Ex/590Em. After that, we subjected the same cell transfection array to a caspase-3/7 assay, Hoechst staining, and a cell-direct real-time reverse transcriptase-polymerase chain reaction (RT-PCR) assay.
Measurement of caspase activity in vitro
Cells were incubated with the Caspase-Glo 3/7 Reagent (Promega) for one hour at room temperature, and the luminescence was then measured.
Hoechst staining
Cells on a cell transfection array were washed with phosphatebuffered saline (PBS), and a fixative and staining solution was added (4% paraformaldehyde, 0.1% Triton X-100, 1 μg/mL Hoechst 33342 in PBS). Twenty minutes after incubation, cells were washed with PBS. We determined the number of apoptotic cells was in three microscopic fields of each well by fluorescence microscopy.
Transfection of siRNA
We carried out transfection of HT-29 cells with siRNA using Liopfectamine 2000 (Invitrogen), according to the manufacturer’s protocol. We plated HT-29 cells 24 hours before transfection, and we then transfected the cells, which were grown to 50% confluence, with 40 nM siRNAs.
Real-time RT-PCR
We purified total RNA from cells and tumor tissues with an RNeasy Mini Kit and RNase-Free DNase Set (QIAGEN, Hilden, Germany), and produced cDNAs with an ExScript RT reagent Kit (Takara Bio, Shiga, Japan). We then subjected cDNA samples to real-time PCR using SYBR Premix Ex Taq (Takara) and specific primers as follows: for PSMA7, forward, 5′-CAAGTGGAGTACGCGCAGGA-3′; reverse, 5′-CTGCAGTTTGGCCACTGACTTC-3′; for RAN, forward, 5′-AAGTTGTCATGGACCCAGCTTTG-3′; reverse, 5′-GCTGGGCTCCAGCTTCATTC-3′. We carried out the reactions using the 7300 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). We normalized gene-expression levels by 18S rRNA or GAPDH.
Cell-direct real-time RT-PCR
We used FastLane Cell cDNA Kit and QuantiTect SYBR Green PCR Kit (QIAGEN). We lysed cells in a well of cell transfection array and synthesized the first-strand cDNA. The cDNA template was then directly subjected to real-time RT-PCR using specific primers.
siRNA treatment in vivo
We performed animal experiments in compliance with the guidelines of the Institute for Laboratory Animal Research, National Cancer Center Research Institute of Japan. We subcutaneously injected 5 × 106 HT-29 cells into athymic nude mice (six-week-old females; CLEA Japan, Tokyo, Japan). When the HT-29 tumor grew to approximately 5–6 mm in diameter, we injected mice with 1 nmol siRNA by intratumoral injection. We harvested tumor tissues for analysis of mRNA and apoptosis at 24 and 72 hours after treatment, respectively.
TUNEL technique
We harvested tumor tissues 72 hours after administration of siRNA and prepared frozen sections. We then performed TUNEL (TdT-mediated dUTP nick-end labeling) staining using an In Situ Cell Death Detection Kit, Fluorescein (Roche Diagnostics, Basel, Switzerland), according to the manufacturer’s protocol. DNA strand breaks in apoptotic cells can be labeled by the addition of fluorescein dUTP using terminal deoxynucleotidyl transferase (TdT). The nuclei were stained with DAPI. We determined the number of fluorescein-positive cells in three microscopic fields of each section by fluorescence microscopy.
Human samples
The study protocol for clinical samples was approved by the Institutional Review Board of Osaka University Medical School (Osaka, Japan), and written informed consent was obtained from each patient. We obtained total RNA from tumor tissues and normal adjacent tissues (FirstChoice Human Tumor/Normal Adjacent Tissue RNA) from Ambion (Austin, TX, USA).
cDNA micro-array analysis
We performed cDNA micro-array analysis using AceGene (DNA Chip Research Inc., Yokohama, Japan) according to the manufacturer’s instructions (http://www.dna-chip.co.jp/thesis/AceGeneProtocol.pdf) to obtain an expression profile of human colorectal cancer.21 As a standard normal control reference, a mixture of total RNA extracted from normal colorectal tissues was used. We synthesized cDNA from total RNA of normal colorectal tissues and colorectal tumor tissues, and labeled cDNA with Cy3 for normal colorectal tissue, and Cy5 for colorectal tumor tissues, respectively. After hybridization of cDNA and array, the array was scanned, and Cy5/Cy3 ratios were log2-transformed to compare levels of mRNA expression in tumor and normal tissues.
Statistical analysis
We conducted statistical analysis using the analysis of variance with Student’s t-test. We considered a P value of 0.05 or less as a significant difference.
Results
RNAi-based screening for identification of molecular targets
To identify molecular targets for therapy, we conducted a study of RNAi-induced gene knockdown in HT-29 human colon cancer cells. The strategy for target identification is schematically shown in Figure 1. We performed gene expression profiling of 191 subjects with colorectal tumors and selected 97 genes whose expression was elevated in human colorectal cancer tissues by rank order of mRNA expression (Table 2). We used the siRNAs specific to these genes which are siRNA pools composed of four distinct siRNA species targeting different sequences of the same target transcript for RNAi-based screening. We transfected HT-29 cells with the siRNAs using reverse transfection-based cell transfection array. To evaluate the efficiency of the cell transfection array, we used GAPDH siRNA against the gene encoding GAPDH (glyceraldehyde-3-phosphate dehydrogenase). GAPDH siRNA transfer caused an approximate 75% reduction of the GAPDH mRNA expression in HT-29 cells relative to the control nontargeting siRNA (data not shown).
Figure 1.
Schematic representation of the strategy for targets identification by RNAi-based reverse genetics in vitro and in vivo. First, we carried out a gene expression profiling of human colorectal tumor tissues and selected genes whose expression was elevated. We performed a functional screening of genes by a cell transfection array to test the efficacy of a specific siRNA related to apoptosis induction in human colorectal cancer. Subsequently, the siRNAs against candidate genes were applied to an in vivo animal tumor model. Finally, we identified new molecular targets for drug and RNAi-based colorectal cancer therapy.
Abbreviation: siRNA, small interfering RNAs.
Table 2.
The list of 97 genes elevated in human colorectal tumor tissues
| No | Gene | Description | Accession no. | Log2 ratio (tumor/normal) |
|---|---|---|---|---|
| 1 | COL1A1 | Collagen, type I, α1 | NM_000088 | 2.137 |
| 2 | SPP1 | Secreted phosphoprotein 1 (osteopontin, bone sialoprotein I, early T-lymphocyte activation 1) | NM_000582 | 1.531 |
| 3 | CCL20 | Chemokine (C-C motif) ligand 20 | NM_004591 | 1.501 |
| 4 | UBD | Ubiquitin D | NM_006398 | 1.392 |
| 5 | TGFBI | Transforming growth factor, β-induced, 68 kDa | NM_000358 | 1.345 |
| 6 | IFITM1 | Interferon induced transmembrane protein 1 (9–27) | NM_003641 | 1.341 |
| 7 | MMP12 | Matrix metallopeptidase 12 (macrophage elastase) | NM_002426 | 1.248 |
| 8 | CEACAM6 | Carcinoembryonic antigen-related cell adhesion molecule 6 (non-specific cross reacting antigen) | NM_002483 | 1.237 |
| 9 | RPS21 | Ribosomal protein S21 | NM_001024 | 1.232 |
| 10 | PFDN4 | Prefoldin 4 | NM_002623 | 1.209 |
| 11 | TIMP1 | TIMP metallopeptidase inhibitor 1 | NM_003254 | 1.191 |
| 12 | NEK6 | NIMA (never in mitosis gene a)-related kinase 6 | NM_014397 | 1.089 |
| 13 | MMP1 | Matrix metallopeptidase 1 (interstitial collagenase) | NM_002421 | 1.079 |
| 14 | S100A11 | S100 calcium binding protein A11 (calgizzarin) | NM_005620 | 1.055 |
| 15 | IFNAR1 | Interferon (α, β and ω) receptor 1 | NM_000629 | 1.031 |
| 16 | CPSF4 | Cleavage and polyadenylation specific factor 4, 30 kDa | NM_006693 | 1.024 |
| 17 | TMEPAI | Transmembrane, prostate androgen induced RNA | NM_020182 | 1.013 |
| 18 | RPL31 | Ribosomal protein L31 | NM_000993 | 0.995 |
| 19 | CPNE3 | Copine III | NM_003909 | 0.988 |
| 20 | UBE2C | Ubiquitin-conjugating enzyme E2C | NM_007019 | 0.979 |
| 21 | NQO1 | NAD(P)H dehydrogenase, quinone 1 | NM_000903 | 0.968 |
| 22 | MYC | V-myc myelocytomatosis viral oncogene homolog (avian) | NM_002467 | 0.966 |
| 23 | LCN2 | Lipocalin 2 (oncogene 24p3) | NM_005564 | 0.959 |
| 24 | PRKAA1 | Protein kinase, AMP-activated, α1 catalytic subunit | NM_006251 | 0.958 |
| 25 | GNGT2 | Guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2 | NM_031498 | 0.932 |
| 26 | PSMA7 | Proteasome (prosome, macropain) subunit, α-type, 7 | NM_002792 | 0.932 |
| 27 | SLC3A2 | Solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2 | NM_002394 | 0.925 |
| 28 | C10ORF137 | Chromosome 10 open reading frame 137 | NM_015608 | 0.917 |
| 29 | CKS2 | CDC28 protein kinase regulatory subunit 2 | NM_001827 | 0.912 |
| 30 | RPS6 | Ribosomal protein S6 | NM_001010 | 0.911 |
| 31 | RPL39 | Ribosomal protein L39 | NM_001000 | 0.910 |
| 32 | COL10A1 | Collagen, type X, α1(Schmid metaphyseal chondrodysplasia) | NM_000493 | 0.910 |
| 33 | MLLT1 | Myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 1 | NM_005934 | 0.896 |
| 34 | GABRD | γ-aminobutyric acid (GABA) A receptor, δ | NM_000815 | 0.895 |
| 35 | COL1A2 | Collagen, type I, α2 | NM_000089 | 0.894 |
| 36 | DKFZP564O0463 | WD repeats and SOF1 domain containing | NM_015420 | 0.894 |
| 37 | RIPK2 | Receptor-interacting serine-threonine kinase 2 | NM_003821 | 0.893 |
| 38 | FOXM1 | Forkhead box M1 | NM_021953 | 0.892 |
| 39 | PIGL | Phosphatidylinositol glycan, class L | NM_004278 | 0.870 |
| 40 | NIN | Ninein (GSK3B interacting protein) | NM_016350 | 0.870 |
| 41 | ATP2B1 | ATPase, Ca++ transporting, plasma membrane 1 | NM_001682 | 0.870 |
| 42 | MIF | Macrophage migration inhibitory factor (glycosylation-inhibiting factor) | NM_002415 | 0.861 |
| 43 | PCYOX1 | Prenylcysteine oxidase 1 | NM_016297 | 0.850 |
| 44 | ARF4 | ADP-ribosylation factor 4 | NM_001660 | 0.843 |
| 45 | MEOX2 | Mesenchyme homeo box 2 (growth arrest-specific homeo box) | NM_005924 | 0.840 |
| 46 | HTR2B | 5-hydroxytryptamine (serotonin) receptor 2B | NM_000867 | 0.836 |
| 47 | HRASLS3 | HRAS-like suppressor 3 | NM_007069 | 0.827 |
| 48 | GYPA | Glycophorin A (includes Ss blood group) | NM_002099 | 0.819 |
| 49 | GDF15 | Growth differentiation factor 15 | NM_004864 | 0.817 |
| 50 | NPHS2 | Nephrosis 2, idiopathic, steroid-resistant (podocin) | NM_014625 | 0.816 |
| 51 | FIBL-6 | Hemicentin 1 | NM_031935 | 0.804 |
| 52 | AKAP8L | A kinase (PRKA) anchor protein 8-like | NM_014371 | 0.787 |
| 53 | SLC12A2 | Solute carrier family 12 (sodium/potassium/chloride transporters), member 2 | NM_001046 | 0.784 |
| 54 | CDK10 | Cyclin-dependent kinase (CDC2-like) 10 | NM_003674 | 0.781 |
| 55 | UFM1 | Ubiquitin-fold modifier 1 | NM_016617 | 0.780 |
| 56 | TBX19 | T-box 19 | NM_005149 | 0.775 |
| 57 | DPEP1 | Dipeptidase 1 (renal) | NM_004413 | 0.775 |
| 58 | NNMT | Nicotinamide N-methyltransferase | NM_006169 | 0.754 |
| 59 | RPS20 | Ribosomal protein S20 | NM_001023 | 0.748 |
| 60 | ZNF84 | Zinc finger protein 84 (hpf2); znf84 | NM_003428 | 0.740 |
| 61 | HIG2 | Hypoxia-inducible protein 2 | NM_013332 | 0.639 |
| 62 | SNAI2 | Snail homolog 2 (Drosophila) | NM_003068 | 0.610 |
| 63 | SLAMF7 | SLAM family member 7 | NM_021181 | 0.604 |
| 64 | RAN | RAN, member RAS oncogene family | NM_006325 | 0.603 |
| 65 | SNAI1 | Snail homolog 1 (Drosophila) | NM_005985 | 0.580 |
| 66 | MRAS | Muscle RAS oncogene homolog | NM_012219 | 0.543 |
| 67 | ARHGEF4 | Rho guanine nucleotide exchange factor (GEF) 4 | NM_015320 | 0.536 |
| 68 | MDK | midkine (neurite growth-promoting factor 2) | NM_002391 | 0.530 |
| 69 | BRAF | Y-raf murine sarcoma viral oncogene homolog B1 | NM_004333 | 0.528 |
| 70 | FBXO11 | F-box protein 11 | NM_012167 | 0.519 |
| 71 | AATF | Apoptosis antagonizing transcription factor | NM_012138 | 0.516 |
| 72 | FIGN | Fidgetin | NM_018086 | 0.506 |
| 73 | MMP9 | Matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) | NM_004994 | 0.497 |
| 74 | VEGFA | Vascular endothelial growth factor | NM_003376 | 0.496 |
| 75 | FBXW5 | F-box and WD-40 domain protein 5 | NM_178225 | 0.481 |
| 76 | LTA | Lymphotoxin α (TNF superfamily, member 1) | NM_000595 | 0.476 |
| 77 | TRAP1 | TNF receptor-associated protein 1 | NM_016292 | 0.455 |
| 78 | LGALS1 | Lectin, galactoside-binding, soluble, 1 (galectin 1) | NM_002305 | 0.453 |
| 79 | RRAS | Related RAS viral (r-ras) oncogene homolog | NM_006270 | 0.441 |
| 80 | MMP10 | Matrix metallopeptidase 10 (stromelysin 2) | NM_002425 | 0.440 |
| 81 | FBXW11 | F-box and WD-40 domain protein 11 | NM_012300 | 0.440 |
| 82 | SAT | Spermidine/spermine N1-acetyltransferase | NM_002970 | 0.440 |
| 83 | RPN2 | Ribophorin II | NM_002951 | 0.400 |
| 84 | RAB4B | RAB4B, member RAs oncogene family | NM_016154 | 0.398 |
| 85 | FETUB | Fetuin B | NM_014375 | 0.395 |
| 86 | ELF4 | E74-like factor 4 (ets domain transcription factor) | NM_001421 | 0.388 |
| 87 | SHAX3 | Chromatin modifying protein 4C | NM_152284 | 0.387 |
| 88 | ECT2 | Epithelial cell transforming sequence 2 oncogene | NM_018098 | 0.384 |
| 89 | HES6 | Hairy and enhancer of split 6 (Drosophila) | NM_018645 | 0.383 |
| 90 | DDEF1 | Development and differentiation enhancing factor 1 | NM_018482 | 0.378 |
| 91 | RHEB | Ras homolog enriched in brain | NM_005614 | 0.375 |
| 92 | CTNND1 | Catenin (cadherin-associated protein), δ1 | NM_001331 | 0.368 |
| 93 | DNMT3B | DNA (cytosine-5-)-methyltransferase 3β | NM_006892 | 0.365 |
| 94 | ASPM | ASP (abnormal spindle)-like, microcephaly associated (Drosophila) | NM_018136 | 0.360 |
| 95 | PCDHA10 | Protocadherin α10 | NM_018901 | 0.349 |
| 96 | VEGFC | Vascular endothelial growth factor C | NM_005429 | 0.347 |
| 97 | RASGRF1 | Ras protein-specific guanine nucleotide-releasing factor 1 | NM_002891 | 0.346 |
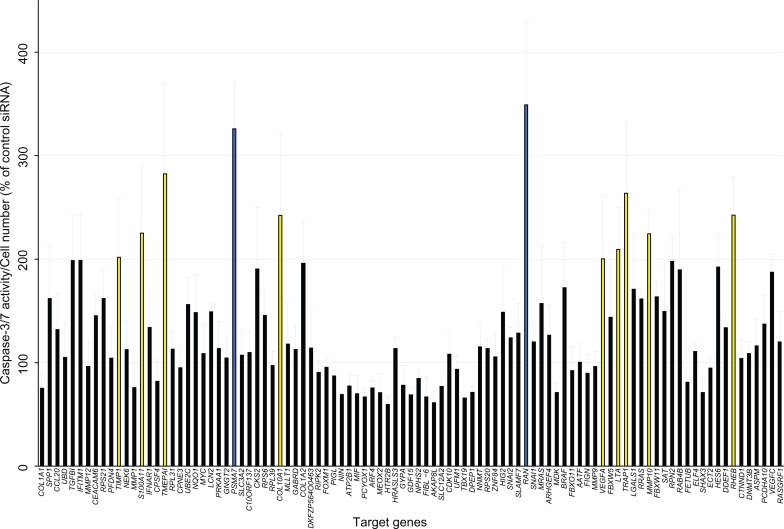
We assessed siRNAs for their ability to induce apoptosis in HT-29 compared with the control nontargeting siRNA. We measured cell viability by resazurin reduction and examined apoptosis by caspase-3/7 activity. Caspase-3/7 activity was normalized by viable cell number (measurement value of resazurin reduction) and Caspase-3/7 activation by siRNAs was compared with control nontargeting siRNA. The results indicated that the downregulation of 11 genes (TIMP1, S100A11, TMEPAI, PSMA7, COL10A1, RAN, VEGFA, LTA, TRAP1, MMP10, and RHEB) resulted in a marked induction of apoptosis in HT-29 cells (caspase-3/7 activity, percentage of nontargeting siRNA > 200, P < 0.05, Figure 2). In particular, the PSMA7 siRNA pool and the RAN siRNA pool strongly enhanced caspase-3/7 activity (caspase-3/7 activity, percentage of nontargeting siRNA > 300, P < 0.001, Figure 2). We validated these results by counting Hoechst-stained cells showing apoptotic nuclear condensation and fragmentation (Figure 3A) and found that there was a significantly higher apoptotic cell death rate in cells given PSMA7 siRNA pool and RAN siRNA pool relative to that in cells given control siRNA (P < 0.01, Figure 3B). At 72 hours after treatment with siRNA, there was substantial cell death induced by the PSMA7 siRNA pool and the RAN siRNA pool compared with the control nontargeting siRNA (Figure 3C).
Figure 2.
Functional screening of genes by RNAi-cell transfection array in cultured colorectal cancer cells. Cell number and caspase-3/7 activity were measured 72 hours after transfection of HT-29 cells. Caspase-3/7 activity was normalized by viable cell number to evaluate apoptosis induction. Control nontargeting siRNA is set to 100% and relative activity is shown. Yellow color bar: relative activity (%) >200, P < 0.05; blue color bar: relative activity (%) >300, P < 0.001. n = 4 per group. Values are mean ± SD.
Abbreviations: SD, standard deviation; siRNA, small interfering RNAs.
Figure 3.
Apoptosis of HT-29 cells transduced with PSMA7 siRNA and RAN siRNA. A) Hoechst staining of cells 72 hours after the transfection of siRNA. Scale bar, 100 μm. The arrows indicate cells with nuclear condensation and fragmentation. B) Numbers of apoptotic cells from (A). The data show the percentage of apoptotic cells. As a control, nontargeting control siRNA was used (n = 4 per group, *P < 0.01). C) Phase contrast micrograph of HT-29 cells 72 hours after the treatment of siRNA. Scale bar, 200 μm. D) Knockdown of mRNA by PSMA7 siRNA pool and RAN siRNA pool using cell transfection array was measured 72 hours after transfection. (n = 5 per group, *P < 0.001). E) Knockdown of mRNA by PSMA7 siRNAs and RAN siRNAs. Expression of PSMA7 mRNA and RAN mRNA was measured 72 hours and 48 hours after transfection, respectively (n = 3 per group, *P < 0.01). As a control, nontargeting siRNA was used. Values are mean ± SD.
Abbreviations: PSMA7, proteasome subunit, α-type, 7; RAN, ras-related nuclear protein; SD, standard deviation; siRNA, small interfering RNAs.
We assessed the efficacy of PSMA7 siRNA pool and RAN siRNA pool for the knockdown of mRNA by celldirect real-time RT-PCR analysis. This analysis revealed that PSMA7 siRNA pool and RAN siRNA pool inhibited the mRNA expression relative to the control nontargeting siRNA, 80% and 60% respectively (Figure 3D). To evaluate the knockdown efficiency of individual siRNAs of the siRNA pool, we performed a liposome-mediated siRNA transfection. PSMA7 siRNA #2 and RAN siRNA #4 most efficiently silenced PSMA expression (70% reduction of mRNA) and RAN expression (70% reduction of mRNA), respectively (Figure 3E). We also confirmed that PSMA7 siRNA #2 and RAN siRNA #4 strongly increased caspase-3/7 activity in HT-29 cells (data not shown). PSMA7 siRNA #2 and RAN siRNA #4 were used in further in vivo experiments.
Thus, downregulation of PSMA7 and RAN expression by siRNA induces apoptosis in colon cancer cells, results that suggest that PSMA7 and RAN might act as a suppressor of apoptosis in colorectal cancer.
Overexpression of PSMA and RAN in colorectal cancer cell lines and tumor tissues
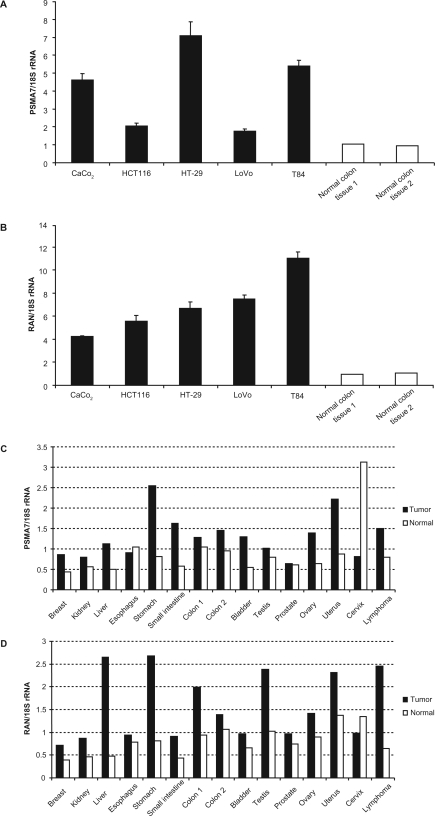
To investigate the role of PSMA and RAN in colorectal cancer, we analyzed the mRNA expression levels of PSMA7 and RAN by real-time RT-PCR. PSMA7 and RAN mRNA were significantly highly expressed in colorectal cancer cell lines Caco-2 (human colorectal adenocarcinoma), HCT116 (human colorectal carcinoma), HT-29 (human colorectal adenocarcinoma), LoVo (human colorectal adenocarcinoma, derived from supraclavicular lymph node metastatic site) and T84 (human colorectal carcinoma, derived from lung metastatic site), compared with normal colon tissues (Figures 4A, 4B). mRNA expression levels of PSMA7 and RAN in human tumoral colon tissues were higher than those of normal adjacent colon tissues (Figures 4C, 4D). These results indicate that PSMA7 and RAN strongly correlated with colorectal tumorigenesis and malignancy.
Figure 4.
PSMA7 and RAN expression in colorectal cancer cell lines and tumor tissues. PSMA7 mRNA and RAN mRNA expression were analyzed by real-time RT-PCR. A) PSMA7 mRNA expression in colorectal cancer cell lines. n = 3 per group. Values are mean ± SD. B) RAN mRNA expression in colorectal cancer cell lines. n = 3 per group. Values are mean ± SD. C) PSMA7 mRNA expression in various tumor tissues (▪, n = 1) and their normal adjacent tissues (□, n = 1). D) RAN mRNA expression in various tumor tissues (▪, n = 1) and their normal adjacent tissues (□, n = 1). mRNA expression was normalized to 18S rRNA. Mean of normal colon tissues is set to 1.
Abbreviations: PSMA7, proteasome subunit, α-type, 7; RAN, ras-related nuclear protein; RT-PCR, reverse transcriptase-polymerase chain reaction; SD, standard deviation.
Furthermore, PSMA7 was overexpressed in other tumor tissues, especially uterus, stomach, and small intestine tumor tissues (Figure 4C). Similarly, RAN was overexpressed in other tumor tissues, in particular uterus, liver, testis, and stomach tumors and lymphoma tissues (Figure 4D). Thus, PSMA7 and RAN were overexpressed in not only colon tumor tissues but also in other tumor tissues, and PSMA7 and RAN might be involved in the development of multiple cancers.
Correlation of PSMA7 and RAN expression with liver and lung metastases in human colorectal cancer
We next investigated the levels of PSMA7 and RAN expression in human colorectal cancers. The results are summarized in Table 3. PSMA7 showed a borderline significance (P = 0.076) for correlation with liver metastasis in colorectal cancer patients. In contrast, no significant correlation between the PSMA7 expression and lung metastasis was found in patients (P = 0.534). Decreased expression of RAN showed a significance (P = 0.023) for correlation with lung metastasis; however, there is no correlation with liver metastasis (P = 0.911). However, to know whether or not the levels of PSMA7 and RAN expression showed correlation with any other clinicopathological features such as depth of invasion, tumor size, lymphatic invasion, or the presence of lymph node metastasis, further analysis is needed.
Table 3.
Correlation of PSMA7 and RAN expression with liver and lung metastases in human colorectal cancer
| No of subjects (n = 191) |
PSMA7 |
RAN |
|||
|---|---|---|---|---|---|
| expression | P-value | expression | P-value | ||
| Liver metastasis | |||||
| Positive | 41 | 1.108 ± 0.116 | 0.0762 | 0.597 ± 0.052 | 0.9112 |
| Negative | 150 | 0.884 ± 0.043 | 0.605 ± 0.031 | ||
| Lung metastasis | |||||
| Positive | 29 | 0.870 ± 0.112 | 0.5336 | 0.460 ± 0.064 | 0.0233 |
| Negative | 162 | 0.943 ± 0.045 | 0.629 ± 0.029 | ||
Note: Values are mean ± standard error.
PSMA7 siRNA and RAN siRNA induce apoptosis in vivo
To extend our in vitro findings and to determine whether PSMA7 and RAN could be effective therapeutic targets for colorectal cancer, we examined the effect of PSMA7 siRNA and RAN siRNA on an animal model of colon tumors by subcutaneously implanting HT-29 cells into mice. We injected the PSMA7 siRNA #2, RAN siRNA #4, or nontargeting control siRNA (1 nmol per tumor) into tumors that had reached 5–6 mm in diameter seven days after inoculation of HT-29 cells. The mRNA levels of PSMA7 and RAN in the tumors given siRNA were measured. mRNA expression was significantly reduced in mouse tumors 24 hours after treatment with PSMA7 siRNA and RAN siRNA, 40% and 30% reduction relative to nontargeting control siRNA, respectively (P < 0.05, Figure 5A).
Figure 5.
Apoptosis induction by PSMA7 siRNA and RAN siRNA treatment in vivo. A) Expression of PSMA7 mRNA and RAN mRNA in HT-29 tumors treated with siRNA. (n = 4 per group, *P < 0.05). B) TUNEL staining of HT-29 tumor tissues treated with siRNA. Scale bar, 20 μm. C) TUNEL-positive cells were counted and represented (n = 3 per group, *P < 0.05). As a control, nontargeting siRNA was used. Values are mean ± SD.
Abbreviations: PSMA7, proteasome subunit, α-type, 7; RAN, ras-related nuclear protein; SD, standard deviation; siRNA, small interfering RNAs.
HT-29 tumors treated with PSMA7 siRNA or RAN siRNA were investigated for apoptotic activity. Three days after siRNA treatment, we performed TUNEL staining which detects apoptotic DNA breaks in situ. TUNEL staining of tumor tissue treated with PSMA7 siRNA and RAN siRNA revealed a significant number of apoptotic cells relative to the number in the nontargeting control siRNA treated tumors (P < 0.05, Figures 5B, 5C). These results indicate that PSMA7 siRNA and RAN siRNA induce apoptotic cell death in vivo.
Discussion
Molecular targeted therapy is one of the most promising approaches in cancer treatment. For cancer researchers it is now possible to understand the molecular mechanisms of the development and progression in many types of cancer. It is expected that many potential new molecular targets will be discovered for cancer therapy. To identify molecular targets for colorectal cancer therapy, we performed a sophisticated strategy using RNAi-based reverse genetics in vitro and in vivo. RNAi is a cellular mechanism for silencing of gene expression. Following the demonstration of RNAi mediated by siRNA in mammalian cells in 2001,22 RNAi has provided new powerful tools for biological research and drug discovery. Additionally, therapeutics based on RNAi offers a new class of pharmaceutical drugs. The inhibition of gene expression through RNAi is applicable to all classes of molecular targets, including the “undruggable targets” of traditional pharmaceutical drugs such as small molecule and protein.
First, we carried out a gene expression profiling of subjects with colorectal tumors and selected 97 genes whose expression was elevated in human colorectal cancer tissues as a screening source of molecular targets. This starting from human disease samples has a great advantage, because cultured cells and disease models cannot faithfully reproduce the pathology of cancer. We then analyzed the function of genes using RNAi in vitro and in vivo. We performed a functional screening of genes by a reverse transfection-based cell transfection array to obtain an unbiased evaluation on the efficacy of a specific siRNA related to apoptosis induction in human colorectal cancer. An unbiased evaluation is important in the discovery of a new and unique target that brings a novel concept of targeted therapy. Our results showed that the siRNA designed for PSMA7 and RAN significantly promoted apoptosis of HT-29 human colon cancer cells. In our next step, we demonstrated in vivo proof-of-concept by the administration of siRNA to a colon tumor model. The in vivo delivery of PSMA7 siRNA and RAN siRNA markedly induced apoptosis in HT-29 tumors in mice. Finally, we identified PSMA7 and RAN as new molecular targets for an RNAi-based colorectal cancer therapy.
PSMA7 (also known as XAPC7) is a proteasome subunit.23 The 26S proteasome consist of a 20S proteasome core and two 19S regulatory subunits.24 The 20S proteasome core is a barrel-shaped structure arranged in four stacked rings. Of these four rings, two end rings are composed of seven α-subunits, and two central rings are composed of seven β-subunits. PSMA7 is one of the seven proteasome α-subunits of 20S proteasome. PSMA7 interacts specifically with hepatitis B virus X protein (HBX), which is important in the life cycle of the hepatitis B virus (HBV).25 In addition, PSMA7 has a principal role in regulating activity of the hepatitis C virus (HCV) internal ribosome entry site (IRES), a function essential for HCV replication.26
The ubiquitin–proteasome pathway is the main extralysosomal system involved in intracellular proteolysis and is critical for the proliferation and survival of all cells.24,27 In particular cancerous cells, the ubiquitin-proteasome pathway plays an integral role in the mechanisms underlying carcinogenesis and metastasis, including cell cycle regulation, apoptosis, and angiogenesis.24,27 The cancer cells exploit the proteasome for their own proliferation through the cell cycle by degrading of cell cycle regulatory proteins. Furthermore, the proteasome regulates apoptotic activity via effects on the pro-oncogenic nuclear factor (NF)-κB pathway, which is a transcriptional activator and activated in many types of tumor.28 Under normal conditions, NF-κB is bound to its inhibitor IκB (IκB) and in an inactive state. The proteasome degrades IκB in response to cellular stresses, and then NF-κB activates transcription of genes for growth factors and apoptosis inhibitors.24,29,30 Therefore, proteasome is an attractive target for cancer therapy. A dipeptidyl boronic acid proteasome inhibitor, bortezomib, suppresses degradation of IκB by proteasome and blocks NF-kB signaling with resultant apoptosis.24,27,29 The efficacy of bortezomib is investigated in various types of malignancies including advanced colorectal cancer.24,31,32 However, the ubiquitin-proteasome pathway is essential for maintenance of cell function in all cells, and the proteasome inhibitor may attack normal cells and cause severe side effects in patients.33,34 A proteasome inhibitor specific for cancer cells should provide a better treatment with minimal side effects. In this study, among proteasome subunits genes, PSMA7 is the only gene listed for genes whose expression is elevated in human colorectal cancer tissues. RNAi-mediated specific inhibition of PSMA7 may be a smart colorectal cancer therapy. Although it is impossible at the moment to judge whether PSMA7 silencing inhibited degradation of IκB by proteasome and blocks NF-kB signaling, in fact, our results indicated that PSMA7 siRNA caused apoptosis in HT-29 colon cells and HT-29 tumor tissues. PSMA7 was overexpressed in not only colon tumor tissues but also in other tumor tissues, which suggests that PSMA7 might be a potential target against various types of cancer.
Moreover, PSMA7 is interesting because of the correlation between its expression and the liver metastasis of colorectal cancer. Our study showed that expression of PSMA7 mRNA was elevated in colorectal tumor tissues from subjects with liver metastasis, although no statistical significance was found. Another research group also reported that overexpression of PSMA7 protein associates with liver metastasis in colorectal cancer.35 PSMA7 may be a predictive marker and a molecular target for liver metastasis from colorectal cancer. Further investigations are needed to confirm the relationship between PSMA7 and colorectal cancer liver metastasis by gene expression profiling on a validation set of colorectal cancer subjects and analysis of PSMA7 expression on a colorectal cancer liver metastatic site at both the mRNA and protein levels.
RAN protein (Ran) is a small GTPase belonging to the Ras superfamily. Ran is essential for the translocation of RNA and proteins through the nuclear pore complex.36,37 GTPase Ran regulates numerous cellular processes by switching between a GTP-bound and GDP-bound form.38 Ran is also critical for the regulation of the cell cycle through mitotic spindle assembly and post-mitotic nuclear envelope assembly.38,39
It is reported that Ran is a suppressor of Bcl-2-associated X protein (Bax), a pro-apoptotic member of the Bcl-2 family of proteins, and that it inhibits apoptosis induced by the anticancer drug paclitaxel.40 Furthermore, it is indicated that silencing of Ran in various tumor cell types causes aberrant mitotic spindle formation, mitochondrial dysfunction, and apoptosis.41,42 Ran is abundantly expressed in most cancer cell lines and cancer tissues.41 This suggests that Ran is associated with malignant transformation and/or the enhanced proliferation of cancer cells. There is a current finding that most tumor cells, but not normal tissues, become dependent on Ran signaling for cell mitosis.41 Targeting the Ran signaling pathway may provide a selective anticancer strategy.
This study showed that RAN mRNA was elevated in colon cancer cell lines and tissues, and in some types of tumor tissue. RAN silencing using RAN siRNA induced apoptosis in HT-29 colon cancer cells and HT-29 tumor tissues. RAN siRNA can specifically inhibit Ran which is a main molecule of RAN signaling, and may be a selective inhibitor against colon cancer.
We identified PSMA7 and RAN as new molecular targets for colorectal cancer therapy using RNAi-based screening in vitro and in vivo. In a further study, we are investigating the antitumor activity of PSMA7 siRNA and RAN siRNA in a colon cancer model and a colon cancer liver metastasis model. Induction of apoptosis is one of the main targets for cancer therapy. The inhibitors of PSMA7 and RAN may provide unique anticancer strategies based on novel mechanisms of action. Since the inhibition of gene expression through RNAi is highly specific and applicable to “undruggable targets”, RNAi-based therapeutics using PSMA7 siRNA and RAN siRNA is a particularly promising approach for cancer treatment.
Acknowledgments
We thank Ms Ayako Inoue for her excellent technical assistances. This work was supported in part by a Grantin- Aid for the Third-Term Comprehensive 10-Year Strategy for Cancer Control of Japan; a Grant-in-Aid for Scientific Research on Priority Areas Cancer from the Ministry of Education, Culture, Sports, Science and Technology, and the Program for Promotion of Fundamental Studies in Health Sciences of the National Institute of Biomedical Innovation (NiBio) of Japan. The authors report no conflicts of interest in this work.
References
- 1.Garcia M, Jemal A, Ward EM, et al. Global Cancer Facts and Figures 2007. Atlanta, GA: American Cancer Society; 2007. pp. 12–14. [Google Scholar]
- 2.National Cancer Center Tokyo, Japan. Cancer statistics in Japan (2008). Available from http://ganjoho.ncc.go.jp/public/statistics/backnumber/2008_en.html Accessed July 2, 2009.
- 3.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 4.Baranda J, Williamson S. The new paradigm in the treatment of colorectal cancer: are we hitting the right target? Expert Opin Investig Drugs. 2007;16(3):311–324. doi: 10.1517/13543784.16.3.311. [DOI] [PubMed] [Google Scholar]
- 5.Köhne CH, Lenz HJ. Chemotherapy with targeted agents for the treatment of metastatic colorectal cancer. Oncologist. 2009;14(5):478–488. doi: 10.1634/theoncologist.2008-0202. [DOI] [PubMed] [Google Scholar]
- 6.Ferrara N, Kerbel RS. Angiogenesis as a therapeutic target. Nature. 2005;438(7070):967–974. doi: 10.1038/nature04483. [DOI] [PubMed] [Google Scholar]
- 7.Kim KJ, Li B, Winer J, et al. Inhibition of vascular endothelial growth factor-induced angiogenesis suppresses tumour growth in vivo. Nature. 1993;362(6423):841–844. doi: 10.1038/362841a0. [DOI] [PubMed] [Google Scholar]
- 8.Hurwitz H, Fehrenbacher L, Novotny W, et al. Bevacizumab plus irinotecan, fluorouracil, and leucovorin for metastatic colorectal cancer. N Engl J Med. 2004;350(23):2335–2342. doi: 10.1056/NEJMoa032691. [DOI] [PubMed] [Google Scholar]
- 9.Wood JM, Bold G, Buchdunger E, et al. PTK787/ZK 222584, a novel and potent inhibitor of vascular endothelial growth factor receptor tyrosine kinases, impairs vascular endothelial growth factor-induced responses and tumor growth after oral administration. Cancer Res. 2000;60(8):2178–2189. [PubMed] [Google Scholar]
- 10.Drevs J, Hofmann I, Hugenschmidt H, et al. Effects of PTK787/ZK 222584, a specific inhibitor of vascular endothelial growth factor receptor tyrosine kinases, on primary tumor, metastasis, vessel density, and blood flow in a murine renal cell carcinoma model. Cancer Res. 2000;60(17):4819–4824. [PubMed] [Google Scholar]
- 11.Mellinghoff I. Why do cancer cells become “addicted” to oncogenic epidermal growth factor receptor? PLoS Med. 2007;4(10):1620–1622. doi: 10.1371/journal.pmed.0040321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henson ES, Gibson SB. Surviving cell death through epidermal growth factor (EGF) signal transduction pathways: implications for cancer therapy. Cell Signal. 2006;18(12):2089–2097. doi: 10.1016/j.cellsig.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 13.Cunningham D, Humblet Y, Siena S, et al. Cetuximab monotherapy and cetuximab plus irinotecan in irinotecan-refractory metastatic colorectal cancer. N Engl J Med. 2004;351(4):337–345. doi: 10.1056/NEJMoa033025. [DOI] [PubMed] [Google Scholar]
- 14.Saltz L, Easley C, Kirkpatrick P. Panitumumab. Nat Rev Drug Discov. 2006;5(12):987–988. doi: 10.1038/nrd2204. [DOI] [PubMed] [Google Scholar]
- 15.Taylor K, Micha D, Ranson M, Dive C. Recent advances in targeting regulators of apoptosis in cancer cells for therapeutic gain. Expert Opin Investig Drugs. 2006;15(6):669–690. doi: 10.1517/13543784.15.6.669. [DOI] [PubMed] [Google Scholar]
- 16.Frenzel A, Grespi F, Chmelewskij W, Villunger A. Bcl2 family proteins in carcinogenesis and the treatment of cancer. Apoptosis. 2009;14(4):584–596. doi: 10.1007/s10495-008-0300-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klasa RJ, Gillum AM, Klem RE, Frankel SR. Oblimersen Bcl-2 antisense: facilitating apoptosis in anticancer treatment. Antisense Nucleic Acid Drug Dev. 2002;12:193–213. doi: 10.1089/108729002760220798. [DOI] [PubMed] [Google Scholar]
- 18.Newsom-Davis T, Prieske S, Walczak H. Is TRAIL the holy grail of cancer therapy? Apoptosis. 2009;14(4):607–623. doi: 10.1007/s10495-009-0321-2. [DOI] [PubMed] [Google Scholar]
- 19.Marini P, Denzinger S, Schiller D, et al. Combined treatment of colorectal tumours with agonistic TRAIL receptor antibodies HGS-ETR1 and HGS-ETR2 and radiotherapy: enhanced effects in vitro and dosedependent growth delay in vivo. Oncogene. 2006;25(37):5145–5154. doi: 10.1038/sj.onc.1209516. [DOI] [PubMed] [Google Scholar]
- 20.Honma K, Iwao-Koizumi K, Takeshita F, et al. RPN2 gene confers docetaxel resistance in breast cancer. Nat Med. 2008;14(9):939–948. doi: 10.1038/nm.1858. [DOI] [PubMed] [Google Scholar]
- 21.Komori T, Takemasa I, Yamasaki M, et al. Gene expression of colorectal cancer: preoperative genetic diagnosis using endoscopic biopsies. Int J Oncol. 2008;32(2):367–375. [PubMed] [Google Scholar]
- 22.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411(6836):494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 23.Gerards WL, de Jong WW, Boelens W, Bloemendal H. Structure and assembly of the 20S proteasome. Cell Mol Life Sci. 1998;54(3):253–262. doi: 10.1007/s000180050147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adams J. The development of proteasome inhibitors as anticancer drugs. Cancer Cell. 2004;5(5):417–421. doi: 10.1016/s1535-6108(04)00120-5. [DOI] [PubMed] [Google Scholar]
- 25.Huang J, Kwong J, Sun EC, Liang TJ. Proteasome complex as a potential cellular target of hepatitis B virus X protein. J Virol. 1996;70(8):5582–5591. doi: 10.1128/jvi.70.8.5582-5591.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krüger M, Beger C, Welch PJ, Barber JR, Manns MP, Wong-Staal F. Involvement of proteasome alpha-subunit PSMA7 in hepatitis C virus internal ribosome entry site-mediated translation. Mol Cell Biol. 2001;21(24):8357–8364. doi: 10.1128/MCB.21.24.8357-8364.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoeller D, Dikic I. Targeting the ubiquitin system in cancer therapy. Nature. 2009;458(7237):438–444. doi: 10.1038/nature07960. [DOI] [PubMed] [Google Scholar]
- 28.Karin M. Nuclear factor-B in cancer development and progression. Nature. 2006;441(7092):431–436. doi: 10.1038/nature04870. [DOI] [PubMed] [Google Scholar]
- 29.Richardson PG, Mitsiades C, Hideshima T, Anderson KC. Proteasome inhibition in the treatment of cancer. Cell Cycle. 2005;4(2):290–296. [PubMed] [Google Scholar]
- 30.Palombella VJ, Rando OJ, Goldberg AL, Maniatis T. The ubiquitinproteasome pathway is required for processing the NF-kappa B1 precursor protein and the activation of NF-kappa B. Cell. 1994;78(5):773–785. doi: 10.1016/s0092-8674(94)90482-0. [DOI] [PubMed] [Google Scholar]
- 31.Cusack JC, Jr, Liu R, Houston M, et al. Enhanced chemosensitivity to CPT-11 with proteasome inhibitor PS-341: implications for systemic nuclear factor-kappaB inhibition. Cancer Res. 2001;61(9):3535–3540. [PubMed] [Google Scholar]
- 32.Pitts TM, Morrow M, Kaufman SA, Tentler JJ, Eckhardt SG. Vorinostat and bortezomib exert synergistic antiproliferative and proapoptotic effects in colon cancer cell models. Mol Cancer Ther. 2009;8(2):342–349. doi: 10.1158/1535-7163.MCT-08-0534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Voortman J, Giaccone G. Severe reversible cardiac failure after bortezomib treatment combined with chemotherapy in a non-small cell lung cancer patient: a case report. BMC Cancer. 2006;6:129. doi: 10.1186/1471-2407-6-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perfetti V, Palladini G, Brunetti L, et al. Bortezomib-induced paralytic ileus is a potential gastrointestinal side effect of this first-in-class anticancer proteasome inhibitor. Eur J Gastroenterol Hepatol. 2007;19(7):599–601. doi: 10.1097/MEG.0b013e32811ebffe. [DOI] [PubMed] [Google Scholar]
- 35.Hu XT, Chen W, Wang D, et al. The proteasome subunit PSMA7 located on the 20q13 amplicon is overexpressed and associated with liver metastasis in colorectal cancer. Oncol Rep. 2008;19(2):441–446. [PubMed] [Google Scholar]
- 36.Kuersten S, Ohno M, Mattaj IW. Nucleocytoplasmic transport: Ran, beta and beyond. Trends Cell Biol. 2001;11(12):497–503. doi: 10.1016/s0962-8924(01)02144-4. [DOI] [PubMed] [Google Scholar]
- 37.Pemberton LF, Paschal BM. Mechanisms of receptor-mediated nuclear import and nuclear export. Traffic. 2005;6(3):187–198. doi: 10.1111/j.1600-0854.2005.00270.x. [DOI] [PubMed] [Google Scholar]
- 38.Joseph J. Ran at a glance. J Cell Sci. 2006;119(Pt 17):3481–3484. doi: 10.1242/jcs.03071. [DOI] [PubMed] [Google Scholar]
- 39.Hetzer M, Gruss OJ, Mattaj IW. The Ran GTPase as a marker of chromosome position in spindle formation and nuclear envelope assembly. Nat Cell Biol. 2002;4(7):E177–E184. doi: 10.1038/ncb0702-e177. [DOI] [PubMed] [Google Scholar]
- 40.Woo IS, Jang HS, Eun SY, et al. Ran suppresses paclitaxel-induced apoptosis in human glioblastoma cells. Apoptosis. 2008;13(10):1223–1231. doi: 10.1007/s10495-008-0247-0. [DOI] [PubMed] [Google Scholar]
- 41.Xia F, Lee CW, Altieri DC. Tumor cell dependence on Ran-GTP-directed mitosis. Cancer Res. 2008;68(6):1826–1833. doi: 10.1158/0008-5472.CAN-07-5279. [DOI] [PubMed] [Google Scholar]
- 42.Morgan-Lappe SE, Tucker LA, Huang X, et al. Identification of Ras-related nuclear protein, targeting protein for xenopus kinesin-like protein 2, and stearoyl-CoA desaturase 1 as promising cancer targets from an RNAi-based screen. Cancer Res. 2007;67(9):4390–4398. doi: 10.1158/0008-5472.CAN-06-4132. [DOI] [PubMed] [Google Scholar]