Abstract
The genetic pathways that partition the developing nervous system into functional systems are largely unknown. The engrailed (En) homeobox transcription factors are candidate regulators of this process in the dorsal midbrain (tectum) and anterior hindbrain (cerebellum). En1 mutants lack most of the tectum and cerebellum and die at birth, whereas En2 mutants are viable with a smaller cerebellum and foliation defects. Our previous studies indicated that the difference in phenotypes is due to the earlier expression of En1 as compared with En2, rather than differences in protein function, since knock-in mice expressing En2 in place of En1 have a normal brain. Here, we uncovered a wider spectrum of functions for the En genes by generating a series of En mutant mice. First, using a conditional allele we demonstrate that En1 is required for cerebellum development only before embryonic day 9, but plays a sustained role in forming the tectum. Second, by removing the endogenous En2 gene in the background of En1 knock-in alleles, we show that Drosophila en is not sufficient to sustain midbrain and cerebellum development in the absence of En2, whereas En2 is more potent than En1 in cerebellum development. Third, based on a differential sensitivity to the dose of En1/2, our studies reveal a genetic subdivision of the tectum into its two functional systems and the medial cerebellum into four regions that have distinct circuitry and molecular coding. Our study suggests that an ‘engrailed code’ is integral to partitioning the tectum and cerebellum into functional domains.
Keywords: Cerebellum, Foliation, Tectum, Patterning, En1, En2, Mouse
Introduction
The genetic regulation of patterning processes that regulate the size, cellular differentiation and morphology of regions of the developing nervous system is fundamental to establishing functional circuits that control different behaviors, emotions and basic bodily functions. The embryonic brain region that gives rise to the midbrain [mesencephalon (mes)] and anterior hindbrain [rhombomere (r1)] is an ideal model system for studying these genetic pathways in vertebrates. Anterior-posterior (A-P) patterning of the midbrain (Mb) and anterior hindbrain is orchestrated by an organizing center in the isthmus located between the mes and r1. Fgf8 is the key isthmic organizer molecule that acts between embryonic day (E) 8.5 and 13 to regulate the expression of genes that direct Mb and r1 development (Wurst and Bally-Cuif, 2001; Zervas et al., 2005). Expression of the engrailed transcription factors (En1 and En2) before E13 is regulated by Fgf8, and En1/2 are crucial for mes/r1 development. It has been challenging to determine the full spectrum of functions of each En gene in mouse because there is an early loss of the mes/r1 in En1 mutants, and the two genes have overlapping functions. It is also unclear whether the two En proteins have equivalent functions in brain development.
Following specification of the mes/r1 region during neural tube closure, the mouse tectum and cerebellum (Cb) develop from the dorsal mes and r1, respectively (Zervas et al., 2005). The tectum of the Mb forms as a layered structure that is divided morphologically and functionally into the anterior superior colliculus and posterior inferior colliculus that process visual and auditory information, respectively. Although expansion of the tectum along the A-P axis is tightly linked to the level of isthmic organizer signaling, the molecular basis of differential allocation of the inferior and superior colliculi is not understood. The Cb is the center for motor control. Differentiated cells of the mouse Cb begin to be generated at E10.5 and form a multi-laminar structure consisting of the deep nuclei surrounded by a dense layer of granule cells, a monolayer of Purkinje cells and an outer molecular layer. The granule cell precursors form a proliferative external granule layer at E13.5 and then migrate past the Purkinje cell layer to form the inner granule layer (IGL) from birth until postnatal day (P) 14. Beginning at E17.5, fissures form in a stereotyped manner and generate a highly foliated Cb. In terms of how early A-P patterning could influence the final structure of the Cb, it is important to note that a morphogenetic rotation of dorsal r1 transforms the A-P axis of r1 into the medial-lateral (M-L) axis of the Cb primordium by E12.5 (Sgaier et al., 2005) (see Fig. 8). Globally, the adult Cb is subdivided into a medial vermis and two lateral hemispheres, with the vermis divided along the A-P axis by 8-10 folia in different inbred mouse strains (referred to as I-X) and the hemispheres divided by 6 folia (Larsell, 1952). Preservation of the general pattern of folia across mammals suggests that there is an evolutionarily conserved genetic program that patterns folia of the Cb (Altman and Bayer, 1997; Herrup and Kuemerle, 1997).
Fig. 8. An ‘En code’ can account for genetic partitioning of the midbrain and cerebellum vermis into functionally related domains.
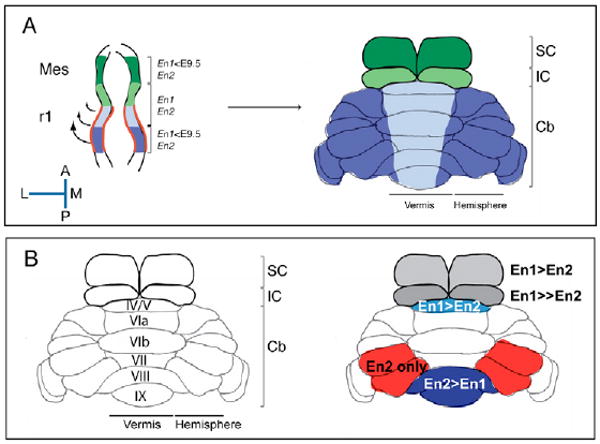
(A) Schematic illustrating the domains (color coded) within the mouse early neural tube (left) along the A-P axis that express either or both of the En genes, and the regions of the adult cerebellum and midbrain (right) that they give rise to owing to a rotation of the neural tube. The red-outlined region delineates rhombomere 1. (B) Schematic illustrating the normal gene dosage requirement for En1 and En2 in sustaining development of distinct domains of the tectum and Cb. Note that based on a sensitive knock-in assay, En2 protein appears to be more active throughout the vermis than En1. Thus, the gene dosage effects are likely to reflect differences in En1 and En2 gene expression. Cb, cerebellum; Mes, mesencephalon; SC, superior colliculus; IC, inferior colliculus; A, anterior; P, posterior; M, medial; L, lateral.
The mouse En1 and En2 genes provide a unique tool for gaining access to the genetic regulation of Cb and tectum patterning. The dynamic expression patterns of the En genes (see Fig. 1) and their mutant phenotypes reflect each successive stage of Cb and tectum development (Joyner, 1996). En1 is first expressed in the mes/r1 at E8.5, ∼12 hours before En2, and is later expressed in the absence of En2 in a number of other tissues. En1-null mutant mice die at birth and have an almost complete deletion of the Mb and Cb owing to tissue loss by E9.5 (Wurst et al., 1994), which is caused, at least in part, by cell death (Chi et al., 2003). Thus, En1 is required for the initial establishment of the mes/r1 region. By contrast, En2-null mutants have a mild phenotype – they are viable and have defects limited to growth of the Cb and patterning of particular folia (Joyner et al., 1991; Millen et al., 1994). An overlap in En gene function has been demonstrated by the complete absence of the tectum and Cb in En1;En2 double mutants (Liu and Joyner, 2001; Simon et al., 2004), and a rescue of the En1 mutant brain phenotype when En1 is replaced with En2 using gene targeting (Hanks et al., 1995). Surprisingly, we found that Drosophila en also can rescue the En1 mutant brain defects in knock-in mouse mutants, although en cannot rescue other defects (Hanks et al., 1998). An important question is whether En1 has any later roles in tectum and Cb patterning, as has been suggested by the Cb phenotype of En1-null mutants that survive on a C57BL/6 genetic background (Bilovocky et al., 2003), and the degree to which such functions overlap with En2.
Fig. 1. Dynamic expression of En1lacZ and En2tau-lacZ in the mes/r1 of mouse embryos.
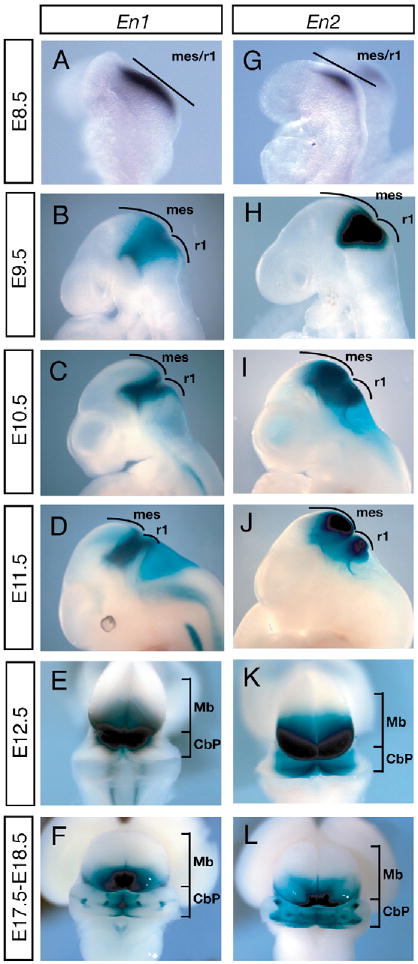
En1 (A) and En2 (G) mRNA expression detected by whole-mount RNA in situ hybridization. (B-F) En1-lacZ and (H-L) En2-tau-lacZ expression at the indicated stages detected by β-gal analysis. En1 is induced earlier and becomes more restricted than En2 by E10.5. Staining in the fourth ventricle in I and D is a result of probe trapping. CbP, cerebellar primordium; mes, mesencephalon; Mb, midbrain; r1, rhombomere 1.
In order to study the temporal requirement for En1 in mes/r1 development, we generated a conditional mutant allele of En1. We find that if En1 is removed at ∼E9, only the posterior tectum is depleted, and two copies of En2 are required to sustain Cb development in these conditional En1 mutants. We next compared the function of Drosophila and mouse En proteins in the mouse brain using a sensitive genetic assay. We provide evidence that En2 is more potent at supporting Cb development than En1, and demonstrate that Drosophila En cannot rescue the En1 mutant brain defects in the absence of endogenous En2. Curiously, our analysis of knock-in mutants and En1/2 double-null mutants uncovered that both genes are preferentially required in particular functional domains of the tectum and cerebellum. We propose an ‘En code’ that divides the tectum and Cb into functional systems based on the dose of En required for the development of each domain.
Materials and Methods
Generation of En1flox knock-in mice and conditional ablation of En1 in r1
The En1flox targeting construct was produced by subcloning a 6.0 kb 5′ BamHI En1 sequence into the BamHI site of pPNTfrt-Neo-frt-loxP to generate pPNTfrt-Neo-frt-loxP+En1-5′ arm. A 2.7 kb BamHI-XbaI fragment of En1 including most of exon 2 was subcloned into the XbaI site of the pGEM-11ZF vector to generate pGEM-11ZF+En1-3′ arm. A loxP sequence was inserted into the KpnI site within the 3′ UTR of En1 and the En1-loxP-3′ arm was released by XbaI and SalI digestion and subcloned into the HindIII site of pPNTfrt-Neo-frt-loxP+En1-5′ arm to generate the final targeting vector.
The En1flox targeting construct was linearized with SalI and electroporated into W4 embryonic stem (ES) cells (Auerbach et al., 2000) as described previously (Matise et al., 2000). Clones were screened by Southern blot analysis using 5′ external and 3′ internal probes to identify targeted clones (see Fig. S1 in the supplementary material). One positive clone was obtained and injected into C57BL/6 blastocysts to generate ES cell chimeric mice (Papaioannou and Johnson, 2000). Chimeric mice were mated with Black Swiss mice to generate En1flox-neo/+ mice. The neo cassette was removed by mating En1flox-neo/+ mice with hACTB-Flpe mice (Rodriguez et al., 2000), which expresses Flpe broadly under the control of the human β-actin promoter. The wild-type (337 bp) and En1flox (380 bp) alleles were detected by PCR with the following primers: En1flox1A, 5′-GCCAAACTGCTTACGACCG-3′; En1flox1B, 5′-TGGGTGGGTAGAGAAGAGGC-3′.
Mes/r1-specific En1 conditional mutant mice (En1flox/Cre) were generated by crossing En1flox/+ mice with En1Cre/+ mice. En1Cre/Cre and En1flox/Cre mice were generated within the same litter by crossing En1flox/Cre mice with En1Cre/+ mice. En1flox/+;En2−/+ were bred to En2−/+ mice to generate En1flox/+;En2−/− and En1Cre/+;En2−/+ were bred to En1flox/+;En2−/− mice to generate En1flox/Cre;En2−/+ and En1flox/Cre;En2−/− mice. To eliminate the possibility that the observed mild phenotype in En1flox/Cre mice was due to a genetic background effect, we generated En1Cre/Cre-null mutants in the same litters as En1flox/Cre mice by mating En1flox/Cre mice to En1Cre/+ mice. We screened a total of 39 mice from seven separate litters in which the expected frequency of all genotypes is 25%. Whereas nine En1flox/Cre mice were found, only one En1Cre/Cre mouse that survived to adulthood was observed (2.6%).
Breeding and genotyping of En1/En2 double mutants
En2ntd (Millen et al., 1994), En2tau-lacZ (Sgaier et al., 2005), En1lki (Hanks et al., 1995), En1Cre (Kimmel et al., 2000) or En1CreERT1 (Sgaier et al., 2005) mutant alleles were used as En2- or En1-null alleles on an outbred Swiss Webster (SW) genetic background. Various combinations of En1- and En2-null alleles were interbred to generate the required genotypes. En12ki (Hanks et al., 1995) and En1Denki (Hanks et al., 1998) mice were bred with En2ntd and En2tau-lacZ, respectively, on an outbred SW genetic background to generate the required genotypes. For embryonic analysis, noon of the day on which a vaginal plug was detected was designated as E0.5. Genotyping was carried out by PCR. The R26R, En1Cre and En1CreERT1 alleles were detected as previously described (Li et al., 2002; Sgaier et al., 2005; Soriano, 1999). The primers used for genotyping the En2 wild-type and En2ntd alleles were: 1, 5′-TGCTCTTTGACGCTTCGGTG-3′; 2, 5′-CCTTGGATGGAGTGCTCAAAGC-3′; and 3, 5′-TCATGCTGGAGTTCTTCGCC-3′. PCR with primers 1 and 2 detected a 300 bp wild-type allele, whereas primers 1 and 3 detected a 500 bp En2ntd mutant allele. The primers used to genotype En1 wild-type, En12ki and En1Denki alleles were: A, 5′-AGCTGCACCGCACCACCAAC-3′; B, 5′-GCACACAAGAGCGAGGCAGC-3′; C, 5′-CCCTGTGCCTTCGCTGAGG-3′; D, 5′-TGCCTGGCGCCTGTAGGACC-3′; and E, 5′-TTGTAGGGTAATGGGGCTGGG-3′. PCR with primers A and B detected the 232 bp En1 wild-type allele, whereas primers C and D detected the 230 bp En12ki allele, and primers E and C detected the 280 bp En1Denki allele.
Histological analysis, β-gal histochemistry and RNA in situ hybridization
Tissue processing, RNA in situ hybridization and β-galactosidase (β-gal) analysis were performed as described on the Joyner website (http://www.mskcc.org/mskcc/html/75282.cfm) using Fgf8 (Crossley and Martin, 1995), Fgf17 (Xu et al., 1999), Spry1 (courtesy of Gail Martin), Otx2 (Simeone, 1993), Gbx2 (Bouillet, 1995), Wnt1 (Parr, 1993), En1 (Joyner and Martin, 1987) antisense RNA probes.
Fate mapping
En1CreERT1/+;En2−/+;R26R/R26R adult males were bred with 5- to 6-week-old En2−/+ females and tamoxifen (Sigma T-5648) administered at 5 mg per 40 g of body weight via gavage at 18.00 h of E10.5 (Sgaier et al., 2005).
Results
En1 is not required after ∼E9.0 for development of the Cb and superior colliculus
Our finding that En2 can fully rescue the En1 mutant phenotype in En1En2ki knock-in mice (Hanks et al., 1995) indicates that the two En proteins are functionally interchangeable and that the brain defects in En1 and En2 single mutants arise from cells that express only one En gene at critical time points in development. To address the question of whether En2 can compensate for En1 after the mes/r1 is specified, we first delineated the dynamic expression patterns of the En genes using En1lacZ (Hanks et al., 1995; Matise and Joyner, 1997) and En2tau-lacZ (Sgaier et al., 2005) knock-in alleles. En1-lacZ expression was first detected at the two-somite stage broadly spanning the mes/r1 region (Fig. 1A) and then progressively narrowed around the mes/r1 junction (isthmic organizer) (Fig. 1B-D). En2-tau-lacZ expression commenced at the five-somite stage in a subdomain of the En1-positive region (Fig. 1G) and quickly broadened (Fig. 1G-J). Within the bilateral wing-like structure of the E12.5 cerebellar primordium (CbP), En1-lacZ was restricted to the medial-most region (vermis anlage), whereas En2-tau-lacZ was expressed in all but the most-lateral CbP (Fig. 1E,K). Similarly, in the Mb, En2-tau-lacZ expression extended more rostrally than En1-lacZ (into the anlage of the superior colliculus). Furthermore, expression of both En genes formed a mirror image double gradient across the mes/r1 region, with highest levels in the isthmus. Starting at E15.5, En1 and En2 expression changed to a sagittal striped pattern along the M-L axis of the CbP, owing to regional downregulation (Fig. 1F,L) (Millen et al., 1995). Thus, the En genes have dynamic gene expression patterns, but after ∼E9 (including postnatal stages; data not shown) the En1 expression domain appears to be encompassed within the En2 expression domain.
The expression analysis of En1 and En2 suggests that En1 might only be required in the mes/r1 before ∼E9. To determine whether this is the case, we generated a conditional mutant allele of En1 (En1flox) in which the coding sequences in the second exon are flanked with loxP sites (see Fig. S1 in the supplementary material). Germline chimeras were bred to Black Swiss mice to avoid rescue of the En1 mutant phenotype by the C57BL/6 background (Bilovocky et al., 2003). In order to delete En1 at ∼E9.0, we combined the En1flox allele with the null En1Cre knock-in allele (Kimmel et al., 2000). Within the mes/r1 region, Cre function is first detected at the five-somite stage and is active in all mes/r1 cells by 15 somites (E9.0) (Li et al., 2002). In En1flox/Cre mice, the second exon of the En1flox allele thus begins to be deleted shortly after its expression is induced, and En1 is expressed for only approximately 24 hours (Fig. 2A).
Fig. 2. En1 is required prior to ∼E9.0 to pattern the anterior cerebellum and after E9.0 to form the posterior midbrain.
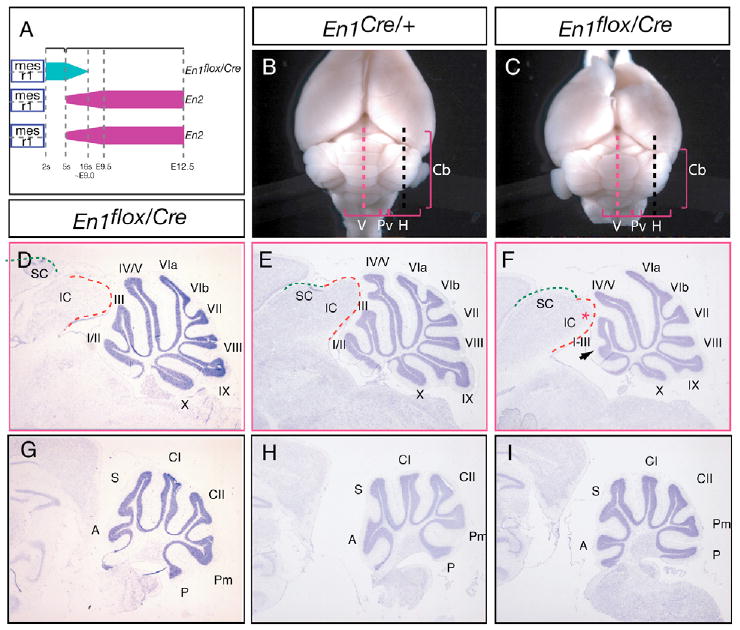
(A) Schematic illustrating the expression profile of En alleles in the mes/r1 region of En1flox/Cre mice. Note that only one functional En1 allele is present because the En1-Cre allele is an En1-null. (B,C) Dorsal view of posterior adult brains of (B) En1Cre/+ and (C) En1flox/Cre mice. (D-I) Cresyl Violet-stained sagittal sections of the brains were taken at the level of (D-F) the vermis and (G-I) hemispheres as indicated in B,C by the red and black dashed lines, respectively. E,H and F,I are sections taken from brains in B and C, respectively. Arrow in F indicates the predominant phenotypes of fusion of vermis folia I-II and III and the red asterisk indicates a slight truncation of the inferior colliculus. In some En1flox/Cre mice, a normally foliated Cb was also seen (D,G). H, hemisphere; V, vermis; Pv, paravermis; Cb, cerebellum; IC and SC are the inferior and superior colliculus, respectively outlined by red and green dashed lines. Vermis and hemisphere folia are indicated by roman numerals (I-X) and letters (A, S, CI, CII, Pm, P), respectively.
Indeed, En1flox/Cre mice were found to be viable and survive to adulthood. En1flox/Cre mutants had a limb phenotype similar to the rare En1 mutants that survive (Loomis et al., 1996) (data not shown). The brains of all but one adult En1flox/Cre mouse analyzed (n=8) appeared grossly normal in whole-mount (Fig. 2B,C). The one mouse that was different had a partial deletion of the Mb and Cb (data not shown). Analysis of sagittal sections of the remaining En1flox/Cre mice revealed that the inferior colliculus (posterior tectum) was partially truncated (in all eight) (Fig. 2E,F and Table 1; compare also with Fig. 3B,I). In addition, five of the seven En1flox/Cre mice had a mild foliation defect in the anterior vermis (medial Cb), and the overall size of the vermis was slightly smaller than normal. The fissure between the anterior-most folia (I/II and III) either failed to form (in two of five) (Fig. 2F), or was shallower than normal (in three of five) in these mutants. Of significance, in two of the seven mutants analyzed, the fissure between folia I/II and III appeared as deep as in wild-type brains (Fig. 2D,E). One likely possibility for the variable rescue in the Cb is that in the two En1flox/Cre mutants that had a normal Cb, En1 was ablated at a slightly later stage. Interestingly, in all eight of the En1flox/Cre mice analyzed, the superior colliculus and the hemispheres (lateral Cb) appeared normal (Fig. 2D-I). Thus, our analysis of the requirement for En1 after ∼E9 demonstrates that two copies of En2 are sufficient to support Cb development, but despite being expressed in a broader domain of the tectum than En1, En2 alone is not able to fully regulate inferior colliculus development.
Table 1. Summary of En1/2 and En1 conditional mutant phenotypes.
| Phenotype | ||||||||
|---|---|---|---|---|---|---|---|---|
| Genotype | Cb subregions | |||||||
| En1 alleles | En2 alleles | SC | IC | Cb size | aV (I-V) | pV (VIII) | pH (CII/PM) | |
| −/− | −/− | X | X | X | ||||
| −/− | −/+ | |||||||
| flox/Cre | −/− | |||||||
| Denki/Denki | −/− | X/√ | X | X | ||||
| flox/Cre | −/+ | √√ | X | X | ||||
| −/− | +/+ | √√/√ | X | (√) | ||||
| −/+ | −/− | √√√ | √/X | √ | √ | √/X | √√* | |
| Denki/+ | −/− | √√√ | √√ | √√ | √√ | X | √√* | |
| (ψ) Denki/Denki | +/− | √√√ | √√ | √√ | √√ | X | √√√ | |
| 2ki/− | −/− | √√√ | √√ | √√ | √√√ | √ | √√ | |
| +/+ | −/− | √√√ | √√√ | √√ | √√√ | √ | √√ | |
| flox/Cre | +/+ | √√√ | √ | √√√ | √√/√√√ | √√√ | √√√ | |
| −/+ | −/+ | √√√ | √√√ | √√√ | √√ | √√ | √√√ | |
| 2ki/2ki | −/− | √√√ | √√√ | √√ | √√√ | √√√ | √√ | |
| 2ki/2ki | −/+ | √√√ | √√√ | √√√ | √√√ | √√√ | √√√ | |
| 2ki/2ki | +/+ | |||||||
| Denki/Denki | +/+ | |||||||
| +/+ | −/+ | |||||||
| −/+ | +/+ | |||||||
| +/+ | +/+ | |||||||
En1/2 alleles: +, wild type; −, null. SC/IC: √√√, normal superior/inferior colliculus; √√, reduced in size; √, little formed; X, not formed. Cb size: √√√, normal; √√, ∼1/3 overall reduction; √, ∼1/2 overall reduction; (√), only the most lateral tissue remains; X, not formed. Anterior vermis (aV): √√√, normal folia I-V; √√, fusion and smaller folia I-III; √, fusion and smaller folia I-V. Posterior vermis (pV): √√√, folium VIII is associated normally with folium VII; √√, folium VIII is positioned between lobule VII and IX; √, folium XIII is smaller and associated with folium IX; X, little or no folium VIII. Posterior hemisphere (pH): √√√, crusII and paramedian folia are normal; √√, crusII and paramedian folia are fused; √√*, one mutant had a partial fissure separating crusII and paramedian. ψ, denotes the mutants that survived into adulthood and that were analyzed.
Fig. 3. Differential requirement of En genes for mouse midbrain and cerebellar development.
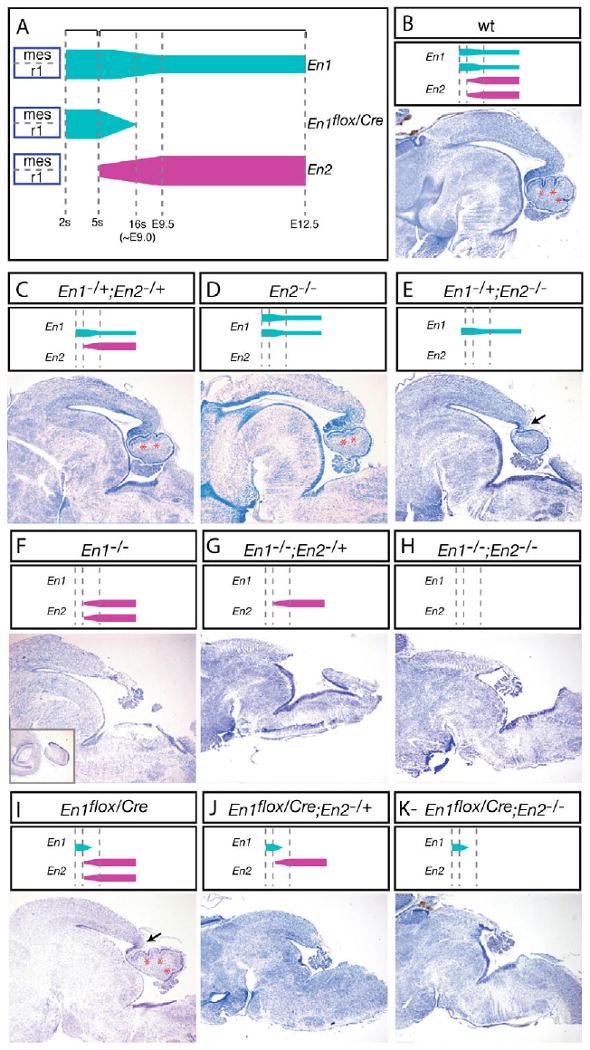
(A) Schematic illustrating the detailed expression profile of each En allele (En1, En2 and En1flox/Cre) within the mes/r1 region. (B-K) Cresyl Violet-stained mid-sagittal sections of E18.5 brains with the corresponding genotypes and all the functional En allele profiles indicated at the top. Asterisks indicate forming fissures. Black arrows indicate truncated inferior colliculus. The inset in F shows the lateral Cb tissue that forms in the En1−/− mutants.
One copy of En2 is not sufficient to support development of the remaining inferior colliculus or the Cb when En1 is deleted by E9
To determine whether one copy of En2 is sufficient to support development of the superior colliculus and Cb when En1 is removed at ∼E9, we removed one copy of En2 on the En1flox/Cre background. Strikingly, En1flox/Cre;En2−/+ mutant mice were not found at weaning (42 mice were analyzed from five litters of a cross between EnCre/+;En2−/+ and Enflox/+;En2−/+ mice). We therefore analyzed the phenotypes of En1flox/Cre embryos lacking one En2 allele at E18.5, and compared them with En1−/− and En1−/−;En2−/− double mutants (Fig. 3). As expected, at E18.5, the vermis of En1flox/Cre;En2+/+ mice was either normal or slightly delayed in forming folia (Fig. 3B,I and Table 1). By contrast, En1flox/Cre;En2−/+ mice (Fig. 3J) displayed a complete deletion of the Cb that was very similar to En1−/−;En2−/+ (Fig. 3G) and En1−/−;En2−/− (Fig. 3H) mutant embryos and more severe than En1−/−;En2+/+ mutants (Fig. 3F) that had some lateral Cb tissue remaining (see also Table 1). However, unlike En1−/−;En2−/+ (Fig. 3G) and En1−/−;En2−/− mutants (Fig. 3H), which have no tectum, in En1flox/Cre;En2−/+ mutant mice (Fig. 3J) some superior colliculus tissue remained, similar to the phenotype of En1−/−;En2+/+ mice (Fig. 3F and Table 1). By contrast, En1flox/Cre;En2−/− mutants had a complete deletion of the Cb and tectum (Fig. 3K and Table 1). In summary, when En1 is expressed until ∼E9.0 (En1flox/Cre), two copies of En2 can support superior colliculus and Cb development and partial development of the inferior colliculus, whereas one copy of En2 is not sufficient to support any development of the inferior colliculus or Cb, and in the absence of all En2 the superior colliculus also does not form.
Particular regions of the Mb and Cb are sensitive to the dose of En1 and En2
Since our analysis of En1 conditional mutants showed that En1 is preferentially required in the inferior colliculus and folia I-III, we further explored the requirement for each mouse En gene in mes/r1 development by analyzing viable En1 and En2 double mutants. Surprisingly, although the Cb of En1−/+ or En2−/+ mice on an outbred background is normal (data not shown), section analysis of En1−/+;En2−/+ double heterozygotes (Fig. 4A,E,C,G) revealed two foliation defects in the vermis. One was a variable but consistent (n=7 of 8) partial fusion of the three anterior-most folia (I-III), similar to the phenotype seen in some En1flox/Cre mutants (Fig. 2F and see Table 1). En1−/+;En2−/+ mice (Fig. 4G) had an additional slight posterior shift in the position of folium VIII that was milder than in En2−/− mutants (Fig. 4F), in which folium VIII is associated with folium IX instead of with VI/VII. Furthermore, there was a general delay in fissure formation in the vermis of E18.5 En1−/+;En2−/+ mutants (Fig. 3B,C), similar to but milder than the delay in En2−/− mutants (Fig. 3D) (Millen et al., 1994).
Fig. 4. Viable En1 and En2 double-mutant adult mice have midbrain and cerebellar defects.
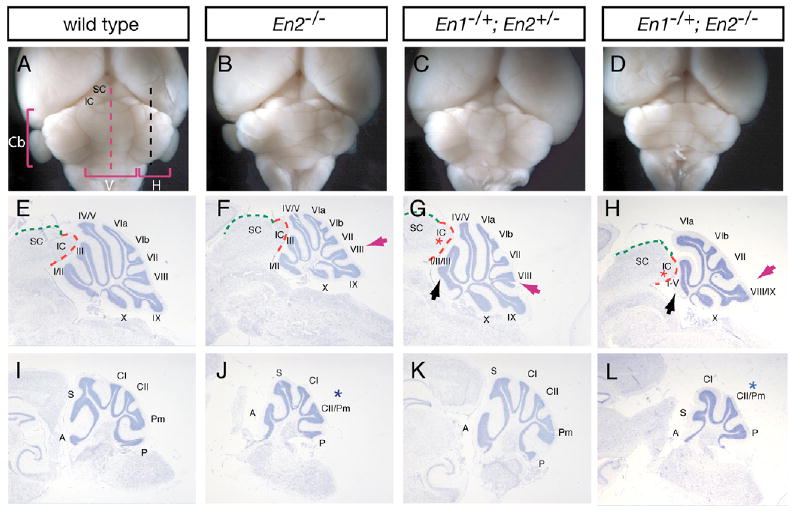
(A-D) Dorsal posterior views of adult brains of (A) wild-type, (B) En2−/−, (C) En1−/+;En2−/+ and (D) En1−/+;En2−/− mutant mice. (E-L) Cresyl Violet-stained sagittal sections of the cerebella shown in A-D at the level of the vermis (E-H) and hemispheres (I-L), as indicated in A by the red and black dashed lines, respectively. Black and red arrowheads indicate the anterior and posterior foliation defects, respectively. Blue and red asterisks indicate hemisphere foliation defects and truncation in the IC, respectively. See Fig. 2 legend for midbrain and cerebellum annotation.
En1−/+;En2−/− mice, which have a smaller Mb and Cb than normal at E18.5 (Simon et al., 2005), were found to survive to adulthood. En1−/+;En2−/− mutants (Fig. 3B,E) had a more severe delay in Cb foliation than En2−/− mutants (Fig. 3D). Consistent with this, the Cb of adult En1−/+;En2−/− mutants (Fig. 4D,H,L) had a variable but greater size reduction and more foliation defects than En2−/− (Fig. 4B,F,J) or En1−/+;En2+/− mice (Fig. 4C,G,K and see Table 1). In the majority of the mutants (six out of eight) the five anterior-most folia (I-V) of the vermis were replaced by a single fold. In addition, folium VIII was more defective than in En2−/− mutants: it was absent or decreased in size and misaligned with folium IX. The inferior colliculus also was partially truncated. In one of the mutants, folia I-V were almost completely lost and fused to a more profoundly truncated tectum (data not shown). One other En1−/+;En2−/− mutant had an even more severe phenotype that included a complete deletion of the vermis and inferior colliculus (data not shown). In contrast to the vermis, although the overall size of the En1−/+;En2−/− hemispheres (Fig. 4I,L) was reduced compared with En2−/− mutants (Fig. 4J), the foliation defect was the same (fusion of the crusII and paramedian folia) or, paradoxically, partially rescued in one mutant. Interestingly, in all the viable En1/2 mutants the relative thickness of the remaining molecular and granule cell layers and Purkinje cell density were normal. Our analysis of viable En1/2 mutants provides additional evidence that the anterior vermis (folia I-V) and inferior colliculus are most sensitive to a reduction in En1, and also reveals redundant functions for En1 and En2 in folia I-V and VIII.
En2 can sustain more extensive Cb vermis development than En1
In order to directly compare the in vivo functions of En1 and En2 proteins, we took advantage of our En1En2ki knock-in mice (Hanks et al., 1995). A sensitive assay for comparing protein function is to combine knock-in alleles with loss-of-function alleles (Wang and Jaenisch, 1997). If the two En proteins have equivalent properties, then removal of En2 in En1En2ki/En2ki mice (En1En2ki/En2ki;En2−/−) should only cause the foliation defects seen in En2−/− mice. Indeed, when expressed in two copies from the En1 locus, En2 is sufficient to support development of the inferior colliculus. Unlike En1flox/Cre;En2+/+ mice, which have a partial truncation of the tectum, the inferior colliculus appeared normal in En1En2ki/En2ki;En2−/− adult or early postnatal mice (Fig. 5A,C and data not shown). Furthermore, consistent with En1 expression being restricted to the primordium of the vermis, as compared with the broader En2 expression in the hemispheres after ∼E9.5, the hemispheres of En1En2ki/En2ki;En2−/− mice (Fig. 5G,I) had the characteristic En2−/− phenotype (Fig. 4J). However, unlike En2−/− mice (Fig. 4F), in which folium VIII is associated with folium IX, in three out of four adult En1En2ki/En2ki;En2−/− mice (Fig. 5A,C), folium VIII was normal. This shows that the vermis foliation defects in En2−/− mice can be rescued by expressing the En2 gene, but not the En1 gene, from the En1 locus. This suggests that En1 and En2 proteins are not equivalent, but rather that En2 activity is either specifically required in the posterior Cb (folium VIII) or more active in the Cb.
Fig. 5. In the absence of En2, expression of mouse En2 but not Drosophila en in place of En1 can rescue mes/r1 defects.
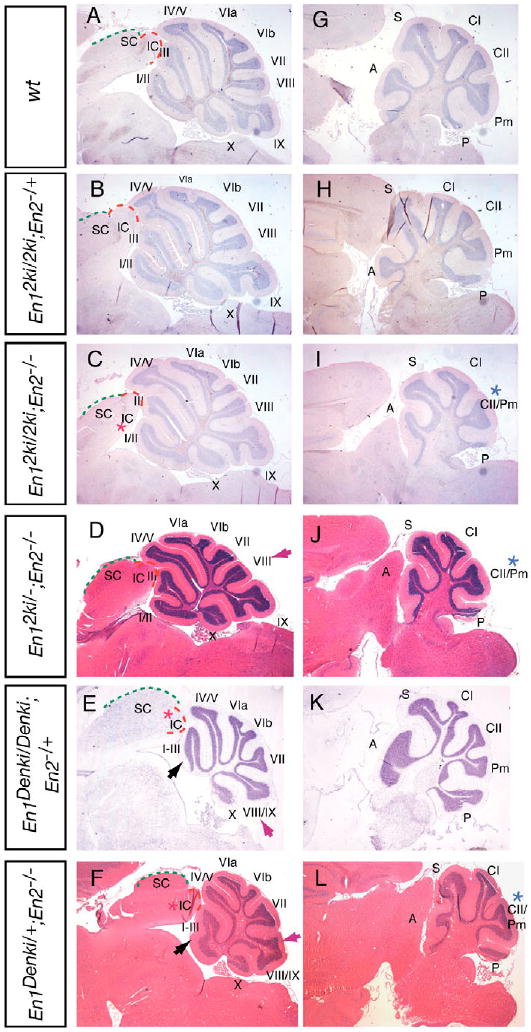
Stained sagittal sections of wild-type (wt) (A,G), En12ki/2ki;En2−/+ (B,H), En12ki/2ki;En2−/− (C,I), En12ki/−;En2−/− (D,J), En1Denki/Denki;En2−/+ (E,K), and En1Denki/+;En2−/− (F,L) at the level of the vermis (A-F) and hemispheres (G-L). A-C,E,G-I,K are stained with Cresyl Violet; D,F,J,L with Hematoxylin and Eosin. For comparison, the En2−/− phenotype is shown in Fig. 4. See Fig. 2 legend for midbrain and cerebellum annotation, and Fig. 4 legend for key to asterisks and arrows.
We further tested whether En2 is generally more potent than En1 by comparing the phenotype of En1−/+;En2−/− mice with En1En2ki/−;En2−/− mice. Strikingly, we found that En1En2ki/−;En2−/− mice (n=3) had a much milder anterior Cb phenotype than En1−/+;En2−/− mutants. The anterior vermis of En1En2ki/−;En2−/− mutants (Fig. 5A,D) had three distinct folia (I/II, III and IV/V) compared with one in En1−/+;En2−/− mice (Fig. 4H). In addition, in the posterior vermis of En1En2ki/−;En2−/− mice (Fig. 5D), folium VIII was only partially displaced toward folium IX, in contrast to En1+/−;En2−/− mutants (Fig. 4H), which have a substantial deletion of folium VIII. This sensitive dosage assay of one copy of En2 or En1 expressed from the En1 locus in the absence of all other En function thus indicates that En2 is generally more active than En1 in regulating Cb development. Finally, the posterior tectum of En1En2ki/−;En2−/− mice (Fig. 5D) was partially truncated (Fig. 4H and data not shown), seemingly less than in En1−/+;En2−/− mutants (Figure 4H and data not shown), indicating that the dose of both En proteins contributes to the truncation of the inferior colliculus in the various En mutants.
Drosophila en expressed in place of En1 cannot support mes/r1 development in the absence of En2
Our finding that one or two copies of En2 expressed from the En1 locus in the absence of endogenous En2 can support tectum and Cb development prompted us to determine whether Drosophila en could do the same. To determine whether Drosophila en, when expressed in place of En1, can sustain mes/r1 development in the absence of En2, we made use of knock-in mice (En1Denki) that express Drosophila en in place of En1 (Hanks et al., 1998). Whereas En1En2ki/En2ki;En2−/− and En1Denki/Denki;En2+/+ mice are viable, we did not detect any En1Denki/Denki;En2−/− mice after birth. Section analysis of E18.5 En1Denki/Denki;En2−/− embryos revealed either the same phenotype as En1−/−;En2−/− embryos (n=1 of 3) or a similar phenotype to En1flox/Cre;En2−/+ mutants (n=2 of 3; data not shown). Interestingly, half the expected number of En1Denki/Denki;En2−/+ mice survived to adulthood, indicating that Drosophila En has some En1-like activity as En1−/−;En2−/+ die at birth. Histological analysis of the brains of three En1Denki/Denki;En2−/+ mice (Fig. 5A,E and Table 1) that survived surprisingly revealed that two of the mutants had a normal tectum and Cb, whereas one had a partial deletion of the posterior tectum and a vermis foliation defect similar to En1+/−;En2−/− mutants (Fig. 4H). The five En1Denki/+;En2−/− mice analyzed had a phenotype that was milder than En1−/+;En2−/− mice, with a distinct IV/V folium (Fig. 5A,F and Fig. 4H). All but one of the five En1Denki/+;En2−/− mutants analyzed had an En2−/− foliation pattern in the hemispheres (Fig. 4J, Fig. 5G,L). These results indicate that Drosophila En function can partially replace normal En1 function in the initiation of normal En2 expression (before ∼E9.0) in En1Denki/Denki;En2+/+ mice, after which mouse En2 contributes a necessary function for continued normal development of mes/r1-derived structures.
Medial-lateral patterning of the Cb appears to be altered in En1−/+;En2−/− mutants
One possible reason for the loss of the posterior tectum and anterior vermis in En1−/+;En2−/− adult mutants is specific loss of the cells that give rise to the these two regions (caudal mes and rostral r1) (Sgaier et al., 2005; Zervas et al., 2004). To determine whether this is the case, we fate mapped the posterior mes and anterior r1 in En1−/+;En2−/− mutants by genetic inducible fate mapping (GIFM) (Joyner and Zervas, 2006) using our null En1CreERT1/+ allele (Sgaier et al., 2005) and the R26R lacZ reporter allele (Soriano, 1999). When tamoxifen (TM) is administered at 18.00 h to E10.5 wild-type (En1CreERT1/+;En2+/+;R26R/+) embryos (marking cells at E11-12), the posterior mes and medial-most domain of the E12.5 CbP are marked (Fig. 6A) and give rise to the vermis and inferior colliculus of the adult, respectively (Fig. 6G,I) (Sgaier et al., 2005). GIFM in En1CreERT1/+;En2−/−;R26R/+ mutants also marked the posterior mes and medial-most domain of the E12.5 CbP (Fig. 6B). A difference that was apparent between the mutants and wild types was that despite an overall reduction in the size of the mes and r1 in the mutants, the size of the initial marked domain in the mes/r1 at E12.5 was similar to that in the wild type, and the regions devoid of marked cells were smaller in the mutants as compared with the wild type (Fig. 6A,B). At E16.5, the size of the marked population of cells in the Cb appeared wider in En1CreERT1/+;En2−/−;R26R/+ embryos (Fig. 6D) than in wild types (Fig. 6C). By contrast, the size of the marked domain in the tectum was smaller in mutants than in wild types (Fig. 6C-F). Similarly, in adult En1CreERT1/+;En2−/−;R26R/+ mutants, the marked domain in the Cb was broader than normal, whereas the size of the marked domain in the tectum was greatly reduced compared with wild types as it was restricted to the remaining inferior colliculus (Fig. 6G-J). The fate mapping results in the mes suggest that in En1/2 mutants, the posterior mes cells marked at E12.5 do not expand normally and this results in a smaller inferior colliculus in the adult. By contrast, because the domain of marked cells in the adult Cb is larger than normal even though the vermis is reduced in size, this suggests that the anterior r1 cells marked at E12.5 are not only retained but contribute to more lateral regions of the vermis than normal. Thus, the loss of tectum and vermis tissue in En1−/+;En2−/− adult mutants is not simply owing to loss of the cells that give rise to the these two regions.
Fig. 6. mes and r1 cells behave differently in En1−/+;En2−/− mutants.
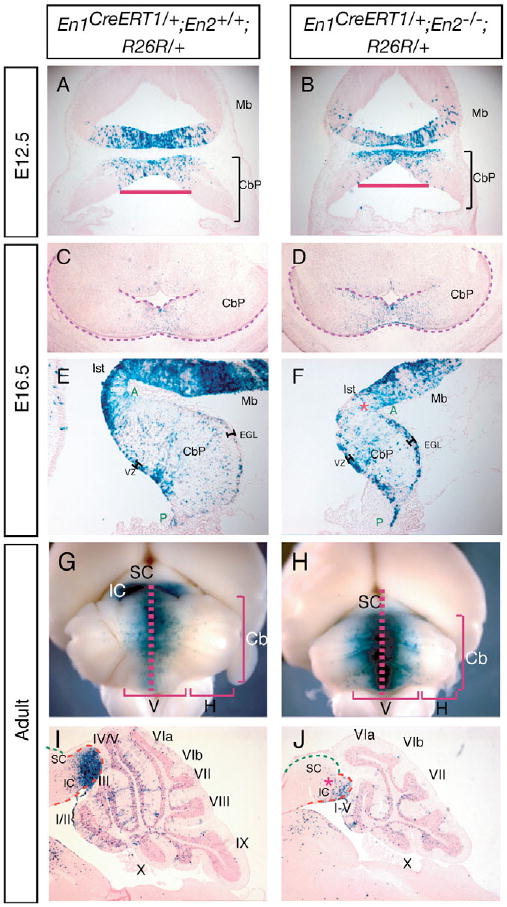
(A-J) Tamoxifen was administered to mouse embryos carrying the indicated alleles at 18.00 h on E10.5. (A,B) Coronal sections of E12.5 brains and (C,D) horizontal and (E,F) midline sagittal sections of E16.5 brains stained for β-gal activity. (G,H) Dorsal views of β-gal-stained adult brains. (I,J) Sagittal sections of adult brains taken at the level of the vermis and stained for β-gal. The domain of marked cells was comparable in the En1CreERT1/+;En2−/− and En1CreERT1/+;R26R/+ embryos at E12.5, but decreased in the tectum and increased in the Cb of En1CreERT1/−;En2−/−;R26R/+ embryos by E16.5. CbP, cerebellar primordium; EGL and associated bar, external granule layer. Ist, isthmus; Mb, midbrain; VZ, ventricular zone; A, anterior; P, posterior; M, medial; L, lateral. Red asterisks indicate truncation of the isthmus/inferior colliculus. CbP is outlined by a purple dashed line in C,D; in I,J, the IC and SC are outlined by dashed red and green lines, respectively. The red bar in A,B indicates the size of the CbP domain with marked cells.
The isthmic organizer is not lost in En2−/− and En1−/+;En2−/− mutants
Since Fgf8 expression is lost in En1/En2 double-homozygous mutant embryos by E9 (Liu and Joyner, 2001), one possible reason for the defects in En2−/− and En1−/+;En2−/− mutants is a disruption of the isthmic organizer (decrease in Fgf signaling). However, our previous whole-mount RNA in situ analysis of En1−/+;En2−/− embryos from the six-somite stage to E9.5 did not reveal any obvious changes in the expression of Fgf8 or mes/r1 morphology (Liu and Joyner, 2001). Based on section analysis, En2−/− embryos had a slight reduction in the size of the Cb by E11.5, and En1−/+;En2−/− mutants had a greater reduction in the CbP and truncation of the tectum (see Fig. 7). We therefore performed RNA in situ analysis at E11.5, a day before Fgf8 expression is normally terminated in the isthmus. Consistent with the fate mapping study, we found that the expression domain of En1 was not obviously altered along the A-P axis in the mutants (Fig. 7G-I). The only obvious difference in the expression domains of Fgf8 and Fgf17 (a related organizer gene) and of the direct target gene Spry1 (Liu et al., 2003) between En2−/− and En1−/+;En2−/− mutants as compared with wild-type mice (En1−/+) was a slight reduction in the size of the domains, which correlated with the reduction in mes/r1 tissue in each mutant (Fig. 7A-F and data not shown). Consistent with retention of organizer activity, the expression domains of two mes genes (Otx2 and Wnt1) and one r1 gene (Gbx2) regulated by Fgf8, were not altered (data not shown).
Fig. 7. Fgf8, Fgf17 and En1 are expressed normally in En2−/− and En1−/+;En2−/− mutant E11.5 mouse embryos.
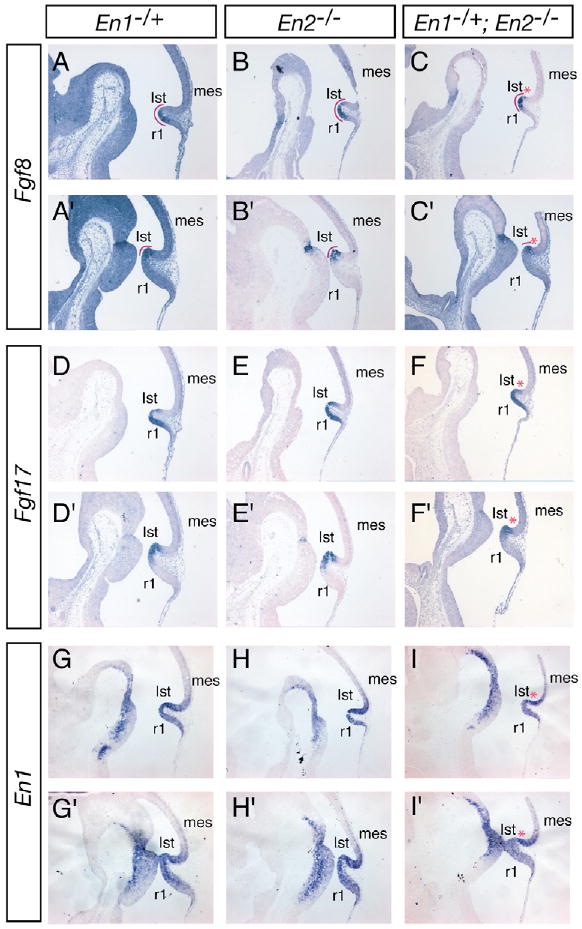
RNA in situ hybridization of Fgf8 (A-C′), Fgf17 (D-F′) and En1 (G-I′) on sagittal sections of En1−/+ (A,D,G), En2−/− (B,E,H) and En1−/+;En2−/− (C,F,I) E11.5 mutant embryos. Upper (A-I) and lower (A′-I′) panels show medial and lateral sections, respectively. The Fgf8/17 expression domains are positioned normally but are slightly reduced in size in mutants (C,F). En1 is expressed normally in mutants (H,I). The isthmus corresponds to the Fgf8 expression domain and is outlined in red. Red asterisks indicate truncation of the isthmus/inferior colliculus. r1, rhombomere 1; Ist, isthmus, mes, mesencephalon.
Discussion
In this study, we have analyzed a series of mouse En conditional knock-in and null mutants to decipher the overlapping and individual functions of the two highly conserved En genes in mes/r1 development. Overall, we found that the inferior colliculus of the tectum and three regions of the Cb are particularly sensitive to the level of En genes (see below and Table 1). Interestingly, the anterior vermis and tectum defects we observed in En1flox/cre, En1Denki/Denki;En2−/+ and En1−/+;En2−/+ mutant mice have similarities to Fgf8−/+;Fgf17−/− mutants (Xu et al., 2000), raising the possibility that a key role of En1/2 is to maintain Fgf8 expression (Liu et al., 2003). We found that Fgf8 expression is maintained as long as one allele of En1 is present, although there are subtle decreases in Fgf8/17 expression. Since En1/2 expression persists after E12.5, when Fgf8 expression is terminated, and En1−/+;En2−/− mutants have a much more severe loss of the tectum and Cb folia than Fgf8−/+;Fgf17−/− mutants, it is possible that En1/2 do not control mes/r1 development solely through regulating Fgf8/17 expression.
En1/2 differentially regulate retention of cells in the mes and r1
By determining the fate of the En1-expressing cells at ∼E11, which normally give rise to the vermis and inferior colliculus, in En1−/+;En2−/− mutants using GIFM, we uncovered an unexpected differential role for En1/2 in regulating growth and survival of cells in the tectum versus the Cb. In En1/2 mutants, the posterior mes cells marked at E12.5 do not expand normally and this results in a smaller inferior colliculus in the adult. By contrast, the anterior r1 cells marked at E12.5 are not only retained but contribute to more lateral regions of the vermis than normal. If the lineage restriction at the mes/r1 border that restricts mes and r1 cells from mixing (Zervas et al., 2004) is disrupted in En1/2 mutants, then it is possible that the population of marked mes cells in En1CreERT1/+;En2−/−;R26R/+ embryos move into r1 and expand the marked population in the Cb. Another possibility is that the precursors of the lateral Cb are selectively lost in the mutants. However, this is not in accordance with our observation that the hemispheres of En1−/+;En2−/− adults are less compromised than the vermis. Although the ultimate overall loss of cells in the mes and r1 of En1−/+;En2−/− mutants could be accounted for by cell death, similar to the situation in En1 mutants (Chi et al., 2003), our fate mapping study shows that it is not as simple as the cells being lost equally on either side of the isthmus.
En1 is required after E9 only for development of the inferior colliculus
Consistent with the En1 expression domain being encompassed by the En2 domain after E9, we found that En1CreERT1/flox conditional mutants are viable and have a normal Cb and superior colliculus. However, despite strong expression of En2 in the posterior mes after E9 when En1 is deleted in these mutants, the inferior colliculus does not develop normally in En1flox/Cre mutants. These results indicate that the En1 protein has a different function from En2, or that En1 and En2 are expressed differently in the tectum after E9. We demonstrated that the latter is the case, because the tectum develops normally when En2 is produced from the En1 locus in the absence of endogenous En2 (En1En2ki/En2ki;En2−/− mice). Based on our expression analysis, the crucial difference must be that En1 is transiently expressed around E9 in a broader domain than En2, or that En1 is later produced at a higher level than En2. Regardless, our studies have uncovered a differential requirement for the two En genes in the superior and inferior colliculi (see Table 1 and Fig. 8A).
En1 and En2 are differentially required in subregions of the Cb
Our analysis of En1/2 double-mutant combinations (null, knock-in and conditional) uncovered additional differential requirements for En1 and En2 in specific regions of the Cb (see Table 1 and Fig. 8B). En1/2 functions are normally uncoupled in the hemispheres as only En2 is required to divide the posterior region into two folds (crusII and paramedian). However, the partial rescue of the hemisphere phenotype in rare En1−/+;En2−/− and En1Denki/+;En2−/− mutants indicates that En1 can support hemisphere development when expressed more laterally than normal. A comparison of the phenotypes of these mice with En1flox/Cre;En2+/+ mutants (which have normal posterior foliation) indicated that En2 plays a greater role than En1 in formation of folium VIII. We demonstrated that this difference is not owing to a difference in gene expression, but instead to a difference in protein activity because the vermis foliation defect seen in En1+/+;En2−/− mutants is rescued in En1En2ki/En2ki;En2−/− mice. Furthermore, En1En2ki/−;En2−/− mice have a milder phenotype than En1−/+;En2−/− mutants. Thus, En2 appears to be more effective in promoting development of the vermis (folia I-V and VIII) than En1. We further discovered that the two En genes act concomitantly to divide the anterior Cb into five folia. En1−/+;En2−/+ double heterozygotes and the majority of En1flox/Cre;En2+/+ mutants have a fusion of the anterior three folia (I-III) and the anterior defect is greatly exaggerated in En1−/+;En2−/− mutants (fusion of folia I-V), despite En2−/− mutants having normal anterior foliation. Since some En1flox/Cre mice have normal anterior folia, this indicates a crucial requirement for expression of En1 only at ∼E9, when En1 expression is fading out in the mutants and En2 is initiating. To our knowledge, this is the first evidence that the pattern of Cb folia can be influenced by genetic events that occur at such an early embryonic stage.
It is revealing to compare the early En1/2 expression patterns and the broad regions of the mes/r1 that differentially require the two genes. Based on our recent fate map of the mes/r1 using GIFM (Sgaier et al., 2005; Zervas et al., 2004), the anterior and posterior mes give rise to the superior and inferior colliculi, respectively (Fig. 8A). Consistent with strong and sustained expression of the En genes in the primordium of the inferior colliculus, this region is most sensitive to the dose of En genes, and in particular to that of En1. However, by using the sensitive assay of En12ki knock-in alleles combined with removal of the endogenous En2 gene, we found that En2 is at least as potent as En1 at promoting inferior colliculus development. Given the transient expression of the En genes in the superior colliculus, it is perhaps surprising that this region is dependent at all on the combination of the two genes. This indicates a requirement for a short burst of En function (En1 or En2) before E9.5. The remaining inferior colliculus tissue in En mutants is likely to correlate with tectum cells that are normally in the low end of the En gradient, suggesting that they are least sensitive to loss of En alleles. There also is a general correlation between the domains of En gene expression and the requirement for each gene in the Cb (vermis versus hemispheres). After E9.5, En1 is only maintained in anterior r1 and the medial Cb primordium (the anlage of the vermis), consistent with no function in the hemispheres. The limit of the En2 expression domain extends more posterior early in r1 and laterally later in the CbP, correlating with a role in the hemispheres. It is not clear, however, why the En genes do not play a major role in the anterior hemispheres or in folia VI/VII and IX/X in the vermis.
An ‘En code’ divides the tectum and Cb into subregions
Taken together, our analysis of a series of En mutants provides evidence that functional domains of the Cb are genetically encoded by the engrailed genes, as specific regions of the tectum and Cb have differential sensitivities to reducing En gene dosage. The phenotypes of multiple mutants point to a genetic division of the tectum into two regions and of the Cb into six. We propose that this represents an ‘En code’ that is used to partition the mes/r1 region into domains that in the adult regulate related neural functions (Fig. 8B). The two functional divisions of the tectum, the inferior and superior colliculi, are delineated based on a temporal requirement for En1 and sensitivity to the overall dose of En protein. Within the vermis of the Cb, the anterior five folia (I-V) and folium VIII are particularly sensitive to a reduction of En genes, and preferentially to En2, thus dividing the vermis into four broad regions (folia I-V, VI/VII, VIII, IX/X). Strikingly, this division of the Cb is very similar to the transverse zones recently proposed based on four different domains of parasagittal gene expression (Armstrong et al., 2005; Ozol et al., 1999). The fact that two independent genetic measures of regionalization of the vermis (mutant phenotypes and gene expression) point to the same subdivisions of the vermis strongly argues that patterning of the folia is fundamental to organization of Cb function. Consistent with this, each transverse zone receives afferent inputs from distinct regions of the spinal cord and/or particular hindbrain nuclei. We predict that, likewise, the division of the hemispheres into regions based on a need for En2 only in two folia (crusII and paramedian) (Fig. 8B) represents genetic partitioning into related functional systems.
Supplementary Material
Acknowledgments
We thank the NYU Transgenic/ES Cell Chimera Facility for the blastocyst injections; Dr P. Soriano for providing the R26R reporter mice; Drs G. Martin, R. Sillitoe, S. Blaess, M. Zervas, J. Li, and Y. Cheng for discussions and comments on the manuscript. This work was supported in part by a grant from NINDS.
Footnotes
Supplementary material: Supplementary material for this article is available at http://dev.biologists.org/cgi/content/full/134/12/2325/DC1
References
- Altman J, Bayer SA. Development of the Cerebellar System in Relation to its Evolution, Structure, and Function. Boca Raton, FL: CRC Press; 1997. [Google Scholar]
- Armstrong CL, Vogel MW, Hawkes R. Development of Hsp25 expression compartments is not constrained by Purkinje cell defects in the Lurcher mouse mutant. J Comp Neurol. 2005;491:69–78. doi: 10.1002/cne.20703. [DOI] [PubMed] [Google Scholar]
- Auerbach W, Dunmore JH, Fairchild-Huntress V, Fang Q, Auerbach AB, Huszar D, Joyner AL. Establishment and chimera analysis of 129SvEv and C57BL/6-derived ES cell lines. Biotechniques. 2000;29:1024–1032. doi: 10.2144/00295st04. [DOI] [PubMed] [Google Scholar]
- Bilovocky NA, Romito-DiGiacomo RR, Murcia CL, Maricich SM, Herrup K. Factors in the genetic background suppress the engrailed-1 cerebellar phenotype. J Neurosci. 2003;23:5105–5112. doi: 10.1523/JNEUROSCI.23-12-05105.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouillet P, Chazaud C, Oulad-Abdelghani M, Dolle P, Chambon P. Sequence and expression pattern of the Stra7 (Gbx-2) homeobox-containing gene induced by retinoic acid in P19 embryonal carcinoma cells. Dev Dyn. 1995;204:372–382. doi: 10.1002/aja.1002040404. [DOI] [PubMed] [Google Scholar]
- Chi CL, Martinez S, Wurst W, Martin GR. The isthmic organizer signal FGF8 is required for cell survival in the prospective midbrain and cerebellum. Development. 2003;130:2633–2644. doi: 10.1242/dev.00487. [DOI] [PubMed] [Google Scholar]
- Crossley PH, Martin GR. The mouse Fgf8 gene encodes a family of polypeptides and is expressed in regions that direct outgrowth and patterning in the developing embryo. Development. 1995;121:439–451. doi: 10.1242/dev.121.2.439. [DOI] [PubMed] [Google Scholar]
- Hanks M, Wurst W, Anson-Cartwright L, Auerbach AB, Joyner AL. Rescue of the En-1 mutant phenotype by replacement of En-1 with En-2. Science. 1995;269:679–682. doi: 10.1126/science.7624797. [DOI] [PubMed] [Google Scholar]
- Hanks MC, Loomis CA, Harris E, Tong CX, Anson-Cartwright L, Auerbach A, Joyner A. Drosophila engrailed can substitute for mouse Engrailed1 function in mid-hindbrain, but not limb development. Development. 1998;125:4521–4530. doi: 10.1242/dev.125.22.4521. [DOI] [PubMed] [Google Scholar]
- Herrup K, Kuemerle B. The compartmentalization of the cerebellum. Annu Rev Neurosci. 1997;20:61–90. doi: 10.1146/annurev.neuro.20.1.61. [DOI] [PubMed] [Google Scholar]
- Joyner AL. Engrailed, Wnt and Pax genes regulate midbrain–hindbrain development. Trends Genet. 1996;12:15–20. doi: 10.1016/0168-9525(96)81383-7. [DOI] [PubMed] [Google Scholar]
- Joyner AL, Martin GR. En-1 and En-2, two mouse genes with sequence homology to the Drosophila engrailed gene: expression during embryogenesis. Genes Dev. 1987;1:29–38. doi: 10.1101/gad.1.1.29. [DOI] [PubMed] [Google Scholar]
- Joyner AL, Zervas M. Genetic fate mapping in mouse: establishing genetic lineages and defining genetic neuroanatomy in the nervous system. Dev Dyn. 2006;253:2376–2385. doi: 10.1002/dvdy.20884. [DOI] [PubMed] [Google Scholar]
- Joyner AL, Herrup K, Auerbach BA, Davis CA, Rossant J. Subtle cerebellar phenotype in mice homozygous for a targeted deletion of the En-2 homeobox. Science. 1991;251:1239–1243. doi: 10.1126/science.1672471. [DOI] [PubMed] [Google Scholar]
- Kimmel RA, Turnbull DH, Blanquet V, Wurst W, Loomis CA, Joyner AL. Two lineage boundaries coordinate vertebrate apical ectodermal ridge formation. Genes Dev. 2000;14:1377–1389. [PMC free article] [PubMed] [Google Scholar]
- Larsell O. The morphogenesis and adult pattern of the lobules and fissures of the cerebellum of the white rat. J Comp Neurol. 1952;97:281–356. doi: 10.1002/cne.900970204. [DOI] [PubMed] [Google Scholar]
- Li JY, Lao Z, Joyner AL. Changing requirements for Gbx2 in development of the cerebellum and maintenance of the mid/hindbrain organizer. Neuron. 2002;36:31–43. doi: 10.1016/s0896-6273(02)00935-2. [DOI] [PubMed] [Google Scholar]
- Liu A, Joyner AL. EN and GBX2 play essential roles downstream of FGF8 in patterning the mouse mid/hindbrain region. Development. 2001;128:181–191. doi: 10.1242/dev.128.2.181. [DOI] [PubMed] [Google Scholar]
- Liu A, Li JY, Bromleigh C, Lao Z, Niswander LA, Joyner AL. FGF17b and FGF18 have different midbrain regulatory properties from FGF8b or activated FGF receptors. Development. 2003;130:6175–6185. doi: 10.1242/dev.00845. [DOI] [PubMed] [Google Scholar]
- Loomis CA, Harris E, Michaud J, Wurst W, Hanks M, Joyner AL. The mouse Engrailed-1 gene and ventral limb patterning. Nature. 1996;382:360–363. doi: 10.1038/382360a0. [DOI] [PubMed] [Google Scholar]
- Matise MP, Joyner AL. Expression patterns of developmental control genes in normal and Engrailed-1 mutant mouse spinal cord reveal early diversity in developing interneurons. J Neurosci. 1997;17:7805–7816. doi: 10.1523/JNEUROSCI.17-20-07805.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matise MP, Auerbach W, Joyner AL. Production of targeted embryonic stem cell clones. In: Hames BD, editor. Gene Targeting: A Practical Approach. New York: Oxford University Press; 2000. pp. 101–132. [Google Scholar]
- Millen KJ, Wurst W, Herrup K, Joyner AL. Abnormal embryonic cerebellar development and patterning of postnatal foliation in two mouse Engrailed-2 mutants. Development. 1994;120:695–706. doi: 10.1242/dev.120.3.695. [DOI] [PubMed] [Google Scholar]
- Millen KJ, Hui CC, Joyner AL. A role for En-2 and other murine homologues of Drosophila segment polarity genes in regulating positional information in the developing cerebellum. Development. 1995;121:3935–3945. doi: 10.1242/dev.121.12.3935. [DOI] [PubMed] [Google Scholar]
- Ozol K, Hayden JM, Oberdick J, Hawkes R. Transverse zones in the vermis of the mouse cerebellum. J Comp Neurol. 1999;412:95–111. [PubMed] [Google Scholar]
- Papaioannou V, Johnson R. Production of chimeras by blastocyst and morula injection of targeted ES cells. In: Hames BD, editor. Gene Targeting: A Practical Approach. New York: Oxford University Press; 2000. pp. 133–175. [Google Scholar]
- Parr BA, Shea MJ, Vassileva G, McMahon AP. Mouse Wnt genes exhibit discrete domains of expression in the early embryonic CNS and limb buds. Development. 1993;119:247–261. doi: 10.1242/dev.119.1.247. [DOI] [PubMed] [Google Scholar]
- Rodriguez CI, Buchholz F, Galloway J, Sequerra R, Kasper J, Ayala R, Stewart AF, Dymecki SM. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat Genet. 2000;25:139–140. doi: 10.1038/75973. [DOI] [PubMed] [Google Scholar]
- Sgaier SK, Millet S, Villanueva MP, Berenshteyn F, Song C, Joyner AL. Morphogenetic and cellular movements that shape the mouse cerebellum; insights from genetic fate mapping. Neuron. 2005;45:27–40. doi: 10.1016/j.neuron.2004.12.021. [DOI] [PubMed] [Google Scholar]
- Simeone A, Acampora D, Mallamaci A, Stornaiuolo A, D'Apice MR, Nigro V, Boncinelli E. A vertebrate gene related to orthodenticle contains a homeodomain of the bicoid class and demarcates anterior neuroectoderm in the gastrulating mouse embryo. Embo J. 1993;12:2735–2747. doi: 10.1002/j.1460-2075.1993.tb05935.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon HH, Thuret S, Alberi L. Midbrain dopaminergic neurons: control of their cell fate by the engrailed transcription factors. Cell Tissue Res. 2004;318:53–61. doi: 10.1007/s00441-004-0973-8. [DOI] [PubMed] [Google Scholar]
- Simon HH, Scholz C, O'Leary DD. Engrailed genes control developmental fate of serotonergic and noradrenergic neurons in mid- and hindbrain in a gene dose-dependent manner. Mol Cell Neurosci. 2005;28:96–105. doi: 10.1016/j.mcn.2004.08.016. [DOI] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Wang Y, Jaenisch R. Myogenin can substitute for Myf5 in promoting myogenesis but less efficiently. Development. 1997;124:2507–2513. doi: 10.1242/dev.124.13.2507. [DOI] [PubMed] [Google Scholar]
- Wurst W, Bally-Cuif L. Neural plate patterning: upstream and downstream of the isthmic organizer. Nat Rev Neurosci. 2001;2:99–108. doi: 10.1038/35053516. [DOI] [PubMed] [Google Scholar]
- Wurst W, Auerbach AB, Joyner AL. Multiple developmental defects in Engrailed-1 mutant mice: an early mid-hindbrain deletion and patterning defects in forelimbs and sternum. Development. 1994;120:2065–2075. doi: 10.1242/dev.120.7.2065. [DOI] [PubMed] [Google Scholar]
- Xu J, Lawshe A, MacArthur CA, Ornitz DM. Genomic structure, mapping, activity and expression of fibroblast growth factor 17. Mech Dev. 1999;83:165–178. doi: 10.1016/s0925-4773(99)00034-9. [DOI] [PubMed] [Google Scholar]
- Xu J, Liu Z, Ornitz DM. Temporal and spatial gradients of Fgf8 and Fgf17 regulate proliferation and differentiation of midline cerebellar structures. Development. 2000;127:1833–1843. doi: 10.1242/dev.127.9.1833. [DOI] [PubMed] [Google Scholar]
- Zervas M, Millet S, Ahn S, Joyner AL. Cell behaviors and genetic lineages of the mesencephalon and rhombomere 1. Neuron. 2004;43:345–357. doi: 10.1016/j.neuron.2004.07.010. [DOI] [PubMed] [Google Scholar]
- Zervas M, Blaess S, Joyner AL. Classical embryological studies and modern genetic analysis of midbrain and cerebellum development. Curr Top Dev Biol. 2005;69:101–138. doi: 10.1016/S0070-2153(05)69005-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


