Abstract
During the preimplantation phase of pregnancy the endometrial stroma differentiates into decidua, a process that implies numerous morphological changes and is an example of physiological transdifferentiation. Here we show that UIII rat endometrial stromal cells cultured in the presence of calf serum acquired morphological features of decidual cells and expressed decidual markers. To identify genes involved in decidualization we compared gene expression patterns of control and decidualized UIII cells using cDNA microarray. We found 322 annotated genes exhibiting significant differences in expression (>3 fold, FDR > 0.005), of which 312 have not been previously related to decidualization. Analysis of overrepresented functions revealed that protein synthesis, gene expression and chromatin architecture and remodeling are the most relevant modified functions during decidualization. Relevant genes are also found in the functional terms differentiation, cell proliferation, signal transduction, and matrix/structural proteins. Several of these new genes involved in decidualization (Csdc2, Trim27, Eef1a1, Bmp1, Wt1, Aes, Gna12, and Men1) are shown to be also regulated in uterine decidua during normal pregnancy. Thus, the UIII cell culture model will allow future mechanistic studies to define the transcriptional network regulating reprogramming of stromal cells into decidual cells.
Keywords: gene expression, microarrays, decidualization, UIII cells
INTRODUCTION
In many mammalian species, endocrine changes during the preimplatation phase as well as signals from the implanting blastocyst lead to extensive modifications of the uterine endometrium that ultimately converts into decidua. The most conspicuous aspect of this tissue transdifferentiation is a transformation of stromal cells into large, polyploidy cells with epithelioid appearance. These decidual cells are characterized by an accumulation of glycogen and lipids in the cytoplasm, numerous lysosomes, large rough endoplasmic reticulum, as well as by extensive cell-to-cell contacts and junctional complexes, including gap junctions, desmosome-like junctions, and tight junctions (Dey et al., 2004). The differentiation of the endometrium into decidual tissue is called the decidual cell reaction (DCR) or decidualization, and it also involves several changes in extracellular components, such as reduction of intercellular space and remodeling of its matrix with a decrease in the number of collagen fibrils (Mulholland et al., 1992).
There is evidence that, in contrast to stromal cells, decidual cells can synthesize prostaglandins (Pgs), hyaluronate, prolactin-like proteins, desmin, vimentin, catechol-O-methyltransferase, ornithine decarboxylase, and alkaline phosphatase (Carson et al., 1987; Gu et al., 1994; Glasser et al., 1986; Heald et al., 1979; Bany et al., 1998). Numerous signaling molecules and factors, including cytokines, homeobox transcription factors, cell-cycle molecules, extracellular matrix remodeling factors and lipid mediators are expressed in the endometrium during decidualization and are crucial to this process (Dey et al., 2004; Wang and Dey, 2006). Primary stromal cells in monolayer culture can acquire some features of decidual differentiation (Vladimirsky et al., 1977; Sananés et al., 1978; Wewer et al., 1998) and express several decidual markers such as Prolactin(Prl)-related protein (dPRP), PRL-like protein-B (PLP-B) (Gu et al., 1994), activin A (Gu et al., 1996), and its binding protein follistatin (Kaiser et al., 1990). However, to date no established cell culture model has been used for studying the molecular mechanisms of decidual transdifferentiation.
UIII cells were described as immortalized stromal cells derived from normal rat uterus that retained progesterone and prolactin receptors and progesterone regulation of cell growth (Cohen et al., 1993). Recently, we found that UIII cells have low levels of PR and ERβ but no ERα. Nevertheless UIII cells proliferate in response to progestins via a crosstalk between PR and ERβ that activates the Erk and the PI3K/Akt signaling pathways (Vallejo et al., 2005). UIII cells have been reported to undergo decidual differentiation in response to arachidonic acid that induces desmin and prolactin expression (Tessier-Prigent et al., 1999). UIII cells have also been shown to respond to medium containing low serum and either progesterone (1 mM), estradiol 17-beta (10 nM), cholera toxin (10 ng/ml) or interleukin-11 (10 ng/ml) through an increase in the expression of a luciferase reporter gene driven by the dPRP promoter (Rider et al., 2005). Here we describe serum-induced in vitro decidualization of UIII cells and characterize the changes in gene expression pattern during this process. In addition to 10 genes already known to participate in decidualization, we identify 312 novel genes that are differentially expressed during in vitro differentiation of UIII cells treated with hormone-deprived calf serum. We verify the differential uterine expression of some of these genes during in vivo decidualization and discuss their possible significance during the transdifferentiation process.
MATERIALS AND METHODS
Animals
Female Sprague Dawley rats (from the IByME, Buenos Aires, Argentina) were used for in vivo validation of microarrays results. The animals were kept under standard conditions. After mating, the presence of sperm in the vaginal smear the following morning was designated as day 1 post coitum (d.p.c). To evaluate the pregnancy outcome, autopsy was performed at 8 d.p.c. and the rats were killed by CO2 asphyxiation. The implantation sites were removed and the material was frozen for mRNA analysis. Control uteri were obtained from non-pregnant female rats. The rats were euthanized under approval protocol of the Animal Experimentation Committee of the IBYME.
Cell culture
UIII cells were maintained in M199 medium supplemented with 10% fetal calf serum (FCS), gentamycin (50 U/ml) at 37°C in humidified 95% air with 5% CO2 (Vallejo et al., 2005). Culture media was changed every 2 days. Cells were cultured in M199 supplemented with 10% dextran–coated charcoal treated FCS (DCC-FCS), and 24 hours later, media was replaced by fresh M199 without serum. After two days in serum-free conditions, media was replaced by fresh M199 with 10% DCC-FCS during the time specified in each figure. Viable cells were counted after 5 days of culture using the 0.1% trypan blue exclusion method. The percentage of cells with enlarged cytoplasm and decidual morphology were counted under light microscope.
Immunofluorescence
Cells were fixed with 3% paraformaldehyde in PBS and permeabilized with 0.01% triton X-100 in PBS (Vallejo et al., 2005). Mouse anti-desmin Dako (dil. 1:100) and Alexa 488 goat anti-Mouse Molecular Probes, The Netherlands (dil. 1:1000) were used. Nucleus was stained with propidium iodide 1mg/ml for 15min. RNA extraction and RT-PCR: In all cases total RNA isolation was performed according to guanidinium thiocyanate-phenol-chloroform extraction single step method (Chomczynski and Sacchi, 1987). To determine mRNA expression levels, cDNA was synthesized from equal amounts of total RNA (2.5µg) (Vallejo et al., 2005). PCR amplification reactions were carried out within the exponential range. All amplification products were routinely checked by gel electrophoresis on a 1.3% agarose gel and then visualized under UV light following staining with 0.05% ethidium bromide to confirm the size of the DNA fragment and that only one product was formed. Amplifications of the cDNA were performed using primers listed in Table S7.
Real-Time PCR
β-actin, GADPH, desmin and Men-1 levels were quantified by Real-Time PCR using the system of detection ABI PRISM 7500. β-actin and GADPH were used as control. The authenticity of the PCR products was verified by melting curve analyses and agarose gel electrophoresis. Fluorescent values were measured at 520 nm. CT values were calculated with 7500 System Software Sequence Detection System 1.3 (Applied Biosystems) and relative quantification was carried out according to User Bulletin Nr2 (Applied Biosystems, 10/2001). Amplification of the cDNA was performed using primers and temperatures listed in Table S7.
PCR Statistical Analysis
Data from desmin and Men-1 gene expression were subjected to ANOVA, followed by Tukey-Kramer Multiple Comparisons Test. Values were considered significant at p< 0.05.
Microarray Analysis
Total RNAs were prepared from UIII cells as indicated for each experiment. [33P]-labeled cDNAs from each sample were hybridized in triplicate on NIA 15K mouse cDNA microarrays (Tanaka et al., 2000). The clone set (15,264 gene features) sequence information is available at the National Institute on Aging web site http://lgsun.grc.nia.nih.gov. About one-half of the genes are novel, with about 90% sequence-verified (Kargul et al., 2001). Only sequence-verified clones are used for the analysis in this paper. We opted for this library in the case of rat cells because this is a more comprehensive set of genes whose embryo and maternal DNA sequences are more representative of the biological processes of our study (Tanaka et al., 2000). We also considered that mouse and rat genome sequences do not bear enough differences to discard their heterogeneous hybridization in benefit of the repertoire of genes tested. Hybridizations and image acquisitions were carried out as previously described (Tanaka et al, 2000). Signal intensities of individual spots were obtained with overlaid grids. Background signal intensities were obtained from media of areas where no DNA was spotted.
Treatment of microarray data and Statistical Analysis
The average of all spots with empty vector material were calculated and used to filter and normalize the data from each membrane hybridization. After transformation of data those clones for which three or more were exact zeroes were no longer considered. For each treatment and replicate, the number of null transformed data was about 70%. The standard deviations were replaced by a “smoothed” value, which is the average of the standard deviations corresponding to genes with similar differences. The smoothed standard deviation was defined as an average from 20 standard deviations. Since normality of the data cannot be assumed, null distribution was obtained by permutations. The Q-Q plots of this “null distribution” show that it can be approximated very precisely by a Student distribution with a small number of degrees of freedom (between 5 and 9, depending on the treatment) (data not shown). Given the p-values, corrected values were obtained by means of the Fold Discovery Rate (FDR) procedure of Benjamini and Hochberg (Bejamini and Hochberg, 1995). Lists of genes up- or down-regulated by at least a factor of 3 and FDR of 0.005 in decidualized cells as compared to control cells were constructed.
Gene Ontology annotation
Gene Ontology (GO) terms significantly over- or under-represented in the group of genes differentially expressed were identified with program GOstat (Beissbarth and Speed, 2004) available at http://gostat.wehi.edu.au/cgi-bin/goStat2.pl. GO terms with p-values < 0.01 after correction for the multiplicity of testing by the Benjamini (FDR) method are reported.
On-line software OntoExpress
Functional profiles using gene ontology terms of regulated genes was further constructed using the on-line software OntoExpress (Khatri et al., 2002; Khatri et al., 2004).
RESULTS
1. Characterization of UIII in vitro decidualization
To induce in vitro differentiation we cultured UIII cells in presence of 10% of DCC-FCS (dextran-coated, charcoal treated calf bovine serum). The cells were plated as previously described (Vallejo et al., 2005) and after two days in serum-free conditions medium containing 10% DCC-FCS was added (day 0) and cells were cultured for 7 days (day 7), changing the media every 2 days of culture. Initially cells grew as a monolayer of polygonal epitheliod cells (Figure 1a), showing some mitotic figures. The kinetic of cell growth shows that the number of cells per culture-well increased significantly up to day 4 of culture in DCC-FCS. Although the cells became confluent after 2 days they continued to grow, forming multilayer structures. After becoming confluent some cells started to increase in size and became binucleated. The percentage of these enlarged cells reached a maximum, 10%, at 5 days (Figure 1b).
Figure 1. UIII cells in vitro decidualization.
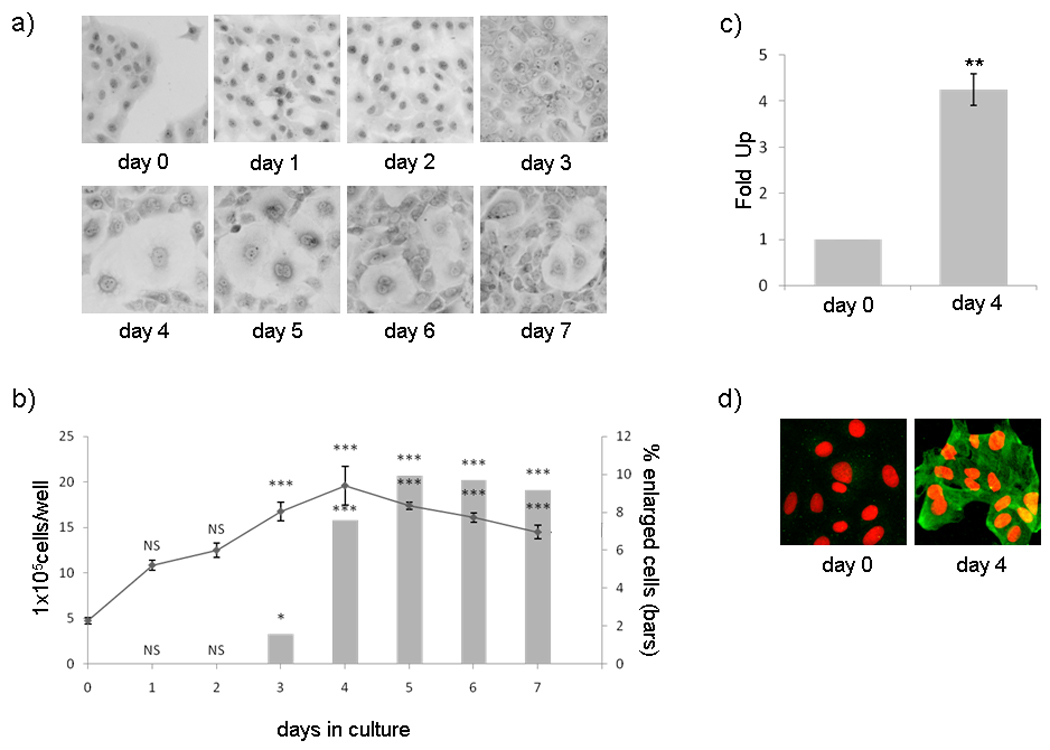
UIII cells were plated in M199 supplemented with 10% DCC-FCS, and 24 hours later, media was replaced by fresh M199 without serum. After two days in serum-free conditions (day 0), cells were cultured in presence of 10% DCC-FCS for 7 days (day 7). a) UIII cells were fixed and stained with Giemsa reagent and observed with a light microscope using a 10× objective. b) Number of total cells/well (line) and percentage of cells with decidual morphology (bars) during 7 days of cultures show the kinetic of cell growth and cell differentiation respectively. Data represent the media ± SEM of 3 independent experiments. NS P>0.05, * P<0.05, ***P<0.001 vs. control (day 0). c) Quantification of desmin mRNA by real-time PCR. Day 4 column shows fold up of desmin mRNA after 4 days in 10% DCC-FCS relative to β-actin mRNA levels. Data represent the media ± SD of three independent experiments with four independent measurements. ** P<0.01 vs. control (day 0). d) Immunofluorescence of desmin protein expression. Day 0 and day 4 cells were fixed, permeabilized and incubated with polyclonal Mouse anti-Desmin and 488 goat anti-Mouse (green). Nucleus was stained with propidium iodide (red).
Expression of the cytoskeleton protein desmin, a marker of in vivo decidualization, increased at the level of mRNAs and protein (Figure 1c and d) after 4 days of culture in the presence of DCC-FCS. Desmin expression already approaches maximum at 3 days of culture around 24 hours after the cells have reached confluence (data not shown). Maximun expression of desmin coincides with the maximum in the morphological changes, but in each experiment all cells were positive for desmin after 3–4 days in culture regardless of their morphological appearance. Therefore, desmin expression seems to be an earlier marker for decidualization than the enlarged cell size or the appearance of binucleated cells and indicates a global decidualization of the culture. We performed RT-PCR assays for dPrl, another marker of decidualization and found that dPrl is not expressed in our condition of culture during in vitro decidualization (data not shown). We also tested the expression of other decidua-related genes (Ccnd1, c-Myc, p21Cip/Kip1, Sod, Gpx, inhA/activinA, Fstl1, Gja1, PrlR, nPR, ERα, ERβ, 11̣β-HSD, 3β̣-HSD, mPRα, pgrm1, desmin) previously reported during in vivo decidualization (Sugino et al., 2006; Riesewijk et al., 2003; Jones et al., 2002; Kashiwagi et al., 2007; Grummer et al., 2004; Tan et al., 2002) showing the expected results (Figure S1).
2. Genes differentially regulated in decidualized cells
To identify genes involved in in vitro decidualization we used microarray analyses of RNAs (Tanaka et al., 2000) from UIII cells cultured in presence of 10% DCC-FCS for 0 and 96 hs. A total of 30% of cDNA probes showed significant hybridization in at least one of the two stages of UIII cells tested. We identified 577 cDNA clones (557 UniGene clusters) with significant differences in expression (≥ 3 fold, and ≤ 0.5 fold; FDR > 0.005) in decidualized versus non-decidualized cells. Of these 336 were overexpressed and 241 were underexpressed in decidualized cells after 96 h of culture (Table S1). Using the full set of protein-encoding genes on the Gene Ontology database we listed 577 IDs including 538 unique genes (data not shown), of which 322 correspond to annotated genes (Table 1 and 2, and Table S1). Only ten of these annotated genes have been previously reported to be regulated in decidual cells (marked with an asterisk in Table 1 and 2). Thus, 312 genes identified in this study have not been previously related to decidualization.
Table 1.
Statistically overrepresented GO terms
| Name | Gene Symbol |
Fold | FDR |
|---|---|---|---|
| Protein synthesis (see table S3 supp material) | |||
| Gene Expression | |||
| cold shock domain containing C2, RNA binding | csdc2 | 68.37 | 0.007 |
| transcription termination factor, RNA polymerase I | ttf1 | 9.69 | 0.002 |
| tripartite motif-containing 27 | trim27 | 9.06 | 0.002 |
| SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 2 |
smarcd2 | 7.53 | 0.002 |
| tripartite motif-containing 28 | trim28 | 7.35 | 0.002 |
| Testis-specific Y-encoded-like protein 2 | tspyl2 | 6.26 | 0.002 |
| heterogeneous nuclear ribonucleoprotein A1 | hnrnpa1 | 6.10 | 0.002 |
| forkhead box N4 | foxn4 | 5.95 | 0.002 |
| zinc finger protein 496 | zkscan17 | 5.89 | 0.002 |
| nuclear transcription factor Y, gamma | nfyc | 5.58 | 0.009 |
| leucine rich repeat (in FLII) interacting protein 1 | lrrfip1 | 5.41 | 0.009 |
| transcription factor CP2-like 1 | tcfcp2l1 | 5.37 | 0.007 |
| far upstream element (FUSE) binding protein 3 | fubp3 | 5.36 | 0.013 |
| timeless homolog (Drosophila) | timeless | 5.17 | 0.009 |
| cleavage stimulation factor, 3' pre-RNA, subunit 1, 50kDa | cstf1 | 4.87 | 0.019 |
| nuclear receptor subfamily 3, group C, member 1 | nr3c1* | 4.84 | 0.019 |
| multiple endocrine neoplasia I | men1 | 4.75 | 0.024 |
| retinoblastoma-like 2 (p130) | rbl2 | 4.69 | 0.019 |
| cold shock domain protein A | csda | 4.39 | 0.025 |
| heterogeneous nuclear ribonucleoprotein C (C1/C2) | hnrnpc | 4.22 | 0.025 |
| Wilms tumor 1 | wt1 | 4.10 | 0.019 |
| mitochondrial ribosomal protein L44 | mrpl44 | 4.06 | 0.023 |
| basic transcription factor 3 | btf3 | 3.79 | 0.024 |
| amino-terminal enhancer of split ( | aes | 3.74 | 0.042 |
| telomeric repeat binding factor (NIMA-interacting) 1 | terf1 | 3.51 | 0.023 |
| histone deacetylase 1 | hdac1 | 3.46 | 0.033 |
| TGFbeta-stimulated clone-22 domain family, member 1 | tsc22d1 | 3.42 | 0.039 |
| chromatin assembly factor 1, subunit B (p60) | chaf1b | 3.26 | 0.043 |
| bromodomain containing 7 | brd7 | 3.24 | 0.043 |
| zinc finger protein 592 | zfp592 | 3.21 | 0.045 |
| adenosine deaminase, RNA-specific, B1 (RED1 homolog rat) | adarb1 | 3.19 | 0.043 |
| NHP2 non-histone chromosome protein 2-like 1 (S. cerevisiae) |
nhp2l1 | 3.16 | 0.043 |
| small nuclear ribonucleoprotein polypeptide E | snrpe | 0.58 | 0.050 |
| tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
tanc2 | 0.58 | 0.042 |
| homeodomain interacting protein kinase 1 | hipk1 | 0.55 | 0.038 |
| alpha thalassemia/mental retardation syndrome X-linked, ATP-dependent helicase |
Atrx | 0.55 | 0.034 |
| MYST histone acetyltransferase (monocytic leukemia) 3 | myst3 | 0.54 | 0.038 |
| TAF9B RNA polymerase II, TATA box binding protein (TBP)- associated factor, 31kDa |
taf9b | 0.54 | 0.038 |
| high mobility group AT-hook 2 | hmga2 | 0.53 | 0.038 |
| polymerase (RNA) III (DNA directed) polypeptide G (32kD) | polr3g | 0.53 | 0.038 |
| TAF6 RNA polymerase II, TATA box binding protein (TBP)- associated factor, 80kDa |
taf6 | 0.51 | 0.039 |
| ankyrin repeat domain 10 | ankrd10 | 0.46 | 0.009 |
| FIP1 like 1 (S. cerevisiae) | fip1l1 | 0.45 | 0.034 |
| down-regulator of transcription 1, TBP-binding (negative cofactor 2) |
dr1 | 0.44 | 0.013 |
| PRP31 pre-mRNA processing factor 31 homolog (S. cerevisiae) |
prpf31 | 0.43 | 0.013 |
| homeobox B3 | hoxb3 | 0.42 | 0.021 |
| activating transcription factor 4 (tax-responsive enhancer element B67) |
atf4 | 0.42 | 0.002 |
| nucleosomal binding protein 1 | nsbp1 | 0.41 | 0.013 |
| SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 5 |
smarca5 | 0.39 | 0.007 |
| suppressor of variegation 3–9 homolog 1 (Drosophila) | suv39h1 | 0.38 | 0.002 |
| TFS2-M domain-containing protein 1 |
a630018 p17rik |
0.37 | 0.002 |
| polymerase (RNA) III (DNA directed) polypeptide B | polr3b | 0.36 | 0.046 |
| HLA-B associated transcript 1 | bat1a | 0.35 | 0.002 |
| RNA binding motif protein 39 | rbm39 | 0.35 | 0.002 |
| excision repair cross-complementing rodent repair deficiency, complementation group 3 (xeroderma pigmentosum group B complementing) |
ercc3 | 0.35 | 0.002 |
| retinoid X receptor, beta | rxrb | 0.33 | 0.002 |
| enhancer of zeste homolog 2 | ezh2 | 0.31 | 0.002 |
| GLI-Kruppel family member GLI2 | gli2 | 0.31 | 0.002 |
| zfpl10 | 0.30 | 0.002 | |
| zinc finger protein 367 | zfp367 | 0.18 | 0.002 |
| zinc finger with KRAB and SCAN domains 1 | zkscan1 | 0.12 | 0.002 |
| Chromosome organization and biogenesis | |||
| heterochromatin protein 1, binding protein 3 | hp1bp3 | 5.98 | 0.002 |
| glutathione peroxidase 4 (phospholipid hydroperoxidase) | gpx4* | 4.35 | 0.024 |
| Structural maintenance of chromosomes 1A | smc1a | 3.75 | 0.024 |
| nucleosome assembly protein 1-like 4 | nap1l4 | 0.51 | 0.034 |
| REC8 homolog (yeast)) | Rec8 | 0.48 | 0.034 |
| nucleolar and spindle associated protein 1 | nusap1 | 0.37 | 0.002 |
| Structural maintenance of chromosomes 3 | Smc3 | 0.32 | 0.002 |
| Structural maintenance of chromosomes 4 | Smc4 | 0.27 | 0.002 |
|
Regulation of anatomical structure morphogenesis/cell shape/epithelial cell development |
|||
| shroom family member 3 | shroom3 | 8.51 | 0.002 |
| dynein, light chain, LC8-type 1 | dynll1 | 4.62 | 0.031 |
| guanine nucleotide binding protein (G protein), alpha 13 | gna13 | 3.51 | 0.023 |
| guanine nucleotide binding protein (G protein) alpha 12 | gna12 | 3.41 | 0.045 |
| craniofacial development protein 1 | cfdp1 | 0.35 | 0.002 |
| gap junction protein, beta 3, 31kDa | gja1* | 0.21 | 0.002 |
| Response to temperature stimulus | |||
| angiotensinogen (serpin peptidase inhibitor, clade A, member 8) |
agt* | 23.24 | 0.002 |
| heat shock 70kDa protein 8 | hspa8 | 5.55 | 0.009 |
| Crystalline, alpha B | cryab | 4.85 | 0.009 |
| heat shock protein 90kDa alpha (cytosolic), class B member 1 |
hsp90ab 1 |
3.11 | 0.046 |
| Latexin | lxn | 0.54 | 0.039 |
Functional gene categories were obtained using GOstat program over a subset of differentially expressed genes (≥ 3 fold and ≤ 0.5 fold, FDR > 0.005) derived from microarray analyses. Protein synthesis, Gene espression, Chromosome organization, regulation of anatomical structure morphogenesis/cell shape/epithelial cell development and response to temperature stimulus were the most representative GO terms. (P-Value Cutoff: 0.1; GO-Cluster Cutoff: -1; Correct Method: Benjamini). Genes in two terms were listed in the category with the highest P-value).
Table 2.
Differentiation and related function terms
| Term function | Gene Name | Gene Symbol |
Fold | FDR |
|---|---|---|---|---|
| 1. Differentiation | ||||
| Tetraspanin 5 | Tspan5 | 48.70 | 0.002 | |
| Fk506-binding protein 52 | Fkbp4* | 6.20 | 0.009 | |
| Forkhead box N4 | Foxn4 | 5.90 | 0.009 | |
| Bone morphogenetic protein 1 | Bmp1 | 4.90 | 0.009 | |
| THO complex 5 | Thoc5 | 4.70 | 0.023 | |
| Wilms tumor homolog 1 | Wt1 | 4.10 | 0.046 | |
| Amino terminus of drosophila enhancer of split groucho |
Aes | 3.70 | 0.024 | |
| Guanine nucleotide-binding protein alpha-12 subunit | Gna12 | 3.40 | 0.002 | |
| Prolactin receptor | PrlR | 3.20 | 0.043 | |
| Epimorphin | Epim | 0.24 | 0.002 | |
| Enhancer of zeste homolog 2 | Ezh2 | 0.31 | 0.002 | |
| Drebrin 1 | Dbn1 | 0.52 | 0.003 | |
| 2.Cell proliferation, cell division or cell cycle (excluding those above or in Table 2) | ||||
| Bcl2 associated atanogene 4/Silencer of death domains | Bag4/SODD | 54.20 | 0.003 | |
| Angiotensin-1 | Agt | 23.20 | 0.003 | |
| S100 calcium-binding protein A6 (calcyclin) | S100a6* | 10.60 | 0.003 | |
| CDC28 protein kinase regulatory subunit 1B | Cks1b | 6.70 | 0.009 | |
| TSPY-like 2 | Tspyl2 | 6.30 | 0.025 | |
| Interferon induced transmembrane protein 3 | Ifitm3 | 5.60 | 0.009 | |
| Heat shock 70kDa protein 8 | Hspa8 | 5.60 | 0.009 | |
| Timeless homolog (Drosophila) | Timeless | 5.30 | 0.009 | |
| Retinoblastoma-like 2 (p130) | Rbl2 | 4.70 | 0.020 | |
| cyclin Y | Ccny | 3.70 | 0.019 | |
| Ribosomal protein S27 (Wdr79) | Rps27 | 3.60 | 0.003 | |
| Telomeric repeat binding factor (NIMA-interacting) 1 | Terf1 | 3.50 | 0.002 | |
| Inhibitor of growth family member 4 | Ing4 | 3.20 | 0.040 | |
| integral membrane protein 2B | Itn2b | 6.80 | 0.003 | |
| clusterin | Clu | 5.10 | 0.009 | |
| DNA fragmentation factor, 40kDa, beta polypeptide (caspase-activated Dnase) |
Dffb | 0.20 | 0.003 | |
| GLI-Kruppel family member GLI2 | Gli2 | 0.31 | 0.003 | |
| craniofacial development protein 1 | Cfdp1 | 0.35 | 0.003 | |
| Growth arrest-specific 1 | Gas1 | 0.40 | 0.007 | |
| ubiquitin-activating enzyme E1C | Ube1c | 0.40 | 0.009 | |
| Collagen, type XVIII, alpha 1 | Col18a1* | 0.56 | 0.043 | |
| Transformation related protein 53 binding protein 2 | Trp53bp2 | 0.56 | 0.043 | |
| 3. Cell communication, signaling, kinase (excluding those above or in Table 2) | ||||
| Rhophilin, Rho GTPase binding protein 2 | Rhpn2 | 25.90 | 0.025 | |
| Protein tyrosine phosphatase, receptor type, F | Ptprf | 12.00 | 0.003 | |
| Protein tyrosine phosphatase, non-receptor type 6 | Ptpn6 | 6.30 | 0.002 | |
| Dickkopf homolog 3 | Dkk3 | 5.60 | 0.046 | |
| Dual specificity phosphatase 16 | Dusp16 | 5.60 | 0.009 | |
| Signal peptidase complex subunit 2 homolog | Spcs2 | 4.70 | 0.023 | |
| Interleukin 11 receptor, alpha chain 2 | Il11ra2* | 3.50 | 0.030 | |
| Phosphoinositide-3-kinase, class 2, alpha polypeptide, |
Pik3c2a | 0.13 | 0.002 | |
| Ankyrin repeat and SOCS box-containing 13 | Asb13 | 0.15 | 0.003 | |
| Zinc finger, AN1-type domain 6 | Zfand6 | 0.19 | 0.002 | |
| Gap junction protein, alpha 1, | Gja1* | 0.21 | 0.002 | |
| Guanine nucleotide binding protein beta polypeptide 2-like1 |
Gnb2l1 | 0.55 | 0.038 | |
| 4. Extracellular matrix | ||||
| decorin | dcn* | 14.50 | 0.019 | |
| biglycan | bgn* | 10.40 | 0.003 | |
| matrilin 2 | matn2 | 7.83 | 0.003 | |
| follistatin-like 1 | fstl1* | 5.80 | 0.034 | |
| ADAM metallopeptidase with thrombospondin type 1 motif, 19 |
adamts19 | 0.26 | 0.000 | |
| collagen, type III, alpha 1 | col3a1 | 0.29 | 0.003 | |
| glycosylphosphatidylinositol specific phospholipase D1 |
gpld1 | 0.45 | 0.034 | |
Functional gene categories were obtained using Onto-express program over Up and Down differentially expressed gene (≥ 3 fold and ≤ 0.5 fold, FDR > 0.005) derived from microarray analyses.
Reported in decidualization processes.
3. Global functional classification
For global functional classification, functional annotations were derived from the NIA publicly available database, http://lgsun.grc.nia.nih.gov. We assigned one function to each up- or down-regulated gene in decidualized cells. The functions were apoptosis, cell cycle, DNA replication, energy/metabolism, heat shock/stress, matrix/structural proteins, protein synthesis/translational control, signal transduction, transcription/chromatin, and unknown. The percentage distribution for all differentially regulated genes in these functions is represented in Table S2 which also indicates up-regulated and down-regulated genes in decidualized cells. We found that protein synthesis (14%), signal transduction (6.5%), matrix/structural proteins (5.5%) and transcription/chromatin (4.5%) were the functions encompassing more differentially regulated genes (Table S2). The number of genes involved in protein synthesis with up-regulation after differentiation was higher than the number of genes down regulated after this event (56 up-regulated, 27%, vs. 6, 2%, down-regulated). Conversely, gene numbers related to signal transduction (19 up-regulated vs. 18 down-regulated), matrix/structural proteins (18 up-regulated vs. 14 down-regulated) and transcriptional regulation and chromatin remodeling (16 up-regulated vs. 9 down-regulated) showed a similar amount of positively and negatively regulated genes (Table S2).
3 a. Overrepresented functions
In a complementary approach we mapped the group of genes differentially expressed using the GOstat program (Beissbarth and Speed, 2004).
Again we found protein synthesis related functions as the most overrepresented functions (P-value: 1.65e-11) (Table S3). Among them there are 41 ribosomal protein genes.
Four additional non-related terms were identified as significantly over-represented functions (Table 1). The second term gene expression (P-value: 6.82e-05) included 102 genes, 41 of which were already listed in protein synthesis. Of the remaining 61 genes, 32 were up regulated and 29 were down regulated (Table 1). Within these 61 genes, if we limit the search to the subset of GO hierarchy that contains the keyword chromatin, we found that the most over-represented was the function establishment or maintenance of chromatin architecture, with 14 genes (P-value: 0.0734). This group includes 8 up regulated genes (Ttf1, Smarcd2, Trim28, Tspyl2, Nr3c1, Men1, Rbl2, and Hdac1) and 6 down regulated genes (Atrx, Myst3, Hmga2, Nsbp1, Smarca5, Ezh2, and Gli2 (Table 1 and Table S4). Within this group of genes we found 26 genes related to the keyword transcription factor (Table S5).
The third overrepresented term Chromosome organization and biogenesis (P-value: 0.206) contains term functions related to chromatin modification and nucleosome assembly. In this term there were 15 genes, of which 7 were already listed in gene expression term (Smarcd2, Men1, Terf1, Suv39h1, Myst3, Ttf1, Hmga2). Of the other 8 genes, 3 were up regulated (Hp1bp3, Gpx4, Smc1a) and 5 were down regulated (Nap1l4, Rec8, Nusap1, Smc3, and Smc4).
The fourth overrepresented terms were regulation of anatomical structure morphogenesis/cell shape/ epithelial cell development (P-value: 0.041, P-value: 0.041, and P-value: 0.0739) with 8 genes. The last overrepresented term was response to temperature stimulus (P-value: 0.0934) with 5 genes.
3 b. Differentiation and related functions
To identify genes that had been reported in cell differentiation or connected processes (i.e. cell proliferation, cell division, cell cycle, signaling and extracellular matrix), we used the online program Onto/Express (http://vortex.cs.wayne.edu/projects.html). This program produced an output of 235 function terms in our query of 199known genes (Table S6). The function terms related with differentiation and related processes are listed in Table 2. We found 12 genes associated with differentiation, 18 genes related to cell proliferation, cell division or cell cycle, 18 genes related to signaling terms and 9 genes related to extracellular matrix.
4. Experimental validation of microarray results
To validate results obtained with a robust statistical methodology (Material and Methods) we selected four up regulated (Figure 2) and four down regulated genes (Figure 3), to test by RT-PCR. We selected genes well characterized in decidualization and new genes, one of them the most upregulated and the others connected with the function of chromatin modification, which is a process of our interest. Three of these 8 genes, Fstl1 (Kaiser et al., 1990), PrlR (Gu et al., 1996) and Gja1 (Grummer et al., 2004) have been previously reported to be expressed during decidualization. The other five, Eef1a1, Ovgp1, Tspan5, Myst3 and Suv39h1, have not been previously described in decidualized cells.
Figure 2. Experimental Microarrays validation of up-regulated genes.
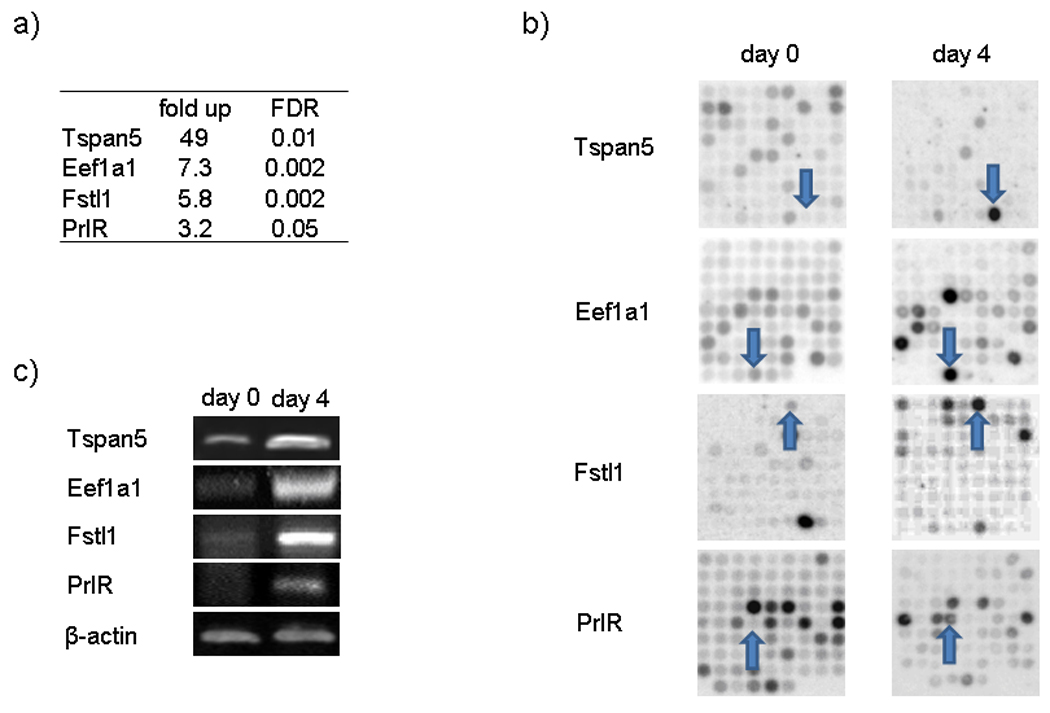
UIII cells were cultured as it was described in Figure 1. After two days in serum-free conditions (day 0, non-decidualized cells), cells were cultured in presence of 10% DCC-FCS for 4 days (day 4, decidualized cells). Day 0 and day 4 cells were resuspended and RNA was extracted for microarray and RT-PCR experiments. a) Table with microarray data of selected genes. Genes up-regulated by at least a factor of 3 and FDR of 0.005 in decidualized cells as compared to control cells were selected. b) Membrane scanning of microarrays of selected genes. c) RT-PCR of selected genes at day 0 (non-decidualized cells) and day 4 (decidualized cells).
Figure 3. Experimental Microarrays validation of down-regulated genes.
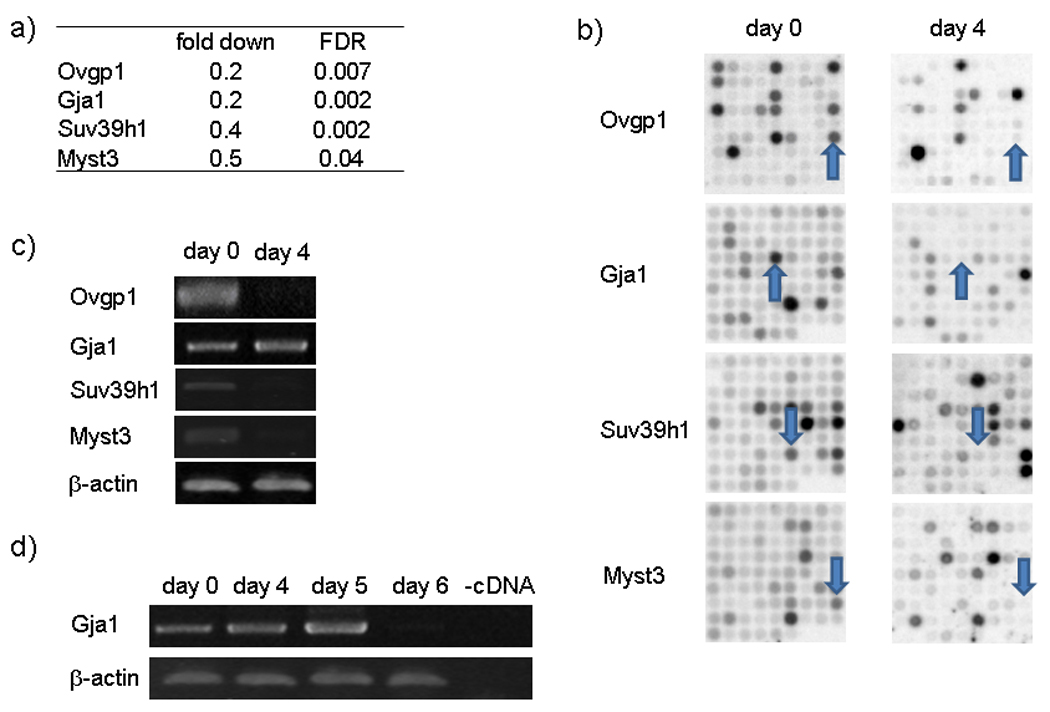
UIII cells were cultured as it was described in Figure 1. After two days in serum-free conditions (day 0, non-decidualized cells), cells were cultured in presence of 10% DCC-FCS for 4 days (day 4, decidualized cells). Day 0 and day 4 cells were resuspended and RNA was extracted to microarray and RT-PCR experiments. a) Table with microarray data of selected genes. Genes down-regulated by at least a factor of 3 and FDR of 0.005 in decidualized cells as compared to control cells were selected. b) Membrane scanning of microarrays of selected genes. c) RT-PCR of selected genes at day 0 (non-decidualized cells) and day 4 (decidualized cells). d) Gja1 RT-PCR six days kinetic expression.
The mean of fold changes in mRNAs hybridization after decidualization and their statistic parameter FDR obtained from three independent experiments are shown in Figure 2a and 3a. Representative spots for each gene are shown in Figure 2b and 3b. The Gja1 mRNA was not significantly expressed at day 4, but a consistent decrease of their mRNA was observed at 6 days of culture (Figure 3d). Taken together, the results show that the changes observed for these eight genes in the microarray analysis were confirmed by RT-PCR (Figure 2c and 3c), supporting the reliability of the array data.
5. In vivo validation of gene expression changes during in vitro decidualization
To evaluate the physiological relevance of the gene expression changes observed during UIII cell differentiation we have performed PCR studies on RNA from non-pregnant uterus and decidual 8 d.p.c. rat tissues. For this study we selected seventeen genes that change expression in differentiated UIII cells, including Cdsdc2 (68.4 fold), Trim27 (9.06 fold), Ef-1 (7.27 fold), Fkbp4 (6.18 fold), Fstl1 (5.8 fold), Bmp1 (4.90 fold), Men1 (4.75 fold), Aes (4.74 fold), Wt1 (4.10 fold), Gna12 (3.40 fold), Myst3 (0.54 fold), Suv39h1 (0.38 fold), Epim (0.24 fold) Ovpg1 (0.21); and 3β-HSD, Prl and desmin not included in the array. Of the seventeen genes tested, seven are within the overrepresented GO term Gene Expression and another seven are within the GO term Differentiation.
To study the in vivo changes of these genes we performed semiquantitative PCR analysis of mRNA levels in rat decidua for Csdc2, Trim27, Fkbp4, Fstl1, Bmp1, Wt1, Aes, Gna12, Myst3, Suv39h1, Epim, Ovpg1, 3β-HSD, and desmin are shown in Figure 4a and in Figure S2. The up-regulation of Fkbp4 and downregulation of Myst3, Suv39h1, Epim and Ovgp1 found by the gene array in decidualized cells and verified by PCR was not seen in the 8dpc decidua. In contrast, Fkbp4 is decreased and Myst3, Suv39h1, Epim and Ovpg1 are increased in 8dpc decidual tissue. The results showed that the mRNAs of Csdc2, Trim27, Eef1a1, Fstl1, Bmp1, Wt1, Aes, Gna12, Men1, 3β-HSD and desmin from 8dpc decidual tissue behave as in decidualized UIII cells (Figure 4a and b). The quantitative analysis of Men1 mRNA using real-time PCR also showed an increase of 2.06 ± 0.56 fold in decidual tissue from 8 d.p.c. rat uterus as compared to non-pregnant uterus (Figure 4b). Thus, gene expression data from whole decidua of 8 d.p.c. implantation sites exhibited similar changes to those observed in decidualized UIII cells in eleven out of seventeen genes (65% of the tested genes). These findings make the results of the UIII microarrays biologically meaningful.
Figure 4. In vivo gene expression of genes found during in vitro UIII differentiation.
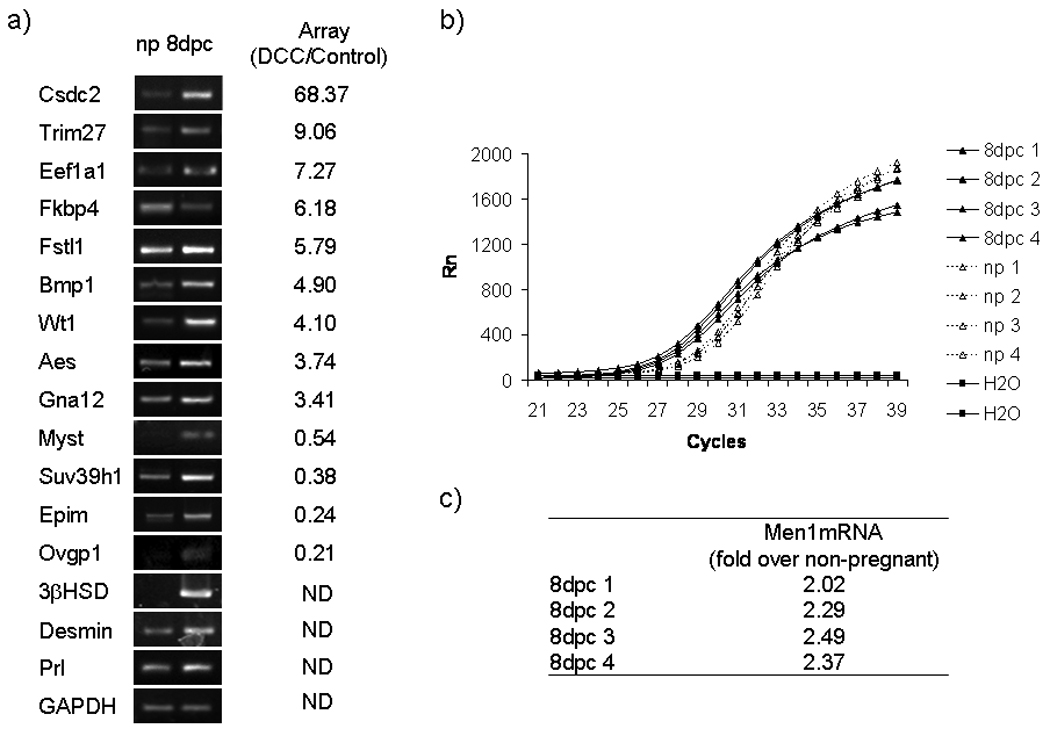
a) Total RNA from non-pregnant uterus (np) and implantation sites from 8 d.p,c uterus (8 pdc) were extracted for RT-PCR. The figure shows ethidium bromide-stained gels of RT-PCR products for: Cdsdc2, Trim27, Eef1a1, Fkbp4, Fstl1, Bmp1, Wt1, Aes, Gna12, Myst3, Suv39h1, Epim, Ovpg1, β-HSD, desmin, Prl and GAPDH. Fold changes from microarray data of selected genes are shown. ND: not determined. b) Quantitation of Men1 mRNA in non-pregnant (np) and 8 d.p.c. in uterus. The figure shows fluorescence intensity pattern of real time PCR products for Men1 from four independent samples of non-pregnant mRNA (np), four independent samples of 8 d.p.c mRNA (8dpc) and two samples without template (H2O). c) Table shows fold up of Men1mRNA from 8 d.p.c. over non-pregnant relative to GAPDH mRNA levels. Data represent the media of four independent experiments.
DISCUSSION
We find that UIII cells behave as a homogeneous population and that all cells express the decidual marker protein desmin after 4 days of culture in the presence of DCC-FCS. This makes UIII cells suitable for global gene expression analysis. We found similar morphological changes as well as changes in the expression of other decidua-related genes (Ccnd1, c-Myc, p21Cip/Kip1, Sod, Gpx, inhA/activinA, Fstl1, Gja1, PrlR, nPR, ERα, ERβ, 11̣β-HSD, 3β-HSD, mPRα, pgrm1, desmin) as previously reported during in vivo decidualization (Sugino et al., 2006; Riesewijk et al., 2003; Jones et al., 2002; Kashiwagi et al., 2007; Grummer et al., 2004; Tan et al., 2002). Moreover, we describe a set of new genes that exhibit characteristic changes in expression as determined by cDNA arrays and validated in physiological uterine decidua. The subset of genes validated includes Csdc2, Trim27, Eef1a1, Fstl1, Bmp1, Wt1, Aes, Gna12, Men1, 3β-HSD and desmin. We therefore assume that the values obtained by microarrays represent real changes in mRNA levels of UIII. Another set of genes detected by microarrays and validated in in vitro samples, Myst3, Suv39h1, Epim and Ovgp1, did not show a similar behavior in in vivo decidua. This suggests that decidualization induced by steroid hormone deprived FCS with cultured stromal cells may not precisely mimic the decidualization observed in uterus under the influence of ovarian hormones. This may be due to the absence of the hormonal stimuli or to the lack of other cellular components present in the endometrium, such as epithelial cells and infiltrating blood cells.
Regarding the physiological meaning of the decidualization protocol with DCC-FCS, which is a media free of steroid hormones, we think that since EGF activates a crosstalk with steroid hormone receptors in cells derived from human breast cancer (MCF-7) and human prostate cancer (LNCaP) (Migliaccio et al., 2006), it is conceivable that EGF or other growth factors acting through a similar pathway could be at work in serum induced decidualization. Moreover, treatment with EGF further increases desmin expression in presences of 10% DCC-FBS in UIII cells (Vallejo et al., unpublished data).
In addition to desmin, we have tested dPrl as a marker of decidualization to support the fact that our model reproduces known changes involved in in vivo decidualization. While the marker was expressed in decidua from 8dpc, we did not detect dPrl in UIII cells after 4 days in culture with 10% DCC (data not shown). The expression of dPrl has been reported in response to medroxyprogesterone acetate (MPA), but not in cells decidualized under influence of progesterone and estradiol (Matsumoto, 2009). There is evidence that dPrl appeared at later time points, in presence of FBS (Prigent-Tessier et al., 2001). As we are interested in early changes in gene expression that may precede the appearance of dPrl we have not studied the late stages of decidualization.
In the following we will briefly discuss some of the genes we found differentially expressed in UIII cells cultured for 4 days in the presence of 10% DCC-FCS. We will start by genes related to the overrepresented Gene Ontology (GO) terms protein synthesis that we will not describe in detail, and gene expression that includes chromatin dynamics and transcription factor activity. Then we will move on to more interesting genes related to differentiation or similar processes (cell proliferation, cell cycle, cell division and embryogenesis), and extracellular matrix, two processes strictly connected with decidualization. In each case we will first mention the genes that have already been related to decidualization and continue to discuss the genes for which this relationship has not been previously established.
Genes involved in protein synthesis
It has been reported that there are two periods of rapid RNA biosynthesis in the uterus during early pregnancy (Heald and O’Grady, 1970), one on the 2nd day after pregnancy and the other on the 4th day. The second increase in RNA synthesis coincides with the beginning of the decidual differentiation of endometrial stromal cells. The significant overrepresentation of genes that were encompassed in GO terms protein synthesis and gene expression during in vitro decidualization of UIII cells correlates with the reported increase in RNA synthesis in the in vivo differentiation of endometrial cells. In particular the increase in mRNAs for ribosomal proteins and ribosome related factors, indicates a need for increased protein synthesis that may well be connected to the increase in size of the decidual cells and to their enhanced secretory activity. As the genes in this functional class have no other specific functions, we will not discuss them any further.
Genes related to gene expression
This category includes 61 genes of which 14 are related to chromatin structure and dynamics and 26 encode transcription factors. We will discuss below validated genes only, which were selected because of the interest of their specific functions in decidualization. Only one of them, Wt1, has been previously related to decidual differentiation (Makrigiannakis et al, 2001). The Wt1 gene is expressed in human endometrial stromal cells (ESCs). Its mRNA and protein levels remain constant in the proliferative and secretory phase of the menstrual cycle. Wt1 mRNA and protein expression increases significantly in ESCs when these cells differentiate into decidual cells. Here we found a similar increase of Wt1 during UIII differentiation, possibly connected to the decrease of receptors for both IGF-I and II and induced endometrial stromal arrest. Such restriction of IGF action is consistent with the proposed role of decidualization- induced IGFBP-1 during ESC differentiation (Giudice et al., 1992). Wt1 also suppresses growth factors encoding genes such as macrophage colony-stimulating factor (M-CSF), and TGF-β1 (Harrington et al., 1993; Dey et al., 1994). In the group of genes encompassing a differentiation function we found positive changes in other protein that interact with M-CSF as negative regulator or macrophage differentiation, Thoc5 (THO complex 5, up 4.7 fold in decidualized cells over non-decidualized) (Tamura et al., 1999). M-CSF provides critical regulation of prostaglanding production in human cultured endometrial stromal cells (Wang et al., 2006) and its regulation could imply a modulation of the activity M-CSF/prostaglandin cytokine axis in endometrial cells. The other six (Csdc2, Trim27, Men1, Aes, Myst3 and Suv39h1) have not been previously described in decidualization.
Csdc2
(Cold shock domain c2, up 68.4 fold), belongs to the so-called Y-box-proteins, which form a highly conserved family of RNA binding proteins present in all organisms, from bacteria to humans. The Y-box proteins exhibit single-stranded (ss) DNA and/or ssRNA-binding activity. Several members of the family have been suggested to couple the nuclear history of a transcript to its translational fate. Csdc2 is expressed at high levels in brain cells and might be a good candidate to regulate the synthesis of specific proteins in response to extracellular stimuli (Derrigo et al., 2000 ).
Trim27
(tripartite motif-containing 27, up 9.06-fold), contains a TRIM motif that includes three zinc-binding domains, a RING, a B-box type 1 and a B-box type 2, and a coiled-coil region. Trim27 localizes to the nuclear matrix and interacts with the enhancer of polycomb that represses gene transcription. It is also thought to be involved in the differentiation of male germ cells (Reymond et al., 2001).
Men1
(menin, up 4.8-fold) gene encodes a tumor suppressor protein associated with a histone methyltransferase complex (Yokoyama et al., 2004). Men1 is localized in the nucleus and inhibits transcriptional activation by JunD (Agarwal et al., 1999). Men1, is a key epigenetic player as a tumor suppressor in cancer pathogenesis (Yokoyama et al., 2008) and increases consistently in rat uterine decidual tissue.
Aes
(amino terminal enhancer of split, up 3.7-fold) encodes a protein with high homology to the amino terminus of Drosophila enhancer of split groucho, a negative regulator of transcription during neural differentiation (Paroush et al., 1994), and is involved in actin filament organization to generate distinct cellular morphologies (Kiger et al., 2003). Aes like Dbn1 (debrin) and Rhpn (Rhophilin, Rho GTPase binding protein 2) might be involved in the actin cytoskeleton changes that accompany morphological changes observed during decidualization.
Myst3
(MYST histone acetyltransferase 3, down 0.543-fold) is a histone acetyltransferase involved in inhibition of the differentiation of M1 myeloid cells into monocytes/macrophages (Kitabayashi et al., 2001) and Suv39h1 (suppressor of variegation 3–9 homolog 1 Drosophila, down 0.381-fold) is a histone-lysine N-methyltransferase involved in gene silencing through chromatin modification in somatic cells (O’Carroll et al., 2000). Suv39h1 had been reported to play an important role in the transition from proliferation to differentiation of muscle cells, preventing gene expression trough interaction with MyoD (Mal, 2006). UIII decidualization is a suitable system to perform mechanistic studies on the connection between chromatin modifiers and cell differentiation.
Genes related to cell differentiation
Of the 12 genes related to differentiation, Fkbp4 (Tranguch et al., 2005), Wt1 (Makrigiannakis et al., 2001) and PrlR (Gu et al., 1996) have been previously reported in decidual cells. Wt1 has been discussed above in gene expression related genes. The other 9 genes in this class (Tspan5, Aes, Bmp1, Foxn4, Gna12, Thoc5, Epim, Ezh2, Dbn1) have not been previously related to the decidualization process.
Bmp1
(Bone morphogenetic protein 1, up 4.9-fold) encodes a secreted astacin metalloprotease that cleaves the COOH-propeptide of procollagen I, II and III. BMP1 plays key roles in regulating the formation of the extracellular matrix, so BMP1 could play an important role in matrix modification in the decidualization process (Suzuki et al., 1996).
Gna12
(Guanine nucleotide-binding protein alpha-12 subunit, up 3.4-fold), also known as G12, encodes a member of a G12 subfamily of G heterotrimeric proteins. The activated forms of G12 and G13 -G 12Q229L and G 13Q229L- were found to cause transformation of fibroblasts, to activate the JNK pathway, to activate the serum response element, and to regulate different isoforms of Na+H+ exchangers (Prasad et al., 1995). It has been demonstrated that G13 plays a role in blood vessel development (Gu et al, 2002). Regulation of Gna12 mRNA suggests that G12 might participate in the generation of new blood vessels in uterine decidua.
Epim
(epimorphin, down 0.24-fold) encodes the morphogenic protein epimorhin that is involved in cell sorting for branching, in mammary gland alveolar hyperplasia, in stem cell patterning of reproductive tissues like ovarian and testis, and in remodeling processes (Radisky et al., 2003; Von Schalburg et al., 2006). Epim increases slightly during the pre-adhesion phase of implantation, diminishes during the post-adhesion phase, and gathers new strength when outgrowth occurs. An active role for Epim has been proposed in the outgrowth of blastocyst (Qin et al., 2005).
If the concept of differentiation is enlarged to include related terms such as cell proliferation, cell division, cell cycle, signaling and extracellular matrix, we found another 35 genes showing altered expression during decidualization of UIII cells. Only 4 of them -Col18a1 (Pollheimer et al., 2004), Il11ra (White et al., 2004; Li et al., 2008); S100a6 (Farnsworth et al., 1998), Gja1 (Grummer et al., 2004) - have been previously reported in decidual cells.
Gja1
(Gap junction protein alpha 1, up 5-fold) encodes a component of gap junctions also known as connexin-43, which is involved in cell-cell contacts. Connexin-43 is found in heart, liver and peripheral nervous tissue and is enhanced during in vivo decidualization (Grummer et al., 2004). Gja1 expression is reduced during decidualization of UIII cells, suggesting that decidualization induced by steroid hormone deprived FCS may not be as complete as observed in uterus under the influence of ovarian hormones. Underexpression of Gja1 is also an early marker of breast cancer and seminoma development (Roger et al., 2004) and its repression in UIII cells treated with DCC-FCS may have to do with the enhanced cell proliferation in monolayer culture with reduced cell-cell contacts.
The other most regulated genes in this class, which have not been related to decidualization nor validated, are Bag4 and Agt.
Bag4
(Bcl2 associated atanogene 4/Silencer of death domains, up 54.2-fold) is a member of the BAG1-related protein family with an anti-apoptotic role (Knee et al., 2001). Bag1 and Bag4 are regulators of heat shock protein 70 kDa (Hsp70/Hsc70) family proteins that interacts with steroid hormone receptors (Knee et al., 2001). Bag4 binds to the death domain of Tumor necrosis factor receptor type 1 in the absence of TNF (Jiang et al., 1999). The high increase in the expression of this gene during decidualization could provide a protection against apoptosis during the implantation period.
The up-regulation of Agt (angiotensinogen, up 23.2-fold) during decidualization adds evidence of a distinct renin-angiotensin-aldosterone system in the uterus with a multiplicity of roles unrelated to its primary functions (Hassan et al., 2000).
Genes related to extracellular matrix remodeling
Our main focus was on identifying genes and mechanisms triggering differentiation rather than on changes that result from differentiation, i.e. extracellular matrix remodeling. However, changes in extracellular matrix were detected in our study. Among genes related to extracellular matrix, Dcn (decorin, up 14.4-fold) (San Martin et al., 2003), Bgn (biglycan, up 10.4-fold) (San Martin et al., 2003) and Fstl1 (Kaiser et al., 1990) have been previously described as expressed in decidual cells and can be related to extracellular matrix reorganization necessary during decidua development.
Fstl1
(follistatin like protein1, up 5.8-fold) encodes a protein like follistatin. Fstl1 binds Activin A, antagonizes binding to its membrane receptor in decidual cells (Arai et al., 2003), and thus interferes with Activin A promoted decidualization. Decidual cell secreted Activin-A, Inhibin and Follistatin, orchestrate a sophisticated autocrine regulation of differenciation (Kaiser et al., 1990). Increased expression of IGF and activin A neutralizing factors (i.e., HtrA1 and Fstl3) is higher in uterine decidua than in artificially induced deciduomata, correlating with a reduction in the number of mitosis, tissue growth, and mitogenic signaling (Kashiwagi et al. 2007). Since our objective was to develop a cell culture model for studying the physiological decidual reaction, we did not compare in detail our microarray data from UIII cells with pseudopregnant deciduomas. However, our results and those of Giudice’s (Kao et al., 2002) support the notion that the follistatin-like protein family is induced during decidualization independently of the presence of the implanting embryo.
CONCLUSIONS
The changes in expression of Fstl1, PrlR, Wt1, FKbp4, Col18a1, Gja1, 2Il11ra2, S100a6, Dcn, and Bgn observed by microarray analysis are compatible with previously reported findings during in vivo decidualization and Csdc2, Trim27, Eef1a1, Fstl1, Bmp1, Wt1, Aes, Gna12, 3βHSD and desmin in vivo validated genes support the usefulness of the UIII cell line as a cell culture model for studying the decidual reaction.
The observed changes in expression of genes not previously related to the decidual reaction are of potential relevance. Changes in genes involved in epigenetic pathways and chromatin remodeling point to the relevance of this level of gene regulation for the implementation of the decidual reaction. The UIII cell culture model will allow mechanistic studies to define the transcriptional network regulating reprogramming of stromal cells into decidual cells by knockdown and Chip-on-chip experiments.
Supplementary Material
Acknowledgments
This research was supported in part by the Intramural Research Program of the NIH, National Institute on Aging; by grants to P.S. from Agencia Nacional de Promoción Científica y Tecnológica (PICT 5-34086) Argentina, Universidad de Buenos Aires (Proyecto UBACYT 2004–2007, Código X318), Consejo Nacional de Investigaciones Científicas y Técnicas (PIP 2005–2006); and by grants to M.B. from CRG, DURSI, and MEC (BFU2006-10693). P.S. is an established investigator from the Consejo Nacional de Investigaciones Científicas y Técnicas.
Footnotes
Summary Sentence: The transcriptome analysis of in vitro decidualized cells reveals changes in genes involved in chromatin remodeling that may have novel functional significance of gene regulation in in vivo decidualization.
REFERENCES
- Agarwal SK, Guru SC, Heppner C, Erdos MR, Collins RM, Park SY, Saggar S, Chandrasekharappa SC, Collins FS, Spiegel AM, Marx SJ, Burns AL. Menin Interacts with the AP1 Transcription Factor JunD and Represses JunD-Activated Transcription. Cell. 1999;96:143–152. doi: 10.1016/s0092-8674(00)80967-8. [DOI] [PubMed] [Google Scholar]
- Arai KY, Tsuchida K, Uehara K, Taya K, Sugino H. Characterization of rat follistatin-related gene: effects of estrous cycle stage and pregnancy on its messenger RNA expression in rat reproductive tissues. Biol Reprod. 2003;68(1):199–206. doi: 10.1095/biolreprod.102.008565. [DOI] [PubMed] [Google Scholar]
- Bany BM, Zhang X, Kennedy TG. Effects of epidermal growth factor and interleukin-1alpha on plasminogen activator secretion and decidualization in rat endometrial stromal cells. Biol Reprod. 1998;59(1):131–135. doi: 10.1095/biolreprod59.1.131. [DOI] [PubMed] [Google Scholar]
- Beissbarth T, Speed TP. GOstat: find statistically overrepresented Gene Ontologies within a group of genes. Bioinformatics. 2004;20(9):1464–1465. doi: 10.1093/bioinformatics/bth088. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J Roy Stat Soc B. 1995;57:289–300. [Google Scholar]
- Carson DD, Dutt A, Tang JP. Glycoconjugate synthesis during early pregnancy: hyaluronate synthesis and function. Dev Biol. 1987;120(1):228–235. doi: 10.1016/0012-1606(87)90120-5. [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenolchloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Cohen H, Pageaux JF, Melinand C, Fayard JM, Laugier C. Normal rat uterine stromal cells in continuous culture: characterization and progestin regulation of growth. Eur J Cell Biol. 1993;61:116–125. [PubMed] [Google Scholar]
- Dey BR, Sukhatme VP, Roberts AB, Sporn MB, Rauscher FJ, 3rd, Kim SJ. Repression of the transforming growth factor-beta 1 gene by the Wilms' tumor suppressor WT1 gene product. Mol Endocrinol. 1994;8(5):595–602. doi: 10.1210/mend.8.5.8058069. [DOI] [PubMed] [Google Scholar]
- Derrigo M, Cestelli A, Savettieri G, Di Liegro I. RNA-protein interactions in the control of stability and localization of messenger RNA. Int J Mol Med. 2000;5(2):111–123. [PubMed] [Google Scholar]
- Dey SK, Lim H, Das SK, Reese J, Paria BC, Daikoku T, Wang H. Molecular cues to implantation. Endocr Rev. 2004;25:341–373. doi: 10.1210/er.2003-0020. [DOI] [PubMed] [Google Scholar]
- Farnsworth RL, Talamantes F. Calcyclin in the mouse decidua: expression and effects on placental lactogen secretion. Biol Reprod. 1998;59(3):546–552. doi: 10.1095/biolreprod59.3.546. [DOI] [PubMed] [Google Scholar]
- Glasser SR, Julian J. Intermediate filament protein as a marker of uterine stromal cell decidualization. Biol Reprod. 1986;35(2):463–474. doi: 10.1095/biolreprod35.2.463. [DOI] [PubMed] [Google Scholar]
- Giudice LC, Dsupin BA, Irwin JC. Steroid and peptide regulation of insulin-like growth factor-binding proteins secreted by human endometria stromal cells is dependent on stromal differentiation. J Clin Endocrinol Metab. 1992;75:1235–1241. doi: 10.1210/jcem.75.5.1385468. [DOI] [PubMed] [Google Scholar]
- Grummer R, Hewitt SW, Traub O, Korach KS, Winterhager E. Different regulatory pathways of endometrial connexin expression: preimplantation hormonal-mediated pathway versus embryo implantation-initiated pathway. Biol Reprod. 2004;71(1):273–281. doi: 10.1095/biolreprod.103.024067. [DOI] [PubMed] [Google Scholar]
- Gu JL, Müller S, Mancino V, Offermanns S, Simon MI. Interaction of G alpha(12) with G alpha(13) and G alpha(q) signaling pathways. Proc Natl Acad Sci USA. 2002;99(14):9352–9357. doi: 10.1073/pnas.102291599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y, Soares MJ, Srivastava RK, Gibori G. Expression of decidual prolactin-related protein in the rat decidua. Endocrinology. 1994;135(4):1422–1427. doi: 10.1210/endo.135.4.7925104. [DOI] [PubMed] [Google Scholar]
- Gu Y, Srivastava RK, Clarke DL, Linzer DI, Gibori G. The decidual prolactin receptor and its regulation by decidua-derived factors. Endocrinology. 1996;137(11):4878–4885. doi: 10.1210/endo.137.11.8895360. [DOI] [PubMed] [Google Scholar]
- Harrington MA, Konicek B, Song A, Xia XL, Fredericks WJ, Rauscher FJ., 3rd Inhibition of colony-stimulating factor-1 promoter activity by the product of the Wilms' tumor locus. J Biol Chem. 1993;268(28):21271–21275. [PubMed] [Google Scholar]
- Hassan E, Creatsas G, Mastorakos G, Michalas S. Clinical implications of the ovarian/endometrial renin-angiotensin-aldosterone system. Ann New York Acad Sci. 2000;900:107–118. doi: 10.1111/j.1749-6632.2000.tb06221.x. [DOI] [PubMed] [Google Scholar]
- Heald PJ, O’Grady JE. The uptake of 3H-uridine into nucleic acids of rat uterus during early pregnancy. Biochem J. 1970;117:65–71. doi: 10.1042/bj1170065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Woronicz JD, Liu W, Goeddel DV. Prevention of Constitutive TNF Receptor 1 Signaling by Silencer of Death Domains. Science. 1999;283:543–546. doi: 10.1126/science.283.5401.543. [DOI] [PubMed] [Google Scholar]
- Jones RL, Salamonsen LA, Findlay JK. Potential roles for endometrial inhibins, activins and follistatin during human embryo implantation and early pregnancy. Trends Endocrinol Metab. 2002;13(4):144–150. doi: 10.1016/s1043-2760(01)00559-8. [DOI] [PubMed] [Google Scholar]
- Kaiser M, Gibori G, Mayo KE. The rat follistatin gene is highly expressed in decidual tissue. Endocrinology. 1990;126(5):2768–2770. doi: 10.1210/endo-126-5-2768. [DOI] [PubMed] [Google Scholar]
- Kao LC, Tulac S, Lobo S, Imani B, Yang JP, Germeyer A, Osteen K, Taylor RN, Lessey BA, Giudice LC. Global gene profiling in human endometrium during the window of implantation. Endocrinology. 2002;143(6):2119–2138. doi: 10.1210/endo.143.6.8885. [DOI] [PubMed] [Google Scholar]
- Kargul GJ, Dudekula DB, Qian Y, Lim MK, Jaradat SA, Tanaka TS, Carter MG, Ko MS. Verification and initial annotation of the NIA mouse 15K cDNA clone set. Nat Genet. 2001;28(1):17–18. doi: 10.1038/ng0501-17. [DOI] [PubMed] [Google Scholar]
- Kashiwagi A, DiGirolamo CM, Kanda Y, Niikura Y, Esmon CT, Hansen TR, Shioda T, Pru JP. The Postimplantation Embryo Differentially Regulates Endometrial Gene Expression and Decidualization. Endocrinology. 2007;148(9):4173–4184. doi: 10.1210/en.2007-0268. [DOI] [PubMed] [Google Scholar]
- Khatri P, Bhavsar P, Bawa G, Draghici S. Onto-Tools: an ensemble of web-accessible, ontology-based tools for the functional design and interpretation of high-throughput gene expression experiments. Nucleic Acids Research. 2004;32(web server issue):W449–W456. doi: 10.1093/nar/gkh409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatri P, Draghici S, Ostermeier GC, Krawetz SA. Profiling gene expression using onto-express. Genomics. 2002;79(2):266–270. doi: 10.1006/geno.2002.6698. [DOI] [PubMed] [Google Scholar]
- Kiger AA, Baum B, Jones S, Jones MR, Coulson A, Echeverri C, Perrimon N. A functional genomic analysis of cell morphology using RNA interference. J Biol. 2003;2(4):27. doi: 10.1186/1475-4924-2-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitabayashi I, Aikawa Y, Nguyen LA, Yokoyama A, Ohki M. Activation of AML1-mediated transcription by MOZ and inhibition by the MOZ–CBP fusion protein. EMBO J. 2001;20(24):7184–7196. doi: 10.1093/emboj/20.24.7184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knee DA, Froesch BA, Nuber U, Takayama S, Reed JC. Structure-Function Analysis of Bag1 Proteins. J Biol Chem. 2001;276:12718–12724. doi: 10.1074/jbc.M010841200. [DOI] [PubMed] [Google Scholar]
- Li F, Devi YS, Bao L, Mao J, Gibori G. Involvement of Cyclin D3, CDKN1A (p21),and BIRC5 (Survivin) in Interleukin 11 Stimulation of Decidualization in Mice. Biol Reprod. 2008;78(1):127–133. doi: 10.1095/biolreprod.107.063313. [DOI] [PubMed] [Google Scholar]
- Makrigiannakis A, Coukos G, Mantani A, Prokopakis P, Trew G, Margara R, Winston R, White J. Expression of Wilms’ Tumor Suppressor Gene (WT1) in Human Endometrium: Regulation through Decidual Differentiation. The Journal of Clinical Endocrinology & Metabolism. 2001;86(12):5964–5972. doi: 10.1210/jcem.86.12.8074. [DOI] [PubMed] [Google Scholar]
- Mal AK. Histone methyltransferase Suv39h1 represses MyoD-stimulated myogenic differentiation. EMBO J. 2006;25(14):3323–3334. doi: 10.1038/sj.emboj.7601229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto K, Yamauchi N, Watanabe R, Oozono S, Kubota K, Nishimura K, Wood C, Soh T, Kizaki K, Hattori MA. In vitro decidualization of rat endometrial stromal cells. Cell Tissue Res. 2009;335(3):575–583. doi: 10.1007/s00441-008-0734-1. [DOI] [PubMed] [Google Scholar]
- Migliaccio A, Castoria G, Di Domenico M, Ciociola A, Lombardi M, De Falco A, Nanayakkara M, Bottero D, De Stasio R, Varricchio L, Auricchio F. Crosstalk between EGFR and extranuclear steroid receptors. Ann N Y Acad Sci. 2006;1089:194–200. doi: 10.1196/annals.1386.006. [DOI] [PubMed] [Google Scholar]
- Mulholland J, Aplin JD, Ayad S, Hong L, Glasser SR. Loss of collagen type VI from rat endometrial stroma during decidualization. Biol Reprod. 1992;46(6):1136–1143. doi: 10.1095/biolreprod46.6.1136. [DOI] [PubMed] [Google Scholar]
- O'Carroll D, Scherthan H, Peters AH, Opravil S, Haynes AR, Laible G, Rea S, Schmid M, Lebersorger A, Jerratsch M, Sattler L, Mattei MG, Denny P, Brown SD, Schweizer D, Jenuwein T. Isolation and Characterization of Suv39h2, a Second Histone H3 Methyltransferase Gene That Displays Testis-Specific Expression. Mol Cell Biol. 2000;20(24):9423–9433. doi: 10.1128/mcb.20.24.9423-9433.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paroush Z, Finley RL, Jr, Kidd T, Wainwright SM, Ingham PW, Brent R, Ish-Horowicz D. Groucho is required for Drosophila neurogenesis, segmentation, and sex determination and interacts directly with hairy-related bHLH proteins. Cell. 1994;79(5):805–815. doi: 10.1016/0092-8674(94)90070-1. [DOI] [PubMed] [Google Scholar]
- Pollheimer J, Bauer S, Huber A, Husslein P, Aplin JD, Knöfler M. Expression pattern of collagen XVIII and its cleavage product, the angiogenesis inhibitor endostatin, at the fetal-maternal interface. Placenta. 2004;25(10):770–779. doi: 10.1016/j.placenta.2004.03.003. [DOI] [PubMed] [Google Scholar]
- Prasad MV, Dermott JM, Heasley LE, Johnson GL, Dhanasekaran N. Activation of Jun kinase/stress-activated protein kinase by GTPase-deficient mutants of G alpha 12 and G alpha 13. J Biol Chem. 1995;270(31):18655–18659. doi: 10.1074/jbc.270.31.18655. [DOI] [PubMed] [Google Scholar]
- Prigent-Tessier A, Barkai U, Tessier C, Cohen H, Gibori G. Characterization of a rat uterine cell line, U(III) cells: prolactin (PRL) expression and endogenous regulation of PRL-dependent genes; estrogen receptor beta, alpha(2)-macroglobulin, and decidual PRL involving the Jak2 and Stat5 pathway. Endocrinology. 2001;142(3):1242–1250. doi: 10.1210/endo.142.3.8004. [DOI] [PubMed] [Google Scholar]
- Qin J, Takahashi Y, Isuzugawa K, Imai M, Yamamoto S, Hirai Y, Imakawa K. Regulation of embryo outgrowth by a morphogenic factor, epimorphin, in the mouse. Mol Reprod Dev. 2005;70(4):455–463. doi: 10.1002/mrd.20225. [DOI] [PubMed] [Google Scholar]
- Radisky DC, Hirai Y, Bissell MJ. Delivering the message: epimorphin and mammary epithelial morphogenesis. Trends Cell Biol. 2003;13(8):426–434. doi: 10.1016/s0962-8924(03)00146-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond A, Meroni G, Fantozzi A, Merla G, Cairo S, Luzi L, Riganelli D, Zanaria E, Messali S, Cainarca S, Guffanti A, Minucci S, Pelicci PG, Ballabio A. The tripartite motif family identifies cell compartments. EMBO J. 2001;20(9):2140–2151. doi: 10.1093/emboj/20.9.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rider V, Patapova T, Dai G, Soares MJ. Stimulation of a rat uterine stromal cell line in culture reveals a molecular switch for endocrine-dependent differentiation. J Endocrinol. 2005;184:119–127. doi: 10.1677/joe.1.05957. [DOI] [PubMed] [Google Scholar]
- Riesewijk A, Martín J, van Os R, Horcajadas JA, Polman J, Pellicer A, Mosselman S, Simón C. Gene expression profiling of human endometrial receptivity on days LH 2 versus LH 7 by microarray technology. Mol Hum Reprod. 2003;9:253–264. doi: 10.1093/molehr/gag037. [DOI] [PubMed] [Google Scholar]
- Roger C, Mograbi B, Chevallier D, Michiels JF, Tanaka H, Segretain D, Pointis G, Fenichel P. Disrupted traffic of connexin 43 in human testicular seminoma cells: overexpression of Cx43 induces membrane location and cell proliferation decrease. J Pathol. 2004;202:241–246. doi: 10.1002/path.1509. [DOI] [PubMed] [Google Scholar]
- San Martin S, Soto-Suazo M, De Oliveira SF, Aplin JD, Abrahamsohn P, Zorn TM. Small leucine-rich proteoglycans (SLRPs) in uterine tissues during pregnancy in mice. Reproduction. 2003;125:585–595. doi: 10.1530/rep.0.1250585. [DOI] [PubMed] [Google Scholar]
- Sananès N, Weiller S, Baulieu EE, Le Goascogne C. In vitro decidualization of rat endometrial cells. Endocrinology. 1978;103(1):86–95. doi: 10.1210/endo-103-1-86. [DOI] [PubMed] [Google Scholar]
- Sugino N, Kashida S, Takiguchi S, Nakamura Y, Kato H. Induction of superoxide dismutase by decidualization in human endometrial stromal cells. Mol Hum Reprod. 2006;2:178–184. doi: 10.1093/molehr/6.2.178. [DOI] [PubMed] [Google Scholar]
- Suzuki N, Labosky PA, Furuta Y, Hargett L, Dunn R, Fogo AB, Takahara K, Peters DM, Greenspan DS, Hogan BL. Failure of ventral body wall closure in mouse embryos lacking a procollagen C-proteinase encoded by Bmp1, a mammalian gene related to Drosophila tolloid. Development. 1996;122(11):3587–3595. doi: 10.1242/dev.122.11.3587. [DOI] [PubMed] [Google Scholar]
- Tamura T, Mancini A, Joos H, Koch A, Hakim C, Dumanski J, Weidner KM, Niemann H. FMIP, a novel Fms-interacting protein, affects granulocyte/macrophage differentiation. Oncogene. 1999;18(47):6488–6495. doi: 10.1038/sj.onc.1203062. [DOI] [PubMed] [Google Scholar]
- Tan J, Raja S, Davis MK, Tawfik O, Dey SK, Das SK. Evidence for coordinated interaction of cyclin D3 with p21 and cdk6 in directing the development of uterine stromal cell decidualization and polyploidy during implantation. Mech Dev. 2002;111(1–2):99–113. doi: 10.1016/s0925-4773(01)00614-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka TS, Jaradat SA, Lim MK, Kargul GJ, Wang X, Grahovac MJ, Pantano S, Sano Y, Piao Y, Nagaraja R, Doi H, Wood WH, 3rd, Becker KG, Ko MS. Genome-wide expression profiling of mid-gestation placenta and embryo using a 15,000 mouse developmental cDNA microarray. Proc Natl Acad Sci USA. 2000;97(16):9127–9132. doi: 10.1073/pnas.97.16.9127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessier-Prigent A, Willems R, Lagarde M, Garrone R, Cohen H. Arachidonic acid induces differentiation of uterine stromal to decidual cells. Eur J Cell Biol. 1999;78:398–406. doi: 10.1016/S0171-9335(99)80082-X. [DOI] [PubMed] [Google Scholar]
- Tranguch S, Cheung-Flynn J, Daikoku T, Prapapanich V, Cox MB, Xie H, Wang H, Das SK, Smith DF, Dey SK. Cochaperone immunophilin FKBP52 is critical to uterine receptivity for embryo implantation. Proc Natl Acad Sci USA. 2005;102(40):14326–14331. doi: 10.1073/pnas.0505775102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallejo G, Ballaré C, Baranao JL, Beato M, Saragueta P. Progestin activation of nongenomic pathways via cross talk of progesterone receptor with estrogen receptor beta induces proliferation of endometrial stromal cells. Mol Endocrinol. 2005;19:3023–3037. doi: 10.1210/me.2005-0016. [DOI] [PubMed] [Google Scholar]
- Vladimirsky F, Chen L, Amsterdam A, Zor U, Lindner HR. Differentiation of decidual cells in cultures of rat endometrium. J Reprod Fertil. 1977;49(1):61–68. doi: 10.1530/jrf.0.0490061. [DOI] [PubMed] [Google Scholar]
- Von Schalburg KR, McCarthy SP, Rise ML, Hutson JC, Davidson WS, Koop BF. Expression of morphogenic genes in mature ovarian and testicular tissues: potential stem-cell niche markers and patterning factors. Mol Reprod Dev. 2006;73(2):142–152. doi: 10.1002/mrd.20359. [DOI] [PubMed] [Google Scholar]
- Wang H, Dey SK. Roadmap to embryo implantation: clues from mouse models. Nat Rev Genet. 2006;7(3):185–199. doi: 10.1038/nrg1808. [DOI] [PubMed] [Google Scholar]
- Wang H, Wen Y, Polan ML, Boostanfar R, Feinman M, Behr B. Regulation of cyclooxygenase activity in cultured endometrial stromal cells by granulocyte-macrophage colony-stimulating factor. Fertil Steril. 2006;85 Suppl 1:1118–1122. doi: 10.1016/j.fertnstert.2005.09.040. [DOI] [PubMed] [Google Scholar]
- Wewer UM, Damjanov A, Weiss J, Liotta LA, Damjanov I. Mouse endometrial stromal cells produce basement-membrane components. Differentiation. 1986;632(1):49–58. doi: 10.1111/j.1432-0436.1986.tb00555.x. [DOI] [PubMed] [Google Scholar]
- White CA, Robb L, Salamonsen LA. Uterine extracellular matrix components are altered during defective decidualization in interleukin-11 receptor alpha deficient mice. Reprod Biol Endocrinol. 2004;2:76. doi: 10.1186/1477-7827-2-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama A, Cleary ML. Menin Critically Links MLL Proteins with LEDGF on Cancer-Associated Target Genes. Cancer Cell. 2008;14:36–46. doi: 10.1016/j.ccr.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama A, Wang Z, Wysocka J, Sanyal M, Aufiero DJ, Kitabayashi I, Herr W, Cleary ML. Leukemia Proto-Oncoprotein MLL Forms a SET1-Like Histone Methyltransferase Complex with Menin To Regulate Hox Gene Expression. Mol Cell Biol. 2004;24:5639–5649. doi: 10.1128/MCB.24.13.5639-5649.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


