Abstract
Electron spin-echo envelope modulation (ESEEM) spectroscopy is a well-established technique for the study of naturally occurring paramagnetic metal centers. The technique has been used to study copper complexes, hemes, enzyme mechanisms, micellar water content, and water permeation profiles in membranes, among other applications. In the present study, we combine ESEEM spectroscopy with site-directed spin labeling (SDSL) and X-ray crystallography in order to evaluate the technique's potential as a structural tool to describe the native environment of membrane proteins. Using the KcsA potassium channel as a model system, we demonstrate that deuterium ESEEM can detect water permeation along the lipid-exposed surface of the KcsA outer helix. We further demonstrate that 31P ESEEM is able to identify channel residues that interact with the phosphate head group of the lipid bilayer. In combination with X-ray crystallography, the 31P data may be used to define the phosphate interaction surface of the protein. The results presented here establish ESEEM as a highly informative technique for SDSL studies of membrane proteins.
Electron spin-echo envelope modulation (ESEEM) is a pulsed electron paramagnetic resonance (EPR) technique that monitors nuclear magnetic resonance (NMR) transitions indirectly through EPR transitions (1). In the experiment, the electron spin-echo amplitude is monitored as it is modulated by the nuclear spin of nearby atoms at characteristic electron-nuclear double resonance (ENDOR) frequencies (2). ESEEM has been widely used for the study of paramagnetic metal centers in metalloproteins (3-6). Typically, ESEEM spectroscopy has been utilized to study metal binding, coordination, radical formation, and metallo-enzyme mechanisms. In addition, ESEEM spectroscopy has been used to investigate D2O permeation profiles in membranes (7, 8) and to study membrane insertion of antimicrobial peptides (9, 10). Recently, ESEEM has also been used to study the major light harvesting membrane protein complex of green plants (LHCIIb) in detergent micelles (11).
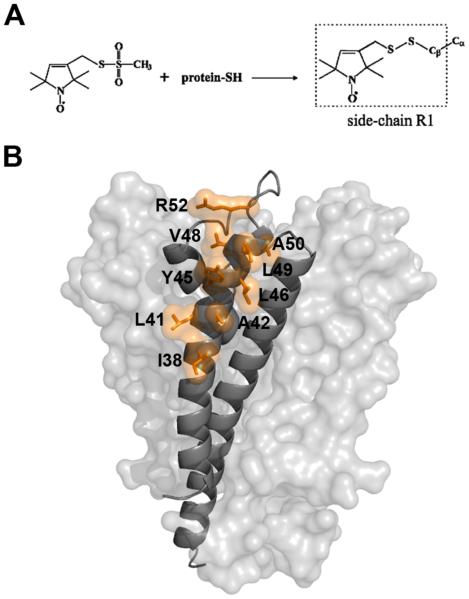
The technique's requirement of a paramagnetic center can be satisfied by site-directed spin labeling (SDSL) (12-15). SDSL involves the introduction of a nitroxide at a location of interest in the protein through molecular engineering (Figure 1A). SDSL has been used to probe structure in a variety of membrane protein systems, including bacteriorhodopsin, rhodopsin, porins, and potassium channels (12-15). Most applications of SDSL have utilized continuous wave (CW) EPR techniques and include nitroxide scanning applications with the goal of determining the underlying secondary structure of a sequence of interest, the measurement of molecular distance distributions between pairs of nitroxides, and the detection of conformational changes during function. In addition, CW-EPR can provide a measure of polarity surrounding the nitroxide side chain (16, 17). The polarity effects are mediated through the degree and direction of hydrogen bonding of the nitroxide with the nearby environment and are measured by determining the A and g tensor elements of the nitroxide (18). However, the identity of the hydrogen-bonding partner of the nitroxide typically remains unknown.
Figure 1.
Site directed spin labeling of KcsA. (A) Reaction of the methanethiosulfonate reagent with cysteine to generate the side chain designated R1. (B) Labeled sites in KcsA. A KcsA monomer is shown as a grey ribbon with the rest of the channel depicted as a van der Waals surface. The side chains of labeled sites are depicted as orange sticks and van der Waals surfaces.
Alternatively, the polarity and accessibility of the nitroxide environment can be determined by CW-EPR using collision studies (19, 20). In membrane proteins, these experiments typically involve exposing labeled and reconstituted protein with hydrophilic (NiEDDA) and hydrophobic (oxygen) paramagnetic colliders that provide additional T1 relaxation pathways to the nitroxide thorough Heisenberg spin exchange. These collision studies are limited by the fact that spin exchange requires direct physical contact between the nitroxide and the paramagnetic collider with a collision radius equal to the sum of the effective radii of the two species (20). In contrast, ESEEM is based on through-space anisotropic hyperfine interactions between the nitroxide and nearby nuclear spins. The amplitude of the ESEEM signal depends on both the number of nearby nuclei (N) and the distance between nuclear and electron spins (R), and is proportional to N/R6. For instance, a deuterium nucleus can be detected at a distance of up to 6-8 Å (9). ESEEM, therefore, can determine the atomic environment surrounding a nitroxide without direct physical contact with the interacting species.
In the present study, we applied ESEEM in combination with SDSL in order to determine the native environment of a membrane protein of known structure reconstituted in lipid vesicles. The KcsA potassium channel is well suited for these studies because it is easily over-expressed in E. coli, is well-behaved biochemically, and can be functionally reconstituted in lipid vesicles (21, 22). In addition, the structure of KcsA is known to high-resolution (23), and extensive biophysical studies support the notion that the crystal structure represents the predominant native state of the channel (24-26). Specifically, we determined the water permeation profile along the surface of the outer transmembrane helix of KcsA using D2O as a water substitute and deuterium substrate. We found that deuterium interactions with the outer helix decrease with increasing immersion depth into the membrane. This decrease of water permeation was similar compared with empty lipid vesicles of the same composition. In addition, we identified two KcsA residues that are within interaction distance to the phosphate head group of the membrane, as determined through 31P-nitroxide coupling. The crystal structure of one of these two constructs allows the determination of the phospholipid head group interaction surface of the channel. The data suggest that ESEEM is well suited for structural studies of membrane proteins and can provide key insight into their local native environment.
Materials and Methods
Protein expression, purification, labeling, and reconstitution
KcsA was over-expressed, purified, and spin-labeled as previously described (26-28). Labeled KcsA was further purified by gel filtration on Superdex 200 in a buffer containing 20 mM Tris, pH 8.0, 150 mM KCl, 5 mM decylmaltoside and reconstituted on Sephadex G-50 (both GE Healthcare) into 4:1 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC): 1-palmitoyl-2-oleoyl-sn-glycero-3-[phospho-rac-(1-glycerol)] (POPG) liposomes (Avanti Polar) at a molar lipid: protein ratio of ~1000: 1 by the method of Green and Bell (29). Proteoliposomes were pelleted by centrifugation (20 hours at 400,000 g) and resuspended to a KcsA concentration of ~200 μM with a solution of 50 mM Tris, pD 7.4, 150 mM KCl, and 25% glycerol for cryo-protection. The samples were flash frozen in liquid nitrogen and stored at −80° C until measurement.
Preparation of doped empty vesicles
1-palmitoyl-2-stearoyl-(n-DOXYL)-sn-glycero-3-phosphocholine and 1-palmitoyl-oleoyl-sn-glycero-3-phospho(TEMPO)choline (Avanti Polar) were reconstituted on Sephadex G-50 (GE Healthcare) into 4:1 POPC: POPG liposomes (Avanti Polar) at a molar lipid: spin-labeled lipid ratio of ~1000:1 as described above.
EPR measurements
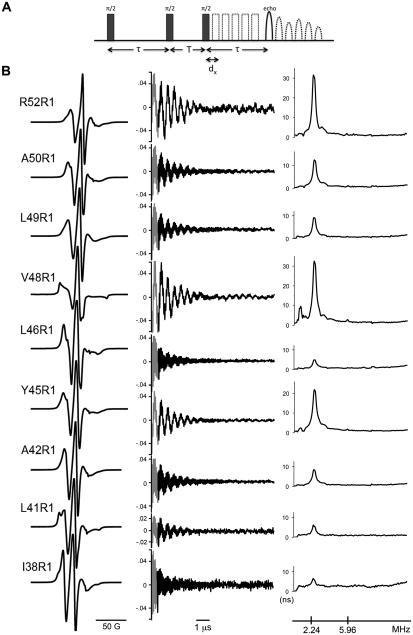
CW-EPR spectra were recorded at room temperature on an X-band EMX spectrometer fitted with an ER4119HS resonator (Bruker Biospin) at a scan width of 200 Gauss. ESEEM samples were flash-cooled in liquid nitrogen and the data were recorded at 80K on an X-band Elexsys-E580 spectrometer fitted with an ER4118X-MS2 resonator (Bruker Biospin). ESEEM data were acquired using a three-pulse sequence (π/2-τ-π/2-T-π/2-τ-echo) with 16 ns π/2 pulses at a frequency corresponding to the centerline of the nitroxide spectrum. A four-step phase cycle was used to suppress unwanted echoes. Timing parameters were adjusted to obtain maximal modulation by specific nuclei: τ = 224 ns for 2H experiments, τ = 252 ns for 31P experiments, and τ = 128 ns for 1H blind experiments. The time-dependent echo amplitudes are shown in Figure S1 and were processed as described (7, 10). The data were normalized by division through a polynomial fit to the underlying T1 echo decay, followed by subtraction of unity. The missing data points were obtained by back-prediction using the LPSVD algorithm (30). The data were further processed by Hamming apodization, zero filling, and Fourier transformation to obtain frequency domain power spectra with spectral densities with dimension of time (10). Three-pulse ESEEM data were simulated (1) with different A and g tensor elements (31, 32) and a single 2H nucleus at 3.2 Å in order to estimate effects due to polarity and nitroxide structure. The error in measurement (<4%) and effects due to polarity and nitroxide structure (<1%) were small in relation to the overall variability between protein samples from different expression batches (<10%). HYSCORE data were acquired using a four-pulse sequence (π/2-τ-π/2-t1-πt2-π/2-τ-echo) with 16 ns π/2 pulses and a 32 ns π pulse. A four-step phase cycle was used to suppress unwanted echoes. The time-dependent echo amplitudes were processed as described above (without back-prediction), followed by 3D Fourier transformation.
Results
Spin-labeling of the KcsA outer helix
Based on the crystal structure of KcsA (23), nine residues (I38, L41, A42, Y45, L46, V48, L49, A50, and R52) were selected along the outer surface of the outer transmembrane helix (Figure 1B). These residues were chosen because they have minimal tertiary interactions with other portions of the protein and are predominately exposed to the outside environment. In addition, all residues are located on the extracellular half of the channel where the structure is known to remain constant independent of the gating state of the channel (33). The inner transmembrane helix was avoided due to incomplete labeling along the region of interest (26, 33). The positions of the nine residues range from a lipid exposed helix surface site in the middle of the membrane (I38) to a solvent exposed loop site located just above the outer helix (R52). Each residue was mutated to cysteine and the introduced thiol was reacted with a nitroxide spin label to form the nitroxide side chain denoted R1 (Figure 1A). The labeled mutant channel was then reconstituted in POPC:POPG lipid vesicles by gel filtration (see Methods). The room temperature CW-EPR spectra at X-band of the selected constructs are shown in Figure 2B. The lineshapes of all nitroxides (except V48R1) are indicative of sites with high side chain mobility and are thus consistent with their external position along the outer helix. In contrast, the lineshape of V48R1 is indicative of an immobilized nitroxide. The crystal structure of KcsA indicates that the immobilization of the R1 side chain at this position probably occurs due to a tertiary contact interaction of the nitroxide with the pore helix of the channel (23). The spectra reported here are thus highly consistent with the known structure of KcsA. Furthermore, the spectra are in agreement with results from earlier studies (25, 27).
Figure 2.
Lineshape and 2H ESEEM data of R1 constructs reconstituted in POPC:POPG vesicles. (A) Schematic of the three-pulse ESEEM experimental design. Three π/2 pulses are separated by the time constants τ and T, with T sequentially delayed by dx increments, resulting in the modulation of the observed spin echo. The time constant τ is adjusted to tune the system for optimal detection of a frequency of interest. (B) Left: X-band first-derivative absorption spectra acquired at room temperature over a scan width of 200 G. Middle: ESEEM data obtained with τ = 224 ns (to optimize detection of 2H) in the presence of D2O. The black trace is the normalized time domain data. The grey segment is the back-calculated dead-time signal (see Methods). Right: frequency domain ESEEM signal. The deuteron (2.24) and phosphorous (5.95) frequencies are indicated in MHz units.
Deuterium ESEEM demonstrates water permeation along the hydrophobic surface of KcsA
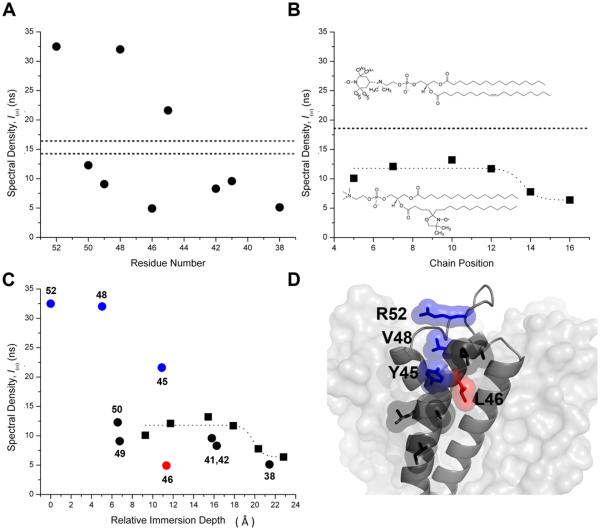
The KcsA constructs were analyzed using the three-pulse ESEEM sequence shown in Figure 2A. D2O was added to the sample in order to isotope label hydrogen derived from water and distinguish it from hydrogen derived from protein and lipid. Figure 2B summarizes the lineshape (left panel) and ESEEM data in both time (middle panel) and frequency domain (right panel) for all sites studied. Proton and deuteron ENDOR frequencies predominate in the ESEEM spectra at all sites. Because the total proton pool in the system arises from multiple sources (protein, lipid, and residual H2O), the proton spectral density at 14.6 MHz cannot unambiguously be assigned to any one source and was not further analyzed. Figure 3A illustrates the amplitude of the deuteron spectral density at 2.2 MHz as a function of residue position using data from all sites studied. The data indicate that although the absolute deuteron spectral density fluctuates significantly along the transmembrane helix, there is a sizeable overall decrease from ~33 ns near the outside surface (R52R1) to ~5 ns near the middle of the membrane (I38R1). In order to place these values into a more meaningful framework, we determined the spectral density at different immersion depths using empty vesicles of the same composition (Figure 3B) doped with trace amounts of spin-labeled phospholipids (lower insert). In these empty vesicles, the deuteron spectral density decreased with an approximately sigmoidal dependence from ~12 ns to ~6 ns as the spin label was moved along the alkyl chain toward the center of the membrane (7).
Figure 3.
KcsA insertion into the membrane preserves the expected water permeation profile. (A) The deuteron spectral density of labeled residues is shown as a function of residue number. Deuterium modulation of the C-terminal mutants F148R1 and L151R1 is represented by the horizontal dashed lines (~14.8 ns and 16.6 ns, respectively). (B) The deuteron spectral density of spin-labeled lipids in POPC:POPG vesicles plotted as a function of chain position. The dotted line is drawn to support the eye. The dashed line is the deuteron spectral density of TEMPO-PC. Top insert: Structure of TEMPOPC. Bottom insert: Structure of 7-doxyl-PC. (C) The labeled lipid (squares) and KcsA (circles) deuteron spectral density data superimposed as a function of relative immersion depth (in Å relative to the Cβ position of R52R1). Data for residues Y45R1, V48R1 and R52R1 are colored blue and L46R1 is colored red. (D) Structural representation of one KcsA subunit depicted as a grey ribbon and the rest of the channel as a van der Waals surface. The studied side chains are depicted in stick representation with accompanying van der Waals surfaces and are colored as in C.
The deuterium modulation was also measured at the level of the lipid head group by means of TEMPO-PC (Figure 3B, upper insert). The spectral density for this interfacial spin-label was ~18 ns (dotted line) and thus significantly higher than at any position along the alkyl chain. Simulation studies show that the different structures of the nitroxide moieties have a negligible impact on the 2H ESEEM data in the current experimental context (data not shown). The data on the empty vesicles are nicely self-consistent and indicate that we can expect decreasing deuteron spectral density as a function of immersion depth, with absolute values ranging from ~18 ns at the water-lipid interface to ~6 ns near the middle of the membrane. We next sought to determine the spectral density of sites on KcsA exposed to bulk water. Two sites known to be located far from the membrane were chosen that had both exhibited exceptionally high NiEDDA accessibility in earlier studies. The sidechains of F148R1 and L151R1 are located on the water-exposed C-terminal domain of KcsA and were shown to have the highest NiEDDA accessibility of any residues in that region of the protein (34). Interestingly, the deuteron spectral density of these two sites (~14.5 ns and ~16.5 ns, dotted lines in Figure 3A) is closer to TEMPO-PC (~18 ns) than to R52R1 (~33 ns). It thus appears that the 2H spectral density of V48R1 and R52R1 is exceptionally high.
It is useful for the direct comparison between lipid and protein data to place all spin labels on approximately the same length scale with regard to immersion depth. For the purposes of this study, the immersion depth of KcsA spin labels was defined as the distance (in the direction normal to the membrane surface) between the Cβ of the residue in question and the Cβ of residue R52. R52 is arbitrarily used as a reference point for zero immersion depth in order to generate only positive values. The immersion depth of lipid spin labels is obtained by modelling the doxyl moiety (Figure 3B, lower inset) onto a well-resolved lipid chain observed in the high resolution structure of KcsA (23) and determining the vertical distance component of the spin-label with respect to R52. The resulting graph (Figure 3C) demonstrates an overall similar depth dependence at some KcsA sites compared to the lipid data. The new depth scale has eliminated some of the original data scatter (Figure 3C) and the only remaining significant outliers are the lower than expected 2H spectral density for residue L46R1 (red), located at an immersion depth of ~11 Å, and the higher than expected 2H spectral densities for Y45R1, V48R1, and R52R1 (blue). What is apparent in the data is that the insertion of KcsA into the membrane did not lead to generally increased water permeation along the surface of the protein when compared to empty vesicles. To the contrary, the lower than expected 2H spectral density of residue L46R1 indicates less water permeation in the vicinity of this residue than at comparable depths in empty vesicles.
31P ESEEM identifies residues interacting with the phosphate of the membrane
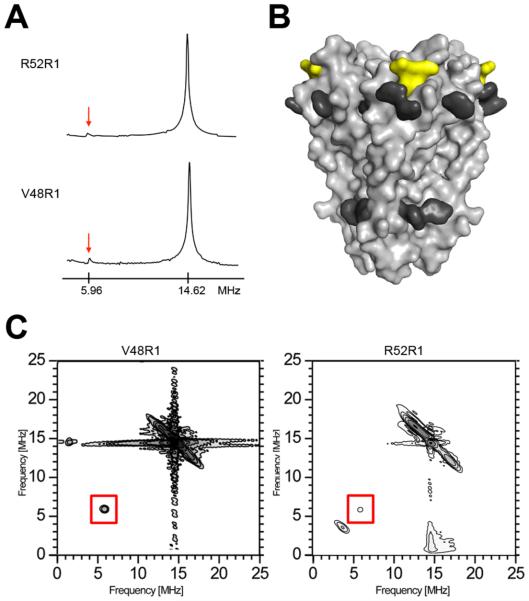
The frequency domain spectra at superficially locates sites, in particular V48R1 and R52R1, show hints of weak 31P modulation at 5.95 MHz that is absent in more lipid-immersed sites (Figure 2B). 31P is the only nucleus in the system that can account for interactions at this frequency (see legend to Figure 2). In order to further confirm the interaction at these two positions, the ESEEM experiment was repeated with a pulse sequence tuned to optimize detection of 31P (Figure 4A). The data confirm 31P modulation near the detection limit, corresponding to a 31P – nitroxide spin-spin distance of ~5 Å (35). The van der Waals surfaces occupied by the two 31P-modulated non-aromatic residues are contiguous and are located above a ring of membrane-exposed aromatic residues in KcsA (Figure 4B), consistent with an interaction between the two side chains and the surrounding phosphate head group layer of the membrane. In addition, one of the interacting residues is R52; arginines are well known to interact closely with the phosphate moiety of membranes (36). The 31P modulation at residues V48 and R52 was further confirmed by a 2D-HYSCORE experiment (Figure 4C) (37). In order to minimize deuteron interference and to clarify the spectra, the HYSCORE samples were prepared in a buffer without added D2O. As expected, both V48R1 and R52R1 spectra show the required diagonal peaks corresponding to 1H and 31P (14.46 and 5.95 MHz, respectively). The off-diagonal splitting of the 1H peak is probably due to interactions of the unpaired electron with nearby protons on the nitroxide ring or with hydrogen-bonded water.
Figure 4.
31P ESEEM of reconstituted KcsA. (A) Frequency domain ESEEM spectra of the KcsA constructs V48R1 and R52R1 using a time constant optimized for 31P modulation (τ= 252 ns). The phosphorous (5.96 MHz) and proton (14.62 MHz) Larmor frequencies are indicated. (B) The phosphorous interaction surface of KcsA is depicted as a yellow van der Waals surface. The side chains of aromatic residues believed to be exposed toward the water-lipid interface are highlighted in dark grey (38). (C) Frequency domain HYSCORE spectra of V48R1 (optimal time constant to observe 31P modulation, τ = 252 ns) and R52R1 (optimal time constant to suppress 1H modulation, τ = 128 ns). The phosphorous Larmor frequency peak at 5.96 MHz is boxed.
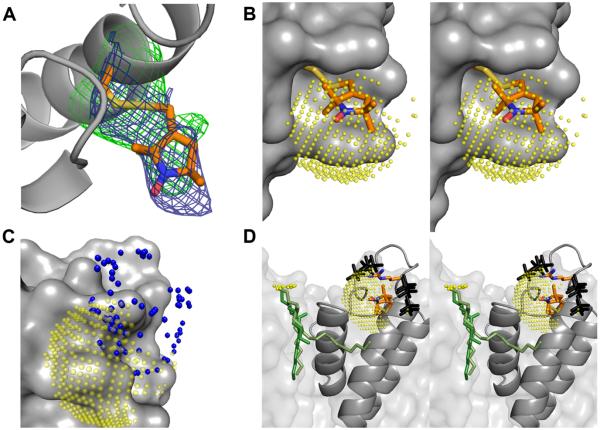
If the precise position of the two nitroxides were known, then the observed 31P spectral densities could be used to approximately determine the position of the interacting phospholipid head group. Given the highly immobile CW-EPR spectrum of residue V48R1 (Figure 2B), it appeared feasible to crystallographically resolve the R1 side chain in this construct. Unfortunately, high quality KcsA-Fab co-crystals could not be grown due to steric interference of the labeled residue with Fab binding (data not shown). Nevertheless, data to an effective resolution of 3.5 Å Bragg spacings was obtained in the absence of a Fab fragment using the original crystallization protocol of the isolated channel (38) (Table S1). Figure 5A shows that the observed electron density for V48R1 clearly determines the position of the R1 side chain.
Figure 5.
The structure of V48R1 and the phosphate interaction surface. (A) KcsA is shown as a grey ribbon and the V48R1 side chain is shown in stick representation with carbon, nitrogen, oxygen, and sulphur atoms colored orange, blue, red, and yellow, respectively. The 2Fo-Fc map is contoured at 1σ (blue) and the Fo-Fc map is contoured at 2.7σ (green). (B) Stereo view of V48R1 with a 5 Å 31P ESEEM interaction surface (translucent yellow spheres) centered on the nitroxide nitrogen. The sphere is sampled at ~0.5 Å and only sterically feasible areas of the sphere are shown. KcsA is shown as a grey van der Waals surface. (C) Possible positions of the R52R1 nitroxide nitrogen based on preferred nitroxide dihedral angles (39) are shown as blue spheres together with the KcsA channel as a grey van der Waals surface and the yellow V48R1 interaction surface. (D) Stereo view of sterically feasible phosphate positions (black) assuming the presence of a R52-phosphate interaction located on the V48R1 interaction surface (yellow), i.e. both V48R1 and R52R1 couple to the same phospholipid. The side chains of R52 and V48R1 as well as a crystallographically observed lipid (two alternative green models, see PDB entries 1K4C and 1R3J) are shown in stick representation. The position of the phosphate head group of the lipid is not resolved. Possible positions of the phosphorous are shown as small yellow spheres.
If it is assumed that the phosphorous modulation at both sites originates from the same 31P nucleus, a reasonable assumption given the relative scarcity of phosphorous in the system under study, then this nucleus must be located at the intersection of the surfaces of two spheres with radius 5 Å, centered at the position of the two nitroxide spins (Figure 5B). However, unlike V48R1, the highly mobile CW-EPR spectrum of R52R1 (Figure 2B) indicates the presence of a highly disordered side chain, meaning that a crystal structure of this construct is unlikely to provide a defined position of the nitroxide moiety. Although the position of the unpaired electron at this site can not be determined precisely, the accessible volume of the side chain can be obtained based on the structure of R1 and the knowledge of its preferred dihedral angles (Figure 5C) (39). If we assume that the R52 side chain closely interacts with the phosphate under native conditions, then the known preferred dihedral angles of arginine, the possible volume of the R52R1 sidechain, the position of the V48R1 spin, and the size and geometry of phosphate jointly constrain the possible position of the phosphate head group (Figure 5D).
Discussion
Deuterium ESEEM
In contrast to the established collision-based EPR techniques for determining membrane exposure (19, 40), ESEEM does not require direct physical access of a collider to the R1 side chain. Physical access of a collider places limits on the types of environments that can be studied by the CW-EPR methodology. This limitation was demonstrated for sites in the internal water-filled cavity of KcsA, which were found to be essentially inaccessible to NiEDDA even though they were located in an aqueous environment (26). The ESEEM approach described here overcomes this limitation by directly determining the deuterium environment surrounding the spin label, independent of any other access parameters. As expected, the two measurements are highly correlated for the structurally unconstrained and solvent exposed residues studied here (Figure S2). The manner in which deuterium is introduced allows for some future technique development. It is conceivable, for instance, that the deuterium can be introduced selectively, such as at limited positions on the alkyl chain of lipids (41) or at specific amino acids on the channel (42). Indeed, studies with spin-labeled peptide inserted into deuterated lipid vesicles have been performed (9, 43). In the present study, deuterium was introduced non-selectively in the form of D2O. Deuterium nuclei are thus present either as part of water or as part of protein or lipid through the mechanism of proton-deuteron exchange. Since proton-deuteron exchange requires D2O access to the site, it is unnecessary to be able to separate the two sources of deuterons for the purposes of this study. The penetration of water into the membrane is followed through the amplitude of deuterium modulation. The amplitude of the deuterium modulation of the electron spin is dependent on both the distance between the interacting nuclei and the spin (R), as well as the number of interacting nuclei (N), and is proportional to N/R6. The high order dependence on distance and the magnetic properties of deuterium ensure that only nuclei within ~6-8 Å can contribute to the deuterium signal (9). Furthermore, the modulation is weighted heavily in favor of the closest interacting nuclei. Therefore, water permeation profiles determined by ESEEM are correlated to the local concentration of D2O and strongly weighted by the deuteron-spin distance distribution. The high order dependence on distance means that different rotameric distributions of R1 along the TM1 helix may affect the measured deuteron spectral density. While sidechains that are immobilized (V48R1) at room temperature are expected to also occupy a single rotamer at low temperature, more mobile sidechains (I38R1, L49R1) sample multiple rotamers at room temperature and are likely to freeze into rotamer distributions with distinct deuterium environments. It is therefore to be expected that, in the limit, highly ordered water surrounding a highly ordered spin label could give rise to exceptionally high spectral densities. The technique may thus more robust when highly mobile nitroxides are studied as they sample a large volume of space, in effect averaging out the local D2O distribution experienced by any one rotameric state.
Water permeation in empty vesicles
It has previously been shown that water permeation into saturated DPPC vesicles can be determined by deuterium ESEEM and that the permeation profile has a sigmoidal, trough-like form (7). The permeation profile of unsaturated POPC: POPG vesicles determined here (Figure 3B) shows a similar overall form, but the point of maximal gradient is shifted towards deeper positions. In POPC:POPG membranes the gradient is highest at approximately chain position 13, whereas the equivalent point occurs at chain position 11.6 in unsaturated DPPC vesicles (7). The higher water penetration into membranes comprised of unsaturated lipids is expected and can be explained by the increased inter-lipid packing distance that exists due to unsaturation. The transition from high water permeation near the outer surface of the lipid bilayer to low water permeation in the middle of the membrane is also affected by temperature. In frozen membranes, the transition point is shifted to slightly deeper chain positions than are observed in fluid membranes (16). Nevertheless, the structure and biophysical properties of membranes are well preserved upon freezing under cryo-protecting conditions (7, 8, 35, 44).
Water permeation along the surface of the outer helix
When an integral membrane protein is inserted into a membrane, a boundary layer of lipids is generated that may have an altered water permeation profile than the more distant parts of the membrane. In the case of KcsA, however, the water permeability along the protein surface does not appear to increase over the level observed in empty vesicles (except for Y45R1, which is located at the water-lipid interface). Therefore, KcsA does not introduce water-accessible crevices along its surface. To the contrary, part of the protein surface may be exposed to less water than would be expected in empty vesicles. Specifically, sites L41R1, A42R1, and L46R1 show lower apparent water penetration than would be expected based on their relative immersion depths. Interestingly, the lowest 2H spectral density is observed for a residue (L46R1) that projects into a groove between two transmembrane helices (Figure 3D). KcsA is known to order lipids in grooves between transmembrane helices (Figure 5D) (23) leading to locally dry protein surface areas. Alternatively, or in addition, a slight bend of the outer helix increases the effective immersion depth of this residue with respect to the other sites studied (Figure 3D). The very low water permeation at site L46R1 is also supported by NiEDDA accessibility data that show a lower than expected accessibility at this site (25).
It remains unclear why the deuterium modulation is so high for V48R1 and R52R1. As we did not exhaustively search for high deuterium modulation, it remains possible that other residues exposed to bulk water could show similarly high deuteron spectral densities. We selected the two specific residues on the C-terminal of KcsA because we reasoned that residues exhibiting exceptionally high NiEDDA collision rates were good candidates to reflect the expected deuterium modulation of a highly water-exposed nitroxide. If this reasoning is sound, then the deuterium modulation of V48R1 and R52R1 is indeed exceptionally high. It is noteworthy that the two nitroxides show very different side chain mobilities at room temperature, arguing against locally ordered water located very close to the side chain as the reason for the high spectral density. Further studies are required to determine whether these densities are indeed exceptional and whether these high values are related to the other aspect that distinguishes these two residues, namely their close interaction with the phosphate head group of a nearby lipid.
Phosphorous ESEEM
Unlike deuteron and proton, 31P is a rare nucleus in the experimental system under study. The only natural source of phosphorous is the phosphate head group of the membrane in which the channel is reconstituted. Based on the geometry of phospholipid packing in membranes, approximately one 31P nucleus is expected per 50 Å2 of membrane surface (45). The modulation depth of the 31P nucleus is smaller than that of 2H (46). As a consequence, the presence of a 31P nucleus can only be detected to a distance of ~5 Å from the nitroxide, whereas a single 2H nucleus can interact up to 6-8 Å (9, 35, 46). Because of the overall scarcity and regular occurrence of phosphorous, it is reasonable to treat phosphorous modulations as if they originated from a single nucleus. The high order dependence on distance and the nuclear properties of 31P limit the number of residues with observable modulation and allow an accurate map of the phosphate interaction surface of KcsA to be made (Figure 4B). The accuracy of the map is further improved if the precise position of the nitroxide side chain can be determined through X-ray crystallography (Figure 5D).
Locating the interacting phosphate head group
The magnitude of the magnetic interaction determined by ESEEM is dominated by the closest distance between nucleus and nitroxide spin. Accordingly, the interaction can be analyzed assuming a single nucleus approximation in order to obtain a lower bound value for the distance of interaction (47). Applying these principles, we conclude that the interacting 31P nucleus is located ~5 Å from both V48R1 and R52R1. What can be learned about the possible location of the interacting phosphate head group relative to the KcsA channel based on these distances? The following procedure was followed in determining the approximate position of the interacting 31P nucleus. First, a sphere with radius 5 Å and origin centered on the nitrogen of the V48R1 nitroxide side chain was calculated. The interacting 31P nucleus must be located on the surface of this sphere. Second, the surface area of the sphere in steric conflict with the channel was eliminated. Figure 5B shows the remaining phosphate interaction surface. Third, the possible interaction surface was further reduced by eliminating those areas that were >5 Å from any feasible R52R1 rotamer (Figure 5C). Fourth, the remaining surface was explored for steric compatibility with regard to a phosphate moiety. A phosphate group was moved along the surface and all areas were eliminated where the phosphate is in steric conflict with the channel irrespective of its orientation. Finally, from this subset of sterically feasible phosphate positions, we chose those that were within reach of the R52 sidechain, assuming a native R52-phosphate interaction. The resulting set of possible phosphate positions is shown in Figure 5D (black models).
Several of these potential phosphate positions are highly compatible with the expected phosphate position based on a crystallographically determined lipid molecule. In the crystal structure of KcsA (PDB entries 1K4C, 1R3J), a lipid molecule binds to the channel in the groove between the inner helix of one subunit and the pore helix of the adjacent subunit. The lipid is resolved in two different orientations (green stick models in Figure 5D). Although the phosphate head group of this lipid could not be observed, the presence of the glycerol backbone in the model allows the possible locations of the nucleus to be determined (small yellow spheres). It is encouraging to note that several of the phosphate positions predicted by our placement are at a similar vertical position as the crystallographically predicted lipid phosphorous nucleus. In addition, the distance of closest approach between the crystallographically determined phosphorous nuclei positions and the modelled phosphate is on the order of ~8 Å, and thus in reasonable agreement with the expected ~7 Å inter-phosphate distance in the membrane (45).
In this study, we have evaluated ESEEM in terms of its potential usefulness to describe the environment surrounding membrane proteins under native conditions. Using electron spin modulation by deuterium, we have demonstrated the water permeation profile along the surface of the KcsA potassium channel. The data support the conclusion that KcsA inserts into the membrane without creating water crevices along its protein surface. The technique may thus prove useful to detect the narrow water crevices that are predicted to occur in some membrane proteins as a result of distorted lipid packing (48, 49). In addition, the ESEEM data identified two residues on KcsA through 31P modulation that are in intimate contact with the phosphate head-group region of the membrane. Based on a crystallographically determined structure of an interacting side chain, the position of the phosphate head-group with respect to the channel could be strongly constrained. The data presented here demonstrate that the technique is well suited to investigate the interaction of membrane proteins with their surrounding lipid environment. This capability may prove particularly useful for the study of those proteins for which lipids are either critical for maintaining native structure, or have been shown to participate in its function (28, 50-52).
Supplementary Material
Acknowledgements
We thank Michael Lenaeus for critically reading the manuscript.
This work was supported by NIH grant GM58568 to A.G and NIH fellowship NS051095 to J.C
Abbreviations
- CW
continuous wave
- DOXYL
4,4-dimethyl-oxazolidine-N-oxyl
- DPPC
dipalmitoylphosphatidylcholine
- ENDOR
electron-nuclear double resonance
- EPR
electron paramagnetic resonance
- ESEEM
electron spin echo envelope modulation
- Fab
fragment- antigen binding
- HYSCORE
hyperfine sub-level correlation
- IPTG
isopropyl β-D-1-thiogalactopyranoside
- KCl
potassium chloride
- MHz
megahertz
- MTSL
S-(2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl methanesulfonothioate
- NiEDDA
nickel(II) ethylenediaminediacetate
- NMR
nuclear magnetic resonance
- POPC
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
- POPG
1-palmitoyl-2-oleoyl-sn-glycero-3-[phospho-rac-[1-glycerol]]
- R1
designation for spin labeled side chain
- SDSL
side-directed spin labeling
- TEMPO-PC
1-palmitoyl-oleoyl-sn-glycero-3-phospho(4-[N,N-dimethyl-N-(2-hydroxyethyl)]ammonium-2,2,6,6-tetramethylpiperidine-1-oxyl)-choline
- TM
transmembrane segment
- Tris
tris(hydroxymethyl)aminomethane
- T1
electron spin-lattice relaxation time.
Footnotes
The coordinates and structure factors of KcsA-V48R1 have been deposited with the Protein Data Bank (accession code 3IFX).
Supporting information available
A table with collection and refinement statistics of KcsA V48R1, a figure showing the unprocessed ESEEM data, a figure comparing the 2H spectral density with collisional CW-EPR data, and additional crystallization and refinement methods. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Schweiger A, Jeschke G. Principles of pulse electron paramagnetic resonance. 1st ed. Oxford University Press; Oxford: 2001. [Google Scholar]
- 2.Hoffman BM. Electron-nuclear double resonance spectroscopy (and electron spin-echo envelope modulation spectroscopy) in bioinorganic chemistry. Proc Natl Acad Sci U S A. 2003;100:3575–3578. doi: 10.1073/pnas.0636464100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mims WB, Peisach J. Assignment of a ligand in stellacyanin by a pulsed electron paramagnetic resonance method. Biochemistry. 1976;15:3863–3869. doi: 10.1021/bi00662a033. [DOI] [PubMed] [Google Scholar]
- 4.Prisner T, Rohrer M, MacMillan F. Pulsed EPR spectroscopy: biological applications. Annu Rev Phys Chem. 2001;52:279–313. doi: 10.1146/annurev.physchem.52.1.279. [DOI] [PubMed] [Google Scholar]
- 5.Hoffman BM. ENDOR of metalloenzymes. Acc Chem Res. 2003;36:522–529. doi: 10.1021/ar0202565. [DOI] [PubMed] [Google Scholar]
- 6.Britt RD, Campbell KA, Peloquin JM, Gilchrist ML, Aznar CP, Dicus MM, Robblee J, Messinger J. Recent pulsed EPR studies of the photosystem II oxygen-evolving complex: implications as to water oxidation mechanisms. Biochim Biophys Acta. 2004;1655:158–171. doi: 10.1016/j.bbabio.2003.11.009. [DOI] [PubMed] [Google Scholar]
- 7.Erilov DA, Bartucci R, Guzzi R, Shubin AA, Maryasov AG, Marsh D, Dzuba SA, Sportelli L. Water concentration profiles in membranes measured by ESEEM of spin-labeled lipids. J Phys Chem B. 2005;109:12003–12013. doi: 10.1021/jp050886z. [DOI] [PubMed] [Google Scholar]
- 8.Bartucci R, Guzzi R, Marsh D, Sportelli L. Intramembrane polarity by electron spin echo spectroscopy of labeled lipids. Biophys J. 2003;84:1025–1030. doi: 10.1016/S0006-3495(03)74918-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carmieli R, Papo N, Zimmermann H, Potapov A, Shai Y, Goldfarb D. Utilizing ESEEM spectroscopy to locate the position of specific regions of membrane-active peptides within model membranes. Biophys J. 2006;90:492–505. doi: 10.1529/biophysj.105.062992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bartucci R, Guzzi R, Sportelli L, Marsh D. Intramembrane water associated with TOAC spin-labeled alamethicin: electron spin-echo envelope modulation by D2O. Biophys J. 2009;96:997–1007. doi: 10.1016/j.bpj.2008.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Volkov A, Dockter C, Bund T, Paulsen H, Jeschke G. Pulsed EPR determination of water accessibility to spin-labeled amino acid residues in LHCIIb. Biophys J. 2009;96:1124–1141. doi: 10.1016/j.bpj.2008.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hubbell WL, Gross A, Langen R, Lietzow MA. Recent advances in site-directed spin labeling of proteins. Curr Opin Struct Biol. 1998;8:649–656. doi: 10.1016/s0959-440x(98)80158-9. [DOI] [PubMed] [Google Scholar]
- 13.Hubbell WL, Cafiso DS, Altenbach C. Identifying conformational changes with site-directed spin labeling. Nat Struct Biol. 2000;7:735–739. doi: 10.1038/78956. [DOI] [PubMed] [Google Scholar]
- 14.Columbus L, Hubbell WL. A new spin on protein dynamics. Trends Biochem Sci. 2002;27:288–295. doi: 10.1016/s0968-0004(02)02095-9. [DOI] [PubMed] [Google Scholar]
- 15.Fanucci GE, Cafiso DS. Recent advances and applications of site-directed spin labeling. Curr Opin Struct Biol. 2006;16:644–653. doi: 10.1016/j.sbi.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 16.Marsh D. Polarity and permeation profiles in lipid membranes. Proc Natl Acad Sci U S A. 2001;98:7777–7782. doi: 10.1073/pnas.131023798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marsh D, Kurad D, Livshits VA. High-field electron spin resonance of spin labels in membranes. Chem Phys Lipids. 2002;116:93–114. doi: 10.1016/s0009-3084(02)00022-1. [DOI] [PubMed] [Google Scholar]
- 18.Griffith OH, Dehlinger PJ, Van SP. Shape of the hydrophobic barrier of phospholipid bilayers. (Evidence for water penetration in biological membranes) J. Membrane Biol. 1974;15:159–192. doi: 10.1007/BF01870086. [DOI] [PubMed] [Google Scholar]
- 19.Altenbach C, Greenhalgh DA, Khorana HG, Hubbell WL. A collision gradient method to determine the immersion depth of nitroxides in lipid bilayers: application to spin-labeled mutants of bacteriorhodopsin. Proc Natl Acad Sci U S A. 1994;91:1667–1671. doi: 10.1073/pnas.91.5.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Altenbach C, Froncisz W, Hemker R, McHaourab H, Hubbell WL. Accessibility of nitroxide side chains: absolute Heisenberg exchange rates from power saturation EPR. Biophys J. 2005;89:2103–2112. doi: 10.1529/biophysj.105.059063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schrempf H, Schmidt O, Kummerlen R, Hinnah S, Muller D, Betzler M, Steinkamp T, Wagner R. A prokaryotic potassium ion channel with two predicted transmembrane segments from Streptomyces lividans. Embo J. 1995;14:5170–5178. doi: 10.1002/j.1460-2075.1995.tb00201.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heginbotham L, Kolmakova-Partensky L, Miller C. Functional reconstitution of a prokaryotic K+ channel. J Gen Physiol. 1998;111:741–749. doi: 10.1085/jgp.111.6.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou Y, Morais-Cabral JH, Kaufman A, MacKinnon R. Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 Å resolution. Nature. 2001;414:43–48. doi: 10.1038/35102009. [DOI] [PubMed] [Google Scholar]
- 24.le Coutre J, Kaback HR, Patel CK, Heginbotham L, Miller C. Fourier transform infrared spectroscopy reveals a rigid alpha-helical assembly for the tetrameric Streptomyces lividans K+ channel. Proc Natl Acad Sci U S A. 1998;95:6114–6117. doi: 10.1073/pnas.95.11.6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perozo E, Cortes DM, Cuello LG. Three-dimensional architecture and gating mechanism of a K+ channel studied by EPR spectroscopy. Nat Struct Biol. 1998;5:459–469. doi: 10.1038/nsb0698-459. [DOI] [PubMed] [Google Scholar]
- 26.Gross A, Columbus L, Hideg K, Altenbach C, Hubbell WL. Structure of the KcsA potassium channel from Streptomyces lividans: a site-directed spin labeling study of the second transmembrane segment. Biochemistry. 1999;38:10324–10335. doi: 10.1021/bi990856k. [DOI] [PubMed] [Google Scholar]
- 27.Gross A, Hubbell WL. Identification of protein side chains near the membrane-aqueous interface: a site-directed spin labeling study of KcsA. Biochemistry. 2002;41:1123–1128. doi: 10.1021/bi015828s. [DOI] [PubMed] [Google Scholar]
- 28.Vamvouka M, Cieslak J, Van Eps N, Hubbell W, Gross A. The structure of the lipid-embedded potassium channel voltage sensor determined by double-electron-electron resonance spectroscopy. Protein Sci. 2008;17:506–517. doi: 10.1110/ps.073310008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Green PR, Bell RM. Asymmetric reconstitution of homogeneous Escherichia coli sn-glycerol-3- phosphate acyltransferase into phospholipid vesicles. J Biol Chem. 1984;259:14688–14694. [PubMed] [Google Scholar]
- 30.Kumaresan R, Tufts DW. Estimating the parameters of exponentially damped sinusoids and pole-zero modeling in noise. Ieee T Acoust Speech. 1982;30:833–840. [Google Scholar]
- 31.Owenius R, Osterlund M, Svensson M, Lindgren M, Persson E, Freskgard PO, Carlsson U. Spin and fluorescent probing of the binding interface between tissue factor and factor VIIa at multiple sites. Biophys J. 2001;81:2357–2369. doi: 10.1016/S0006-3495(01)75882-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marsh D, Toniolo C. Polarity dependence of EPR parameters for TOAC and MTSSL spin labels: correlation with DOXYL spin labels for membrane studies. J Magn Reson. 2008;190:211–221. doi: 10.1016/j.jmr.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 33.Kelly BL, Gross A. Potassium channel gating observed with site-directed mass tagging. Nat Struct Biol. 2003;10:280–284. doi: 10.1038/nsb908. [DOI] [PubMed] [Google Scholar]
- 34.Cortes DM, Cuello LG, Perozo E. Molecular architecture of full-length KcsA: role of cytoplasmic domains in ion permeation and activation gating. J Gen Physiol. 2001;117:165–180. doi: 10.1085/jgp.117.2.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kurshev VV, Kevan L. Electron spin echo modulation studies of doxylstearic acid spin probes in frozen vesicle solutions: interaction of the spin probe with 31P in the surfactant headgroups. J Phys Chem. 1995;99:10616–10620. [Google Scholar]
- 36.Hunte C. Specific protein-lipid interactions in membrane proteins. Biochem Soc Trans. 2005;33:938–942. doi: 10.1042/BST20050938. [DOI] [PubMed] [Google Scholar]
- 37.Hofer P, Grupp A, Nebenfuhr H, Mehring M. Hyperfine sublevel correlation (HYSCORE) spectroscopy - a 2D electron-spin-resonance investigation of the squaric acid radical. Chem. Phys. Lett. 1986;132:279–282. [Google Scholar]
- 38.Doyle DA, Cabral JM, Pfuetzner RA, Kuo A, Gulbis JM, Cohen SL, Chait BT, MacKinnon R. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 39.Tombolato F, Ferrarini A, Freed JH. Dynamics of the nitroxide side chain in spin-labeled proteins. J Phys Chem B. 2006;110:26248–26259. doi: 10.1021/jp0629487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oh KJ, Altenbach C, Collier RJ, Hubbell WL. Site-directed spin labeling of proteins. Applications to diphtheria toxin. Methods Mol Biol. 2000;145:147–169. doi: 10.1385/1-59259-052-7:147. [DOI] [PubMed] [Google Scholar]
- 41.Seelig J, Seelig A. Deuterium magnetic resonance studies of phospholipid bilayers. Biochem Biophys Res Commun. 1974;57:406–411. doi: 10.1016/0006-291x(74)90945-0. [DOI] [PubMed] [Google Scholar]
- 42.Tugarinov V, Kanelis V, Kay LE. Isotope labeling strategies for the study of high-molecular-weight proteins by solution NMR spectroscopy. Nat Protoc. 2006;1:749–754. doi: 10.1038/nprot.2006.101. [DOI] [PubMed] [Google Scholar]
- 43.Gordon-Grossman M, Gofman Y, Zimmermann H, Frydman V, Shai Y, Ben-Tal N, Goldfarb D. A combined pulse EPR and Monte Carlo simulation study provides molecular insight on peptide-membrane interactions. J Phys Chem B. 2009;113:12687–12695. doi: 10.1021/jp905129b. [DOI] [PubMed] [Google Scholar]
- 44.Noethig-Laslo V, Cevc P, Arcon D, Sentjurc M. Hydrophobic barrier in liposomes studied by FT-ESEEM. Orig Life Evol Biosph. 2004;34:237–242. doi: 10.1023/b:orig.0000009843.52972.bd. [DOI] [PubMed] [Google Scholar]
- 45.Engelman DM. Lipid bilayer structure in the membrane of Mycoplasma laidlawii. J Mol Biol. 1971;58:153–165. doi: 10.1016/0022-2836(71)90238-5. [DOI] [PubMed] [Google Scholar]
- 46.Zanker PP, Jeschke G, Goldfarb D. Distance measurements between paramagnetic centers and a planar object by matrix Mims electron nuclear double resonance. J Chem Phys. 2005;122:024515. doi: 10.1063/1.1828435. [DOI] [PubMed] [Google Scholar]
- 47.Halkides CJ, Farrar CT, Larsen RG, Redfield AG, Singel DJ. Characterization of the active site of p21 ras by electron spin-echo envelope modulation spectroscopy with selective labeling: comparisons between GDP and GTP forms. Biochemistry. 1994;33:4019–4035. doi: 10.1021/bi00179a031. [DOI] [PubMed] [Google Scholar]
- 48.Freites JA, Tobias DJ, von Heijne G, White SH. Interface connections of a transmembrane voltage sensor. Proc Natl Acad Sci U S A. 2005;102:15059–15064. doi: 10.1073/pnas.0507618102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Freites JA, Tobias DJ, White SH. A voltage-sensor water pore. Biophys J. 2006;91:L90–92. doi: 10.1529/biophysj.106.096065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.MacKinnon R. Structural biology. Voltage sensor meets lipid membrane. Science. 2004;306:1304–1305. doi: 10.1126/science.1105528. [DOI] [PubMed] [Google Scholar]
- 51.Lee SY, Lee A, Chen J, MacKinnon R. Structure of the KvAP voltage-dependent K+ channel and its dependence on the lipid membrane. Proc Natl Acad Sci U S A. 2005;102:15441–15446. doi: 10.1073/pnas.0507651102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schmidt D, Jiang QX, MacKinnon R. Phospholipids and the origin of cationic gating charges in voltage sensors. Nature. 2006;444:775–779. doi: 10.1038/nature05416. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.