Abstract
Membrane proteins are of great interest to plant physiologists because of their important function in many physiological processes. However, their study is hampered by their low abundance and poor solubility in aqueous buffers. Proteomics studies of non-model plants are generally restricted to gel-based methods. Unfortunately, all gel-based techniques for membrane proteomics lack resolving power. Therefore, a very stringent enrichment method is needed before protein separation. In this study, protein extraction in a mixture of chloroform and methanol in combination with gel electrophoresis is evaluated as a method to study membrane proteins in non-model plants. Benefits as well as disadvantages of the method are discussed. To demonstrate the pitfalls of working with non-model plants and to give a proof of principle, the method was first applied to whole leaves of the model plant Arabidopsis. Subsequently, a comparison with proteins extracted from leaves of the non-model plant, banana, was made. To estimate the tissue and organelle specificity of the method, it was also applied on banana meristems. Abundant membrane or lipid-associated proteins could be identified in both tissues, with the leaf extract yielding a higher number of membrane proteins.
Electronic supplementary material
The online version of this article (doi:10.1007/s00425-010-1121-1) contains supplementary material, which is available to authorized users.
Keywords: Chloroform/methanol extraction, Leaf, Meristem, Non-model, Plant membrane proteomics
Introduction
Membrane proteins play a crucial role in many physiological processes and are therefore of great interest to plant physiologists. Classical two-dimensional electrophoresis (2DE) is the most commonly applied technique to study plant proteins (Carpentier et al. 2005, 2008b; Agrawal and Rakwal 2006; Jorrin et al. 2007). Unfortunately, one main drawback is that 2DE fails to analyze integral membrane proteins with multiple membrane spanning domains (Braun et al. 2007; Rabilloud et al. 2008). Indeed, due to their hydrophobic nature, membrane proteins are difficult to extract and dissolve in aqueous buffers and tend to precipitate during isoelectric focusing (Santoni et al. 2000). The relatively low abundance of membrane proteins is another challenge to the study of membrane proteins (i.e., “the abundance problem”). The presence of more abundant soluble proteins makes the detection and identification of membrane proteins troublesome (Santoni et al. 2000). Therefore, the addition of a subcellular or biochemical fractionation step is essential for a successful membrane proteome study. Commonly used fractionation techniques in plant membrane proteomics include differential centrifugations and salt or alkaline treatments, as reviewed by Ephritikhine et al. (2004).
Proteomics in non-model plant species is currently restricted to protein separation techniques with sufficient resolving power (i.e., gel-based separations) combined with cross-species identification (Wilkins and Williams 1997). In case the resolution is too low, multiple proteins are digested simultaneously resulting in a complex peptide pool in which peptides that are derived from the same protein can no longer be associated with each other (i.e., “the identification problem”). Consequently, a combination of masses of non-related peptides is used for database searching, thereby enhancing the risk of false positive identifications. This is especially true for the shotgun approach where the protein extract is digested prior to separation (Haynes and Roberts 2007). Additionally, proteomics studies on non-model plants require the application of tandem mass spectrometry (MS/MS) to retrieve identifications (Carpentier et al. 2008b). In the MS mode, peptides with a high signal-to-noise ratio (S/N) are selected for further fragmentation to obtain more informative data on the parent ion mass, enlarging the probability of obtaining significant hits. The S/N of a peptide not only depends on the abundance of the protein from where it is derived, but also on its ease to desorb and ionize and on its m/z. For this reason, the peptides with the highest S/N in a complex peptide mixture might be derived from different proteins. In case such intense peptides are less or not informative (i.e., not conserved in a sequenced species), identification will fail since more homologous peptides with a lower S/N will not be selected for MS/MS. However, when the peptide mixture consists of a limited amount of peptides, which are all derived from one protein, the informative peptides with a lower S/N have a bigger chance of being selected for MS/MS, making protein identification possible.
All published methods for membrane proteomics are limited in resolution (Braun et al. 2007). Therefore, a very stringent enrichment step is needed prior to protein separation. A simple strategy to enrich a protein mixture with highly hydrophobic proteins is to extract them in a mixture of chloroform and methanol (C/M). This extraction method was first presented in 1951 by Folch et al. as a way to extract lipids from brain tissue (Folch et al. 1951). Nowadays, the method is still popular as a protein precipitation method (Jiang et al. 2004). It has been observed that some proteins do not precipitate and remain soluble in the mixture of chloroform and methanol. Henriques and Park were the first to characterize C/M-soluble proteins from spinach chloroplast membranes and demonstrated that the C/M-soluble fraction is enriched in proteins containing a high number of hydrophobic amino acids (Henriques and Park 1976). Joyard et al. thoroughly characterized the proteins found soluble in the organic phases of different ratios of chloroform to methanol (Joyard et al. 1982; Seigneurin-Berny et al. 1999). In the following years, the method was applied to study hydrophobic proteins of purified chloroplast envelope (Ferro et al. 2002, 2003) and thylakoid (Friso et al. 2004) membranes, and of mitochondrial (Brugiere et al. 2004), tonoplast (Schmidt et al. 2007) and plasma membranes (Marmagne et al. 2004).
In this study, we evaluate whether C/M extraction is a suitable method to analyze membrane proteins from total cellular lysates of a non-model plant. Chloroform/methanol extraction was combined with one- as well as two- dimensional gel electrophoresis (dSDS). Resolution of both separations was compared. The method was applied on a model (Arabidopsis) and a non-model (banana) plant to demonstrate the difficulties associated with non-model studies. To evaluate organelle specificity, the method was also applied to non-green tissue, namely banana shoot apical meristems (Musa spp.).
Although banana (Musa spp.) is one of the most important food crops in the world, only 1% of the Musa genome is currently sequenced (http://www.Musagenomics.org/). Moreover, banana is only distantly related to sequenced monocots, which makes cross-species identification challenging (Aert et al. 2004; Lescot et al. 2008). Therefore, banana can be considered as a good representative of a non-model plant. The presence of very high levels of oxidative enzymes, phenol compounds and carbohydrates makes the study of the banana proteome even more challenging (Carpentier et al. 2005).
The aim of this study was to demonstrate the difficulties encountered in proteomics studies on non-model plants by comparing results obtained in a model and non-model plant. Since membrane proteins are very interesting, but troublesome to analyze, a method to enrich membrane proteins to facilitate membrane proteomics studies was evaluated. This overview of the merits and disadvantages of C/M extraction estimates its value and is a basis for membrane proteomics studies in non-model plants.
Materials and methods
Plant material
Arabidopsis thaliana var. Columbia plants, kindly provided by F. Rolland (K. U. Leuven, Belgium), were grown using a daily cycle of 12-h light (75 μmol m−2 s−1) at 22°C and 12-h darkness at 18°C and 50% relative humidity. After 2 months, leaves were harvested and immediately frozen in liquid nitrogen. Plantlets of the banana cultivar Mbwazirume (ITC0084) were obtained from the Bioversity International Musa collection at K. U. Leuven, Belgium. Banana plants were grown in the greenhouse with 27° and 20°C of respective day and night temperatures, a 12-h photoperiod and 80% relative humidity. Leaves were collected from 1-year-old plants. Multiple shoot meristem cultures were initiated as described by Strosse et al. (2006) and subsequently maintained on a standard control medium containing 0.09 M sucrose (i.e., Murashige and Skoog medium supplemented with benzylaminopurine). All cultures were kept in the dark at 25°C. After 1 month of culture, meristems were grinded in liquid nitrogen.
Protein extraction
After grinding in liquid nitrogen, 200–400 mg material (fresh weight) was transferred to 1 ml of ice-cold extraction buffer, containing 100 mM Tris–HCl (pH 8.3), 5 mM EDTA.Na2, 100 mM KCl, 1% DTT and complete protease inhibitor cocktail (Roche, Mannheim, Germany). Samples were added to an ice-cold chloroform/methanol mixture in the ratio 1/9 as described by Seigneurin-Berny et al. (1999) and carefully mixed. Different C/M ratios were tested. Samples were incubated on ice for 30 min and centrifuged at 16,000g (4°C) for 1 h. Pellets were washed overnight in ice-cold acetone containing 0.2% DTT. Organic phases were collected and proteins that were soluble in these phases were precipitated overnight at −20°C by addition of 1 ml cold di-ethylether. After centrifugation at 4°C (16,000g for 1 h), pellets were solubilized in 100 mM Tris–HCl (pH 6.8), 4% SDS. Total protein extract was retrieved after trichloroacetic acid (TCA) precipitation. As much as 100 μl of sample was added to 1.9 ml of 10% TCA in ice-cold acetone with 0.2% DTT and precipitated overnight. After 1 h of centrifugation at 16,000g (4°C), pellets were washed with ice-cold acetone containing 0.2% DTT. The pellets were dissolved in the same SDS buffer as the C/M-soluble and insoluble proteins. The protein concentration was measured using a micro-Bradford membrane protein assay as described by Zuo and Lundahl (2000) and samples were stored at −80°C.
Protein separation
After thawing, loading buffer was added up to a final buffer concentration of 3% SDS, 75 mM Tris–HCl (pH 6.8), 15% glycerol, 3.5 M urea, 1% DTT and 0.05% bromophenol blue. Samples were heated at 37°C for 30 min and briefly centrifuged. Equal amounts (40 μg) of proteins were loaded and proteins were separated via SDS PAGE. For dSDS separation of banana leaf proteins, 90 μg of sample was loaded. For all separations, the Laemmli protocol (Laemmli 1970) was used.
One-dimensional separation
For 1D separation, a 10–15.5% hyperbolic gradient gel (18 × 24 cm, 1.5 mm), generated by a 2DE optimizer (NextGen Sciences, Alconbury, UK) was used. The stacking gel consisted of 4% acrylamide (Bio-Rad laboratories, Hercules, CA, USA). Gels were run overnight at 2 W/gel at 12°C.
Two-dimensional separation
For dSDS PAGE, gels were poured manually. The separating gel of the first dimension (18 × 24 cm, 1 mm) consisted of 10% acrylamide, while the second dimension separating gel (26 × 20 cm, 1.5 mm) contained 15% acrylamide. Both stacking gels contained 4% acrylamide. The dSDS protocol was performed as described by Rais et al. (2004) with slight modifications. Urea was omitted from the gel since the identification of highly hydrophobic proteins was desired (Rais et al. 2004). After a first dimension separation, lanes were excised, swollen for 45 min in buffer (100 mM Tris, 0.2% DTT, pH 2.0) and placed on top of the second dimension gels. Gaps between the excised gel lane and the spacer were filled with agarose sealing solution containing 0.5% agarose, 0.002% bromophenol blue and 1× Laemmli buffer. The first dimension was run overnight at 2 W/gel at 12°C. Second dimension gels were run at 20°C. For entering and protein migration through the stacking gel, 2 W/gel was applied. After 1.5 h, the power was increased to 16.7 W/gel.
All gels were stained with G-250 Colloidal Coomassie (Serva, Heidelberg, Germany; Neuhoff et al. 1988).
Image analyses
Gel images were captured with labscan 5 software (GE Healthcare) and analysis of 1D and 2D images was performed using Quantity One (Bio-Rad) and Image Master 2-D platinum (GE healthcare) software, respectively.
Protein identification
After Coomassie blue staining and image analysis, spots were manually picked. In-gel digestion with trypsin and analysis of the tryptic peptides by MALDI TOF-TOF was performed at the “Centre de Recherche Public Gabriel Lipmann” in Luxembourg. For digestion, the Ettan dalt spot handling workstation (GE Healthcare) was used. After reduction and alkylation, gel pieces were washed and desalted first in 50 mM ammonium bicarbonate/50% v/v methanol and subsequently in 75% (v/v) acetonitrile (ACN). After adding 8 μl of a Trypsin Gold solution (5 ng ml−1) in 20 mM ammonium bicarbonate (Promega, Madison, WI, USA) to the dried gel plugs, samples were incubated at 37°C for 6 h. After extraction and drying, the resulting peptides were dissolved in 3 μl of a 50% ACN solution containing 0.1% trifluoroacetic acid (TFA) and 0.7 μl of each well was spotted on disposable MALDI-TOF target plates (Applied Biosystems, Foster City, CA, USA). Spotted peptides were mixed with 0.7 μl of α-cyano-4-hydroxycinnamic acid (7 mg ml−1, 50% ACN/0, 1% TFA) and allowed to air dry.
Mass spectrometrical analyses (MS and MS/MS) were carried out using the Applied Biosystems 4800 Proteomics Analyzer in positive electron mode, externally calibrated using the peptide mass calibration kit, according to the manufacturer’s instructions (Applied Biosystems).
An in-house Mascot platform was used for searching against the NCBI Viridiplantae database and the NCBI Musa EST database. The searching parameters allowed two missed cleavages, a tolerance of 0.50 Da on MS/MS fragments and 100 ppm on precursor mass, as well as carbamidomethylation on cysteine as a fixed modification. Double oxidation and kynurenin formation of tryptophan and oxidation of methionine were allowed as variable modifications. The probability score (Mowse score, Perkins et al. 1999) calculated by the software was used as a criterion for accurate identification. Estimation of false positive rates was made by searching a decoy database with the same search criteria. This decoy database was composed of the proteins present in the NCBI Viridiplantae database and from which the amino acid composition was randomly scrambled. Search against this database gave no significant hits at the MS/MS level.
Prediction methods
Identifications obtained by the NCBI Viridiplantae and MUSA EST search were blasted in batch against the Swissprot database using the blastcl3 tool, which interacts directly with the NCBI BLAST server (http://www.ncbi.nlm.nih.gov/staff/tao/URLAPI/netblast.html). The ProtParam tool of the ExPASy server (http://ca.expasy.org/) was used to calculate the grand average of hydropathicity (GRAVY) score (Kyte and Doolittle 1982) and other parameters such as the theoretical pI and M r. The number of transmembrane domains was calculated by the TMHMM Server v 2.0 (http://protfun.net/services/TMHMM/), Phobius (http://phobius.cbr.su.se/), HMMTOP (http://www.enzim.hu/hmmtop/) (Tusnady and Simon 2001) and the Aramemnon Web site (http://aramemnon.botanik.uni-koeln.de).
Sequence alignments were performed using the ClustalW2 tool of the European Bioinformatics Institute (http://www.ebi.ac.uk/Information/).
Protein location was predicted using the Target P server (http://www.cbs.dtu.dk/services/TargetP/) and functional domains were predicted by Pfam (http://pfam.sanger.ac.uk/). Information on Arabidopsis proteins was obtained from the ARAMEMNON and TAIR (http://www.arabidopsis.org/) Web site. Results were evaluated and updated according to the information found in the plant proteome database (http://ppdb.tc.cornell.edu/).
Results and discussion
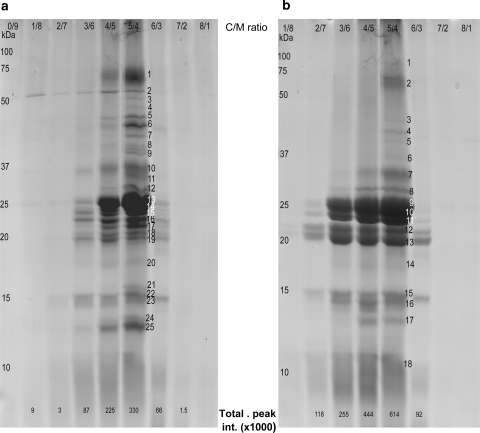
Since C/M extraction was not yet evaluated for use on whole cell lysates, the ratio of chloroform to methanol (ranging from 0/9 to 8/1) yielding the highest amount of proteins was determined for Arabidopsis as well as for banana, as recommended by Rolland et al. (2006). Because the C/M method has a bias toward smaller proteins (Brugiere et al. 2004), the protein patterns of the organic phases of the different ratios were compared using optimized 1D gradient gels (10–15.5%, hyperbolic gradient). As illustrated in Fig. 1, only in the C/M ratios of 3/6 to 6/3, a considerable amount of proteins remained soluble in the organic phase, irrespective of the plant species. Since the 5/4 ratio resulted in the highest number of bands and the highest total peak intensity, this ratio was chosen for further analyses for both Arabidopsis and banana. As predicted by Bligh and Dyer (1959), only one organic phase and a white pellet containing the insoluble proteins was obtained using this ratio. Since also lipids and pigments were extracted in the organic phase, they were further eliminated through protein precipitation with diethylether.
Fig. 1.
Proteins from Arabidopsis (a) or banana (b) leaves, extracted in organic phases of different ratios of chloroform to methanol separated on a 10–15.5% (hyperbolic acrylamide gradient) SDS gel and Coomassie Brilliant Blue stained. Molecular masses (kDa) of a protein standard are indicated on the left; C/M ratios are shown on top. At the bottom, the total peak intensity (×1,000) as calculated by the Quantity One software (Bio-Rad) is given. The numbers are indicative of the bands, which were cut for protein identification. Identifications are given in the Online Resource S2
Chloroform/methanol extraction of leaf proteins of the model plant Arabidopsis to assess the “abundance identification” problem
Resolution: 1D gradient SDS versus dSDS PAGE
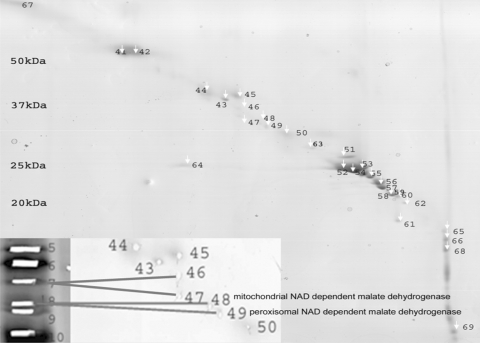
The main constraint for protein identification in non-model plants is the lack of genomic information. Hence, a sufficient separation power to separate proteins from each other is indispensable. For that reason, the resolution of a 1D separation (Fig. 1a, 5/4 lane) was compared with that of a 2D separation (Fig. 2). Double SDS was chosen as 2D technique, since it is considered to be superior to separate and visualize highly hydrophobic proteins in comparison with, for example, benzyldimethyl-n-hexadecylammonium chloride (16-BAC)-gels (Burre et al. 2006). Proteins soluble in 5/4 C/M were separated on a 10–15.5% hyperbolic gradient gel (Fig. 1a). A total of 25 bands were picked; 23 gave rise to identifications and in 48% (11 out of 23) of the bands more than one protein was identified (Online Resource S2). From the dSDS gel, 29 spots were picked, 27 resulted in identifications and only in 19% of the spots (5 out of 27) two or more proteins were identified. This is a good indication that implementing a second dimension improves resolution and will thus simplify the final tryptic digest. Moreover, dSDS allowed the study of protein isoforms like the mitochondrial and peroxisomal form of NAD-dependent malate dehydrogenase (Fig. 2). These were detected in two different spots on the dSDS gel (spots 48 and 49, respectively), while on the 1D gel they were located in one band (band 8).
Fig. 2.
Double SDS gel of proteins from Arabidopsis leaves soluble in 5/4 C/M. As much as 40 μg of extract was loaded. Numbers indicate the number of the spot, corresponding to one or more identified proteins described in Online Resource S1 and S2. Molecular masses of standard proteins are indicated on the left. The gel was Coomassie Brilliant Blue stained. The enlargement in the left lower corner shows that proteins that are detected in one band on the 1D gradient gel (Fig. 1a, band 8) are dispersed in two spots on the 2D gel
However, the application of dSDS also has its disadvantages: the resolution remains limited (Rabilloud et al. 2008) and the introduction of an extra dimension is inevitably associated with loss of proteins. Due to this protein loss and the fact that protein identification depends on protein abundance and peptide intensities, protein identifications after one- or two- dimensional SDS PAGE complemented each other.
In total, the extraction of Arabidopsis leaf proteins in a 5/4 C/M mixture and 1D and dSDS separation yielded 36 different protein identifications (Online Resources S1 and S2). This number is of the same order of magnitude as the 37 proteins identified by Ferro et al. in a study of Arabidopsis chloroplasts (Ferro et al. 2003) and the 31 proteins reported by Brugiere et al. (2004) in a study of the mitochondrial proteome. This underlines that the C/M method is very selective.
As already observed by Henriques and Park (1976) and Brugiere et al. (2004), only proteins with a low M r were present in the C/M-soluble phase. High M r proteins probably precipitate during C/M extraction. Schroder and Hasilik (2006) proposed the addition of a halogenic acid and an extra phase separation to recover these high M r membrane proteins. However, this extra phase separation is associated with additional significant protein losses (Schroder et al. 2007).
Increase in abundance of membrane proteins by chloroform/methanol extraction
One of the major problems in identifying membrane proteins in whole cell lysates is their low abundance compared to water-soluble proteins. It is therefore important to evaluate whether C/M extraction indeed results in an extract that is enriched in membrane proteins and depleted in highly abundant water-soluble proteins. The total protein extract from Arabidopsis leaves was compared with the protein extracts that are soluble and insoluble in C/M (Fig. 3). Some representative proteins were identified as proof of principle (Table 1). The most abundant protein in the total leaf (T) as well as in the C/M insoluble (I) extract was identified as the large chain of Rubisco (Fig. 3, band numbers 1 and 2). It was almost completely absent in the C/M-soluble (S) fraction. The most heavily stained bands of the C/M-soluble fraction contained proteins associated with light-harvesting complex II such as chlorophyll a-/b-binding proteins (band numbers 9 and 10). Those membrane proteins were clearly depleted from the insoluble fraction, but could be identified in the total protein fraction. Other membrane proteins, such as the photosystem II 44-kDa protein (band numbers 3 and 4), could also be identified in the total cell lysate. Less abundant integral membrane proteins such as photosystem II Qb protein (band number 6) could only be detected in the C/M-soluble fraction. This was indicative of an enrichment in integral membrane proteins in the C/M-soluble fraction.
Fig. 3.
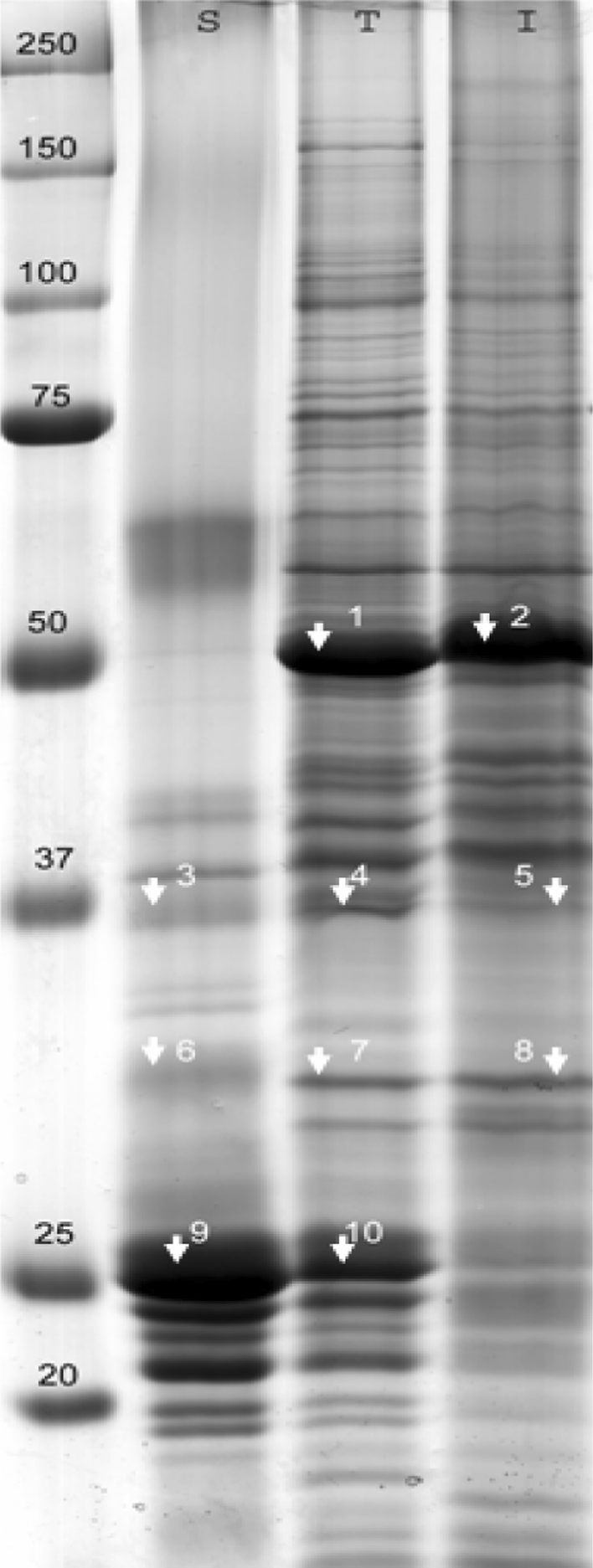
Proteins soluble (S) and insoluble (I) in a 5/4 C/M mixture compared to the total protein content (T) of Arabidopsis leaves on a uniform 10% acrylamide gel. As much as 40 μg of proteins was loaded. Gel was Coomassie Brilliant blue stained. Molecular masses of the protein standard (kDa) are indicated on the left. Some representative bands (numbered) were cut for protein identification (Table 1, Online Resource S2)
Table 1.
Proteins identified in the bands depicted in Fig. 3
| gia | Name | Band number | GRAVY score | TMH | ||
|---|---|---|---|---|---|---|
| C/M sol (S) | Total (T) | C/M insol (I) | ||||
| 7525041 | Rubisco large chain | / | 1 | 2 | −0.272 | 0 |
| 7525029 | Photosystem II 44-kDa protein | 3 | 4 | / | 0.252 | 6 |
| 18420348 | Probable fructose-bisphosphate aldolase 2 | / | 4 | 5 | −0.167 | 0 |
| 8131597 | Photosystem II Qb protein | 6 | / | / | −0.062 | 8 |
| 15240013 | 33-kDa oxygen-evolving protein | / | 7 | 8 | −0.327 | 0 |
| 16374 | Chlorophyll a-/b-binding protein | 9 | 10 | / | −0.056 | 2 |
agi number, name, the band where the protein was identified, GRAVY score and number of transmembrane helices (TMH; determined by TMHMM v2.0) are shown. C/M sol (S) = proteins soluble in a 5/4 C/M mixture; total (T) = total protein extract after TCA precipitation; C/M insol (I) = proteins insoluble in a 5/4 C/M mixture
Protein extraction of a whole cell lysate: chloroform/methanol extraction versus classical 2DE
The main motivation to perform C/M extraction combined with gel electrophoresis was to complement classical 2DE studies and to find an approach to study membrane proteins in non-model plants. To evaluate whether membrane proteins were indeed identified, the number of transmembrane helices (TMH) was determined. Different prediction programs are available of which TMHMMv2.0 is reported to be the best (Moller et al. 2001). The TMHMMv2.0 server predicted that only 22% of the C/M-soluble proteins were transmembrane proteins. The plant-specific membrane database, Aramemnon, uses a consensus of 17 prediction programs and predicted that 58% of the obtained proteins were integral membrane proteins (Schwacke et al. 2003). Using the HMMTOP server (Tusnady and Simon 2001), the number of transmembrane proteins was much higher and reached 72%. These differences clearly demonstrate that setting the threshold of the prediction too strict might result in false negative results, while setting it too broad might result in false positives. Therefore, Aramemnon was preferred. However, in silico predictions should be validated by localization studies, which will be instrumental to improve the algorithms and make them more plant specific.
We compared our results with a published 2DE map of Arabidopsis leaf proteins. Giavalisco et al. (2005) published such a map. Although the authors took special precautions to increase the number of membrane proteins on the 2DE gels, proteins containing more than one transmembrane helix were not identified. As much as 70% of our proteins extracted with C/M could not be found on this 2DE map (Online Resource S1), which confirms that C/M extraction complements classical 2DE and leads to an enrichment of hydrophobic proteins.
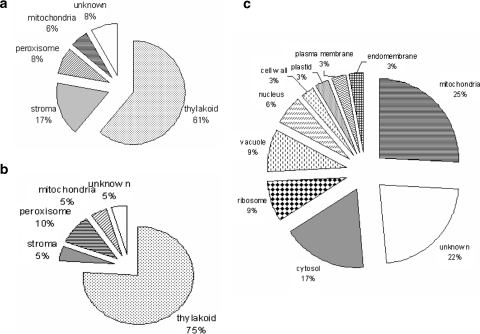
Most of the identified proteins were associated with the chloroplast thylakoid membrane (Fig. 4a) and were involved in photosynthesis (78%). A detailed study of the location and function of the obtained proteins was not performed, as protein functions have already been discussed in previous papers (Ferro et al. 2003; Friso et al. 2004).
Fig. 4.
Location of Arabidopsis leaf (a) or banana leaf (b) and meristem (c) proteins extracted in a 5/4 C/M mixture. Information was retrieved from the Target P server (http://www.cbs.dtu.dk/services/TargetP/) and the plant proteome database (http://ppdb.tc.cornell.edu)
Reproducibility
Because reproducibility of the C/M extraction is essential, a comparison with related publications on plants was made. Henriques and Park (1976) reported that more than 50% of the total C/M-soluble proteins are present in a 25-kDa band. This band was specified further as containing the main component of the light-harvesting chlorophyll–protein complex (LHC). The most abundant proteins found in the C/M fraction are indeed associated with LHC II and have a molecular mass of approximately 25 kDa (Figs. 1, 2, 3). The majority of the 36 identified proteins (28) were previously reported (Online Resource S1), 22 of them in the study of the thylakoid proteome by Friso et al. (2004). The other eight proteins were not previously reported to be C/M soluble, but were not identified as membrane proteins. This might be an indication that a mixture of chloroform and methanol extracts a higher number of non-membrane proteins from a total cellular lysate compared to purified membrane structures.
Chloroform/methanol extraction applied to the non-model plant banana
Extraction of leaf proteins to allow comparison with the model plant Arabidopsis
The above results obtained with Arabidopsis leaf extracts demonstrated that C/M extraction combined with gel electrophoresis (1) yielded more than 50% transmembrane proteins, (2) complemented 2DE studies and (3) was a very stringent and reproducible method. The latter was desirable to cope with the limited resolution of gel-based separation techniques for membrane proteins.
When applying the method on leaf extracts of the non-model plant banana, 20 different proteins were identified using both the 1- and 2D approach (Table 2, Online Resource S2). This corresponds to an average identification rate of 61%, which was comparable to the identification rate in our 2DE analyses of banana (Carpentier et al. 2008a), but which was significantly lower than the 92.5% obtained with Arabidopsis.
Table 2.
Proteins identified after C/M extraction of a whole banana leaf lysate combined with one-dimensional gradient (1D) or dSDS (2D) PAGE
| Accessiona | Closest homolog | M r | pI | TMH | AA/TMH | GRAVY | Location | Spot/band number | ||
|---|---|---|---|---|---|---|---|---|---|---|
| 1D | 2D | 2 DE | ||||||||
| Q40433 | Photosystem I psaH protein (Nicotiana sylvestris) | 15.3 | 9.95 | 0 | / | −0.134 | Chloroplast thylakoid | 44 | ||
| O24045 | Rubisco small subunit (Musa acuminata) | 20.5 | 8.78 | 0 | / | −0.242 | Chloroplast stroma | 17 | 42 | x |
| Q9SUI4 | Photosystem I reaction center subunit XI (Nicotiana attenuata) | 23.1 | 9.85 | 0 | / | 0.31 | Chloroplast thylakoid | 15 | ||
| Q41039 | Lhca4 (Pinus sylvestris) | 26.8 | 7.12 | 0 | / | −0.093 | Chloroplast thylakoid | 21 | ||
| Q94JA2 | Malate dehydrogenase (mitochondrial) (Oryza sativa) | 35.5 | 8.74 | 0 | / | 0.076 | Mitochondrion | 37 | ||
| O49124 | Putative serine-glyoxylate aminotransferase (Fritillaria agrestis) | 44.1 | 7.63 | 0 | / | −0.012 | Others | 3 | 47 | x |
| Q6V8T3 | Chlorophyll a-/b-binding protein type I (Malus × domestica) | 15.6 | 5.05 | 1 | 147 | 0.116 | Chloroplast thylakoid | 22 | ||
| MUSF352TF | Rieske FeS protein precursor (Cytochrome b6-f complex iron-sulfur subunit) | 23.9 | 8.55 | 1 | 225 | −0.08 | Chloroplast thylakoid | |||
| O64450 | Lhcb1*9 (Nicotiana sylvestris) | 28.3 | 5.48 | 1 | 267 | 0.016 | Chloroplast thylakoid | 29 | ||
| P93260 | Glycolate oxidase (Mesembryanthemum crystallinum) | 31.3 | 9.16 | 1 | 75 | 0.011 | Peroxisome | 4 | 36 | |
| Q0ILQ0 | Peroxisomal malate dehydrogenase (Oryza sativa) | 37.4 | 8.09 | 1 | 145 | 0.181 | Peroxisome | 6 | 34 | x |
| Q67HN4 | Cytochrome b-559 alpha subunit (Cartonema philydroides) | 8.6 | 4.75 | 2 | 110 | 0.193 | Chloroplast thylakoid | 45 | ||
| MUSO477TF | Photosystem I reaction center subunit V | 13.2 | 10.47 | 2 | 64 | 0.192 | Chloroplast thylakoid | 44 | ||
| P15192 | Chlorophyll a–b-binding protein type 2 member 2 (Pinus sylvestris) | 16.1 | 4.79 | 2 | 75 | 0.156 | Chloroplast thylakoid | 28 | ||
| P36494 | Chlorophyll a–b-binding protein CP24 (Solanum lycopersicum) | 27.8 | 6.15 | 2 | 131 | 0.085 | Chloroplast thylakoid | 24 | ||
| Q6ZF30 | Putative chlorophyll a-/b-binding protein of LHCII type III (Oryza sativa) | 28.8 | 5.82 | 2 | 133 | 0.025 | Chloroplast thylakoid | 11 | 26 | |
| 1908421A | Light-harvesting complex IIa protein (Hordeum vulgare) | 31.3 | 5.33 | 2 | 143 | −0.149 | Chloroplast thylakoid | 30 | ||
| P05643 | Cytochrome b6/f complex subunit IV (Zea mays) | 17.5 | 6.56 | 3 | 53 | 0.551 | Chloroplast thylakoid | 15 | ||
| Q8HTU2 | Photosystem II D2 protein (Columnea sp. Lindqvist and Albert 30) | 83.2 | 6.66 | 9 | 59 | 0.372 | Chloroplast thylakoid | 1,2,5,6,7 | ||
| Q7YJY8 | Photosystem II D1 protein (Calycanthus floridus) | 82.5 | 6.89 | 11 | 67 | 0.120 | Chloroplast thylakoid | 33 | ||
aSwissprot or MusaEST accession number, the closest protein homolog, physicochemical properties; relative molecular masses (M r, ×1,000), pI, the number of transmembrane helices (TMH, determined by Aramemnon), GRAVY scores and the location are shown. The number of the bands on the gradient gel (Fig. 1b) or the spots on the dSDS gel (gel not shown) of the identified proteins are indicated as well as whether the proteins were also identified on a classical 2DE map of banana leaf proteins (Carpentier et al. 2008a)
Using cross-species identification and the Aramemnon database, 70% of the identified proteins were predicted to be transmembrane proteins. However, as stated above, care should be taken to consider a protein as being a real transmembrane protein, especially when working with non-model organisms and consequently relying on cross-species identification. As in Arabidopsis, most proteins were associated with chloroplast thylakoids and participated in the photosynthesis process.
The majority of the identified proteins (Table 2) could not be located on our previously generated 2DE map of banana leaf proteins (Carpentier et al. 2008a). This map, like most 2DE maps, did not reveal membrane proteins with more than one transmembrane domain (Carpentier et al. 2008a). Only the putative serine-glyoxylate aminotransferase, peroxisomal malate dehydrogenase and Rubisco small subunit were detected using both classical 2DE and C/M extraction combined with gel electrophoresis.
The identification of the Lhcb1*9 isoform demonstrates the need of an increased availability of DNA sequences of more plant species. The plant kingdom is characterized by the existence of many multigene families, which give rise to protein isoforms. In case the protein isoform of a non-model plant slightly differs from that of a model plant, there is a chance that peptides with the highest S/N are derived from a region that differs in amino acid sequence. Indeed, the peptides that led to the identification of Lhcb1*9 differ from the homologous peptides of the five Arabidopsis Lhcb1 isoforms (Online Resource S3).
Extraction of meristem proteins to determine the specificity of C/M extraction
To determine whether C/M preferentially extracts proteins that are associated with a specific organelle, the method was applied on total cellular extracts of shoot apical meristems. In contrast to leaves, meristems are non-green tissues containing undifferentiated, but actively metabolizing, cells. Their cytoplasm contains a large number of cell organelles, a lot of small vacuoles and an endoplasmic reticulum that is associated with ribosomes (Helliot et al. 2003). Proplastids are not yet differentiated to chloroplasts, so a different C/M-soluble proteome was expected in comparison with leaves (Carpentier et al. 2007).
First, the optimal C/M ratio was determined as described above for leaves (data not shown). Again, the 5/4 C/M ratio was selected. Extracted proteins were separated on a 10–15.5% gradient gel. Figure 5 clearly demonstrates that meristem and leaf protein band patterns are highly dissimilar and that the meristem lane contains a higher number of protein bands. In total, 35 non-redundant proteins were identified (Table 3; Online Resource S1 and S2). As could be expected, protein IDs from the meristem cells differed from those found in the leaf extract except for the mitochondrial form of malate dehydrogenase.
Fig. 5.
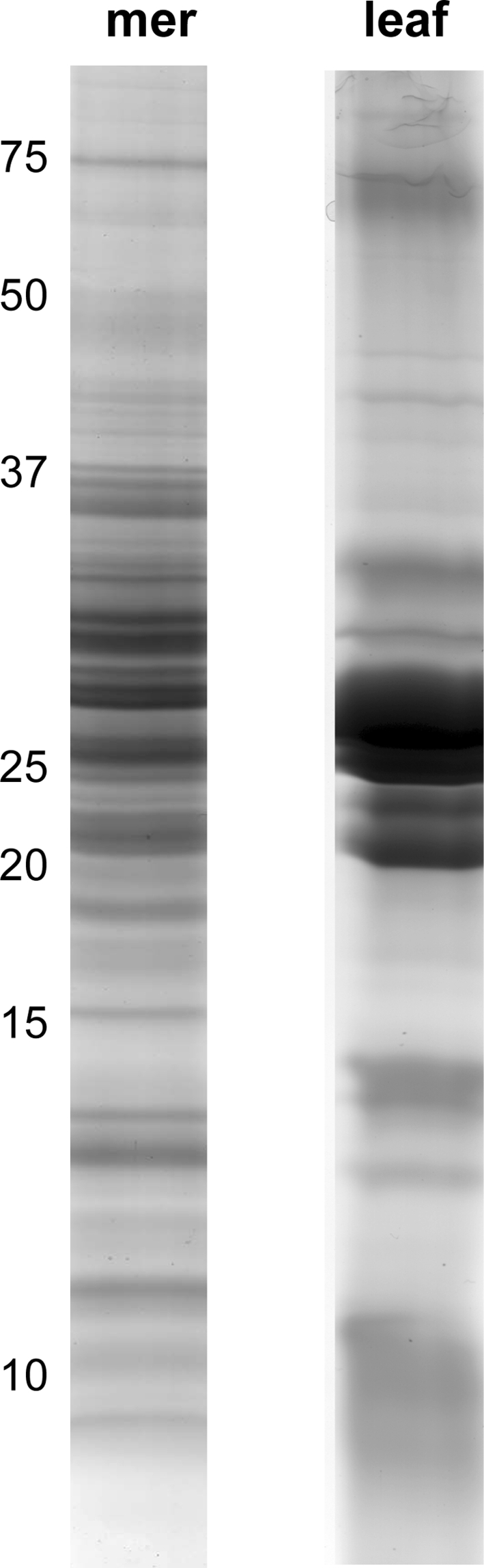
Banana meristem (mer) and leaf (leaf) proteins soluble in a 5/4 C/M mixture separated on a 10–15.5% gradient gel and Coomassie Brilliant Blue stained. Molecular masses (kDa) of a protein standard are indicated on the left
Table 3.
Proteins identified after C/M extraction of a whole banana meristem lysate combined with one-dimensional gradient (1D) SDS PAGE
| Accessiona | Closest homolog | M r | pI | TMH | GRAVY | Location |
|---|---|---|---|---|---|---|
| A7Q777 | Chromosome chr18 scaffold_59, whole genome shotgun sequence (Vitis vinifera) | 39.18 | 5.39 | 1 | −0.153 | Cell wall |
| P38076 | Cysteine synthase (Triticum aestivum) | 34.11 | 5.48 | 0 | 0.07 | Cytosol |
| P48534 | l-ascorbate peroxidase, cytosolic (Pisum sativum) | 27.06 | 5.52 | 0 | −0.332 | Cytosol |
| P29448 | Thioredoxin H-type 1 (Arabidopsis thaliana) | 12.67 | 5.64 | 0 | 0.034 | Cytosol |
| Q5JL11 | Putative soluble inorganic pyrophosphatase (Oryza sativa subsp. Japonica) | 23.62 | 5.88 | 0 | −0.31 | Cytosol |
| O23714 | Proteasome subunit beta type-2-A (Arabidopsis thaliana) | 22.54 | 5.95 | 0 | −0.034 | Cytosol |
| P34921 | Glyceraldehyde-3-phosphate dehydrogenase, cytosolic (Dianthus caryophyllus) | 36.9 | 6.46 | 0 | −0.143 | Cytosol |
| Q9AYP4 | 40S ribosomal protein S10 (Oryza sativa) | 20.26 | 9.76 | 0 | −0.878 | Cytosol-ribosome |
| Q8H0X6 | Cysteine proteinase inhibitor 6 (Arabidopsis thaliana) | 23.47 | 5.66 | 1 | −0.353 | Endomembrane system |
| Q07078 | Heat shock protein 81-3 (Oryza sativa) | 80.18 | 4.98 | 0 | −0.599 | Mitochondrion |
| Q9FWR4 | Glutathione S-transferase DHAR1, mitochondrial (Arabidopsis thaliana) | 23.64 | 5.56 | 0 | −0.173 | Mitochondrion |
| P47922 | Nucleoside diphosphate kinase 1 (Pisum sativum) | 16.46 | 5.94 | 0 | −0.064 | Mitochondrion |
| P17783 | Malate dehydrogenase, mitochondrial (Citrullus lanatus) | 33.24 | 6.26 | 0 | 0.135 | Mitochondrion |
| P27084 | Superoxide dismutase [Mn], mitochondrial precursor (Pisum sativum) | 25.82 | 7.16 | 0 | −0.269 | Mitochondrion |
| Q6K548 | Mitochondrial outer membrane protein porin (Oryza sativa subsp. Japonica) | 29.09 | 7.21 | 18 β | −0.16 | Mitochondrion |
| O48646 | Probable phospholipid hydroperoxide glutathione peroxidase 6, mitochondrial (Arabidopsis thaliana) | 19.61 | 7.85 | 0 | −0.277 | Mitochondrion |
| A5AHP2 | Chromosome chr15 scaffold_37, whole genome shotgun sequence-Putative uncharacterized protein (Vitis vinifera) | 31.95 | 9.61 | 1 | −0.124 | Mitochondrion |
| Q2PF08 | ADP,ATP carrier protein 1, mitochondrial (Trifolium pratense) | 39.9 | 9.84 | 3 | 0.007 | Mitochondrion |
| Q9LT08 | 26S proteasome non-ATPase regulatory subunit 14 (Arabidopsis thaliana) | 34.35 | 6.31 | 0 | −0.243 | Nucleus |
| A7PF22 | Chromosome chr11 scaffold_13, whole genome shotgun sequence (Vitis vinifera) | 15.69 | 6.1 | 1 | 0.113 | Nucleus-cytosol |
| Q7XLR1 | Probable aquaporin PIP2-6 (Oryza sativa) | 29.96 | 9.08 | 5 | 0.483 | Plasma membrane |
| P04907 | Glutathione S-transferase 3 (Zea mays) | 23.72 | 6.06 | 0 | 0.066 | Plastid |
| Q08682 | 40S ribosomal protein Sa-1 (Arabidopsis thaliana) | 32.29 | 5.05 | 0 | −0.316 | Ribosome |
| P59263 | Ubiquitin (Arabidopsis thaliana) | 8.52 | 6.56 | 0 | −0.445 | Ribosome |
| B9IJE3 | Predicted protein (Populus trichocarpa) | 35.87 | 5.55 | 0 | −0.016 | Unknown |
| P49036 | Sucrose synthase 2 (Zea mays) | 92.94 | 6.03 | 0 | −0.282 | Unknown |
| P52578 | Isoflavone reductase homolog (Solanum tuberosum) | 33.85 | 6.16 | 0 | −0.063 | Unknown |
| Q6L4X6 | Os05g0508400 protein (Oryza sativa) | 66.32 | 7 | 0 | −0.448 | Unknown |
| A9PIA4 | Predicted protein-Putative uncharacterized protein (Populus trichocarpa) | 24.28 | 7.02 | 0 | −0.028 | Unknown |
| Q0DIK0 | Os05g0383000 (Oryza sativa) | 17.89 | 7.7 | 1 | −0.222 | Unknown |
| Q39613 | Peptidyl-prolyl cis–trans isomerase (Catharanthus roseus) | 18.28 | 8.36 | 0 | −0.27 | Unknown |
| Q0J8X2 | Os04g0683100 protein (Oryza sativa) | 29.1 | 9.82 | 0 | −0.392 | Unknown |
| P21616 | Pyrophosphate-energized vacuolar membrane proton pump (Phaseolus aureus) | 79.85 | 5.32 | 15 | 0.647 | Vacuole |
| P24091 | Endochitinase B (Nicotiana tabacum) | 31.47 | 8.31 | 0 | −0.322 | Vacuole |
| P07979 | Lichenase (Nicotiana plumbaginifolia) | 37.48 | 9.63 | 1 | −0.128 | Vacuole |
aSwissprot accession number, the closest protein homolog, physicochemical properties; relative molecular masses (M r, ×1,000), pI, the number of transmembrane helices (TMH, determined by Aramemnon), GRAVY scores and the location are shown
Only the most abundant proteins could be identified. Meristematic cells are fast dividing cells, associated with a high level of protein synthesis and consequently a high abundance of ribosomal proteins. Moreover, the expression of proteasome subunit genes is elevated since plant cell division is linked to a timely proteolysis of several cell cycle regulators and a rapid formation and removal of structures such as the spindle apparatus and phragmoplast (Kurepa and Smalle 2008). This implies a high abundance of proteins that are involved in the ubiquitination process and explains the identification of ubiquitin, superoxide dismutase and proteasome subunits. Due to the presence of a high number of small vacuoles in meristem cells, vacuolar proteins such as endochitinase were detected. Also cyclophilin (peptidyl-prolyl cis–trans isomerase) is a highly abundant protein in meristematic tissue (Nuc et al. 2001). Again, mainly low molecular mass proteins were identified and no correlation between the different physico-chemical parameters could be demonstrated.
Most proteins were located in mitochondria (Fig. 4c). However, the location of the identified proteins was more diverse in meristems compared with leaves (Fig. 4b). This is of no surprise since leaf cells are specialized in photosynthesis, while meristem cells are still undifferentiated.
The number of identified membrane proteins was lower compared to leaves (29%). Nevertheless, the pyrophosphate-energized vacuolar membrane proton pump, which contains 15 TMH, could be detected.
We hypothesize that also non-membrane proteins are extracted with a mixture of chloroform and methanol because proteins soluble in C/M are able to associate with lipids or contain a short hydrophobic region as mentioned by Rolland et al. (2006). It was, for example, demonstrated that the cytosolic form of nucleoside diphosphate kinase associates with membranes of a wide variety of intracellular compartments in humans (Mitchell et al. 2009). Also, GAPDH was reported to associate with subcellular membranes in a rather unspecific way (Zinser and Daum 1995). Mitochondrial and chloroplast proteins could still be associated with their transit peptide at the moment of C/M extraction. The monomeric form of malate dehydrogenase was shown to associate with phospholipid vesicles, whereas the native dimer did not (Webster et al. 1980). Also, the phospholipid hydroperoxide glutathione peroxidase is able to bind phospholipds. Glutathione S-transferase contains a hydrophobic binding site because it links glutathione to a hydrophobic substrate to detoxify endo- and xenobiotic compounds (Neuefeind et al. 1997). This hypothesis was also valid for some proteins without a predicted TMH identified in the C/M-soluble fraction of leaves. Aldolases are for example major components of chloroplast plastoglobules, which are lipid-rich structures (Ytterberg et al. 2006). Also, some proteins of the photosynthetic apparatus, like photosystem I psaH and Lhca4 protein, were detected in these globules.
Evaluation of the method
To estimate the value of chloroform/methanol extraction as a method of studying membrane proteins from total cellular lysates, a comparison was made with reports that utilized the same C/M ratio (i.e., 5/4) to study membrane proteins from purified membrane structures Table 4). These data indicate that C/M extraction is more suitable for membrane proteome studies on chloroplasts compared to other organelles. A possible explanation is the presence of highly abundant membrane proteins in chloroplasts compared to other membrane structures in which the proteome is more diverse (e.g., the plasma membrane). The higher number could also be partly explained by the less stringent criteria HMMTOPv2.0 uses for prediction of TMH. Indeed, it predicted 72% of the C/M extracted proteins from Arabidopsis leaves to be transmembrane proteins.
Table 4.
Comparison of the number of membrane proteins identified after chloroform/methanol (5/4) extraction of different samples
| Organellea | Separation method | TMH prediction | NR proteins identified | Membrane proteins | Non-membrane proteins | Percentage of membrane proteins |
|---|---|---|---|---|---|---|
| Spinach chloroplast (envelope) (Ferro et al. 2002) | 1D SDS PAGE | HMMTOP | 53 | 42 | 11 | 79 |
| Arabidopsis chloroplast (envelope) (Ferro et al. 2003) | geLC | HMMTOP | 37 | 34 | 3 | 92 |
| Arabidopsis mitochondria (Brugiere et al. 2004) | geLC | HMMTOP | 31 | 22 | 9 | 71 |
| Arabidopsis plasma membrane (Marmagne et al. 2004) | 1D SDS PAGE and geLC | Aramemnon | 59 | 32 | 27 | 54 |
| Cauliflower vacuoles (tonoplast) pH 4 (Schmidt et al. 2007) | geLC | Aramemnon | 43 | 27 | 16 | 63 |
| Cauliflower vacuoles (tonoplast) pH 6 (Schmidt et al. 2007) | geLC | Aramemnon | 30 | 19 | 11 | 63 |
| Arabidopsis total leaf | 1D and dSDS PAGE | Aramemnon | 36 | 21 | 17 | 58 |
| Banana total leaf | 1D and dSDS PAGE | Aramemnon | 20 | 14 | 6 | 70 |
| Banana total meristems | 1D SDS PAGE | Aramemnon | 35 | 10 | 25 | 29 |
aThe tissue where the extraction was performed on, the method of separating the proteins (geLC: combination of SDS PAGE and liquid chromatography), the prediction program for TMH and the number of identified proteins [total non-redundant (NR), membrane, and non-membrane proteins] are shown. The percentage of membrane proteins was calculated based on these numbers
Extracts from total cellular lysates contain less membrane proteins in comparison with extracts from purified organelles. In studies on Arabidopsis chloroplasts, 42 of 53 (Ferro et al. 2002) and 34 of 37 (Ferro et al. 2003) extracted proteins were identified as integral membrane proteins. In the total leaf extract, 21 of 36 proteins were predicted to contain at least one transmembrane helix. This lower ratio can be explained by the presence of abundant non-membrane proteins in the total extract, which are removed through purification of the organelle. The lower number of identified membrane proteins from meristem samples can be explained by its more diverse proteome with a smaller number of abundant membrane proteins.
We conclude that a 5/4 mixture of chloroform and methanol is able to extract abundant membrane proteins of up to 15 TMH. In combination with SDS PAGE and cross-species identification, it can be considered as a valuable tool to study membrane or membrane-associated proteins of a non-model plant in a reproducible way. However, also abundant non-membrane proteins, probably associated with lipids at the moment of extraction, are retrieved. Regarding its high selectivity, it is recommended to use the method in combination with other extraction methods designed for membrane proteins. Chloroform/methanol extraction can especially be useful in membrane proteome studies of fractions where one protein is highly abundant (e.g., chloroplasts). Indeed, it allows the study of proteins, which are otherwise difficult to analyze, because they are masked by the presence of Rubisco.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
The authors thank Prof. Filip Rolland (K. U. Leuven, Belgium) for the gift of the Arabidopsis plants and Dr. Isabelle Henry for critical reading. Dr. Jenny Renaut, Dr. Kjell Sergeant and Sébastien Planchon from the Centre de Recherche Public Gabriel Lipmann in Luxembourg are acknowledged for protein identifications. This work was supported by the Flanders Research Foundation (FWO Belgium). Dr.S.·C. Carpentier is supported by a postdoctoral fellowship of the same foundation.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Abbreviations
- 2DE
Classical two-dimensional electrophoresis
- C/M
Chloroform/methanol
- MS
Mass spectrometry
- S/N
Signal-to-noise ratio
- (d)SDS PAGE
(Double) sodium dodecylsulfate polyacrylamide electrophoresis
- TMH
Transmembrane helix
References
- Aert R, Sagi L, Volckaert G. Gene content and density in banana (Musa acuminata) as revealed by genomic sequencing of BAC clones. Theor Appl Genet. 2004;109:129–139. doi: 10.1007/s00122-004-1603-2. [DOI] [PubMed] [Google Scholar]
- Agrawal GK, Rakwal R. Rice proteomics: a cornerstone for cereal food crop proteomes. Mass Spectrom Rev. 2006;25:1–53. doi: 10.1002/mas.20056. [DOI] [PubMed] [Google Scholar]
- Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- Braun RJ, Kinkl N, Beer M, Ueffing M. Two-dimensional electrophoresis of membrane proteins. Anal Bioanal Chem. 2007;389:1033–1045. doi: 10.1007/s00216-007-1514-6. [DOI] [PubMed] [Google Scholar]
- Brugiere S, Kowalski S, Ferro M, Seigneurin-Berny D, Miras S, Salvi D, Ravanel S, D’herin P, Garin J, Bourguignon J, Joyard J, Rolland N. The hydrophobic proteome of mitochondrial membranes from Arabidopsis cell suspensions. Phytochemistry. 2004;65:1693–1707. doi: 10.1016/j.phytochem.2004.03.028. [DOI] [PubMed] [Google Scholar]
- Burre J, Beckhaus T, Schagger H, Corvey C, Hofmann S, Karas M, Zimmermann H, Volknandt W. Analysis of the synaptic vesicle proteome using three gel-based protein separation techniques. Proteomics. 2006;6:6250–6262. doi: 10.1002/pmic.200600357. [DOI] [PubMed] [Google Scholar]
- Carpentier SC, Witters E, Laukens K, Deckers P, Swennen R, Panis B. Preparation of protein extracts from recalcitrant plant tissues: an evaluation of different methods for two-dimensional gel electrophoresis analysis. Proteomics. 2005;5:2497–2507. doi: 10.1002/pmic.200401222. [DOI] [PubMed] [Google Scholar]
- Carpentier SC, Witters E, Laukens K, Van Onckelen H, Swennen R, Panis B. Banana (Musa spp.) as a model to study the meristem proteome: acclimation to osmotic stress. Proteomics. 2007;7:92–105. doi: 10.1002/pmic.200600533. [DOI] [PubMed] [Google Scholar]
- Carpentier SC, Coemans B, Podevin N, Laukens K, Witters E, Matsumura H, Terauchi R, Swennen R, Panis B. Functional genomics in a non-model crop: transcriptomics or proteomics? Physiol Plant. 2008;133:117–130. doi: 10.1111/j.1399-3054.2008.01069.x. [DOI] [PubMed] [Google Scholar]
- Carpentier SC, Panis B, Vertommen A, Swennen R, Sergeant K, Renaut J, Laukens K, Witters E, Samyn B, Devreese B. Proteome analysis of non-model plants: a challenging but powerful approach. Mass Spectrom Rev. 2008;27:354–377. doi: 10.1002/mas.20170. [DOI] [PubMed] [Google Scholar]
- Ephritikhine G, Ferro M, Rolland N. Review: plant membrane proteomics. Plant Physiol Biochem. 2004;42:943–962. doi: 10.1016/j.plaphy.2004.11.004. [DOI] [PubMed] [Google Scholar]
- Ferro M, Salvi D, Riviere-Rolland H, Vermat T, Seigneurin-Berny D, Grunwald D, Garin J, Joyard J, Rolland N. Integral membrane proteins of the chloroplast envelope: identification and subcellular localization of new transporters. Proc Natl Acad Sci USA. 2002;99:11487–11492. doi: 10.1073/pnas.172390399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferro M, Salvi D, Brugiere S, Miras S, Kowalski S, Louwagie M, Garin J, Joyard J, Rolland N. Proteomics of the chloroplast envelope membranes from Arabidopsis thaliana. Mol Cell Proteomics. 2003;2:325–345. doi: 10.1074/mcp.M300030-MCP200. [DOI] [PubMed] [Google Scholar]
- Folch J, Ascoli I, Lees M, Meath JA, LeBaron FN. Preparation of lipid extracts from brain tissue. J Biol Chem. 1951;191:833–841. [PubMed] [Google Scholar]
- Friso G, Giacomelli L, Ytterberg AJ, Peltier J-B, Rudella A, Sun Q, van Wijk KJ. In-depth analysis of the thylakoid membrane proteome of Arabidopsis thaliana chloroplasts: new proteins, new functions, and a plastid proteome database. Plant Cell. 2004;16:478–499. doi: 10.1105/tpc.017814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giavalisco P, Nordhoff E, Kreitler T, Kloppel KD, Lehrach H, Klose J, Gobom J. Proteome analysis of Arabidopsis thaliana by two-dimensional gel electrophoresis and matrix-assisted laser desorption/ionisation—time of flight mass spectrometry. Proteomics. 2005;5:1902–1913. doi: 10.1002/pmic.200401062. [DOI] [PubMed] [Google Scholar]
- Haynes PA, Roberts TH. Subcellular shotgun proteomics in plants: looking beyond the usual suspects. Proteomics. 2007;7:2963–2975. doi: 10.1002/pmic.200700216. [DOI] [PubMed] [Google Scholar]
- Helliot B, Swennen R, Poumay Y, Frison E, Lepoivre P, Panis B. Ultrastructural changes associated with cryopreservation of banana (Musa spp.) highly proliferating meristems. Plant Cell Rep. 2003;21:690–698. doi: 10.1007/s00299-002-0537-x. [DOI] [PubMed] [Google Scholar]
- Henriques F, Park RB. Compositional characteristics of a chloroform–methanol-soluble protein fraction from spinach chloroplast membranes. Biochim Biophys Acta. 1976;430:312–320. doi: 10.1016/0005-2728(76)90087-6. [DOI] [PubMed] [Google Scholar]
- Jiang L, He L, Fountoulakis M. Comparison of protein precipitation methods for sample preparation prior to proteomic analysis. J Chromatogr. 2004;1023:317–320. doi: 10.1016/j.chroma.2003.10.029. [DOI] [PubMed] [Google Scholar]
- Jorrin JV, Maldonado AM, Castillejo MA. Plant proteome analysis: a 2006 update. Proteomics. 2007;7:2947–2962. doi: 10.1002/pmic.200700135. [DOI] [PubMed] [Google Scholar]
- Joyard J, Grossman A, Bartlett S, Douce R, Chua N. Characterization of envelope membrane polypeptides from spinach chloroplasts. J Biol Chem. 1982;257:1095–1101. [PubMed] [Google Scholar]
- Kurepa J, Smalle JA. Structure, function and regulation of plant proteasomes. Biochimie. 2008;90:324–335. doi: 10.1016/j.biochi.2007.07.019. [DOI] [PubMed] [Google Scholar]
- Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during assembly of head of bacteriophage-t4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lescot M, Piffanelli P, Ciampi AY, Ruiz M, Blanc G, Leebens-Mack J, Da Silva FR, Santos CMR, D’hont A, Garsmeur O, Vilarinhos AD, Kanamori H, Matsumoto T, Ronning CM, Cheung F, Haas BJ, Althoff R, Arbogast T, Hine E, Pappas GJ, Sasaki T, Souza MT, Miller RNG, Glaszmann JC, Town CD (2008) Insights into the Musa genome: syntenic relationships to rice and between Musa species. BMC Genomics 9: Artn 58 [DOI] [PMC free article] [PubMed]
- Marmagne A, Rouet M-A, Ferro M, Rolland N, Alcon C, Joyard J, Garin J, Barbier-Brygoo H, Ephritikhine G. Identification of new intrinsic proteins in Arabidopsis plasma membrane proteome. Mol Cell Proteomics. 2004;3:675–691. doi: 10.1074/mcp.M400001-MCP200. [DOI] [PubMed] [Google Scholar]
- Mitchell KAP, Szabo G, Otero AD. Direct binding of cytosolic ndp kinases to membrane lipids is regulated by nucleotides. Biochim Biophys Acta Mol Cell Res. 2009;1793:469–476. doi: 10.1016/j.bbamcr.2008.12.009. [DOI] [PubMed] [Google Scholar]
- Moller S, Croning MDR, Apweiler R. Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics. 2001;17:646–653. doi: 10.1093/bioinformatics/17.7.646. [DOI] [PubMed] [Google Scholar]
- Neuefeind T, Huber R, Dasenbrock H, Prade L, Bieseler B. Crystal structure of herbicide-detoxifying maize glutathione s-transferase-i in complex with lactoylglutathione: evidence for an induced-fit mechanism. J Mol Biol. 1997;274:446–453. doi: 10.1006/jmbi.1997.1402. [DOI] [PubMed] [Google Scholar]
- Neuhoff V, Arold N, Taube D, Ehrhardt W. Improved staining of proteins in polyacrylamide gels including isoelectric-focusing gels with clear background at nanogram sensitivity using Coomassie Brilliant Blue G-250 and R-250. Electrophoresis. 1988;9:255–262. doi: 10.1002/elps.1150090603. [DOI] [PubMed] [Google Scholar]
- Nuc K, Nuc P, Slomski R. Yellow lupine cyclophilin transcripts are highly accumulated in the nodule meristem zone. Mol Plant Microbe Interact. 2001;14:1384–1394. doi: 10.1094/MPMI.2001.14.12.1384. [DOI] [PubMed] [Google Scholar]
- Perkins DN, Pappin DJC, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Rabilloud T, Chevallet M, Luche S, Lelong C. Fully denaturing two-dimensional electrophoresis of membrane proteins: a critical update. Proteomics. 2008;8:3965–3973. doi: 10.1002/pmic.200800043. [DOI] [PubMed] [Google Scholar]
- Rais I, Karas M, Schagger H. Two-dimensional electrophoresis for the isolation of integral membrane proteins and mass spectrometric identification. Proteomics. 2004;4:2567–2571. doi: 10.1002/pmic.200400829. [DOI] [PubMed] [Google Scholar]
- Rolland N, Ferro M, Ephritikhine G, Marmagne A, Ramus C, Brugiere S, Salvi D, Seigneurin-Berny D, Bourguignon J, Barbier-Brygoo H, Joyard J, Garin J. A versatile method for deciphering plant membrane proteomes. J Exp Bot. 2006;57:1579–1589. doi: 10.1093/jxb/erj162. [DOI] [PubMed] [Google Scholar]
- Santoni V, Molloy M, Rabilloud T. Membrane proteins and proteomics: un amour impossible? Electrophoresis. 2000;21:1054–1070. doi: 10.1002/(SICI)1522-2683(20000401)21:6<1054::AID-ELPS1054>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Schmidt UG, Endler A, Schelbert S, Brunner A, Schnell M, Neuhaus HE, Marty-Mazars D, Marty F, Baginsky S, Martinoia E. Novel tonoplast transporters identified using a proteomic approach with vacuoles isolated from cauliflower buds. Plant Physiol. 2007;145:216–229. doi: 10.1104/pp.107.096917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder B, Hasilik A. A protocol for combined delipidation and subfractionation of membrane proteins using organic solvents. Anal Biochem. 2006;357:144–146. doi: 10.1016/j.ab.2006.05.035. [DOI] [PubMed] [Google Scholar]
- Schroder B, Wrocklage C, Pan C, Jager R, Kosters B, Schafer H, Elsasser HP, Mann M, Hasilik A. Integral and associated lysosomal membrane proteins. Traffic. 2007;8:1676–1686. doi: 10.1111/j.1600-0854.2007.00643.x. [DOI] [PubMed] [Google Scholar]
- Schwacke R, Schneider A, Van Der Graaff E, Fischer K, Catoni E, Desimone M, Frommer WB, Flugge UI, Kunze R. Aramemnon, a novel database for Arabidopsis integral membrane proteins. Plant Physiol. 2003;131:16–26. doi: 10.1104/pp.011577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seigneurin-Berny D, Rolland N, Garin J, Joyard J. Differential extraction of hydrophobic proteins from chloroplast envelope membranes: a subcellular-specific proteomic approach to identify rare intrinsic membrane proteins. Plant J. 1999;19:217–228. doi: 10.1046/j.1365-313X.1999.00511.x. [DOI] [PubMed] [Google Scholar]
- Strosse H, Schoofs H, Panis B, Andre E, Reyniers K, Swennen R. Development of embryogenic cell suspensions from shoot meristematic tissue in bananas and plantains (Musa spp.) Plant Sci. 2006;170:104–112. doi: 10.1016/j.plantsci.2005.08.007. [DOI] [Google Scholar]
- Tusnady GE, Simon I. The Hmmtop transmembrane topology prediction server. Bioinformatics. 2001;17:849–850. doi: 10.1093/bioinformatics/17.9.849. [DOI] [PubMed] [Google Scholar]
- Webster KA, Freeman KB, Ohki S. Hydrophobic interaction between the monomer of mitochondrial malate-dehydrogenase and phospholipid-membranes. Biochem J. 1980;186:227–233. doi: 10.1042/bj1860227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins MR, Williams KL. Cross-species protein identification using amino acid composition, peptide mass fingerprinting, isoelectric point and molecular mass: a theoretical evaluation. J Theor Biol. 1997;186:7–15. doi: 10.1006/jtbi.1996.0346. [DOI] [PubMed] [Google Scholar]
- Ytterberg AJ, Peltier JB, Van Wijk KJ. Protein profiling of plastoglobules in chloroplasts and chromoplasts. A surprising site for differential accumulation of metabolic enzymes. Plant Physiol. 2006;140:984–997. doi: 10.1104/pp.105.076083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinser E, Daum G. Isolation and biochemical characterization of organelles from the yeast, Saccharomyces cerevisiae. Yeast. 1995;11:493–536. doi: 10.1002/yea.320110602. [DOI] [PubMed] [Google Scholar]
- Zuo SS, Lundahl P. A micro-Bradford membrane protein assay. Anal Biochem. 2000;284:162–164. doi: 10.1006/abio.2000.4676. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.