Abstract
Maternally expressed gene 3 (MEG3) is an imprinted gene highly expressed in the human pituitary. However, MEG3 expression is lost in human gonadotroph-derived pituitary adenomas and most human tumor cell lines. Expression of MEG3 in tumor cells results in growth suppression, p53 protein increase, and activation of p53 downstream targets. The MEG3 gene encodes a noncoding RNA of approximately 1700 nucleotides. There are 12 different MEG3 gene transcripts, generated by alternative splicing. They contain the common exons 1-3 and exons 8-10, but each uses one or more exons 4-7 in a different combination in the middle. MEG3 isoform expression patterns are tissue and cell type specific. Functionally, each isoform stimulates p53-mediated transactivation and suppresses tumor cell growth. We analyzed the secondary RNA folding structure of each MEG3 isoform, using the computer program mfold. All MEG3 RNA isoforms contain three distinct secondary folding motifs M1, M2, and M3. Deletion analysis showed that motifs M2 and M3 are important for p53 activation. Furthermore, a hybrid MEG3 RNA, containing a piece of artificially synthesized sequence different from the wild type but folding into a similar secondary structure, retained the functions of both p53 activation and growth suppression. These results support the hypothesis that a proper folding structure of the MEG3 RNA molecule is critical for its biological functions. This study establishes for the first time the structure-function relationship of a large noncoding RNA and provides a first look into the molecular mechanisms of the biological functions of a large noncoding RNA.
Multiple MEG3 non-coding RNA isoforms exist and their functions are dependent upon unique secondary folding structures.
The Maternally expressed gene 3 (MEG3) is the human homolog of mouse Gtl2, a maternally expressed imprinted gene first identified by gene trapping (1,2,3). We previously observed that MEG3 is highly expressed in the normal human pituitary, including normal gonadotroph cells; however, MEG3 expression is lost in the vast majority of human pituitary tumors derived from gonadotroph cells (4). Furthermore, expression of MEG3 in human cancer cell lines results in growth suppression, accumulation of p53 protein, and activation of p53 downstream targets (4,5). The MEG3 gene product functions as a noncoding RNA (5). Although a TATA- and CCAAT-box was found in its promoter and the RNA transcripts from this gene contain poly(A) tails, making it a target gene of RNA polymerase II, MEG3 does not encode a protein. As we have shown, although the functions of MEG3 required its transcription, a mutant MEG3 from a cDNA containing no translatable open reading frame still retained the full functions (5).
Multiple RNA isoforms transcribed from mouse Gtl2 have been reported (3). We also identified several MEG3 cDNA isoforms by screening a human fetal liver cDNA library (4,5). Because MEG3/Gtl2 is a single-copy gene, these isoforms are transcribed from the same gene but generated by alternative splicing, using different exons in the middle of the RNA. However, several questions remain unanswered, including the number of all isoforms, the expression pattern of each isoform, and whether there are functional differences between them. Importantly, previous studies with ribozymes have shown that particular folding structures within these RNA molecules are important for their self-splicing function (6,7), and distinct RNA folding structures are important for the function of steroid receptor RNA coactivator (8). However, the relationship between MEG3 RNA structure and function is unknown. Here we report the identification of 12 human MEG3 cDNA isoforms and a comparison of their expression and function. Using the RNA secondary structure prediction program mfold, we investigated the relationship between MEG3 secondary RNA folding and p53 activating function.
Materials and Methods
RT-PCR, cloning, and sequence analysis
Total RNA was extracted from a normal human pituitary (obtained from autopsy performed at the Massachusetts General Hospital), a human GH-secreting pituitary tumor (obtained from surgery performed at the Massachusetts General Hospital), human normal fibroblast WI38, and adrenal NCI-H295 cells (both obtained from American Type Culture Collection, Manassas, VA) using TRIzol reagent (Invitrogen Life Technologies, Inc., Carlsbad, CA), and reverse transcribed using the RT System from Promega Corp. (Madison, WI) according to the manufacturers’ protocols. The study was approved by the Institutional Review Board of Partners HealthCare. PCR was performed with the primers ISO forward, (5′-ATG AGA GCA ACC TCC TAG GGT TGT TGT GAG-3′) and ISO reverse, (5′-CCC GCC AGG AAG AAGN ACT TGG GTC CGG-3′) using the following conditions: 94 C for 2 min, 94 C for 30 sec, 60 C for 30 sec, and 72 C for 1 min for 40 cycles and 72 C for 10 min. In addition to the four samples mentioned above, a human fetal liver cDNA library (purchased from CLONTECH, Palo Alto, CA) was also used for PCR. The PCR products were purified by gel extraction and cloned into pCR4-TOPO vector (Invitrogen). Two hundred fifty to three hundred ten clones from each tissue or cell type were examined by DNA sequence analysis.
Cell culture, transfection, reporter assays, and 5-bromo-2′-deoxyuridine (BrdU) incorporation assays
HCT116 cells were kindly provided by Dr. Bert Vogelstein (Johns Hopkins University, Baltimore, MD) and maintained in McCoy 5A medium conditioned with 10% fetal bovine serum. Derived from human colon cancer, this cell is an ideal cell line for our study because it does not express endogenous MEG3 but expresses p53 protein and contains functional p53-related signaling pathways. Furthermore, there is a p53-null HCT116 cell line available, which allowed us to address the specificity of p53-related function as we previously described (5). Each MEG3 isoform cDNA was cloned into pCI-neo or pCMS-d2EGFP as previously described (4). For the reporter assays, cells in a 12-well plate were transfected with Mirus TransIT-LT1 reagent (Mirus Bio, Madison, WI) overnight, each well with plasmid DNAs containing 50 ng p53-Luc, 0.1 μg pCMVβ, and others as indicated. The reporter assays were performed as previously described (9). The BrdU incorporation assays were performed as previously described (5). Briefly, cells in six-well plates were transfected with pCMS-d2EGFP vector containing a MEG3 isoform cDNA, using Mirus TransIT-LT1 reagent (Mirus Bio). This expression vector expresses both green fluorescent protein (GFP) protein and a MEG3 isoform RNA. After BrdU labeling, cells were fixed and stained with anti-BrdU antibody. The labeled cells were examined under a microscope. The number (100–200) of cells with GFP expression was counted. Within these GFP-positive cells, the number of cells with active DNA synthesis, as indicated by the presence of BrdU, was also recorded. The percentage of the BrdU-positive cells within the total GFP-positive cells was calculated as a labeling index.
mfold
The potential secondary structure of each MEG3 RNA isoform was predicted using mfold at the Rensselaer Bioinformatics Web Server (http://www.bioinfo.rpi.edu/applications/mfold/).
Motif deletion and replacement
The deletion of each secondary folding motif M1, M2, and M3 was generated by standard molecular cloning techniques. First, two EcoRV restriction sites were generated in MEG3 cDNA by site-directed mutagenesis, flanking the sequence to be deleted. Next, the resultant cDNA construct was digested with EcoRV and religated, thus generating the deletion mutation, designated MEG3-dM1, MEG3-dM2, and MEG3-dM3, respectively. In MEG3-dM1, 114 nucleotides were deleted, from position 59 to 173. In MEG3-dM2, 419 nucleotides were deleted, from position 469 to 888. In MEG3-dM3, 275 nucleotides were deleted, from position 1067 to 1342. To generate the hybrid MEG3 cDNA with one branch in M2 replaced with an unrelated sequence, we first deleted nucleotide sequence from position 542 to 698 and also generated an EcoRV site to replace this sequence of 157 nucleotides. We chose this region because according to mfold, this region forms a simple stem-loop structure, which is easy to mimic by an unrelated sequence. Two nucleotide sequences were synthesized at the Massachusetts General Hospital DNA Core Facility: 5′-GGCCG ATATC CCTTC CTGCG TAATC CGCAG GTCAC GCACC TGGCG GTTCG CCTTC GGTCC GTACC GCGGA CCTTC GCGTT CCGCC GCGAC CGAG-3′ and 5′-GCGCG ATATC CGCGT AGCCC GTCGG TCACG TGGCG ACAGA GCTGT CGGTT TGGGA AAGAC CGACC TTGAC GGCTC GGTCG CGGCG GAACG CGAAG GTCCG CG-3′. The last 30 nucleotides in these two sequences are complementary to each other. The two nucleotides were mixed, heated at 90 C for 15 min and cooled down slowly at room temperature to align to each other. The aligning mix was extended by PCR at the following conditions: 72 C for 10 min, followed by 1 min at 95 C, 5 min at 60 C, 10 min at 72 C for 10 cycles and then followed by 72 C for 20 min. After PCR, the products were digested with EcoRV, gel purified, and ligated with the MEG3 cDNA in which the nucleotides 542–698 were deleted. The resultant construct was designated as MEG3-RM2B1. For BrdU incorporation assays, cDNA of MEG3-dM1, MEG3-dM2, MEG3-dM3, and MEG3-RM2B1 each was cloned into pCMS-d2EGFP vector and used in BrdU incorporation assays as described above.
Northern blots
Expression vector for each MEG3 isoform (0.5 μg) was used to transfect HCT116 cells in a 100-mm culture dish using Mirus TransIT-LT1 reagent (Mirus Bio) according to the manufacturer’s protocol. Forty-eight hours after transfection, total RNA was extracted from the cells using TRIzol reagent (Invitrogen), and Northern blots were performed as previously described (5), using 2.5 μg of total RNA for each lane.
Results
Identification of human MEG3 RNA isoforms
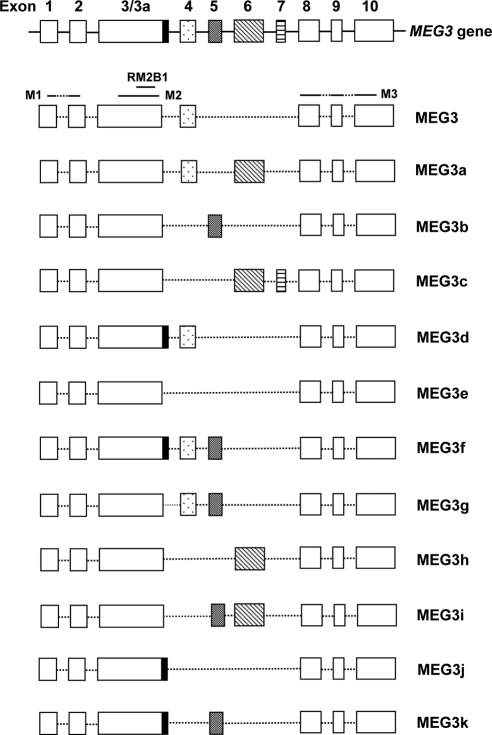
The human MEG3 gene contains 10 exons (Fig. 1). During the screening of a human fetal liver library, we found four MEG3 cDNA isoforms, designated as MEG3a, MEG3b, MEG3c, and MEG3d. Sequence comparison revealed that these four isoforms, along with the original MEG3 found in human EST library (2), all contain the common exons 1, 2, 3, 8, 9, and 10. However, each isoform contains a different combination of exons 4-7 (Fig. 1). RT-PCR was performed using a pair of primers, located within exon 3 and exon 8, respectively, to amplify MEG3 cDNA from several human cell and tissue samples. The amplified cDNA fragments were examined by sequence analysis. From more than 1300 cDNA sequences, we identified a total of 12 MEG3 cDNA isoforms (Fig. 1), named MEG3 (the first reported expressed sequence tag sequence) (2), MEG3a (4), and MEG3b through MEG3k (GenBank Accession no. MEG3b, GQ183494; MEG3c, GQ183495; MEG3d, GQ183496; MEG3e, GQ183497; MEG3f, GQ183498; MEG3g, GQ183499; MEG3h, GQ183500; MEG3i, GQ183501; MEG3j, GQ183503; and MEG3k, GQ183503). The schematic representation of each isoform is demonstrated in Fig. 1.
Figure 1.
The schematic representation of molecular structures for the human MEG3 gene and its mature isoform transcripts. Each box represents an exon. Exon 3a is an alternative exon that contains 40 extra nucleotides at the 3′-end of exon 3. The regions corresponding to folding motifs M1, M2, M3, and RM2B1 are indicated.
MEG3 RNA isoform expression pattern
The RT-PCR described above was performed using five human samples, which have been shown to express MEG3 in our previous studies (Refs. 4 and 10 and our unpublished data): a normal pituitary, a fetal liver, normal fibroblast WI38 cells, adrenal NCI-H295 cells, and a GH-secreting pituitary tumor. Thus, sequence analysis identified different MEG3 isoforms and their expression pattern in each tissue or cell type. From each sample, 250–310 MEG3 cDNA fragments were analyzed by sequencing. We found that only human fetal liver contains all 12 MEG3 isoforms. Normal pituitary tissue expresses MEG3, MEG3b, MEG3d, MEG3e, and MEG3g. Fibroblast WI38 cells express MEG3, MEG3b, MEG3d, MEG3e, and MEG3f. Adrenal NCI cells express MEG3, MEG3b, MEG3e, and MEG3j. The GH-secreting pituitary tumor expresses MEG3, MEG3a, MEG3d, MEG3e, and MEG3g. In all samples, MEG3 is the most abundant, accounting for 40–86% of all MEG3 isoforms expressed. These results are summarized in Table 1.
Table 1.
MEG3 isoform expression in different human tissue and cell types
| MEG3 (%) | MEG3a (%) | MEG3b (%) | MEG3c (%) | MEG3d (%) | MEG3e (%) | MEG3f (%) | MEG3g (%) | MEG3h (%) | MEG3i (%) | MEG3j (%) | MEG3k (%) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Normal pituitary | 79 | 0 | 12 | 0 | 1 | 5 | 0 | 3 | 0 | 0 | 0 | 0 |
| Fibroblast WI38 | 68 | 0 | 3 | 0 | 1 | 26 | 2 | 0 | 0 | 0 | 0 | 0 |
| Fetal liver | 40 | 1 | 31 | 1 | 6 | 5 | 1 | 10 | 1 | 1 | 1 | 1 |
| GH tumor | 86 | 1 | 0 | 0 | 9 | 2 | 0 | 2 | 0 | 0 | 0 | 0 |
| Adrenal NCI-H295 | 85 | 0 | 2 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 3 | 0 |
MEG3 RNA isoform function
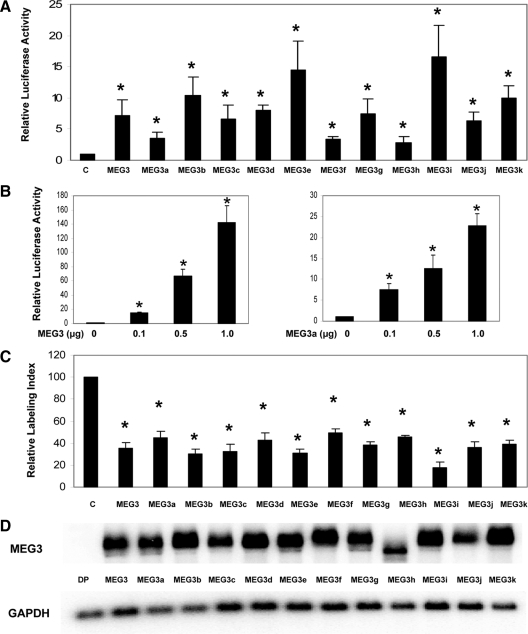
We next examined two functions of MEG3 isoforms: stimulation of p53-mediated transactivation and suppression of cell proliferation. First, each isoform was cloned into the expression vector pCI-neo and used in reporter assays to test its ability to stimulate p53-mediated transactivation. As shown in Fig. 2A, in HCT116 cells, even a small amount of each MEG3 isoform (the ratio of MEG3 isoform to reporter is 1:5) caused significant enhancement of p53-mediated reporter expression. However, activities vary among MEG3 isoforms: MEG3a, MEG3f, and MEG3h are weak activators, which stimulate p53-mediated transactivation by approximately 3.5-fold at this ratio, whereas MEG3e and MEG3i are strong activators with an activity approximately 5 times stronger than that of the weak activators such as MEG3a, MEG3f, and MEG3h, stimulating p53-mediated transactivation by more than 15-fold under the same conditions. A dose-dependent activity was observed for each MEG3 isoform in this assay. When the amount of MEG3 isoform used in reporter assays increased, each isoform was capable of stimulating p53-mediated transactivation by 20- to greater than 100-fold. Representative results using MEG3 and MEG3a are shown in Fig. 2B.
Figure 2.
A, Stimulation of p53-mediated transactivation by MEG3 isoforms in reporter assays in HCT116 cells. For each activator (each MEG3 isoform or blank pCI-neo vector as a control), 10 ng of plasmid DNA were used. *, P < 0.01 compared with the control. B, A dose-dependent stimulation of p53-mediated transactivation by MEG3 (left panel) or MEG3a (right panel). For p53-Luc reporter, 50 ng were used in all experiments. The luciferase activity observed from the cotransfection of pCI-neo was designated as 1. Values are mean ± sd from six experiments for each construct. *, P < 0.01 compared with the control. C, Suppression of DNA synthesis by MEG3 isoforms. The expression vector for each MEG3 isoform or blank vector as a control was transfected into HCT116 cells, and BrdU incorporation assays were performed. The relative labeling index from the blank vector control was designated as 100. Values are mean ± sd from four experiments for each construct. *, P < 0.01 compared with the control. D, MEG3 isoform RNA expression in transfected cells measured by Northern blot. For control, a MEG3 expression vector without a promoter (DP) is used. GAPDH, Glyceraldehyde-3-phosphate dehydrogenase.
Second, we tested the ability of each MEG3 isoform to suppress cell proliferation in HCT116 cells. Each MEG3 isoform was cloned into the vector pCMS-d2EGFP, which allows expression of both GFP protein and MEG3 RNA isoform. The resultant construct was used to transiently transfect HCT116 cells, and BrdU incorporation assays followed. As shown in Fig. 2C, each MEG3 isoform was able to suppress DNA synthesis in transfected cells by at least 50%. Cell growth suppression function among MEG3 isoforms was less variable than p53 stimulation. Therefore, each MEG3 isoform was capable of both stimulating p53-mediated transactivation and suppressing DNA synthesis in HCT116 cells.
To investigate whether the functional difference in stimulating p53-mediated transactivation observed among MEG3 isoforms is due to the differences in the expression level and/or RNA stability of each isoform, we used Northern blot to examine RNA expression level of each MEG3 isoform in transfected HCT116 cells. As shown in Fig. 2D, there is no major difference in the RNA expression levels of MEG3 isoforms, indicating that in transfected cells, the expression and stability of each MEG3 isoform were comparable with each other.
Folding structures of MEG3 RNA isoforms
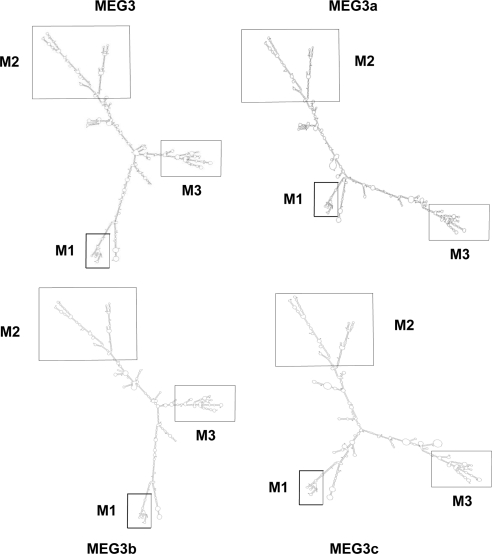
We have shown previously that MEG3 and its isoforms function as noncoding RNAs (5). Such a large RNA molecule containing approximately 1700 nucleotides must form a compact folding structure to carry out its function. As a first step in investigating the mechanisms by which MEG3 noncoding RNA stimulates p53-mediated transactivation and suppresses cell proliferation, we analyzed the potential folding structures of MEG3 RNA isoforms using the widely used computer program mfold (11,12). Using thermodynamic methods, mfold predicted the potential secondary structure of each MEG3 RNA isoform (see the predicted secondary folding structures for MEG3, MEG3a, MEG3b, and MEG3c in Fig. 3). The folding structures for the other isoforms can be found in the Supplemental Folding Structures (MEG3a–j) published on The Endocrine Society’s Journals Online web site at http://endo.endojournals.org. As predicted by mfold, each MEG3 RNA isoform contains three conserved branch motifs, designated as M1, M2, and M3 (Fig. 3).
Figure 3.
Predicted RNA secondary folding structure of MEG3, MEG3a, MEG3b, and MEG3c, determined by mfold. The structures for additional MEG3 RNA isoforms can be found in the Supplemental Folding Structures. The three conserved motifs found in all isoforms, M1, M2, and M3, are indicated.
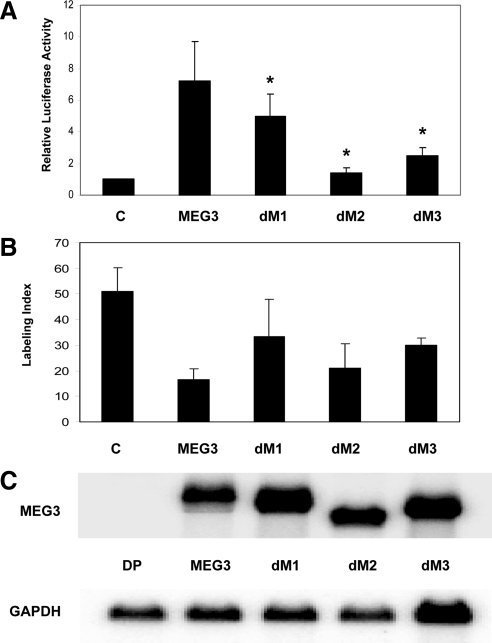
To test the functional importance of these conserved motifs, we generated MEG3 mutants in which each motif, M1, M2, or M3, was deleted, respectively. mfold predicted that these mutations did not change the overall secondary structure of the whole MEG3 RNA molecule. Rather, only local folding structure corresponding to each motif was removed (data not shown). Each of the mutants, designated as MEG3-dM1, MEG3-dM2, and MEG3-dM3, respectively, was tested for its ability to stimulate p53-mediated transactivation in reporter assays and suppress cell proliferation in BrdU incorporation assays. As shown in Fig. 4A, compared with the wild-type MEG3, deletion of M1 results in a reduction in the activity of p53 stimulation only by approximately 30% (P < 0.01), whereas deletion of M3 reduces this activity by approximately 67% (P < 0.01). In contrast, deletion of M2 almost completely abolishes this activity (P < 0.01), indicating that this structural motif is critical for MEG3 RNA to stimulate p53-mediated transactivation. However, in BrdU incorporation assays, deletion of M1 reduced growth suppression function by approximately 49% (P < 0.01) compared with the wildtype; deletion of M3 reduced growth suppression function by approximately 41% (P < 0.01); but deletion of M2 reduced growth suppression function only by approximately 14% (P = 0.11) (Fig. 4B). Northern blot showed that the RNA expression levels for these mutants in transfected cells were very similar to each other (Fig. 4C), indicating that the impaired functions in these mutants were not due to lowered expression level or decreased RNA stability.
Figure 4.
A, Stimulation of p53-mediated transactivation by MEG3 deletion mutants dM1, dM2, and dM3, in the reporter assays in HCT116 cells. For each activator (each MEG3 deletion mutant, wild-type MEG3, or blank pCI-neo vector as a control), 10 ng of plasmid DNA were used. For p53-Luc reporter, 50 ng were used. The luciferase activity observed from the cotransfection of pCI-neo was designated as 1. Values are mean ± sd from six experiments for each construct. *, P < 0.01 compared with the wild type. B, Suppression of DNA synthesis by MEG3 deletion mutants. The expression vector for MEG3, each deletion mutant or blank vector as a control was transfected into HCT116 cells, and BrdU incorporation assays were performed. Values are mean ± sd from four experiments for each construct. C, Expression of MEG3 and deletion mutants in transfected cells measured by Northern blot. For control, a MEG3 expression vector without a promoter (DP) is used. GAPDH, Glyceraldehyde-3-phosphate dehydrogenase.
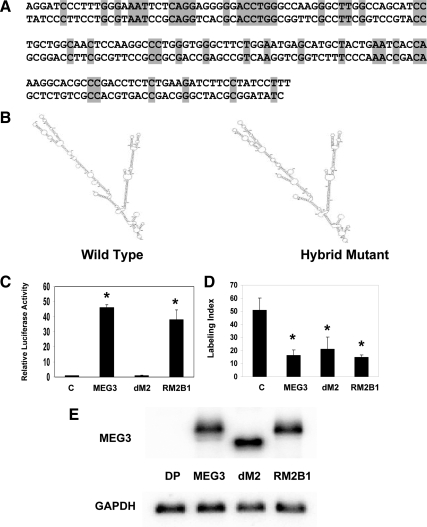
After identifying the functional importance of motif M2 for p53-mediated transactivation, we investigated whether its folding structure plays an essential role for this function. Therefore, we generated a hybrid cDNA for MEG3, named MEG3-RM2B1, in which approximately half of the M2 sequence, a total of 157 nucleotides corresponding to the MEG3 sequence from 542 to 698, was replaced by an artificially synthesized sequence bearing no resemblance to the original wild-type sequence (Fig. 5A). This region forms a simple stem-loop structure, which is easy to mimic by an unrelated sequence. Sequence alignment analysis confirmed that there is no homology between the original MEG3 sequence and this artificially synthesized sequence. However, according to mfold, this artificial hybrid MEG3-RM2B1 RNA molecule has a similar secondary folding structure to the wild-type MEG3 (Fig. 5B). When this hybrid MEG3-RM2B1 cDNA is used in functional assays, it shows strong activity to enhance transcription from a p53-dependent promoter and to suppress DNA synthesis, similar to that of wild-type MEG3 (Fig. 5, C and D; there is no statistically significant difference between the wild-type MEG3 and the hybrid MEG3-RM2B1). Its expression level in the transfected cells is similar to that of the wild-type MEG3 and the mutant MEG3-dM2 (Fig. 5E). These results clearly demonstrate that the folding structure of MEG3 RNA is more important to its biological function than its primary sequence.
Figure 5.
A, Comparison of the wild-type primary sequence in MEG3 cDNA (top) from position 542 to 698, composed of the major folding branch in M2, and the artificially synthesized primary cDNA sequence (bottom), which replaces the original sequence in MEG3-RM2B1. B, The folding structure of M2 in wild-type MEG3 RNA (left) and that of the hybrid MEG3-RM2B1 (right). C, Stimulation of p53-mediated transactivation by MEG3-RM2B1. For each activator, 250 ng of plasmid DNA were used. The luciferase activity observed from the cotransfection of pCI-neo was designated as 1. Values are mean ± sd from six experiments for each construct. *, P < 0.01 compared with the control. D, Suppression of DNA synthesis by MEG3-RM2B1. The expression vector for MEG3, MEG3-dM2, MEG3-RM2B1, or blank vector as a control was transfected into HCT116 cells, and BrdU incorporation assays were performed. Values are mean ± sd from four experiments for each construct. *, P < 0.01 compared with the control. E, Expression of MEG3, dM2, and RM2B1 in transfected cells measured by Northern blot. For control, a MEG3 expression vector without a promoter (DP) is used. GAPDH, Glyceraldehyde-3-phosphate dehydrogenase.
Discussion
In a representational difference analysis comparing the gene expression profile between normal human pituitary and gonadotroph-derived pituitary adenomas, we identified a small piece of MEG3 cDNA that is present in the normal human pituitary but absent in this pituitary tumor type (4,13). By screening a human fetal liver cDNA library, we identified four MEG3 cDNA isoforms (4,5). This finding prompted us to investigate the presence of other MEG3 cDNA isoforms and their expression patterns and investigate whether the function of MEG3 mRNA was dependent on its secondary folding structure. Sequence analysis of more than 3000 MEG3 cDNA clones from five different human tissue and cell types revealed a total of 12 MEG3 cDNA isoforms, as we report here. However, the original MEG3 first identified as an expressed sequence tag sequence of mouse Gtl2 homolog (2) is the most abundantly expressed transcript. It should be pointed out that we used only one normal human pituitary and one human GH-secreting pituitary tumor as the starting material because of the overwhelming labor intensiveness of such a study. Nevertheless, using a pool of samples from different individuals may well generate a more comprehensive isoform distribution with physiological relevance, and such a study would be important to do.
Schuster-Gossler et al. (3) reported molecular cloning of Gtl2, which is the mouse homolog of human MEG3. In their report, mouse Gtl2 contained nine exons. However, when the published Gtl2 cDNA sequence was used to align with the most updated mouse genome sequence, we found that there are 10 exons in the mouse Gtl2 gene: the alternative exon 6a and 6b reported by Schuster-Gossler et al. (3) are actually two different exons according to the most recent mouse genome sequence data. We found that human MEG3 also contains 10 exons. Therefore, the overall gene structure of mouse Gtl2 and human MEG3 are very similar. Schuster-Gossler et al. (3) also identified several alternatively spliced Gtl2 cDNA isoforms, generated by differential usage of middle exons 4, 5, 6, and 7. Again, the pattern of alternative splicing in mouse Gtl2 and human MEG3 is well conserved. However, there has been no publication to date that has thoroughly examined mouse Gtl2 isoforms and their expression patterns. In this study, we identified 12 human MEG3 isoforms by examination of more than 1300 MEG3 cDNA clones obtained from five different tissue and cell types and investigated their expression patterns.
Although numerous studies have been published focusing on the imprinting control of the Dlk1/Gtl2 (DLK1/MEG3 in human) gene cluster (14,15,16,17,18,19), the biological function of mouse Glt2 itself has not been investigated. In humans, we first reported the antiproliferative and p53-stimulating functions of MEG3 and MEG3a and provided experimental evidence to show that MEG3 functions as a noncoding RNA (4,5). In this study, we found that all MEG3 isoforms have the same ability to stimulate p53-mediated transactivation and suppress DNA synthesis in HCT116 cells. Interestingly, although suppression of DNA synthesis by each MEG3 isoform is comparable, stimulation of p53-mediated transactivation varies between different MEG3 isoforms. These data suggest that these two functions may be independent of each other. This hypothesis is confirmed by our experiments using MEG3 deletion mutants. For example, MEG3-dM1 functions almost like the wide-type in stimulation of p53-mediated transactivation, but its ability to suppress DNA synthesis is severely impaired. In contrast, in MEG3-dM2, the function to stimulate p53-mediated transactivation is completely lost, but its function in growth suppression is only slightly affected. Therefore, it is clear that different folding structures in MEG3 RNA molecules are involved in different biological functions.
We observed that all 12 MEG3 isoforms are present in human fetal liver, but only a few are present in adult tissue and cell lines, suggesting a difference in splicing mechanism between adult and fetal tissues. The difference in the abundance of different MEG3 isoforms is probably related to the dynamic feature of the coupled transcription-splicing process. As the large RNA precursor for mature MEG3 is transcribed, it could be folded into different secondary and tertiary structures, thus allowing for differential splicing. However, certain folding structures are either more stable or easier to form, resulting in the increased abundance of particular mature isoforms. We have shown that the RNA levels for MEG3 isoforms in the transfected cells were very similar to each other, indicating that there is no major difference in RNA stabilities among different MEG3 isoforms. Therefore, the abundance of a particular isoform is probably due to the particular efficiency in RNA processing.
To begin to understand the molecular mechanisms for the biological functions of MEG3 RNA, we analyzed the potential secondary folding structure of each MEG3 RNA isoform using the most widely used computer program mfold (11,12). This program predicted the presence of three folding structures, or motifs, in all MEG3 RNA isoforms. Deletion analysis showed that one of these motifs, M2, is critical for the stimulation of p53-mediated transactivation by MEG3 RNA. To confirm that this particular folding structure is important for this function, we generated a hybrid MEG3 RNA molecule in which half of the primary sequence in this M2 motif was replaced by an entirely unrelated, artificially synthesized sequence. However, according to mfold, this hybrid RNA molecule has a secondary folding structure similar to the wild-type MEG3 RNA. Indeed, this hybrid MEG3 RNA is fully functional in both stimulating p53-mediated transcription and suppressing cell proliferation. Therefore, for the first time, we have shown that RNA molecules with partially different primary sequences but similar folding structures can function similarly. For proteins, it is well known that a proper tertiary structure is critical for protein functions. Our data show that similarly, the folding structures of large noncoding RNA molecules are involved in their functions. Like proteins, we can assume that large functional noncoding RNA will further fold into tertiary structures based on their secondary structures, and their biological functions will ultimately depend on the correct tertiary structures. Further investigations, including crystallography and RNA-protein interactions, will reveal more molecular details about the functional mechanisms in this new field of the biology of noncoding RNAs.
Supplementary Material
Acknowledgments
We thank Drs. Peter Ansell and Ali Mahta for technical support.
Footnotes
This work was supported by National Institutes of Health Grant R01DK40947, The Guthart Family Foundation, and the Jarislowsky Foundation.
Present address for Y.W.: Kunming Primate Research Center and Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming, Yunnan 650223, China.
Present address for Y.Z.: Department of Anesthesiology, The First Affiliated Hospital of Kunming Medical College, 295 Xichang Road, Kunming, Yunnan 650032, China.
Disclosure Summary: The authors have nothing to disclose.
First Published Online December 23, 2009
Abbreviations: BrdU, 5-Bromo-2′-deoxyuridine; GFP, green fluorescent protein.
References
- Schuster-Gossler K, Simon-Chazottes D, Guenet JL, Zachgo J, Gossler A 1996 Gtl2lacZ, an insertional mutation on mouse chromosome 12 with parental origin-dependent phenotype. Mamm Genome 7:20–24 [DOI] [PubMed] [Google Scholar]
- Miyoshi N, Wagatsuma H, Wakana S, Shiroishi T, Nomura M, Aisaka K, Kohda T, Surani MA, Kaneko-Ishino T, Ishino F 2000 Identification of an imprinted gene, Meg3/Gtl2 and its human homologue MEG3, first mapped on mouse distal chromosome 12 and human chromosome 14q. Genes Cells 5:211–220 [DOI] [PubMed] [Google Scholar]
- Schuster-Gossler K, Bilinski P, Sado T, Ferguson-Smith A, Gossler A 1998 The mouse Gtl2 gene is differentially expressed during embryonic development, encodes multiple alternatively spliced transcripts, and may act as an RNA. Dev Dyn 212:214–228 [DOI] [PubMed] [Google Scholar]
- Zhang X, Zhou Y, Mehta KR, Danila DC, Scolavino S, Johnson SR, Klibanski A 2003 A pituitary-derived MEG3 isoform functions as a growth suppressor in tumor cells. J Clin Endocrinol Metab 88:5119–5126 [DOI] [PubMed] [Google Scholar]
- Zhou Y, Zhong Y, Wang Y, Zhang X, Batista DL, Gejman R, Ansell PJ, Zhao J, Weng C, Klibanski A 2007 Activation of p53 by MEG3 non-coding RNA. J Biol Chem 282:24731–24742 [DOI] [PubMed] [Google Scholar]
- Doudna JA, Cech TR 2002 The chemical repertoire of natural ribozymes. Nature 418:222–228 [DOI] [PubMed] [Google Scholar]
- Toor N, Keating KS, Pyle AM 2009 Structural insights into RNA splicing. Curr Opin Struct Biol 19:260–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanz RB, Razani B, Goldberg AD, O'Malley BW 2002 Distinct RNA motifs are important for coactivation of steroid hormone receptors by steroid receptor RNA activator (SRA). Proc Natl Acad Sci USA 99:16081–16086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Mehta KR, Choi AP, Scolavino S, Zhang X 2003 DNA damage-induced inhibition of securin expression is mediated by p53. J Biol Chem 278:462–470 [DOI] [PubMed] [Google Scholar]
- Gejman R, Batista DL, Zhong Y, Zhou Y, Zhang X, Swearingen B, Stratakis CA, Hedley-Whyte ET, Klibanski A 2008 Selective loss of MEG3 expression and intergenic differentially methylated region hypermethylation in the MEG3/DLK1 locus in human clinically nonfunctioning pituitary adenomas. J Clin Endocrinol Metab 93:4119–4125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews DH, Sabina J, Zuker M, Turner DH 1999 Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J Mol Biol 288:911–940 [DOI] [PubMed] [Google Scholar]
- Zuker M 2003 Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31:3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Sun H, Danila DC, Johnson SR, Zhou Y, Swearingen B, Klibanski A 2002 Loss of expression of GADD45γ, a growth inhibitory gene, in human pituitary adenomas: implications for tumorigenesis. J Clin Endocrinol Metab 87:1262–1267 [DOI] [PubMed] [Google Scholar]
- Croteau S, Charron MC, Latham KE, Naumova AK 2003 Alternative splicing and imprinting control of the Meg3/Gtl2-Dlk1 locus in mouse embryos. Mamm Genome 14:231–241 [DOI] [PubMed] [Google Scholar]
- da Rocha ST, Tevendale M, Knowles E, Takada S, Watkins M, Ferguson-Smith AC 2007 Restricted co-expression of Dlk1 and the reciprocally imprinted non-coding RNA, Gtl2: implications for cis-acting control. Dev Biol 306:810–823 [DOI] [PubMed] [Google Scholar]
- Kagami M, Sekita Y, Nishimura G, Irie M, Kato F, Okada M, Yamamori S, Kishimoto H, Nakayama M, Tanaka Y, Matsuoka K, Takahashi T, Noguchi M, Tanaka Y, Masumoto K, Utsunomiya T, Kouzan H, Komatsu Y, Ohashi H, Kurosawa K, Kosaki K, Ferguson-Smith AC, Ishino F, Ogata T 2008 Deletions and epimutations affecting the human 14q32.2 imprinted region in individuals with paternal and maternal upd(14)-like phenotypes. Nat Genet 40:237–242 [DOI] [PubMed] [Google Scholar]
- Schmidt JV, Matteson PG, Jones BK, Guan XJ, Tilghman SM 2000 The Dlk1 and Gtl2 genes are linked and reciprocally imprinted. Genes Dev 14:1997–2002 [PMC free article] [PubMed] [Google Scholar]
- Takada S, Paulsen M, Tevendale M, Tsai CE, Kelsey G, Cattanach BM, Ferguson-Smith AC 2002 Epigenetic analysis of the Dlk1-Gtl2 imprinted domain on mouse chromosome 12: implications for imprinting control from comparison with Igf2–H19. Hum Mol Genet 11:77–86 [DOI] [PubMed] [Google Scholar]
- Takada S, Tevendale M, Baker J, Georgiades P, Campbell E, Freeman T, Johnson MH, Paulsen M, Ferguson-Smith AC 2000 Δ-Like and gtl2 are reciprocally expressed, differentially methylated linked imprinted genes on mouse chromosome 12. Curr Biol 10:1135–1138 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.