Abstract
STAT3 is a latent cytoplasmic transcription factor responsive to cytokine signaling and tyrosine kinase oncoproteins by nuclear translocation when tyrosine phosphorylated. We report that malignant transformation by activated Ras is impaired without STAT3, in spite of the inability of Ras to drive STAT3 tyrosine phosphorylation or nuclear translocation. Moreover, STAT3 mutants that cannot be tyrosine phosphorylated, are retained in the cytoplasm, or cannot bind DNA nonetheless supported Ras-mediated transformation. Unexpectedly, STAT3 was detected within mitochondria, and exclusive targeting of STAT3 to mitochondria without nuclear accumulation facilitated Ras transformation. Mitochondrial STAT3 sustained altered glycolytic and oxidative phosphorylation activities characteristic of cancer cells. Thus, in addition to its nuclear transcriptional role, STAT3 regulates a metabolic function in mitochondria, supporting Ras-dependent malignant transformation.
Signal Transducers and Activators of Transcription (STATs) mediate cellular differentiation, proliferation, survival, and immune function (1). Tyrosine phosphorylated STATs undergo phosphotyrosine-SH2 domain interactions, leading to nuclear translocation and expression of target genes (2). STAT3 mediates acute phase responses to cytokines such as interleukin 6 (3–6) and has been linked to cancer development (7). Deregulated tyrosine kinase oncoproteins can target constitutive STAT3 phosphorylation (8), resulting in gene expression associated with tumor cell survival and proliferation. Augmented STAT3 activity is associated with multiple human tumors and STAT3 inhibition can mediate tumor regression (9). However, additional STAT3 functions inconsistent with functioning solely as a transcription factor have been described (10).
Ablation of STAT3 impairs malignant transformation by protein tyrosine kinase oncoproteins, such as anaplastic lymphoma kinase (ALK) and v-Src (11, 12). To determine whether oncogenes that lack tyrosine kinase activity also require STAT3, we examined cellular transformation by activated Ras (H-RasV12, where V12 indicates valine-12). Although previous evidence suggested Ras transformation was independent of STAT3 (7, 8), we found the ability of H-Ras to cause cell growth in soft agar, like that of v-src, was impaired without STAT3 (Fig. 1A). Similarly, growth of Ras-transformed tumors in mice was abrogated without STAT3 (Fig. 1B). Reconstitution of STAT3-null cells with mutated STAT3 lacking its tyrosine phosphorylation site (Y705F) restored growth of H-RasV12 expressing tumors (Fig. 1B). Lack of a STAT3 tyrosine phosphorylation requirement for tumor formation by H-RasV12 is distinct from the phosphorylation requirement for v-Src transformation (11) but is consistent with absence of detectable tyrosine-phosphorylated STAT3 in Ras-transformed cells (fig. S1A).
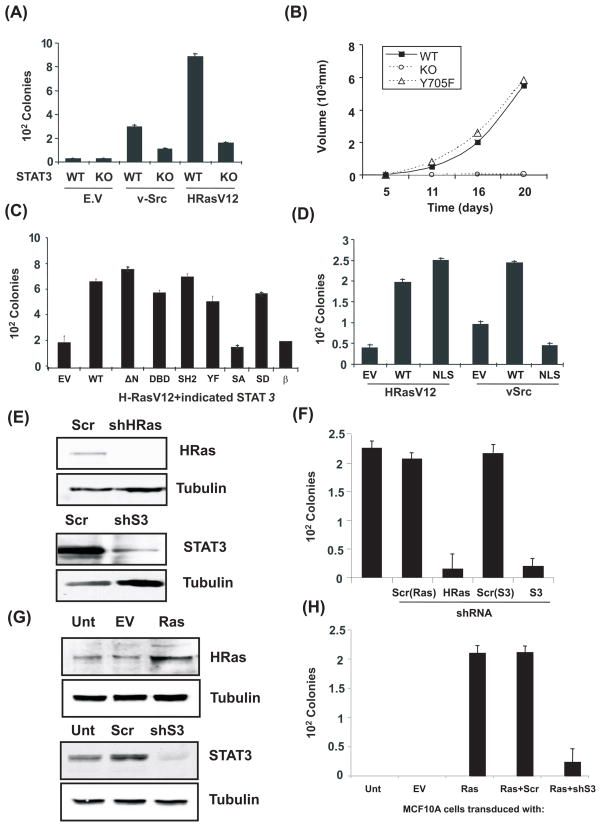
Figure 1. STAT3 is essential for Ras transformation but not as a transcription factor.
(A) Colony formation of wild type (WT) or STAT3-deficient (KO) cells with or without v-Src or H-RasV12 plated in soft agar. (B) Average tumor volume formed by H-RasV12-expressing STAT3-null cells transduced with empty vector (KO), wild type (WT) or Y705F-mutant STAT3 (Y705F) and injected into Balb/cnu/nu mice (5 mice per group). (C) Average colony formation of H-RasV12-expressing STAT3-deficient cells stably transduced with empty vector (EV), wild type STAT3 (WT), or STAT3 mutants: N-terminal 132 amino acid deletion (ΔN), DNA binding domain VVV461-463AAA (DBD), SH2 domain R609K (SH2), tyrosine phosphorylation site Y705F (YF), serine phosphorylation site S727A and S727D (SA and SD), and the STAT3β isoform (β). (D) Average colony formation of H-RasV12 or v-Src-expressing cells transduced with empty vector (EV), wild type STAT3 (WT), or STAT3 with a mutated nuclear localization signal (NLS). (E) Depletion of H-Ras or STAT3 protein by shRNA in T24 human bladder carcinoma cells compared to scrambled shRNA control (Scr). (F) Average colony formation of T24 cells depleted for STAT3 or H-Ras. (G) Expression of H-Ras and STAT3 in human breast epithelial MCF10A cells transduced with empty vector (EV) or H-RasV12 (Ras) and transfected with shRNA to STAT3 (shS3) or scrambled control (Scr). (H) Average colony formation of MCF10A cells with and without H-RasV12 and STAT3. Error bars indicate SD.
To further evaluate STAT3 structural requirements for Ras transformation, we reconstituted STAT3-null cells with STAT3 mutants (Fig. 1C). Versions of STAT3 inert for cytokine-activated transcription or v-Src transformation supported Ras transformation. STAT3 N-terminal, DNA-binding, SH2 domains, and tyrosine phosphorylation site were not required. However, mutation of the serine phosphorylation site in the C-terminus (S727A) or deletion of the C-terminal domain (STAT3β) abrogated cooperation between STAT3 and Ras. Replacement of S727 with phosphorylation-mimetic aspartate (S727D) restored transformation (Fig. 1C), suggesting S727 phosphorylation is required.
The C-terminus of STAT proteins and phosphorylation of S727 are implicated in transcriptional activity (13), but lack of a requirement for STAT3 DNA binding or SH2 domains implied that STAT3-mediated transcription was not necessary for Ras transformation. We tested whether STAT3 nuclear translocation is required to augment Ras transformation. We mutated the two nuclear localization sequences (NLS) in STAT3 (14), causing cytoplasmic retention (fig. S2). Cytoplasmic-restricted STAT3 restored transformation by Ras but not v-Src (Fig. 1D). These data are consistent with dual functions of STAT3 in transformation. The tyrosine kinase oncoprotein v-Src required tyrosine phosphorylated, transcriptionally-competent nuclear STAT3 for transformation, but activated Ras required non-transcriptional, non-nuclear STAT3.
We tested whether STAT3 is required for transformation of human cells. We ablated STAT3 expression by shRNA knockdown in the T24 cell line derived from a spontaneous H-Ras-transformed human bladder cell carcinoma (15). Ablation of STAT3 expression impaired T24 colony formation to the same extent as shRNA-ablation of Ras expression (Fig. 1E–F). Similarly, transformation of normal human mammary epithelial cell line MCF10A by H-RasV12 depended on expression of STAT3 (Fig. 1G–H). Notably, neither T24 nor MCF10A cells required STAT3 for proliferation or survival during adherent growth.
Activated Ras signals primarily through stimulation of MAPK, PI3K, and RalGDS pathways (16). We interrogated Ras signaling biochemically and by using Ras-effector domain mutants (fig. S1). Ras-mediated activation of RalA was the only effector function showing dependence on STAT3, but activation of this pathway did not explain STAT3-dependence of Ras-mediated transformation (fig. S3). Since STAT3 has been documented in discrete cytoplasmic bodies (17), we fractionated cytosol into plasma membrane, organelle-free cytoplasmic, and organelle fractions, and probed for STAT3. STAT3 was detected in all three fractions, with greatest amounts in cytosol (S100) but distinct presence in the organelle fraction, which was mostly mitochondria (Fig. 2A). Mitochondrial STAT3 was also detected in primary liver tissue, non-transformed MCF10A mammary epithelial cells, and T24 bladder carcinoma cells (fig. S4). We fractionated STAT3 knockout cells reconstituted with mutant forms of STAT3. Presence of STAT3 in mitochondria did not require tyrosine phosphorylation or intact SH2 or DNA binding domains, mirroring the lack of requirement of these domains for Ras-transformation (Fig. 2B).
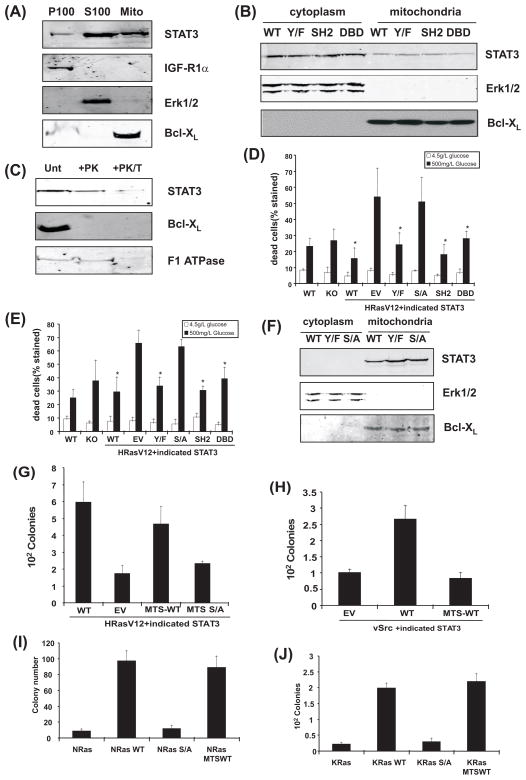
Figure 2. Localization of STAT3 to mitochondria supports Ras transformation.
(A) Fractionation of cells mechanically disrupted in absence of detergent and separated into P100 (plasma membrane), S100 (organelle-free cytosol), and mitochondrial fractions. Samples probed for STAT3, IGF1-R1α, or Bcl-XL, as indicated. (B) Distribution of STAT3 mutants expressed in H-RasV12-transduced cells. Erk1/2 and Bcl-XL expression verified fraction purity. (C) Protease-sensitivity of proteins associated with mitochondrial membranes in absence (unt) or presence of proteinase K (+PK) or proteinase K and Triton X-100 (+PK/T). (D) Survival of H-RasV12 expressing cells following glucose depletion. Survival of STAT3-null (KO) or wild type cells expressing H-RasV12 (WT) or STAT3-null cells expressing H-RasV12 reconstituted with empty vector (EV) or STAT3 mutants after growth in low or high glucose medium, as indicated. Asterisks indicate statistically significant differences (p<.05 by student’s t-test) of individual conditions relative to empty vector. (E) Survival of cells exposed to 2% oxygen. (F) Mitochondrially-targeted wild type and mutant STAT3 protein expression. (G) Colony formation by H-RasV12-, (H) v-Src-, and (I, J) N- and K-Ras-expressing cell lines.
To further assess STAT3 mitochondrial localization, we tested its protease sensitivity with or without detergent. Proteins associated with outer mitochondrial membranes are expected to be protease-sensitive, whereas internal proteins are degraded only following membrane disruption with detergent (18). Incubation of purified mitochondria with proteinase K readily removed Bcl-X expressed on the outer mitochondrial membrane, but STAT3 and F1 ATPase, an internal mitochondrial protein, were retained unless mitochondria were treated with both protease and detergent (Fig. 2C). Therefore, like F1 ATPase, STAT3 appears to be an internal mitochondrial protein.
Given the presence of STAT3 in mitochondria and the dependence of tumor cells for glucose due to high glycolytic activity (19), we probed for metabolic defects in RasV12-expressing cells engineered to express STAT3 mutants. Activated Ras increased dependence on high glucose medium in absence of STAT3 (Fig. 2D). Reconstitution of cells with STAT3 suppressed this sensitivity, and the structure-function relationship between STAT3 and cell survival mirrored that for transformation. That is, tyrosine phosphorylation and intact SH2 and DNA binding domains were not required for protection from cell death but S727 was essential (Fig. 2D). Similarly, RasV12-expressing cells lacking STAT3 were more sensitive to glucose restriction when cultured in reduced oxygen concentrations, requiring S727 for protection but not other STAT3 domains implicated in cytokine signaling (Fig. 2E). In contrast, cells with or without functional STAT3 were equally viable when cultured in presence of high glucose, demonstrating that mitochondrial STAT3 is not essential for normal cell viability.
To determine if mitochondrial STAT3 was sufficient to support Ras-transformation, we reconstituted STAT3-null cells with STAT3 artificially targeted exclusively to mitochondria. We transfected Ras-expressing STAT3-null cells with wild type or mutant versions of STAT3 fused with a mitochondrial targeting sequence (MTS) derived from cytochrome c oxidase subunit VIII, an integral mitochondrial protein (20). Cytoplasmic and mitochondrial extracts from stably transfected cells confirmed exclusive presence of STAT3 in mitochondria, with approximately equal abundance of wild type and tyrosine- or serine-phosphorylation-deficient mutants (Fig. 2F). STAT3 expressed exclusively in mitochondria supported anchorage-independent growth, but S727 was required (Fig. 2G). In contrast, mitochondrially-restricted STAT3 did not support v-Src-driven anchorage-independent growth (Fig. 2H), consistent with our previous finding that v-src requires nuclear functions of STAT3 (11). Cellular transformation by activated N- or K-Ras also required STAT3, and impaired transformation without STAT3 was reversed by STAT3 artificially restricted to mitochondria, again dependent on S727 (Fig. 2I–J and S5). Mitochondrially-restricted STAT3 supported growth of Ras-transformed cells as solid tumors in vivo, which could not grow without STAT3 (fig. S6). Thus, all three major human Ras oncoproteins depended on mitochondrial STAT3 for full transforming potential.
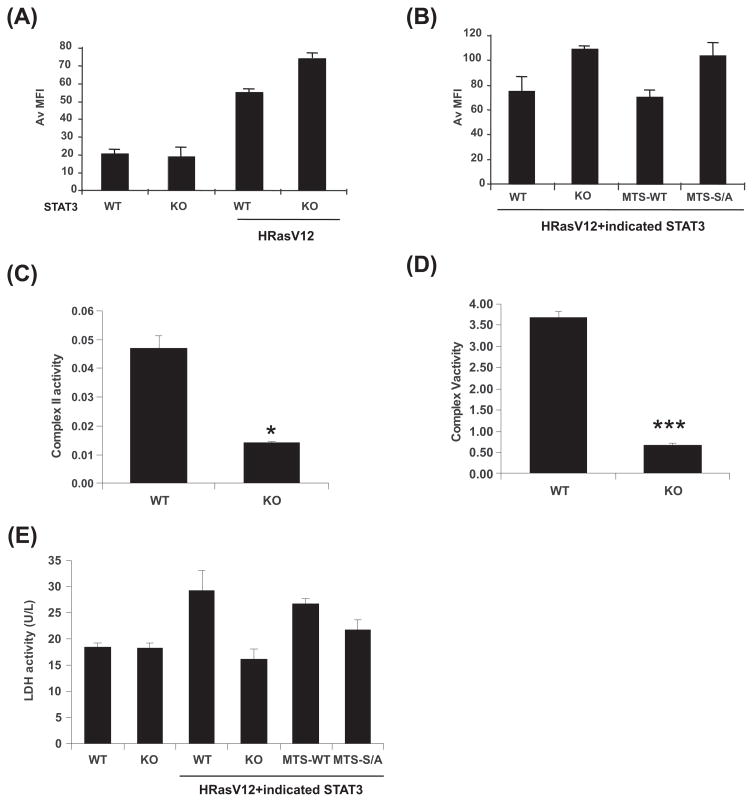
We detected no STAT3 requirement for mitochondrial formation, maintenance, or gene expression (fig. S6A–C). However, loss of STAT3 led to a 50% reduction in cellular ATP levels (fig. S6D) and altered sensitivity to mitochondrial inhibitors (fig. S6E–G). Therefore, we examined mitochondrial function in the presence and absence of STAT3. Transformed cells exhibited increased mitochondrial membrane potential (Fig. 3A), considered a characteristic of cancer cells (21). Although normal cells exhibited no dependence on STAT3 for mitochondrial membrane potential, absence of STAT3 reproducibly accentuated the increased polarization of H-RasV12-expressing cells (Fig. 3A) and this accentuated polarization was reverted by mitochondrially-restricted STAT3 expression (Fig. 3B). Like transformation, this mitochondrial STAT3 function required S727. Since mitochondrial membrane potential is maintained by oxidative phosphorylation, we measured activities of each enzyme complex of the electron transport chain (Fig. 3C–D and fig. S8). Mitochondrial function was impaired without STAT3, with significantly reduced activity of succinate oxidoreductase (complex II) and ATP synthase (complex V). Additionally, lactate dehydrogenase activity was increased in transformed cells, and this increased activity required mitochondrial STAT3, largely dependent on S727 (Fig. 3E).
Figure 3. Mitochondrial STAT3 augments electron transport chain activity.
(A) Mitochondrial membrane potential measured with TMRE and recorded as mean fluorescence intensity (MFI) by flow cytometry in wild type (WT) or STAT3-deficient (KO) cells with or without H-RasV12. (B) Mitochondrial membrane potential of H-RasV12-expressing cells with wild type (WT), vector (KO), mitochondrially-targeted wild type (MTS-WT) or S727A-mutant STAT3. Activities of electron transport chain complexes II (C) and V (D) compared between H-RasV12-expressing wild type (WT) or STAT3-deficient (KO) cells. Unit activity of individual complexes normalized to citrate synthase activity from equivalent numbers of mitochondria. Statistical significance with p value <0.05* or <0.001*** by student’s t-test. (E) Lactate dehydrogenase activity in wild type (WT), STAT3-deficient (KO), and H-RasV12-expressing wild type (WT), STAT3-deficient (KO), mitochondrially-targeted wild type (MTS-WT), or mutant STAT3 (MTS-S/A) expressing cells. Results are means of 3 replicates. Error bars indicate SD.
These data demonstrate a transformation-specific function for mitochondrial STAT3, in addition to its previously characterized nuclear roles. Although previous data implicated a Ras-STAT3 axis in transformation, those cases were in the context of activated tyrosine kinases, such as NPM-ALK (12), RET (22), or autocrine cytokine signaling (23), requiring STAT3 function in the nucleus. Mitochondrial STAT3 appears to contribute to Ras-dependent cellular transformation by augmenting electron transport chain activity, particularly that of complexes II and V, accompanied somewhat paradoxically by shifted energy production to favor fermentation. Impaired complex V activity may contribute to a build up of protons, causing enhanced mitochondrial membrane polarization without STAT3. STAT3 loss increased sensitivity to the small molecule inhibitors of oxidative phosphorylation, rotenone and antimycin A (fig. S4C, D), but protected cells from the proton channel inhibitor oligomycin (fig. S4E). The reduced sensitivity of STAT3-null cells to oligomycin underscores that STAT3 is not simply a general survival factor for transformed cells but rather has specific effects on mitochondrial function. In spite of transformation-specific functions of mitochondrial STAT3 documented here, STAT3 also accumulated in mitochondria of non-transformed cells and primary tissues (fig. S4) and recently was shown to modulate respiration in mouse heart tissue (24). Therefore, the metabolic shift important for tumor growth mediated by mitochondrial STAT3 may reflect exploitation of a normal function. If so, mitochondrial STAT3 function could provide an attractive target for therapeutic approaches to cancer.
Supplementary Material
References and Notes
- 1.Levy DE, Lee CK. J Clin Invest. 2002;109:1143. doi: 10.1172/JCI15650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levy DE, Darnell JE., Jr Nat Rev Mol Cell Biol. 2002;3:651. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- 3.Zhong Z, Wen Z, Darnell JE. Science. 1994;264:95. doi: 10.1126/science.8140422. [DOI] [PubMed] [Google Scholar]
- 4.Raz R, Durbin JE, Levy DE. J Biol Chem. 1994;269:24391. [PubMed] [Google Scholar]
- 5.Lütticken C, et al. Science. 1994;263:89. doi: 10.1126/science.8272872. [DOI] [PubMed] [Google Scholar]
- 6.Akira S, et al. Cell. 1994;77:63. doi: 10.1016/0092-8674(94)90235-6. [DOI] [PubMed] [Google Scholar]
- 7.Bromberg JF, et al. Cell. 1999;98:295. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- 8.Yu CL, et al. Science. 1995;269:81. doi: 10.1126/science.7541555. [DOI] [PubMed] [Google Scholar]
- 9.Inghirami G, et al. Cell Cycle. 2005;4:1131. doi: 10.4161/cc.4.9.1985. [DOI] [PubMed] [Google Scholar]
- 10.Gao SP, Bromberg JF. Sci STKE. 2006;2006:pe30. doi: 10.1126/stke.3432006pe30. [DOI] [PubMed] [Google Scholar]
- 11.Schlessinger K, Levy DE. Cancer Res. 2005;65:5828. doi: 10.1158/0008-5472.CAN-05-0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiarle R, et al. Nat Med. 2005;11:623. doi: 10.1038/nm1249. [DOI] [PubMed] [Google Scholar]
- 13.Schindler C, Levy DE, Decker T. J Biol Chem. 2007;282:20059. doi: 10.1074/jbc.R700016200. [DOI] [PubMed] [Google Scholar]
- 14.Reich NC, Liu L. Nat Rev Immunol. 2006;6:602. doi: 10.1038/nri1885. [DOI] [PubMed] [Google Scholar]
- 15.Taparowsky E, et al. Nature. 1982;300:762. doi: 10.1038/300762a0. [DOI] [PubMed] [Google Scholar]
- 16.Mitin N, Rossman KL, Der CJ. Curr Biol. 2005;15:R563. doi: 10.1016/j.cub.2005.07.010. [DOI] [PubMed] [Google Scholar]
- 17.Sehgal PB. Semin Cell Dev Biol. 2008;19:329. doi: 10.1016/j.semcdb.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lorenz H, Hailey DW, Lippincott-Schwartz J. Nat Methods. 2006;3:205. doi: 10.1038/nmeth857. [DOI] [PubMed] [Google Scholar]
- 19.Warburg O. Science. 1956;123:309. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 20.Rizzuto R, et al. J Biol Chem. 1989;264:10595. [PubMed] [Google Scholar]
- 21.Fantin VR, St-Pierre J, Leder P. Cancer Cell. 2006;9:425. doi: 10.1016/j.ccr.2006.04.023. [DOI] [PubMed] [Google Scholar]
- 22.Plaza-Menacho I, et al. J Biol Chem. 2007;282:6415. doi: 10.1074/jbc.M608952200. [DOI] [PubMed] [Google Scholar]
- 23.Gao SP, et al. J Clin Invest. 2007;117:3846. doi: 10.1172/JCI31871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wegrzyn J, et al. Science. 2009;323:793. doi: 10.1126/science.1164551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.We thank A. Pellicer, L. Feig, C. Der, R. Jove, S. Watowich and M. Philips for gifts of reagents and helpful discussions. This work was supported by NIH grant R01AI28900 and NYSTEM Foundation.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.