Abstract
Dyslexia is a hereditary neurological disorder that manifests as an unexpected difficulty in learning to read despite adequate intelligence, education, and normal senses. The prevalence of dyslexia ranges from 3 to 15% of the school aged children. Many genetic studies indicated that loci on 6p21.3, 15q15-21, and 18p11.2 have been identified as promising candidate gene regions for dyslexia. Recently, it has been suggested that allelic variants of gene, DYX1C1 influence dyslexia. In the present study, exon 2 and 10 of DYX1C1 has been analyzed to verify whether these single nucleotide polymorphisms (SNPs) influence dyslexia, in our population. Our study identified 4 SNPs however, none of these SNPS were found to be significantly associated with dyslexia suggesting DYX1C1 allelic variants are not associated with dyslexia.
Keywords: Candidate gene, chromosome, dyslexia, DYX1C1
Introduction
Developmental dyslexia is a hereditary neurological disorder that manifests as a persistent difficulty in learning to read and spell in children with otherwise normal intellectual functioning and educational opportunities.[1] Prevalence of dyslexia in school children has been found to be 3-17.5%.[2] The tendency of dyslexia to run in families has become clear since its earliest descriptions and modern family and twin studies indicate that heritability is 50-60%.[3] Molecular linkage studies have indicated different chromosomal areas harboring dyslexia candidate genes on chromosomes 1, 2, 3, 6, 15, and 18.[4–9] However, characterization of candidate genes for dyslexia is still in the infancy. One of the possible candidate genes, which influence dyslexia, is DYX1C1 which is near DYX1 locus on chromosome 15q21. Sequence analysis of DYX1C1 shows eight single nucleotide polymorphisms (SNPs), of them two SNPs, -3G>A and 1249G>T are functionally important and influences dyslexia.[10]
Because SNPs are inherited and do not change much from generation to generation, analysis of SNPs is essential for finding genes that predispose people to more common conditions in which inheritance patterns are complex and also it will have a wide range of applications for developing diagnostic, therapeutic, and preventative strategies. Since dyslexia is a major educational problem, there is a need for detailed genetic analysis to find out the genes which are responsible for dyslexia which in turn will provide simple diagnostic tools to ease the clinicians for early evaluation of the disorder and treatment. In the present investigation, an attempt has been made to verify whether allelic variants of DYX1C1 are responsible for dyslexia in our population.
Materials and Methods
Dyslexic children were ascertained through special schools for learning disabled as well as from regular schools of Karnataka state. Following tests were used for the diagnosis: a) Teacher rating: Rutter's Proforma A and B[11] was used to get the teachers rating on children's academic performance as well as the presence of behavioral and emotional problems to eliminate those with severe behavioral/ emotional problems if they are primary causes of poor academic achievement. b) Raven's (Colored) Progressive Matrices (RCPM/RPM) was used to ascertain that those children with poor reading/writing are not below normal in their intellectual/reasoning function.[12] c) Graded reading and spelling tests were administered to ascertain that they were behind at least by two grade norms in reading as required by the operational definition of dyslexia. In addition to the above-mentioned criteria, school examination marks and clinical certificates issued from institutes such as National Institute of Mental Health and Neurosciences, Bangalore and All India Institute of Speech and Hearing, Mysore were used as supportive evidences. The age range of the subjects was 8-17 years. Individuals who had no history of reading, spelling, or other academic difficulties were selected as control subjects.
Genomic DNA was isolated from peripheral blood of 51 control subjects and 52 dyslexic subjects by phenol-chloroform DNA isolation method and subjected for screening of SNPs and mutations in the exon 2 as well as 10 of DYX1C1 gene. Using exon 2 flanking intronic primers, a total of 477bp were amplified in all the subjects and also using exon 10 flanking intronic primers, a total of 698bp were amplified. The amplified products were subjected for Sanger's DNA sequencing.
Results
At-164 position of exon 2 of DYX1C1, C to T transversion was observed in one dyslexic proband and none of the controls showed this polymorphism at this site [Figure 1]. At –3 position of exon 2, G to A polymorphism was found in 4 controls and 7 dyslexics [Figure 2]. At 1249 position of 10th exon G to T SNP was found in three dyslexic cases and none of the control sample showed this polymorphism [Figure 3]. At 1259 position of 10th exon, C to G polymorphism was found in 4 controls and 8 dyslexic cases [Figure 4]. All the polymorphisms were in both exons 2 and 10 were heterozygous except the SNP at 1259 position of exon 10. Allele frequencies of these SNPs in dyslexics and control subjects are presented in Table 1. However, chi squire test shows no significant P values for these SNPs [Table 1]. Comparison of DYX1C1 allele frequencies observed in UK, Finnish, and present study is given in Table 2.
Figure 1.
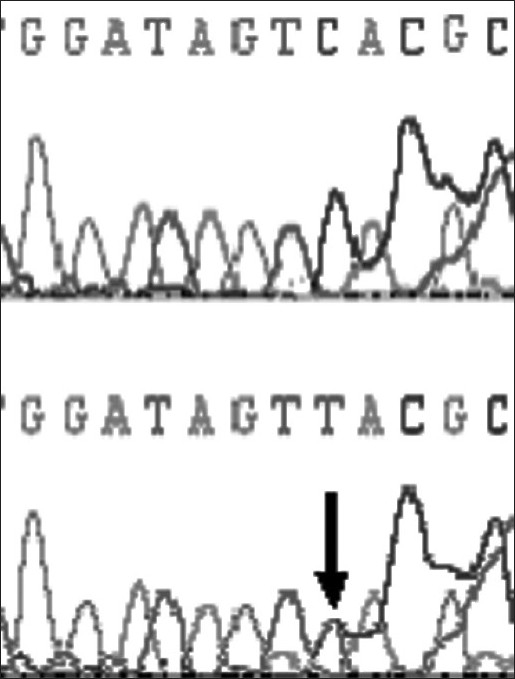
Chromatogram of control sample showing normal sequence and dyslexic sample showing heterozygote (C-T) at -164 position of exon 2 of DYX1C1 gene (arrow indicates the site)
Figure 2.
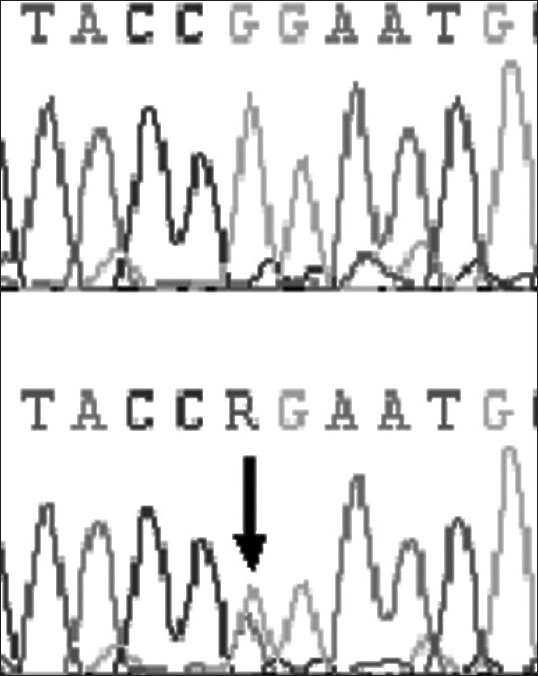
Chromatogram of dyslexic sample showing normal and another sample showing heterozygote (G-A) at -3 position of exon 2 of DYX1C1 gene (arrow indicates the site). R indicatesG/A
Figure 3.
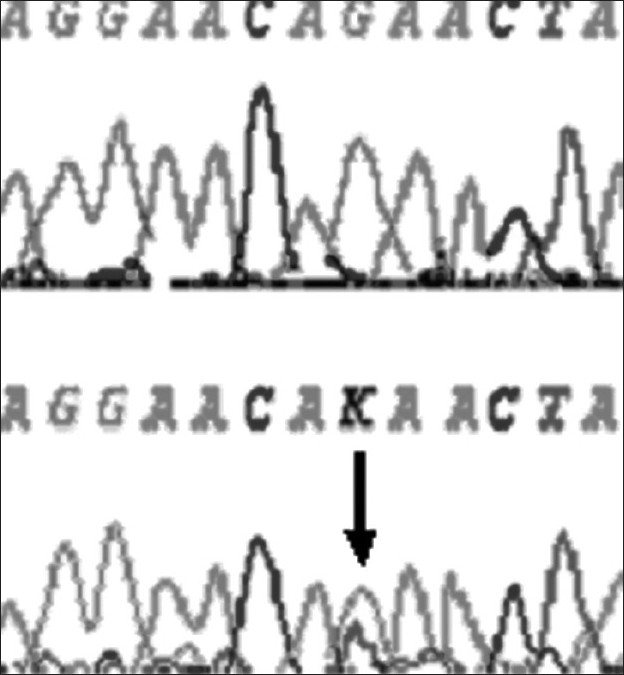
Chromatogram of dyslexic sample showing normal and another sample showing heterozygote (G-T) at 1249 position of exon 10 of DYX1C1 gene (arrow indicates the site). K indicates G/T
Figure 4.
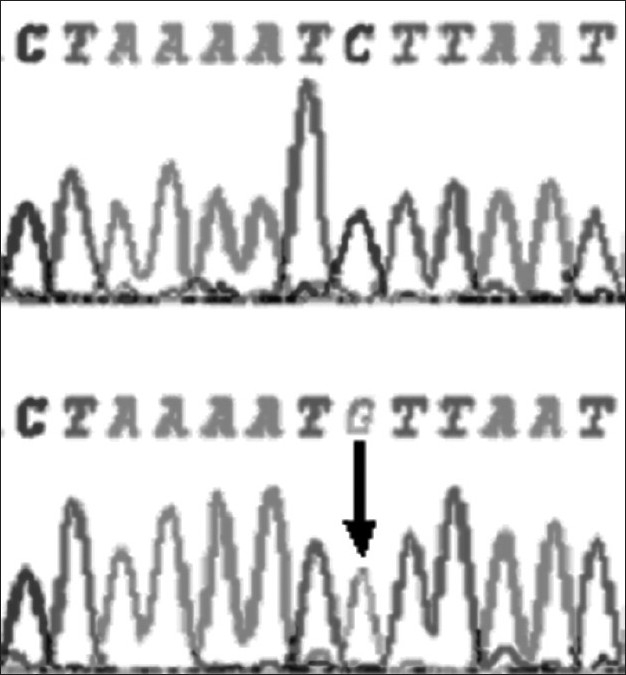
Chromatogram of dyslexic sample showing normal and another sample showing C-G SNP (homozygote) at 1259 position of exon 10 of DYX1C1 gene (arrow indicates the site)
Table 1.
Single nucleotide polymorphisms observed in 2nd and 10th exon in 52 dyslexic cases and 51 controls subjects
| Polymorphism | Exon | % of allele frequency | Chi-square value | P value | |
|---|---|---|---|---|---|
| Dyslexics | Controls | ||||
| -164 C>T | 2 | 0.96 | 0 | 0.986 | 0.421 |
| -3 G>A | 2 | 6.73 | 3.92 | 0.804 | 0.370 |
| 1249 G>T | 10 | 2.88 | 0 | 2.986 | 0.084 |
| 1259 C>G | 10 | 7.84 | 3.9 | 1.335 | 0.248 |
Table 2.
Comparison of DYX1C1 allele frequencies observed in UK, Finland, and present study
| Sequence | Coding | UK allele (%) | Finnish | Present |
|---|---|---|---|---|
| variant | change | allele (%) | study (%) | |
| −164C-T | (5′-UTR) | T (1.56) | T (1.0–6.4) | T (0.96) |
| −3G-A | (5′-UTR) | A (6.37) | A (2.5–8.3) | A (6.73) |
| 1249G-T | Glu417X | T (9.63) | T (5–13.2) | T (2.88) |
| 1259C-G | Ser420Cys | G (9.80) | G (2–10) | G (7.84) |
Discussion
Many genetic studies on dyslexia have identified specific chromosomal loci for different dyslexia related phenotypes which suggest many genes are contributing to the predisposition of dyslexia.[13] One of the possible candidate genes which influence dyslexia is DYX1C1. It consists of 10 exons and codes for 420 amino acid protein which is expressed in brain, lung, kidney, and testis. In the brain, it is expressed in white matter glial cells and cortical neuronal cells. Eight SNPs are located in DYX1C1, of them two SNPs -3G>A and 1249C>T was reported to be associated with dyslexia. -3G>A is located in the binding sequence of the transcription factors and it was reported that transcriptional activator, Elk-1 has been associated with learning in rats. SNP 1249C>T brings a functional effect by truncating the protein. Thus, it has been suggested that both SNPs are functionally important and influences dyslexia.[10] To verify whether these SNPs are unique across the language, in the present study, exon 2 and exon 10 of DYX1C1 were amplified and sequenced for SNPs. Our study identified 4 SNPs however, none of these SNPS were found to be significantly associated with dyslexia. Marino et al.[14] suggested unitary hypothesis of biological basis of dyslexia. If so, the genes responsible for dyslexia should be universal however, in our study it was found that SNPs, -3G-A and 1249C-T are not functionally important to manifest dyslexia. Reports from Italian and UK population also suggest DYX1C1 allelic variants are not associated with dyslexia.[15–16] Cellular function of DYX1C1 is not known so far hence, DYX1C1 cannot be considered as the candidate gene for dyslexia. Since dyslexia is a complex cognitive disability that affects different aspects of reading related skills which are coordinated by visual, motor, cognitive, and language areas of the brain, it is obvious that dyslexia results from many genetic variants. Most recently a new gene, ROBO1 is reported near the DYX5 locus on chromosome 3p which was disrupted due to a translocation t(3;8)(p12;q11), in a dyslexic patient. ROBO1 is a neuronal axon guidance receptor gene involved in brain development and thus an attractive candidate gene for dyslexia. Two functional copies of ROBO1 is required in brain development to acquire normal reading development and dyslexia may be caused by partial haplo-insufficiency for ROBO1.[17] Another candidate gene for dyslexia is DCDC2 which is located in the DYX2 locus and DCDC2 localizes to the regions of the brain where fluent reading occurs.[18] Though reports of candidate genes of dyslexia are accumulating none of the studies are replicated so far.
Dyslexia is a complex cognitive disability that affects different aspects of reading related skills which are coordinated by visual, motor, cognitive, and language areas of the brain. Thus, dyslexia can result from deviation of normal anatomy and function of those areas in the brain.[9] Studies have identified loci on 6p21.3, 15q15-21, and 18p11.2 as promising candidate gene regions.[8,19–20] Identification of specific risk genes on these regions would help in early diagnosis and once genes have been identified, the study of their gene products and areas of the brain in which they are expressed can shed light on the neurobiological basis of dyslexia.
Acknowledgments
We thank all the dyslexic and control individuals participated in the study. Thanks are due to the Chairman, department of studies in Zoology. PS is grateful to University of Mysore for UPG-SRF.
Footnotes
Source of Support: Nil
Conflict of Interest: None declared.
References
- 1.Shaywitz SE. Dyslexia. N Engl J Med. 1998;338:307–12. doi: 10.1056/NEJM199801293380507. [DOI] [PubMed] [Google Scholar]
- 2.Shaywitz SE, Shaywitz BA, Pugh KR, Fulbright RK, Constable RT, Mencl WE. Functional disruption in the organization of the brain for reading in Dyslexia. Proc Natl Acad Sci. 1998;95:2636–41. doi: 10.1073/pnas.95.5.2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DeFries JC, Alarcon M. Genetics of specific reading disability. Ment Retard Dev Disabil Res Rev. 1996;2:39–47. [Google Scholar]
- 4.Cardon LR, Smith SD, Fulker DW, Kimberling WJ, Pennington BF, DeFries JC. Quantitative trait locus for reading disability on Chromosome 6. Science. 1994;266:276–9. doi: 10.1126/science.7939663. [DOI] [PubMed] [Google Scholar]
- 5.Schulte-Körne G, Nöthen MM, Muller-Myhsok B, Cichon S, Vogt IR, Propping P, et al. Evidence for linkage of spelling disability to chromosome 15. Am J Hum Genet. 1998;63:279–82. doi: 10.1086/301919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fagerheim T, Raeymaekers P, Tonnessen FE, Pedersen M, Tranebjaerg L, Lubs HA. A new gene (DYX3) for Dyslexia is located on chromosome 2. J Med Genet. 1999;36:664–9. [PMC free article] [PubMed] [Google Scholar]
- 7.Nopola-Hemmi J, Myllyluoma B, Haltia T, Taipale M, Ollikainen V, Ahonen T, et al. A dominant gene for developmental dyslexia on chromosome 3. J Med Genet. 2001;38:658–64. doi: 10.1136/jmg.38.10.658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fisher SE, Francks C, Marlow AJ, MacPhie LI, Newbury DF, Cardon LR, et al. Independent genome-wide scans identify a chromosome 18 quantitative-trait locus influencing dyslexia. Nat Genet. 2002;30:86–91. doi: 10.1038/ng792. [DOI] [PubMed] [Google Scholar]
- 9.Saviour P, Ramachandra NB. Biological basis of dyslexia: A maturing perspective. Curr Sci. 2006;90:168–75. [Google Scholar]
- 10.Taipale M, Kaminen N, Nopola-Hemmi J, Haltia T, Myllyluoma B, Lyytinen H, et al. A candidate gene for developmental dyslexia encodes a nuclear tetratricopeptide repeat domain protein dynamically regulated in brain. Proc Natl Acad Sci. 2003;100:11553–8. doi: 10.1073/pnas.1833911100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rutter M. Children's behavior questionnaire for completion by teachers: Preliminary findings. J Child Psychol Psychiatry. 1967;8:1–11. doi: 10.1111/j.1469-7610.1967.tb02175.x. [DOI] [PubMed] [Google Scholar]
- 12.Rao SN, Reddy IK. Indian norms of Revan's A, AB, B form. J Psychol Studies. 1968;13:2. [Google Scholar]
- 13.Londin ER, Meng H, Gruen JR. A transcription map of the 6p22.3 reading disability locus identifying candidate genes. BMC Genomics. 2003;4:25–30. doi: 10.1186/1471-2164-4-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marino C, Giorda R, Vanzin L, Nobile R, Lorusso M, Baschirotto C, et al. A locus on 15q15-15qter influences dyslexia: Further support form a transmission / disequilibrium study in an Italian speaking population. J Med Genet. 2004;41:42–6. doi: 10.1136/jmg.2003.010603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scerri TS, Fisher SE, Francks C, MacPhie IL, Paracchini S, Richardson AJ, et al. Putative functional alleles of DYX1C1 are not associated with dyslexia susceptibility in a large sample of sibling pairs from the UK. J Med Genet. 2004;41:853–7. doi: 10.1136/jmg.2004.018341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bellini G, Bravaccio C, Calamoneri F, Cocuzza DM, Filorillo P, Gagliano A, et al. No evidence for association between dyslexia and DYX1C1 functional variants in a group of children and adolescents from Southern Italy. J Mol Neurosci. 2005;27:311–4. doi: 10.1385/jmn:27:3:311. [DOI] [PubMed] [Google Scholar]
- 17.Hannula-Jouppi K, Kaminen-Ahola N, Taipale M, Eklund R, Nopola-Hemmi J, Kaariainen H, et al. The axon guidance receptor gene ROBO1 is a candidate gene for developmental dyslexia. PLOS Genet. 2005;1:467–74. doi: 10.1371/journal.pgen.0010050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meng H, Smith SD, Hager K, Keld M, Liu J, Olson RK, et al. DCDC2 is associated with reading disability and modulates neuronal development in the brain. Proc Natl Acad Sci. 2005;102:17053–8. doi: 10.1073/pnas.0508591102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grigorenko EL, Wood FB, Meyer MS, Pauls DL. Chromosome 6p influences different Dyslexia- related processes: Further confirmation. Am J Hum Genet. 2000;66:715–23. doi: 10.1086/302755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morris DW, Robinson L, Turic D, Duke M, Webb V, Milham C, et al. Family-based association mapping provides evidence for a gene for reading disability on chromosome 15q. Hum Mol Gene. 2000;9:843–8. doi: 10.1093/hmg/9.5.843. [DOI] [PubMed] [Google Scholar]


