Abstract
Gene transcription is associated with local changes in chromatin, both in nucleosome positions and in chemical modifications of the histones. Chromatin dynamics has mostly been studied on a single-gene basis. Those genome-wide studies that have been made primarily investigated steady-state transcription. However, three studies of genome-wide changes in chromatin during the transcriptional response to heat shock in the budding yeast Saccharomyces cerevisiae revealed nucleosome eviction in promoter regions but only minor effects in coding regions. Here, we describe the short-term response to nitrogen starvation in the fission yeast Schizosaccharomyces pombe. Nitrogen depletion leads to a fast induction of a large number of genes in S. pombe and is thus suitable for genome-wide studies of chromatin dynamics during gene regulation. After 20 min of nitrogen removal, 118 transcripts were up-regulated. The distribution of regulated genes throughout the genome was not random; many up-regulated genes were found in clusters, while large parts of the genome were devoid of up-regulated genes. Surprisingly, this up-regulation was associated with nucleosome eviction of equal magnitudes in the promoters and in the coding regions. The nucleosome loss was not limited to induction by nitrogen depletion but also occurred during cadmium treatment. Furthermore, the lower nucleosome density persisted for at least 60 min after induction. Two highly induced genes, urg1+ and urg2+, displayed a substantial nucleosome loss, with only 20% of the nucleosomes being left in the coding region. We conclude that nucleosome loss during transcriptional activation is not necessarily limited to promoter regions.
Gene transcription is usually accompanied by changes in both nucleosome positions and histone modifications (Lee et al. 2004; Li et al. 2007; Jiang and Pugh 2009). Three studies have monitored the genome-wide nucleosome occupancy before and after the transcriptional changes induced by heat shock or glucose in the budding yeast Saccharomyces cerevisiae (Lee et al. 2004; Shivaswamy et al. 2008; Zawadzki et al. 2009). These studies revealed nucleosome eviction in the promoter regions rather than broader genome-wide changes. In addition, genome-wide studies measuring steady-state transcription have been performed in budding yeast and in the fission yeast Schizosaccharomyces pombe (Lee et al. 2004; Wiren et al. 2005; Sinha et al. 2006; Walfridsson et al. 2007). These studies reveal nucleosome depletion in both the promoters and in the coding regions of certain highly active genes. The studies in fission yeast revealed that the depletion was dependent on two nucleosome remodelers of the CHD (chromodomain helicase DNA binding) subfamily, Hrp1 and Hrp3, and the histone chaperone Nap1 (Walfridsson et al. 2007). Steady-state investigations have also shown that actively transcribed genes in S. pombe are highly acetylated on histone H3, but not on histone H4 (Sinha et al. 2006). Other studies of chromatin changes during transcription have been done for single genes. One example is the highly induced GAL1 gene in S. cerevisiae, which is up-regulated at least 1000-fold in the combined absence of glucose and presence of galactose (Johnston and Davis 1984; Nehlin et al. 1991). During induction of this gene, massive nucleosome eviction occurs both in the promoters and in the coding regions (Schwabish and Struhl 2004). Furthermore, this nucleosome eviction was shown to be dependent on histone H3 acetylation by the histone acetyltransferase Gcn5 (Govind et al. 2007). Another example is oxidative stress in S. cerevisiae, where a prominent nucleosome loss is detected after 10–15 min both in the promoter and in the coding region of the STL1 gene (Mas et al. 2009). The nucleosome depletion in the coding region is, however, transient, and already after 30 min, the nucleosome occupancy is back to the level before induction.
Genes are not randomly scattered throughout the eukaryotic genome; frequently coregulated genes are found in clusters (Lee and Sonnhammer 2003; de Wit and van Steensel 2009). One example is the gene cluster in S. cerevisiae that contains GAL1, GAL10, and GAL7 (Johnston and Davis 1984). Other examples of coregulated gene clusters are the genes that are induced early during the sexual program in S. pombe (Mata et al. 2002). The sexual cycle in fission yeast is induced by nitrogen starvation and is driven by a complex transcriptional program (Mata et al. 2002, 2007). Thus, there are several waves of transcription during conjugation and the following meiosis. Some of the genes that are induced within 1 h of nitrogen starvation colocalize within five different clusters. Four of these clusters reside next to the subtelomeric regions of chromosomes I and II, while the fifth cluster is located in the middle of the left arm of chromosome I (Mata et al. 2002). Moreover, it has been noted that upon deletion of the cen1 centromere, neocentromeres can only be formed in the two subtelomeric clusters of genes that are induced during nitrogen starvation, but nowhere else on chromosome I (Ishii et al. 2008). These results could indicate the presence of a specific chromatin structure in these regions, or alternatively that these are the only two regions on chromosome I that are sufficiently large to accommodate a centromere, where there are no genes that are essential for vegetative growth. The cluster in the middle of chromosome I, Chr1, and the gene cluster near the left telomere of chromosome I, Tel1L, also change location during nitrogen starvation. The two clusters normally reside at the nuclear periphery, in the proximity of the nucleolus (Alfredsson-Timmins et al. 2009). This localization is dependent on the histone deacetylase (HDAC) Clr3. However, within 20 min after the onset of nitrogen starvation, there is a pronounced transcription-dependent change in the subnuclear localization of these two gene clusters, which move away from the nuclear membrane and the nucleolus, and instead are found in the center of the nucleus.
The key regulator of entry into the sexual program in S. pombe is Ste11 (Sugimoto et al. 1991; Mata and Bahler 2006; Xue-Franzen et al. 2006). Induction of Ste11 and entry into the sexual life cycle is induced by nitrogen starvation, and is dependent on the mitogen activated protein kinase (MAPK) Sty1 and on the transcription factors Atf1 and Rst2 (Takeda et al. 1995; Shiozaki and Russell 1996; Higuchi et al. 2002). Within 1 h after the onset of nitrogen starvation, around 250 genes are up-regulated or down-regulated, but it is not clear whether all these early induced genes are dependent on Sty1 and Atf1 or Rst2 (Mata et al. 2002). Sty1 and Atf1 are also involved in gene regulation during many different types of stresses such as oxidative, cadmium, heat, osmotic, and DNA damage stress (Shiozaki and Russell 1996; Chen et al. 2003). A set of 140 genes that are up-regulated at least twofold in at least four of these types of stresses has been defined as the core environmental stress response (CESR)–induced genes. Only 11 genes could be identified as CESR-repressed using the same criteria (Chen et al. 2003).
In order to gain a deeper understanding of chromatin alterations early during a transcriptional perturbation, we have made a genome-wide study of the chromatin changes that occur in S. pombe within 20 min after the onset of nitrogen depletion, measuring both nucleosome occupancy and histone acetylation levels. We also made a genome-wide study of transcription before and after nitrogen removal, using the same time points. We found that genes that are involved in pyrimidine salvage and nucleotide catabolism are up-regulated early during nitrogen depletion, whereas genes that are involved in ribosome biogenesis are down-regulated. Surprisingly, unlike previous studies of genome-wide chromatin changes during a transcriptional change (Lee et al. 2004; Shivaswamy et al. 2008; Zawadzki et al. 2009), we saw nucleosome loss of equal magnitude in the promoters and coding regions of the up-regulated genes. The nucleosome eviction was accompanied by an increase in H3AcK9, but not in H4AcK5 levels. The nucleosome reduction in the coding region also occurred for genes induced by cadmium treatment. Two genes in the Chr1 cluster, urg1+ and urg2+, showed a particularly pronounced nucleosome loss in the coding regions 20 min after nitrogen depletion, with only 20% of the nucleosomes remaining.
Results
Genes involved in nucleotide catabolism are up-regulated early after the onset of nitrogen starvation
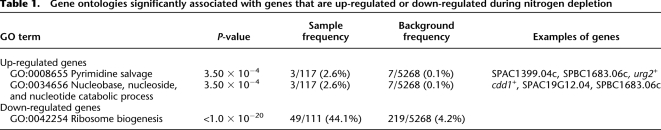
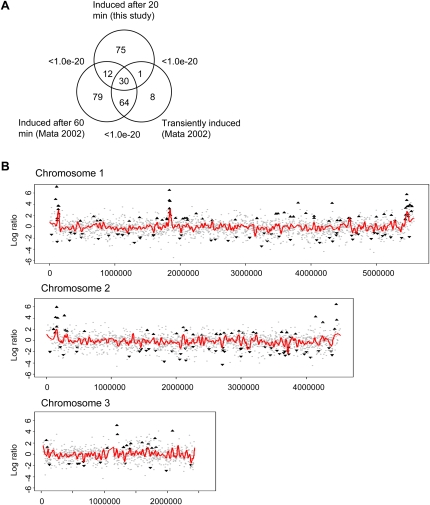
Expression profiling was used to find out which genes were up-regulated early, that is, within 20 min after the onset of nitrogen starvation. In total, 118 genes were significantly up-regulated (more than twofold, P < 0.05), and we found that they were enriched for genes involved in pyrimidine salvage and nucleotide catabolic processes (Table 1; Supplemental Table S3). The 111 genes that were significantly down-regulated were enriched for genes involved in ribosome biogenesis. We compared our gene set with the one obtained by Mata et al. (2002) and found a significant overlap with the genes that were up-regulated within 60 min after nitrogen depletion in that study (P < 1.0 × 10−20, Fisher's exact test) (Fig. 1A). In addition, there was also a significant overlap between the genes that were up-regulated in our study and genes that were defined as transient by Mata et al. (2002), meaning that the genes are up-regulated when nitrogen is depleted, but then return to their lower basal level of transcription (P < 1.0 × 10−20, Fisher's exact test) (Fig. 1A). When the changes in expression of all genes were plotted genome-wide, it was evident that the three gene clusters on chromosome I that are up-regulated after 60 min of nitrogen starvation (Mata et al. 2002) were up-regulated already after 20 min (Fig. 1B; Supplemental Fig. S1). The clusters were named Chr1, Tel1L, and Tel1R, and the genes up-regulated in these clusters are listed in Supplemental Table S4.
Table 1.
Gene ontologies significantly associated with genes that are up-regulated or down-regulated during nitrogen depletion
Figure 1.
Overview of gene expression changes after nitrogen starvation. (A) Overlap between genes that were up-regulated 20 min after the onset of nitrogen starvation (this study) and genes that were up-regulated after 60 min or defined as transiently induced genes (Mata et al. 2002). P-values were calculated using Fisher's exact test. (B) Genomic distribution of up-regulated and down-regulated genes. The x-axis shows the position of the gene in base pairs from the left end of each chromosome, and the y-axis shows the change in gene expression log2 ratio after 20 min of nitrogen starvation. (▲) Significantly up-regulated genes; (▼) significantly down-regulated genes. (Red lines) A loess curve fitted to the gene expression log2 ratios.
Chromatin changes after nitrogen depletion
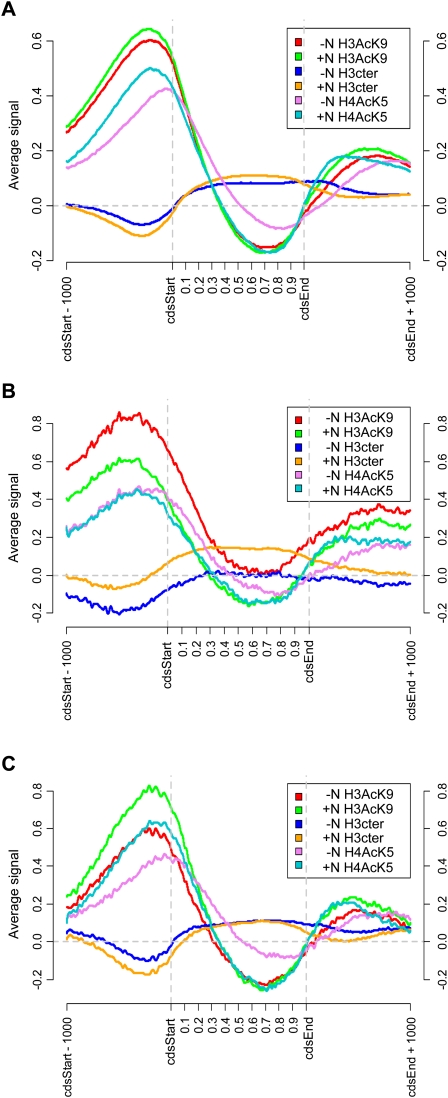
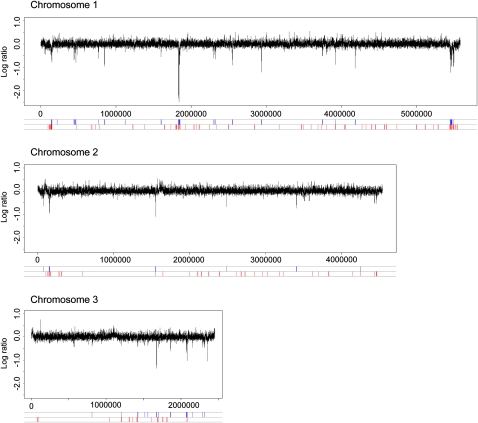
Genome-wide changes in the chromatin during nitrogen depletion were monitored by chromatin immunoprecipitation chip analysis using Affymetrix tiling microarrays (ChIP-chip) with 20-bp resolution. The genome-wide changes in nucleosome occupancy were determined by performing triplicate ChIP-chip measurements with an antibody against the unmodified C terminus of histone H3 (H3Cter). Changes in histone modifications were examined by performing triplicate ChIP-chip analyses with an antibody against histone H3 acetylated at lysine 9 (H3AcK9) or against histone H4 acetylated at lysine 5 (H4AcK5). To get a better overview of the changes during nitrogen removal, we aligned all genes (Fig. 2A), genes that were significantly up-regulated (Fig. 2B), and genes that were significantly down-regulated (Fig. 2C), and plotted the average chromatin profiles (footprints) for each group of genes. There were no major effects on nucleosome occupancy or histone acetylation when all genes were considered as a group (Fig. 2A). When the chromatin footprint of the 118 genes that were up-regulated during nitrogen depletion was plotted, it was evident that nucleosomes were lost to the same extent in both the promoters and in the coding regions (Fig. 2B). The nucleosome occupancy was lowest, 500 bp, before the start codon in the presence of nitrogen, and thus the promoter region also had the lowest nucleosome density after induction. However, the nucleosome loss was highly significant not only in the promoters (−500 to 0 bp, P = 4.2 × 10−17, Wilcoxon rank-sum test), but also in the coding regions (0 to 1000 bp, P = 6.2 × 10−14, Wilcoxon rank-sum test). There was also a significant increase in H3AcK9 in both the promoter regions (P < 1.0 × 10−20, Wilcoxon rank-sum test), with a peak around 300–400 bp upstream of the start codon, and in the coding regions (P = 1.4 × 10−10, Wilcoxon rank-sum test). In addition, there was a small change in the H4AcK5 level in the promoters (P = 1.2 × 10−7, Wilcoxon rank-sum test), which was not seen in the coding regions. The 111 genes that were down-regulated had a decrease in the histone acetylation modifications that we monitored (H3AcK9: P = 5.8 × 10−18, Wilcoxon rank-sum test; and H4AcK5: P = 4.9 × 10−7, Wilcoxon rank-sum test) in the promoters, but in contrast, there were no significant changes in H3AcK9 or H4AcK5 in the coding regions (Fig. 2C). When the changes in nucleosome density were plotted over the whole genome, it was clear that the regions where the gene clusters that are regulated by nitrogen are localized had a substantial nucleosome loss 20 min after the onset of nitrogen starvation (Fig. 3). A detailed examination of the up-regulated genes in the Tel1R cluster revealed that the nucleosome profiles within each gene were similar in the presence and in the absence of nitrogen, though shifted toward less nucleosome occupancy after nitrogen depletion (Fig. 4A). Thus, the nucleosome eviction was detected in a broad region rather then restricted to the promoter regions. The changes in acetylation levels at H3AcK9 and H4AcK5 were also plotted for the entire genome (Supplemental Figs. S2, S3). These plots showed extensive changes in H3AcK9, especially within the gene clusters that were up-regulated after nitrogen removal, whereas the changes in H4AcK5 were less pronounced, which was consistent with the results in Figure 2. In conclusion, there was an overall loss of nucleosomes both in the promoters and in the coding regions of genes that were up-regulated during nitrogen depletion. The nucleosomes that were left had an increase in H3AcK9 and to a lesser extent in H4AcK5 modification.
Figure 2.
Averages over different groups of genes of H3, H3AcK9, and H4AcK5 profiles before and after nitrogen starvation. (A) All genes in the genome; (B) up-regulated genes; (C) down-regulated genes. The positions relative to the coding regions are shown on the x-axis. The genes in each group were aligned so that the starts and stops of the open reading frames would coincide. Since the open reading frames have different lengths, relative positions were used inside the coding regions. In addition, flanking regions of 1000 bp were included on each side. The y-axis shows the average signal (on a log2 scale) for each combination of antibody and condition (plus or minus nitrogen).
Figure 3.
Differences in the histone H3 occupancy over the entire genome, before and after nitrogen starvation. The genomic position in base pairs from the left end of each chromosome is shown on the x-axis and the H3 occupancy log2 ratio on the y-axis. Regions with a significant histone H3 depletion 20 min after nitrogen starvation are shown in blue at the bottom; up-regulated genes are shown in red.
Figure 4.
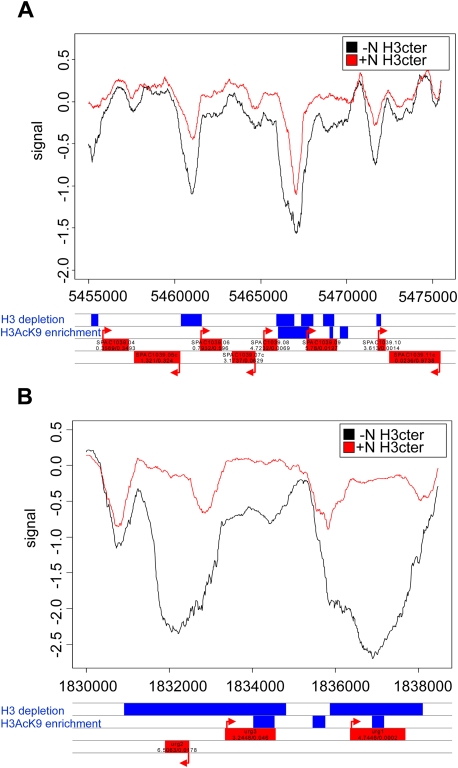
Chromatin profiles over selected regions. The histone H3 binding to the DNA before and after nitrogen depletion is shown. (A) The cluster in the Tel1R region. (B) The region including the urg1+, urg2+, and urg3+ genes. Individual genes are shown in red below the profiles, and regions with significant H3 depletion or significant H3AcK9 enrichment in blue.
Pronounced nucleosome loss in the Chr1 cluster after nitrogen depletion
The Chr1 cluster contains a group of genes that are regulated by uracil, cadmium, and nitrogen, namely, urg1+, urg2+, and urg3+ (Mata et al. 2002; Chen et al. 2003; Watt et al. 2008). When monitored using gene expression microarrays the urg1+ gene was induced 26-fold after 20 min of nitrogen removal and 522-fold after 60 min (this study; Mata et al. 2002). The urg2+ gene was up-regulated 90-fold after 20 min and 396 times after 60 min. These two highly induced genes show a very pronounced nucleosome loss (≥80%) after 20 min of nitrogen depletion (Fig. 4B). Furthermore, the nucleosome loss in these two genes was even greater in their coding regions than in their promoters. In contrast, the urg3+ gene, which was less strongly up-regulated (9.5 times after 20 min and 76 times at 60 min), only lost ∼40% of its nucleosomes (Fig. 4B).
Since the RSC complex was shown to be important for both transcriptional activation and nucleosome remodeling in response to stress in S. cerevisiae (Mas et al. 2009), we tested whether induction of urg1+, urg2+, and SPAC1039.02 (a gene in the Tel1R cluster) was dependent on snf21+ by using a temperature-sensitive snf21-36 mutation (Yamada et al. 2008). A gene that is not regulated by nitrogen starvation, dis2+, was included as a control. We found that all three genes were still induced by nitrogen withdrawal at 35°C, indicating that Snf21 (and thus presumably the RSC complex) was dispensable for this induction. The nucleosome remodelers Hrp1 and Hrp3 have previously been implicated in nucleosome eviction from both the promoters and the coding regions of highly expressed genes (Walfridsson et al. 2007). We therefore investigated whether Hrp1 and Hrp3 are required for nucleosome removal after nitrogen depletion. To this end, we made a ChIP experiment using the H3Cter antibody, comparing nucleosome densities before and after nitrogen withdrawal in the wild type and in a strain lacking Hrp1 and Hrp3. We examined the promoters and coding regions of several up-regulated genes from the gene clusters Chr1, Tel1L, and Tel1R. There was no difference between the nucleosome loss in the wild-type strain and in the hrp1Δ hrp3Δ strain, and hence we conclude that the CHD nucleosome remodelers were not involved in nucleosome eviction during nitrogen removal (data not shown). We proceeded to examine the effects of several other nucleosome remodelers on induction of the urg1+ gene. We found that deletions of the mit1+, fft3+, and snf22+ genes had no effects. Moreover, the urg1+ gene was still induced in a strain lacking the mst2+ gene and at the restrictive temperature in a strain with a ts mutation in the mst1+ gene (Gomez et al. 2008), indicating that the two HATs of the MYST subfamily that are encoded by these genes are not responsible (at least not alone) for the increase in histone acetylation during gene activation by nitrogen depletion (data not shown). In summary, there was a pronounced nucleosome loss throughout the strongly induced urg1+ gene in response to nitrogen depletion, but the lack of effects of all mutations tested so far suggests that this loss was likely dependent on several redundant nucleosome remodelers and/or other mechanisms that remain to be identified.
Nucleosome loss was persistent and also detected after Cd2+ induction
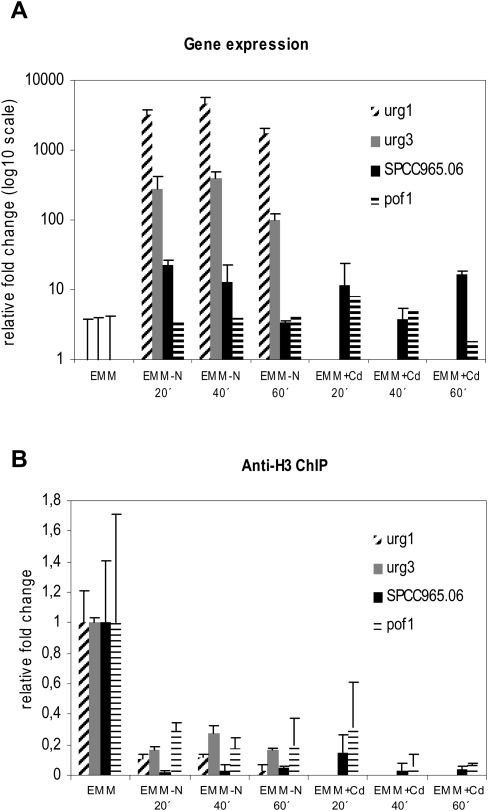
To further elucidate whether the nucleosome loss during gene induction was limited to nitrogen starvation or a common feature during dynamic changes in gene expression in S. pombe, we treated the cells with cadmium. Cadmium treatment in rich media has been shown to up-regulate around 400 genes in S. pombe, including SPCC965.06 and pof1+ (Chen et al. 2003). We treated cells either with cadmium or nitrogen starvation in minimal medium (EMM) and made a subsequent ChIP using antibodies against histone H3 to monitor nucleosome density. At the same time, we also did RNA preparations to monitor gene expression before and after the two inductions. The two genes that we examined, SPCC965.06 and pof1+, were moderately up-regulated by cadmium, and also by nitrogen depletion (five to 10 times) (Fig. 5A). Significantly, the nucleosome loss in the coding region was evident when SPCC965.05 and pof1+ were induced both by nitrogen starvation and with cadmium treatment (Fig. 5B).
Figure 5.
Gene expression and nucleosome occupancy at different time points after nitrogen depletion and treatment with Cd2+. Cells were grown in EMM and starved for nitrogen or treated with 0.5 mM CdSO4, and samples were taken out for RNA preparation or ChIP at time points indicated. (A) Gene expression monitored by real-time PCR and normalized to expression of dis2+ and to the signal obtained for EMM media. Data are represented as mean + SD. (B) ChIP analysis of H3 association with the open reading frame of indicated genes measured by real-time PCR. Data were normalized to the input (nonimmunoprecipitated) material and the signal obtained for EMM media. Data are represented as mean + SD.
Nucleosome loss from the coding regions could either be a transient effect that was associated with the induction process or a more stable effect that would continue as long as the gene was transcribed. To distinguish between these two possibilities, we monitored both gene expression and nucleosome occupancy in the coding regions at different time points after treatment. We removed the nitrogen source or treated the cells with Cd2+ for 20, 40, and 60 min and investigated the nucleosome occupancy as well as monitored the gene expression for the urg1+, urg3+, SPCC965.06, and pof1+ genes (Fig. 5). It was evident that the nucleosome levels did not return to the levels before the gene induction, following either treatment, and hence was not a transient event during activation, but remained during transcription of the genes. We conclude that the nucleosome loss that we detected in the coding region of induced genes was not limited to nitrogen depletion and remained 60 min after induction, and thus appears to be a general feature of transcription in S. pombe.
Overlap between nitrogen regulated and CESR genes
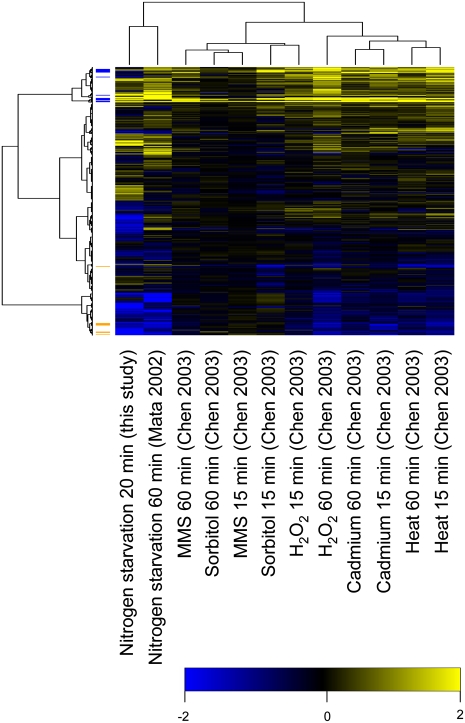
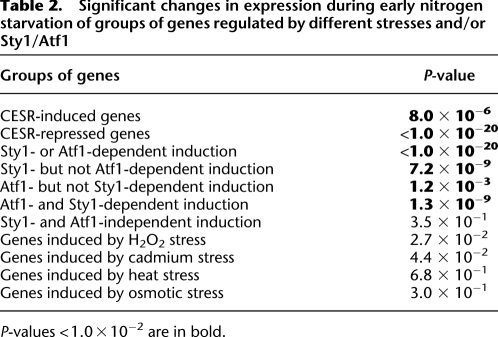
In order to identify similarities in gene expression patterns, which might indicate that similar signaling pathways and/or mechanisms for gene regulation could be involved, we used hierarchical clustering to compare our expression microarray data with published microarray data obtained under different conditions. As expected, we found that our data clustered together with data obtained after 60 min of nitrogen starvation (Mata et al. 2002). We further note that the data from all other types of stresses examined—DNA damage (MMS), osmotic stress (sorbitol), oxidative stress (H2O2), heat stress, and heavy metal (cadmium) stress after 15 and 60 min, respectively (Mata et al. 2002; Chen et al. 2003)—clustered together, indicating that these responses are more similar to each other than to the response to nitrogen depletion (Fig. 6). Still, when the data were clustered in the second dimension, that is, on genes, it was clear that there were also certain similarities between nitrogen depletion and the other stresses. Thus, the CESR-induced genes (Chen et al. 2003) as a group were significantly up-regulated also after 20 min of nitrogen withdrawal (P = 8.0 × 10−6, Wilcoxon rank-sum test compared to all other genes) (see Table 2; Fig. 6). However, the up-regulation was modest for most genes, as only three CESR genes (SPCC338.12, SPAC97.13c, and SPBC23G7.06c) were up-regulated more than twofold in our data. Finally, we note that the genes defined as being CESR-repressed (Chen et al. 2003) were also significantly down-regulated in our data set (P < 1.0 × 10−20, Wilcoxon rank-sum test) (Table 2; Fig. 6).
Figure 6.
Hierarchical clustering of stress response gene expression data. The clustering includes 4145 genes (rows) under 12 different stress conditions (columns). Colors indicate gene expression log ratios compared to relevant controls; (yellow) up-regulation; (blue) down-regulation. The 20-min nitrogen starvation data are from the present investigation and the other data are from Mata et al. (2002) and Chen et al. (2003). At left, CESR-induced genes are labeled in blue, and CESR-repressed genes in orange.
Table 2.
Significant changes in expression during early nitrogen starvation of groups of genes regulated by different stresses and/or Sty1/Atf1
P-values < 1.0 × 10−2 are in bold.
Overlap between nitrogen-regulated genes and genes regulated by Sty1 and/or Atf1
Stress response genes, like the genes that are induced after oxidative or osmotic stress, are regulated by the MAP kinase Sty1 and/or the transcription factor Atf1 (Shiozaki and Russell 1996; Chen et al. 2003). Moreover, Sty1 and Atf1 are also needed for conjugation and meiosis, processes that are induced by nitrogen starvation (Takeda et al. 1995; Shiozaki and Russell 1996). Consistent with this, we found that the Sty1/Atf1-dependent genes (Chen et al. 2003) as a group were up-regulated after 20 min of nitrogen removal (P < 1.0 × 10−20, Wilcoxon rank-sum test) (see Table 2). The strong correlation between the nitrogen-regulated genes and genes that are regulated by Atf1 and/or Sty1 during other types of stresses prompted us to test for Atf1/Sty1 dependency in the response to nitrogen depletion, using strains deleted for atf1+ or sty1+. Two of the genes in the Chr1 cluster, urg1+ and urg2+, are known to be dependent on Atf1 for their up-regulation during cadmium stress (Chen et al. 2003). However, in a RT-PCR experiment, these two genes were up-regulated after 20 min of nitrogen removal also in the strains lacking Atf1 or Sty1. Moreover, two oxidative stress-induced genes that are dependent on Sty1, SPAC1399.01c and SPAC11D3.17 (Chen et al. 2003), were still up-regulated during nitrogen depletion in a strain lacking Sty1 (data not shown).
We proceeded to investigate whether Sty1 or Atf1 are physically associated with genes that are up-regulated during nitrogen depletion by using ChIP analysis with myc epitope-tagged Atf1 and Sty1 proteins. There was a slight enrichment of both Sty1 and Atf1 in the promoters of urg1+ and urg2+; however, this enrichment was not affected by nitrogen depletion (data not shown). A search for known binding sites and/or response elements of Atf1, Rst2, and Sty1 (Takeda et al. 1995; Higuchi et al. 2002; Smith et al. 2007) revealed no enrichment of such elements in either the promoters or the coding regions of the nitrogen-regulated genes. Furthermore, de novo searches for new sequence motifs (see Methods) did not reveal any significantly enriched motifs in these genes.
In conclusion, whereas we see a statistically significant correlation between the genes that were up-regulated early during nitrogen starvation and genes that are up-regulated during other stress conditions, and which are dependent on Sty1 and/or Atf1, direct evidence for a general role for Sty1 and/or Atf1 in the early response to nitrogen depletion remains elusive.
Correlations between genome-wide chromatin changes and the transcriptional responses to nitrogen depletion
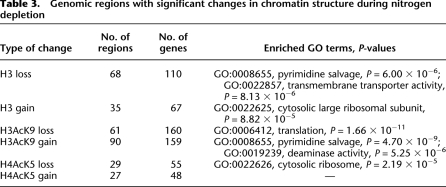
Regions on the chromosomes that exhibited significant chromatin changes early during nitrogen starvation were identified (Table 3). There were 68 regions with significant reduction of histone H3 during nitrogen depletion, and 110 genes were associated with these regions (Fig. 3). Significantly, these regions were enriched for genes involved in pyrimidine salvage and transmembrane transport (Table 3). This was consistent with our finding that the set of genes that were up-regulated after nitrogen depletion also was enriched for genes involved in pyrimidine salvage (Table 1). Thus, there was an overlap between the two groups (23 genes, P = 2.1 × 10−16, Wilcoxon rank-sum test). Furthermore, we found that regions that gain nucleosomes early during nitrogen starvation were enriched for genes encoding ribosomal proteins, which was consistent with our finding that genes involved in ribosome biogenesis were down-regulated during nitrogen depletion (Tables 1, 2). Interestingly, we further found that regions with a significant increase in H3AcK9 levels also were enriched for genes involved in pyrimidine salvage (Table 3; Supplemental Fig. S2). Finally, we found that regions that lose histone acetylation were enriched for genes involved in translation (for H3AcK9) and genes encoding ribosomal proteins (for H4Ac5) (Table 3; Supplemental Figs. S2, S3).
Table 3.
Genomic regions with significant changes in chromatin structure during nitrogen depletion
We also explicitly checked the correlation between changes in nucleosome occupancy, acetylation levels, and gene expression (Supplemental Fig. S4). When all genes were considered as a group, there was a negative correlation coefficient of −0.22 (P < 1.0 × 10−20) between changes in gene expression and changes in nucleosome occupancy, and a positive correlation coefficient of 0.23 (P < 1.0 × 10−20) between changes in gene expression and changes in H3AcK9 and to a smaller extent with changes in H4AcK4 (0.039, P = 3.3 × 10−3). Examination of only the up-regulated genes made these patterns clearer; then the correlation between nucleosome occupancy and gene expression was −0.54 (P = 3.6 × 10−8), while the correlation between H3AcK9 levels and gene expression was 0.60 (P = 2.0 × 10−9). When we considered only the genes annotated as being involved in “pyrimidine metabolism,” there was a correlation coefficient of −0.81 (P = 2.2 × 10−3) between nucleosome occupancy and gene expression and a correlation of 0.69 (P = 5.9 × 10−3) between gene expression and changes in H3AcK9 levels. In conclusion, there was a clear correlation between the changes in the chromatin structure and changes in expression for up-regulated genes. On the other hand, for the down-regulated genes, we found no significant correlation between changes in gene expression and changes in nucleosome occupancy (correlation coefficient −0.043, P = 0.33). Changes in acetylation levels and expression levels were barely significant for H3AcK9 (correlation −0.16, P = 0.044) and H4AcK5 (−0.18, P = 0.029). Finally, when we examined a large group of genes that were down-regulated during nitrogen depletion, namely, the ribosomal genes, there was no correlation between changes in gene expression and changes in neither nucleosome occupancy or histone acetylation levels (Supplemental Fig. S4).
Changes in nucleosome occupancy and histone acetylation correlate both with known gene functions and with changes in gene expression
The aligned chromatin profiles for individual genes were clustered on the basis of differences in nucleosome profiles before and after nitrogen removal, in order to reveal if any groups of genes undergo similar changes in nucleosome occupancy. We used hierarchical clustering on nucleosome occupancy profiles, from 500 bp upstream of the start codon to 500 bp into the open reading frame. Based on the resulting dendrogram, all genes were divided into seven groups, which we named H3:1 to H3:7 (Supplemental Fig. S5A). Groups H3:1, H3:2, and H3:3 were characterized by nucleosome loss both in the promoters and in the coding regions. These three groups contained many of the genes that were up-regulated 20 min after nitrogen starvation. Groups H3:1 and H3:2, which had a smaller reduction in nucleosome occupancy, contained many genes found in the telomeric clusters on chromosome I. Group H3:3, a small group of genes with a larger reduction in nucleosome occupancy, included genes found in the telomeric clusters as well as in the Chr1 cluster. This group was also enriched for genes involved in nucleotide metabolism (Supplemental Fig. S5B,C). In addition, group H3:1 contained genes involved in amino acid transmembrane transport (Supplemental Fig. S5C). A second clustering was done including only the genes that were differentially expressed during nitrogen withdrawal (Supplemental Fig. S6).
A similar hierarchical clustering was also made for changes in H3AcK9 profiles in response to nitrogen depletion, from which six groups of genes could be identified. One of these groups, H3AcK9:6, had an increase in H3AcK9 both in the promoters and in the coding regions, and many of the genes in the Chr1 cluster and in the subtelomeric clusters on chromosome 1 were found within this group of genes (Supplemental Fig. S7). Group H3AcK9:6 was also enriched for genes involved in pyrimidine base metabolism (Supplemental Fig. S7C). Group H3AcK9:4 contained genes where the H3AcK9 level increased in the promoters but not in the coding regions, and this group also contained genes involved in the cellular response to stress (Supplemental Fig. S7C). Clustering of the H5AcK5 profiles resulted in eight groups. Group H4AcK5:8 showed an increase in acetylation levels both in the promoters and in the coding regions, and included several genes from the telomeric clusters and the Chr1 cluster (Supplemental Fig. S8). Group H4AcK5:8 was enriched for genes involved in nitrogen compound metabolic processes (Supplemental Fig. S8B,C). Finally, there was a significant overlap between the groups of genes that showed nucleosome loss and those that showed significant changes in histone modification levels; thus H3:1 and H3AcK9:5 were strongly correlated (P < 1.0 × 10−20, Fisher's exact test) as well as H3:3 and H3AcK9:6 (P < 1.0 × 10−20, Fisher's exact test) and also H3:3 and H4AcK5:8 (P < 1.0 × 10−20, Fisher's exact test).
A de novo search for DNA sequence motifs in the different groups of genes that were identified by clustering on the chromatin profiles revealed several motifs that were significantly enriched (Supplemental Fig. S9). One motif, GTGACTG, has been described previously as being associated with genes encoding ribosomal proteins and was enriched in cluster H3AcK9:2 (Supplemental Fig. S7), which indeed contains many genes encoding ribosomal proteins (Gasch et al. 2004). The other motifs shown in Supplemental Figure S9 have not been described previously. Many of the motifs are AT-rich and may reflect the fact that AT-rich sequences facilitate nucleosome depletion (Segal and Widom 2009). We conclude that hierarchical clustering of genes based on the changes in their chromatin profiles after nitrogen depletion resulted in several groups of genes that are significantly enriched for certain Gene Ontology (GO) terms, and, furthermore, that several novel DNA motifs whose function(s) remain to be elucidated are associated with these groups of genes.
Discussion
Nucleosome eviction from genes that were induced during nitrogen depletion
The ChIP-chip analysis using antibodies against histone H3 and two differentially acetylated histones provides us with snapshots of the state of the chromatin during a major transcriptional change that affects a large number of genes. For the genes that were up-regulated during nitrogen depletion, we could detect a decrease in nucleosome occupancy both in the promoter, coding, and terminating regions (Fig. 2B). Surprisingly, the nucleosomes were lost to the same extent in promoter and coding regions. When the nucleosome profiles before and after nitrogen depletion in the affected genes were compared, for example, for the genes in the Tel1R cluster, it was evident that the nucleosome distribution over the promoter and coding regions of a given gene were quite similar, but shifted downward (toward fewer nucleosomes) after nitrogen depletion (Fig. 4A). This indicates that nucleosome remodelers were active throughout the genes, and not only in the promoters. This finding differ from the conclusions drawn by Shivaswamy et al. (2008), who suggested that chromatin remodeling in response to a transcriptional stimulus is associated with the eviction or reposition of one to two nucleosomes in the promoter region rather than broader region-wide changes. In fact, the highly up-regulated genes urg1+ and urg2+ showed an even higher nucleosome loss in their coding regions than in their promoters (Fig. 4B).
However, studies of single genes have revealed substantial nucleosome loss in the coding regions of some highly induced genes such as the GAL genes in S. cerevisiae. Thus, when the GAL genes are fully induced, only ∼10% of the nucleosomes remain bound to the promoter regions and 30% to the coding regions (Schwabish and Struhl 2004). In addition, nucleosome eviction from the GAL1 gene was shown to be preceded by acetylation of histone H3, mediated by the histone acetyltransferase Gcn5 (Govind et al. 2007). Moreover, when a time-course study was made of certain genes that are induced during osmotic stress in S. cerevisiae, a prominent, but transient nucleosome loss within the coding regions was detected early, within 10–15 min, after induction (Mas et al. 2009). However, after 30 min, the nucleosome profiles in the coding regions were more or less back to the levels seen prior to the onset of osmotic stress. In the promoters of these genes, nucleosome occupancy also decreased initially, but unlike the coding region, they remained at a lower level as compared to the uninduced state. In contrast, our results suggest that nucleosome loss in S. pombe is not a transient event during induction of genes, but a change of state that remains also 60 min after induction by either nitrogen depletion or addition of cadmium and presumably persists as long as the gene is being transcribed (Fig. 5).
It should be emphasized that the prominent nucleosome loss that we detected in the coding regions of genes that were induced by nitrogen depletion was a general phenomenon and not limited to a few highly expressed genes. Furthermore, our results with cadmium induction show that the effect is not limited to induction by nitrogen depletion (Fig. 5). Unlike the situation in S. cerevisiae, nucleosome loss in S. pombe therefore seems to be a general feature of transcribed genes. Further studies on the dynamics of nucleosome occupancy as well as histone modifications in the coding regions of regulated genes are needed to understand the biological significance of this nucleosome depletion.
H3AcK9 modifications were associated with up-regulated genes
We detected a strong increase in the levels of H3AcK9 and a minor increase in the levels of H4AcK5 during gene activation, and this increase was seen throughout the genes (Fig. 2B). This is consistent with previous studies of steady-state transcription in fission yeast, where a strong correlation was found between strongly expressed genes and high H3AcK9 acetylation levels (Wiren et al. 2005; Sinha et al. 2006). We could also identify a number of regions throughout the genome with significantly different levels of nucleosome occupancy as well as H3AcK9 or H4AcK5 modification levels in the absence and in the presence of nitrogen. These regions correlated well with genes that were significantly up-regulated or down-regulated during nitrogen depletion, as revealed by our microarray experiment. Thus, regions with changes usually found in active chromatin, that is, nucleosome depletion and an increase in H3AcK9 acetylation, contained genes that were up-regulated during nitrogen removal, such as the genes involved in pyrimidine salvage (Table 3).
Genome-wide expression profile after nitrogen depletion
Genes that we found to be up-regulated early, after 20 min, of nitrogen starvation correlated well with the genes previously described as being up-regulated after 60 min of nitrogen starvation (Fig. 1A; Mata et al. 2002). The 118 up-regulated genes were enriched for genes involved in pyrimidine salvage and nucleotide catabolism (Table 1). This is not surprising since nucleotides are rich in nitrogen, and their salvage therefore is presumably important during nitrogen starvation. The 111 genes that were significantly down-regulated were enriched for genes involved in ribosome biogenesis. The latter group of genes is down-regulated at an early stage during most stress conditions investigated so far. It is likely that it simply reflects the fact that the cells transiently cease to grow as they are adjusting to a new environment (Chen et al. 2003, 2008; Asp et al. 2008).
The gene clusters observed by Mata et al. (2002), which comprise genes that are up-regulated during nitrogen starvation and which are located in the subtelomeric areas and in the middle of the left arm of chromosome I, were also evident after 20 min of nitrogen starvation (Fig. 1B). Moreover, there was a nonrandom distribution of nitrogen-regulated genes throughout the genome, with certain areas, like the left arm of chromosome III, being completely devoid of up-regulated genes. When the genome-wide transcription profile after 20 min of nitrogen removal was clustered together with the profile from 60 min of nitrogen starvation as well as with the profiles induced by other types of stresses (Fig. 5; Mata et al. 2002; Chen et al. 2003), it was evident that the profile seen during nitrogen removal was different from those seen during other types of stresses. Thus, the transcription profiles seen during nitrogen depletion formed a distinct outgroup, with all the other stress response profiles being more similar to each other than to the nitrogen depletion profiles.
Interestingly, in spite of this, there was a statistically significant overlap between the genes that were up-regulated during nitrogen removal and genes defined as CESR-induced genes by virtue of being up-regulated at least twofold in four out of five types of stresses (Table 2; Chen et al. 2003). Conversely, the CESR-repressed genes, as a group, were also significantly down-regulated during early nitrogen starvation. Moreover, two genes in the Chr1 cluster, urg1+ and urg2+, which also are induced by cadmium stress and which are dependent on Atf1 in that case (Chen et al. 2003), were not Atf1-dependent in the nitrogen depletion context (data not shown). This suggests a differential type of regulation, which was independent of Atf1, for at least some of the genes that are up-regulated early during nitrogen starvation. The correlation between nitrogen-regulated genes and genes that are Atf1- and/or Sty1-dependent in their responses to other types of stresses further suggests that a common set of stress-induced genes exists that, however, are regulated by several distinct pathways in response to different types of stresses.
Clustering of genes according to nitrogen-dependent changes in their chromatin profiles
The changes in chromatin profiles that we observed during nitrogen depletion could be clustered into several groups of genes that were enriched in specific gene ontology terms, for example, cytosolic large ribosomal subunit, transmembrane transport, and pyrimidine salvage (Supplemental Figs. S5c, S6c, S7c, S8c). Moreover, searches for DNA sequence motifs within these clusters resulted in the identification of several significantly enriched motifs. For example, a previously described motif GTGACTG known to be associated with ribosomal protein genes was enriched in cluster H3AcK9:2, a cluster of genes that show loss in H3AcK9 over both the promoters and the coding regions (Supplemental Figs. S7, S9; Gasch et al. 2004). The other enriched motifs that we found have not previously been described. Many of these motifs are AT-rich and may reflect regions that are particularly susceptible to nucleosome depletion (Segal and Widom 2009). The possible importance of these motifs in nitrogen-dependent gene regulation remains to be elucidated.
Nitrogen depletion in S. pombe as a model for genome-wide studies of chromatin dynamics
In this study, we have established nitrogen depletion in fission yeast as a model for genome-wide studies of the chromatin dynamics during transcriptional activation. In a previous study we detected substantial changes in the positions within the cell nucleus of gene clusters that are induced by nitrogen depletion (Alfredsson-Timmins et al. 2009). The clusters examined thus moved from the nuclear periphery to a more internal localization upon induction. We have now shown that the same gene clusters undergo pronounced nucleosome loss during induction by nitrogen withdrawal. Further studies of the response to nitrogen depletion may help us to better understand the connection between changes in chromatin status and changes in subnuclear localization of nitrogen-regulated genes.
Methods
Strains and media
The yeast strains used in this study are listed in Supplemental Table S1. Media were prepared as previously described (Thon et al. 1999; Mata et al. 2002). Nitrogen depletion or cadmium treatment were performed as follows: log-phase cells grown in Edinburgh minimal media (EMM) were washed in either EMM without NH4Cl or EMM with 0.5 mM CdSO4, prewarmed to 30°C, and then incubated for 20, 40, or 60 min in the same media at 30°C. A control with EMM media was also included.
Expression profiling
RNA was extracted from 2 × 107 yeast cells in biological triplicates. The cDNA synthesis, labeling, and hybridizations to Affymetrix GeneChip Yeast Genome 2.0 Arrays were performed according to standard Affymetrix protocols at the microarray core facility at the Department of Biosciences, Karolinska Institutet, Novum, Huddinge, Sweden. For quantification of the transcripts, cDNA was made using oligo d(T) and the SuperScriptIII First Strand Synthesis SuperMix (Invitrogen). cDNA was used for real-time PCR using qPCR MasterMix Plus for SYBR Green I (Eurogentec) on an Applied Biosystems 7900HT Fast Real-Time PCR system.
ChIP-chip
Chromatin immunoprecipitation experiments using tiling microarrays (ChIP-chip experiments) were performed in biological triplicates (Robyr and Grunstein 2003). Nucleosome ChIPs were carried out using an antiserum against the histone H3 C terminus (Ab1791; Abcam). To 100 μL of crude lysate, either 2 μL of histone H3 antiserum, 1 μL of histone H3AcK9 antiserum (Upstate), or 1 μL of histone H4AcK5 antiserum (Upstate) was added. Amplification of DNA was done according to Robyr and Grunstein (2003) with the following modifications: an extra round of amplification was carried out in the first stage of amplification, and in the second stage of amplification, 12.5 mM dTTP was replaced by 12.5 mM dUTP. Shearing of DNA at dUTP sites, labeling of DNA, and hybridization to a GeneChip S. pombe Tiling 1.0FR Array were performed according to standard Affymetrix protocols at the microarray core facility at the Department of Biosciences, Karolinska Institutet, Novum, Huddinge, Sweden. Real-time PCR quantification of the ChIP:ed DNA was made using the qPCR MasterMix Plus for SYBR Green I kit (Eurogentec) on an Applied Biosystems 7900HT Fast Real-Time PCR system.
Data analysis
Gene expression microarray data were processed using the Affy package (Gautier et al. 2004) from Bioconductor (Gentleman et al. 2004). The GCRMA normalization pipeline (Wu and Irizarry 2004) was used. Genes were tested for differential expression with a moderated t-statistic, with FDR correction to compensate for multiple hypothesis testing (Benjamini and Hochberg 1995; Smyth 2004). A gene was considered to be differentially expressed if its expression differed at least twofold between high and low nitrogen conditions, and if the FDR corrected P-value was below 0.05. The expression data from this study were clustered together with data from Mata et al. (2002) and Chen et al. (2003). Hierarchical clustering was carried out on both the genes and the conditions using a Euclidean distance measure (Ward 1963).
Tiling array data for H3, H3AcK9, and H4AcK5 were first normalized against the amount of input DNA using the Affymetrix TAS software with the bandwidth set to 250 bp, producing signals on a log2 scale. In all the analyses the maps of H3AcK9 and H4AcK5 were corrected for nucleosome loss by subtracting the (log2) signal for H3Cterm.
To identify regions where the H3, H3AcK9, and H4AcK5 signals significantly differed between high and low nitrogen conditions, a procedure adapted from Rada-Iglesias et al. (2008) was used. First, an empirical Bayes algorithm (Smyth 2004) was applied to the combined data, resulting in a B-value for each array probe. Contiguous regions with a significant difference were then defined as windows containing at least 10 probes where the log2-ratio for each probe differed at least 3 SD from the mean log2-ratio and the B-value for each probe was at least 5 SD above the mean B-value. The maximum allowed gap between two probes passing the above criteria was set to 100 bp, and the distance between two different regions was required to be at least 400 bp. Each such contiguous region was functionally annotated with all genes found within 2000 bp of the region.
Promoter regions of all protein-coding genes in the genome were clustered on the differences in the H3, H3AcK9, and H4AcK5 signals before and after nitrogen starvation. Each gene was represented by the log2-ratios in a region starting 500 bp upstream of and ending 500 bp downstream from the start codon. Prior to clustering, the signals were smoothened by applying a sliding window of 25 bp. The genes were clustered by hierarchical clustering (Ward 1963) using a Euclidean distance measure.
The AMIGO program (Carbon et al. 2009) was used to test for enrichment of Gene Ontology annotations. Annotations were considered as significantly enriched if they were associated with at least three genes and had a P-value of at most 1 × 10−3.
De novo motif finding was carried out using Bioprospector (Liu et al. 2001). Bioprospector was run with 100 restarts to search for motifs of lengths 10 and 15 bp, using a bootstrap procedure with 50 randomizations to estimate the significance. Enrichment of predefined motifs within a given set of sequences was assessed using Fisher's exact test. Differential expression of groups of genes was tested for significance using Wilcoxon's rank-sum test. Differences between groups of genes (e.g., between up-regulated genes and other genes) in histone occupancy or histone acetylation levels (i.e., in Fig. 2) were tested for significance by comparing the corresponding log-ratios using Wilcoxon's rank-sum test.
Acknowledgments
Paul Russel, Shiv Grewal, Susan Forsburg, Kunihiro Ohta, and Karl Ekwall are thanked for sending us S. pombe strains. Adam Ameur, Robin Andersson, and Stefan Enroth from LCB are thanked for help with the data analysis. P.B. acknowledges grants 2004-3286 from the Swedish Research Council (VR) and CTS0841 from Carl Tryggvers Foundation. J.K. was supported by grants from the Swedish Foundation for Strategic Research and the Knut and Alice Wallenberg Foundation. H.R. was supported by grants from the Swedish Research Council (VR) and the Swedish Foundation for Strategic Research.
Footnotes
[Supplemental material is available online at http://www.genome.org. The microarray data from this study have been submitted to ArrayExpress (http://www.ebi.ac.uk/microarray-as/ae/) under accession nos. E-TABM-784 and E-TABM-785.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.098558.109.
References
- Alfredsson-Timmins J, Kristell C, Henningson F, Lyckman S, Bjerling P. Reorganization of chromatin is an early response to nitrogen starvation in Schizosaccharomyces pombe. Chromosoma. 2009;118:99–112. doi: 10.1007/s00412-008-0180-6. [DOI] [PubMed] [Google Scholar]
- Asp E, Nilsson D, Sunnerhagen P. Fission yeast mitogen-activated protein kinase Sty1 interacts with translation factors. Eukaryot Cell. 2008;7:328–338. doi: 10.1128/EC.00358-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57:289–300. [Google Scholar]
- Carbon S, Ireland A, Mungall CJ, Shu S, Marshall B, Lewis S. AmiGO: Online access to ontology and annotation data. Bioinformatics. 2009;25:288–289. doi: 10.1093/bioinformatics/btn615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Toone WM, Mata J, Lyne R, Burns G, Kivinen K, Brazma A, Jones N, Bahler J. Global transcriptional responses of fission yeast to environmental stress. Mol Biol Cell. 2003;14:214–229. doi: 10.1091/mbc.E02-08-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Wilkinson CR, Watt S, Penkett CJ, Toone WM, Jones N, Bahler J. Multiple pathways differentially regulate global oxidative stress responses in fission yeast. Mol Biol Cell. 2008;19:308–317. doi: 10.1091/mbc.E07-08-0735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E, van Steensel B. Chromatin domains in higher eukaryotes: Insights from genome-wide mapping studies. Chromosoma. 2009;118:25–36. doi: 10.1007/s00412-008-0186-0. [DOI] [PubMed] [Google Scholar]
- Gasch AP, Moses AM, Chiang DY, Fraser HB, Berardini M, Eisen MB. Conservation and evolution of cis-regulatory systems in ascomycete fungi. PLoS Biol. 2004;2:e398. doi: 10.1371/journal.pbio.0020398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier L, Cope L, Bolstad BM, Irizarry RA. affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez EB, Nugent RL, Laria S, Forsburg SL. Schizosaccharomyces pombe histone acetyltransferase Mst1 (KAT5) is an essential protein required for damage response and chromosome segregation. Genetics. 2008;179:757–771. doi: 10.1534/genetics.107.085779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govind CK, Zhang F, Qiu H, Hofmeyer K, Hinnebusch AG. Gcn5 promotes acetylation, eviction, and methylation of nucleosomes in transcribed coding regions. Mol Cell. 2007;25:31–42. doi: 10.1016/j.molcel.2006.11.020. [DOI] [PubMed] [Google Scholar]
- Higuchi T, Watanabe Y, Yamamoto M. Protein kinase A regulates sexual development and gluconeogenesis through phosphorylation of the Zn finger transcriptional activator Rst2p in fission yeast. Mol Cell Biol. 2002;22:1–11. doi: 10.1128/MCB.22.1.1-11.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii K, Ogiyama Y, Chikashige Y, Soejima S, Masuda F, Kakuma T, Hiraoka Y, Takahashi K. Heterochromatin integrity affects chromosome reorganization after centromere dysfunction. Science. 2008;321:1088–1091. doi: 10.1126/science.1158699. [DOI] [PubMed] [Google Scholar]
- Jiang C, Pugh BF. Nucleosome positioning and gene regulation: Advances through genomics. Nat Rev Genet. 2009;10:161–172. doi: 10.1038/nrg2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M, Davis RW. Sequences that regulate the divergent GAL1–GAL10 promoter in Saccharomyces cerevisiae. Mol Cell Biol. 1984;4:1440–1448. doi: 10.1128/mcb.4.8.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JM, Sonnhammer EL. Genomic gene clustering analysis of pathways in eukaryotes. Genome Res. 2003;13:875–882. doi: 10.1101/gr.737703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CK, Shibata Y, Rao B, Strahl BD, Lieb JD. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat Genet. 2004;36:900–905. doi: 10.1038/ng1400. [DOI] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Liu X, Brutlag DL, Liu JS. BioProspector: Discovering conserved DNA motifs in upstream regulatory regions of co-expressed genes. Pac Symp Biocomput. 2001;2001:127–138. [PubMed] [Google Scholar]
- Mas G, de Nadal E, Dechant R, de la Concepcion ML, Logie C, Jimeno-Gonzalez S, Chavez S, Ammerer G, Posas F. Recruitment of a chromatin remodelling complex by the Hog1 MAP kinase to stress genes. EMBO J. 2009;28:326–336. doi: 10.1038/emboj.2008.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata J, Bahler J. Global roles of Ste11p, cell type, and pheromone in the control of gene expression during early sexual differentiation in fission yeast. Proc Natl Acad Sci. 2006;103:15517–15522. doi: 10.1073/pnas.0603403103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata J, Lyne R, Burns G, Bahler J. The transcriptional program of meiosis and sporulation in fission yeast. Nat Genet. 2002;32:143–147. doi: 10.1038/ng951. [DOI] [PubMed] [Google Scholar]
- Mata J, Wilbrey A, Bahler J. Transcriptional regulatory network for sexual differentiation in fission yeast. Genome Biol. 2007;8:R217. doi: 10.1186/gb-2007-8-10-r217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehlin JO, Carlberg M, Ronne H. Control of yeast GAL genes by MIG1 repressor: A transcriptional cascade in the glucose response. EMBO J. 1991;10:3373–3377. doi: 10.1002/j.1460-2075.1991.tb04901.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Ameur A, Kapranov P, Enroth S, Komorowski J, Gingeras TR, Wadelius C. Whole-genome maps of USF1 and USF2 binding and histone H3 acetylation reveal new aspects of promoter structure and candidate genes for common human disorders. Genome Res. 2008;18:380–392. doi: 10.1101/gr.6880908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robyr D, Grunstein M. Genomewide histone acetylation microarrays. Methods. 2003;31:83–89. doi: 10.1016/s1046-2023(03)00091-4. [DOI] [PubMed] [Google Scholar]
- Schwabish MA, Struhl K. Evidence for eviction and rapid deposition of histones upon transcriptional elongation by RNA polymerase II. Mol Cell Biol. 2004;24:10111–10117. doi: 10.1128/MCB.24.23.10111-10117.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E, Widom J. Poly(dA:dT) tracts: Major determinants of nucleosome organization. Curr Opin Struct Biol. 2009;19:65–71. doi: 10.1016/j.sbi.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiozaki K, Russell P. Conjugation, meiosis, and the osmotic stress response are regulated by Spc1 kinase through Atf1 transcription factor in fission yeast. Genes & Dev. 1996;10:2276–2288. doi: 10.1101/gad.10.18.2276. [DOI] [PubMed] [Google Scholar]
- Shivaswamy S, Bhinge A, Zhao Y, Jones S, Hirst M, Iyer VR. Dynamic remodeling of individual nucleosomes across a eukaryotic genome in response to transcriptional perturbation. PLoS Biol. 2008;6:e65. doi: 10.1371/journal.pbio.0060065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha I, Wiren M, Ekwall K. Genome-wide patterns of histone modifications in fission yeast. Chromosome Res. 2006;14:95–105. doi: 10.1007/s10577-005-1023-4. [DOI] [PubMed] [Google Scholar]
- Smith B, Fang H, Pan Y, Walker PR, Famili AF, Sikorska M. Evolution of motif variants and positional bias of the cyclic-AMP response element. BMC Evol Biol. 2007;7:S15. doi: 10.1186/1471-2148-7-S1-S15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article 3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- Sugimoto A, Iino Y, Maeda T, Watanabe Y, Yamamoto M. Schizosaccharomyces pombe ste11+ encodes a transcription factor with an HMG motif that is a critical regulator of sexual development. Genes & Dev. 1991;5:1990–1999. doi: 10.1101/gad.5.11.1990. [DOI] [PubMed] [Google Scholar]
- Takeda T, Toda T, Kominami K, Kohnosu A, Yanagida M, Jones N. Schizosaccharomyces pombe atf1+ encodes a transcription factor required for sexual development and entry into stationary phase. EMBO J. 1995;14:6193–6208. doi: 10.1002/j.1460-2075.1995.tb00310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thon G, Bjerling KP, Nielsen IS. Localization and properties of a silencing element near the mat3-M mating-type cassette of Schizosaccharomyces pombe. Genetics. 1999;151:945–963. doi: 10.1093/genetics/151.3.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walfridsson J, Khorosjutina O, Matikainen P, Gustafsson CM, Ekwall K. A genome-wide role for CHD remodelling factors and Nap1 in nucleosome disassembly. EMBO J. 2007;26:2868–2879. doi: 10.1038/sj.emboj.7601728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward JH. Hierachical grouping to optimize an objective function. J Am Stat Assoc. 1963;58:236–244. [Google Scholar]
- Watt S, Mata J, Lopez-Maury L, Marguerat S, Burns G, Bahler J. urg1: A uracil-regulatable promoter system for fission yeast with short induction and repression times. PLoS One. 2008;3:e1428. doi: 10.1371/journal.pone.0001428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiren M, Silverstein RA, Sinha I, Walfridsson J, Lee HM, Laurenson P, Pillus L, Robyr D, Grunstein M, Ekwall K. Genomewide analysis of nucleosome density histone acetylation and HDAC function in fission yeast. EMBO J. 2005;24:2906–2918. doi: 10.1038/sj.emboj.7600758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Irizarry RA. Preprocessing of oligonucleotide array data. Nat Biotechnol. 2004;22:656–658. doi: 10.1038/nbt0604-656b. [DOI] [PubMed] [Google Scholar]
- Xue-Franzen Y, Kjaerulff S, Holmberg C, Wright A, Nielsen O. Genomewide identification of pheromone-targeted transcription in fission yeast. BMC Genomics. 2006;7:303. doi: 10.1186/1471-2164-7-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Hirota K, Mizuno K, Shibata T, Ohta K. Essential roles of Snf21, a Swi2/Snf2 family chromatin remodeler, in fission yeast mitosis. Genes Genet Syst. 2008;83:361–372. doi: 10.1266/ggs.83.361. [DOI] [PubMed] [Google Scholar]
- Zawadzki KA, Morozov AV, Broach JR. Chromatin-dependent transcription factor accessibility rather than nucleosome remodeling predominates during global transcriptional restructuring in Saccharomyces cerevisiae. Mol Biol Cell. 2009;20:3503–3513. doi: 10.1091/mbc.E09-02-0111. [DOI] [PMC free article] [PubMed] [Google Scholar]