Abstract
ATRX (alpha thalassemia/mental retardation syndrome X-linked) belongs to the SWI2/SNF2 family of chromatin remodeling proteins. Besides the ATPase/helicase domain at its C terminus, it contains a PHD-like zinc finger at the N terminus. Mutations in the ATRX gene are associated with X-linked mental retardation (XLMR) often accompanied by alpha thalassemia (ATRX syndrome). Although ATRX has been postulated to be a transcriptional regulator, its precise roles remain undefined. We demonstrate ATRX localization at the telomeres in interphase mouse embryonic stem (ES) cells in synchrony with the incorporation of H3.3 during telomere replication at S phase. Moreover, we found that chromobox homolog 5 (CBX5) (also known as heterochromatin protein 1 alpha, or HP1 alpha) is also present at the telomeres in ES cells. We show by coimmunoprecipitation that this localization is dependent on the association of ATRX with histone H3.3, and that mutating the K4 residue of H3.3 significantly diminishes ATRX and H3.3 interaction. RNAi-knockdown of ATRX induces a telomere-dysfunction phenotype and significantly reduces CBX5 enrichment at the telomeres. These findings suggest a novel function of ATRX, working in conjunction with H3.3 and CBX5, as a key regulator of ES-cell telomere chromatin.
The ATRX (alpha thalassemia/mental retardation syndrome X-linked) gene (Picketts et al. 1996) is located on the X chromosome (Xq13). It is a large gene spanning ∼300 kb and contains 36 exons. It encodes at least two alternatively spliced, 10.5-kb mRNA transcripts that differ at their 5′ ends and are predicted to give rise to slightly different proteins of 280 and 265 kDa. It is a member of the SNF2 family of helicase/ATPases that contribute to the remodeling of nucleosome structure (Argentaro et al. 2007). In the C-terminal half of the protein is the helicase/adenosine triphosphatase (ATPase) domain comprising seven highly conserved helicase/ATPase motifs. At the N terminus is the ATRX-DNMT3-DNMT3L (ADD) domain (Argentaro et al. 2007) that is a plant homeodomain (PHD)–like zinc finger with an additional C2-C2 motif. Other proteins that share this motif are DNMT3A, DNMT3B, and DNMT3L, three proteins involved in DNA methylation. PHD fingers are common features of chromatin-related proteins (Nagamine et al. 1997; Lu et al. 1998; Bienz 2006) and function to help tether proteins to chromatin and have a preference for binding the methylated lysine residue of H3. There is also evidence that the ADD domain of ATRX plays a role in DNA binding (Cardoso et al. 2000). In human and mouse cells, ATRX is highly enriched at pericentric heterochromatin and is associated with chromobox homolog 5 (CBX5) (also known as HP1 alpha [heterochromatin protein 1 alpha]), DAXX, MECP2, and promyelocytic leukemia (PML) nuclear bodies (McDowell et al. 1999; Xue et al. 2003; Ishov et al. 2004; Kourmouli et al. 2005; Nan et al. 2007).
The association of ATRX mutations with a reduction in alpha globin synthesis in alpha thalassemia patients suggests that the protein plays a role in the regulation of alpha globin gene expression. However, the pleiotropic ATRX syndrome comprising multiple congenital abnormalities, including profound developmental delay, facial dysmorphism, and genital abnormalities, suggests that ATRX is involved in the regulation of other yet unidentified genes. The complex function of ATRX in the developmental process is also apparent in ATRX-null mice. ATRX-null male mice are not viable, and embryos die around 9.5 days post-coitum (dpc) (Berube et al. 2002, 2005; Garrick et al. 2006). Also, conditional ablation of full-length ATRX in the mouse forebrain results in decreased cortical size (Guerrini et al. 2000), consistent with mental retardation in patients carrying ATRX mutations. It is also interesting that loss of ATRX in ES cells leads to reduced cell-growth and a higher rate of spontaneous differentiation (Garrick et al. 2006), suggesting that ATRX plays a role in controlling embryonic stem (ES) cell proliferation and differentiation, in addition to its function for sister chromatid cohesion and chromosome congression during mitosis (Ritchie et al. 2008).
In mammalian cells, telomeric chromatin contains epigenetic markers characteristic of silenced chromatin such as those found at pericentric heterochromatin (Garcia-Cao et al. 2004; Gonzalo et al. 2005, 2006). Knockout deletion of the histone and DNA methyltransferases (SUV39H1/2, SUV420H1/2, DNMT3A/B, and DNMT1) in the mouse results in defective telomere function, aberrantly increased telomere length, and chromosomal instability, suggesting that these repressive markers are essential for telomere length maintenance and structural integrity.
H3.3 is a universal histone predominantly incorporated into transcription sites and associated with active and open chromatin (Loyola et al. 2006). H3.3 phosphorylated at serine 31 (H3.3S31P) is a mitosis-specific marker enriched in the pericentric heterochromatin (Hake et al. 2005). Recently, we have shown the enrichment of H3.3S31P at metaphase telomeres in mouse ES cells and embryonic germ (EG) cells, but not in nonpluripotent cells or differentiated ES cells (Wong et al. 2009). In addition, we have shown localization of H3.3 at interphase telomeres in mouse ES cells. In ES cells undergoing differentiation, H3.3 level at the telomeres decreases, accompanied by increased association of heterochromatin repressive markers H4K20me3 and H3K9me3. Our findings in ES cells challenge the long-standing notion that the telomere chromatin is “silenced,” and are the first to show the existence of a unique and functionally essential telomere chromatin in ES cells that undergoes dynamic differentiation-dependent remodeling.
In this study, we demonstrate that ATRX localizes to mouse ES cell telomeres during interphase. The association of ATRX with the telomeres is synchronized with the incorporation of H3.3 at the telomeres during telomere-replication at S phase. We show a direct interaction between ATRX and H3.3 by chromatin immunoprecipitation analysis and site-directed mutagenesis of H3.3. RNAi-mediated inhibition of ATRX in mouse ES cells induces an increased telomere-dysfunction phenotype and a decrease in CBX5 binding at the telomeres. We propose that ATRX, through interaction with H3.3 and CBX5, plays a novel function as a key regulator of telomere chromatin in ES cells.
Results
ATRX localizes at telomeres in interphase mouse ES and EG cells
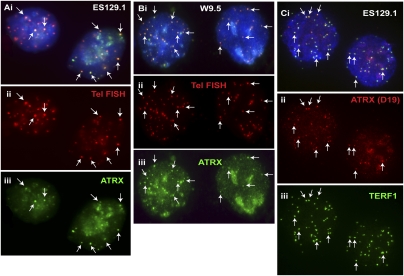
Previous studies have shown that ATRX localizes at pericentric heterochromatin and PML nuclear bodies through interaction with the chromoshadow domain (CSD) of CBX5 and MECP2 and recruitment by DAXX (McDowell et al. 1999; Xue et al. 2003; Ishov et al. 2004; Kourmouli et al. 2005; Nan et al. 2007). Here, we show that ATRX, in addition to its localization at the pericentric heterochromatin (Supplemental Fig. S1A,B), is prominently present at the telomeres in pluripotent mouse ES cell lines ES129.1 (Fig. 1A) and ES-W9.5 (Fig. 1B) during interphase. Specificity of the antiserum was confirmed by competitive immunofluorescence analysis with ATRX peptide (Supplemental Fig. S2A–D). Telomeric localization of ATRX was confirmed by FISH analysis using a telomere-specific TTAGGG probe. As ATRX associates with pericentric heterochromatin and mouse chromosomes are telocentric, a triple staining of telomeres, centromeres (with human CREST antiserum), and ATRX was performed in mouse ES cells to validate ATRX association at the telomeres (Fig. 2A). Finally, immunofluorescence analysis using a different source of anti-ATRX antibody gave the same staining pattern at the telomeres and pericentric heterochromatin in ES cells (Fig. 1C; Supplemental Fig. S1C).
Figure 1.
Enrichment of ATRX at telomeres in mouse ES cell lines, ES129.1 and W9.5. Two sources of antibodies against ATRX (H300 and D19) were used in immunofluorescence analysis. (A,B) In interphase ES129.1 and ES-W9.5 cells, ATRX (Aiii,Biii, green; H300) was found at the telomeres (arrows) and pericentric heterochromatin (for ATRX localization at pericentric heterochromatin, as indicated by centromeric CREST staining, see Supplemental Fig. S1). Telomeric localization of ATRX (Aiii,Biii, green) was confirmed by telomere FISH (Aii,Bii, red). (C) Immunofluorescence analysis using a second source of antibody also showed ATRX (ii, red; D19) enrichment at the telomeres (arrows) in interphase ES129.1 cells, as evident from its colocalization with TERF1 (iii, green).
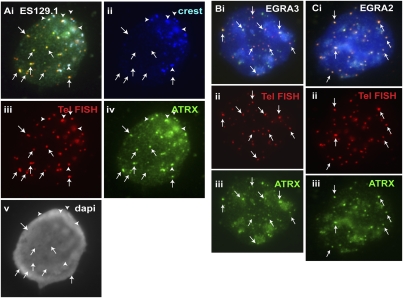
Figure 2.
Cellular distribution of ATRX in mouse ES cell lines ES129.1 and W9.5 and in mouse EG cell lines EGRA2 and EGRA3. (A) In interphase ES129.1 cells, a triple-staining immunofluorescence analysis showed clear colocalization of ATRX (iv, green) with telomere FISH signals (iii, red), without any CREST signal (arrows). ATRX localization at pericentric heterochromatin was indicated by its close proximity to the centromeric CREST staining (ii, blue; arrowheads). (B,C) In interphase EGRA3 and EGRA2 cells, ATRX (Biii,Ciii, green) was similarly enriched at the telomeres (arrows). Telomeric localization of ATRX (Biii,Ciii, green) was confirmed by telomere FISH (Bii,Cii, red).
In somatic mouse and human cells, including mouse NIH3T3 and L cells and human HT1080 and HeLa cells, ATRX mainly accumulated at the pericentric heterochromatin but not at the telomeres (data not shown), suggesting that ATRX localization at ES cell telomeres may potentially be linked to the pluripotent state of the ES cells. To further investigate whether ATRX incorporation at the telomere was associated with cellular pluripotency, we included EG cells in our analysis. We observed distinct ATRX localization at the telomeres in two different mouse EG cell lines, EGRA3 (Fig. 2B) and EGRA2 (Fig. 2C).
ATRX localizes at telomeres during S phase of the cell cycle in ES cells
In our previous study, we have shown that MYC-H3.3 (H3.3 protein with a MYC epitope tag) is incorporated into the telomere chromatin in ES cells during the time of telomere replication and processing (Wong et al. 2009). For the preliminary determination of the timing of ATRX localization at the telomeres, combined 5-bromo-2′-deoxyuridine (BrdU) staining and immunofluorescence analysis was performed on ES129.1 cells (Supplemental Fig. S3A–C). In BrdU-negative, non-S-phase cells, ATRX staining was enhanced at the pericentric heterochromatin (indicated by stronger DAPI staining) but not noticeably at the telomeres, as indicated by the lack of punctate spots that typify telomeric staining. However, in BrdU-positive cells, particularly during mid-to-late S phase, punctate signals indicative of telomeric ATRX staining were significantly increased, suggestive of the binding of ATRX at the telomeres during the period of DNA replication.
In order to clearly determine the timing of ATRX recruitment at the telomeres, we performed immunofluorescence analysis on synchronized ES129.1 cells (arrested at G1/S using a thymidine-block protocol) (Fig. 3). Following the release of cells from G1/S thymidine-induced block, cell-cycle progression was determined by fluorescence-activated cell sorting (FACS) analysis (Supplemental Fig. S3D), which indicated that ES129.1 cells released from G1/S-thymidine block progressed through S phase at 4–6 h post-release, entered M phase at 8 h, and re-entered G1 phase at 10 h. At 0 h, less than 20% of cells showed positive telomeric localization of ATRX. After the initial 2 h post-G1/S release, ATRX level at telomeres became apparent (Fig. 3A; Supplemental Table S1). ATRX recruitment predominantly occurred at mid-to-late S phase, with 74.29% and 82.86% of cells showing five or more positive ATRX binding at the telomeres per cell after 4–6 h of release (Fig. 3B,C; Supplemental Table S1), respectively, in synchrony with the time of telomere replication and processing (Verdun and Karlseder 2006, 2007). At mitosis (after 8 h of release), ATRX dissociated from the telomeres but remained associated with the pericentric heterochromatin (Fig. 3D,E). Similar results were obtained when combined cross-linked chromatin immunoprecipitation (ChIP), and dot-blot assay was used to assess the association of ATRX, MYC-H3.3, TERF1 (telomere binding factor), and histone H4 at the telomeres in these synchronized ES cell population (Supplemental Fig. S4). As previously described (Wong et al. 2009), MYC-H3.3 level at the telomeres increased significantly during S phase and remained prominent as cells entered mitosis. ChIP/dot blot analysis using antibodies against ATRX also showed a clear association of ATRX at the telomeres during S phase, but ATRX level at the telomeres reduced significantly during mitosis. Both TERF1 and histone H4 levels at the telomeres remained high throughout the entire cell cycle, in particular after the entry of cells into S phase. The increases in TERF1 and H4 levels could be attributed to an increase in the activity of telomere replication as cells progressed through S phase. ChIP using rabbit IgG antibody did not result in any significant enrichment of telomeric DNA. Together, these results provided support that ATRX association at the telomeres is coupled with the timing of telomere replication and chromatin assembly in S phase of the cell cycle.
Figure 3.
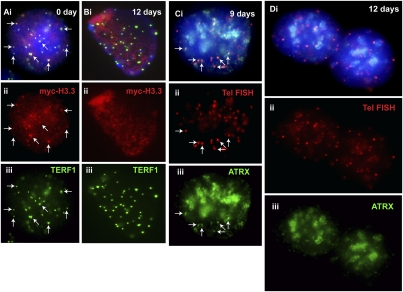
Timing of telomeric loading of ATRX. We previously showed that MYC-H3.3 was loaded at the telomeres in interphase ES129.1 cells during S phase (Wong et al. 2009). Here, ES129.1 cells were similarly synchronized using a thymidine-induced G1/S-block protocol (for FACS analysis, see Supplemental Fig. S3). (A–C) ATRX (iii, green) was present at the telomeres in cells from 2–6 h post-release from G1/S block, as indicated by colocalization with telomere FISH signals (ii, red). (D) By mitosis (8 h post-release), ATRX (iii, green) completely delocalized from the telomeres (ii, red; indicated by telomere FISH signals) but remained at pericentric heterochromatin (arrowheads). (E) Enlarged images of the boxed areas shown in D, showing no ATRX signals at the telomeres (arrows) but strong localization of ATRX (iii, green) at the pericentric regions (arrowheads); note that the telocentric nature of the mouse chromosomes meant an inevitable display of the observed colocalization of telomeric FISH signals with the centromeres at this resolution. At 10 h post-release, the cells re-entered G1 phase of the cell cycle (data not shown).
Cellular differentiation of ES cells leads to decreased ATRX signals at the telomeres
In order to examine if the localization of ATRX at the telomeres was affected by cellular differentiation, we induced differentiation of ES129.1 cells by treatment of leukemia inhibitory factor (LIF) withdrawal and retinoid acid (RA) supplement as previously described (Wong et al. 2009). Prior to differentiation, >90% of these cells expressed the stem-cell surface marker POU5F1 (also known as OCT4). After 3 d of differentiation, less than 50% expressed POU5F1. By 6 d, POU5F1 was barely detectable (data not shown).
We demonstrated the presence of MYC-H3.3 at telomeres in undifferentiated interphase ES 129.1 cells as previously described (Fig. 4A; Wong et al. 2009). MYC-H3.3 signals remained present at the telomeres after 6 and 9 d of induction (data not shown). However, after 12 d, MYC-H3.3 signals at the telomeres were barely detectable (Fig. 4B). In addition, the induction of differentiation of mouse ES cells also led to a significant increase in H4K20me3 levels at the telomeres, as previously observed (Supplemental Fig. S5; Wong et al. 2009).
Figure 4.
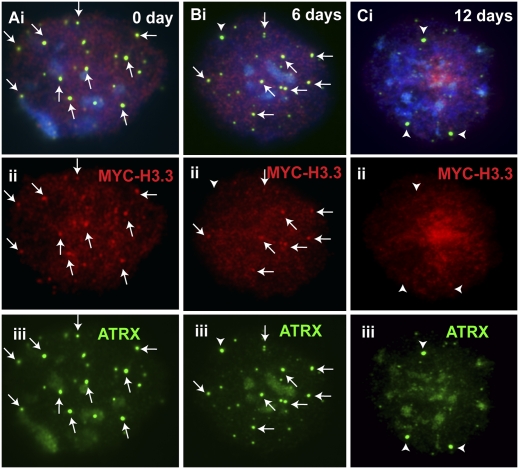
Cellular localization of MYC-H3.3 and ATRX in differentiated ES cells. (A,B) MYC-H3.3 construct was transfected into ES129.1 cells and induced with 1 μM doxycycline for 24 h MYC-H3.3 (Aii, red) colocalized with TERF1 (Aiii, green) at the telomeres (examples shown by the arrows) in undifferentiated (0 day) interphase ES 129.1 cells. MYC-H3.3 and ATRX signals remained apparent at the telomeres after 6 and 9 d of induction (data not shown). However, no enrichment of MYC-H3.3 (Bii, red) was detected at the telomeres (indicated by TERF1 staining; Biii, green) 12 d following the induction of differentiation. (C,D) Likewise, ATRX association at the telomeres was also not noticeably affected by the induction of differentiation for 6 and 9 d (Ciii, green), but ATRX (Diii, green) completely delocalized from the telomeres after 12 d of induction of cellular differentiation.
Like MYC-H3.3, ATRX association at the telomeres was not noticeably affected by the induction of cellular differentiation for 6 and 9 d (Fig 4C; although there was a slight decrease in ATRX telomeric signals after 9 d of differentiation), but it was greatly reduced after 12 d of induction (Fig. 4D). The combined immunofluorescence and quantitative assay confirmed that ATRX staining at the telomeres remained prominent in ES129.1 cells after 6 and 9 d of induction of cellular differentiation (Supplemental Table S2). Telomeric localization of ATRX in these cells was only significantly reduced after 12 d of induction of differentiation, with only about 17.14% of cells showing five or more positive ATRX staining per cell (Supplemental Table S2). It is important to note that the loss of ATRX signals at the telomeres in differentiated ES129.1 cells (12 d post induction of differentiation) was not contributed by a decrease in the number of cells in S phase because these cells were clearly undergoing DNA replication, as evident from the positive BrdU staining (Supplemental Fig. S6).
ATRX colocalizes with H3.3 at the telomeres in ES cells
Both the reported endogenous full-length and truncated ATRX (different proteins of 280 and 265 kDa) are highly conserved between mouse and human. Although the effects of deleting the helicase domain of the full-length ATRX have been reported (Garrick et al. 2006), little is known of the functions of the ADD domain and the truncated ATRX. PHD fingers are a common feature of chromatin-related proteins and are highly specialized methyl-lysine binding domains with a marked preference for H3K4 (1ARTK4). H3.3 has the same protein sequence as H3 (except for five amino acids at S31, A87, I89, G90, and S96), including its K4 residue being a target site for histone methylation (Loyola et al. 2006).
Given the common association of PHD domain with lysine residue on histone H3, we hypothesized that the localization of ATRX at the telomeres was dependent on H3.3 loading at the telomeres. The localization of ATRX and MYC-H3.3 at the telomeres in interphase ES129.1 cells lent further support for this possibility. To investigate such a possibility, we performed immunofluorescence analysis on ES129.1-MYC-H3.3 cells (Fig. 5). In undifferentiated ES129.1 cells, ATRX was clearly colocalized with MYC-H3.3 in ES129.1 cells (Fig. 5A). The colocalization between ATRX and MYC-H3.3 remained prominent after 6 d (Fig. 5B) and 9 d (data not shown) of induction. After 12 d, the costaining of ATRX and MYC-H3.3 was barely detectable (Fig. 5C). These results suggest that ATRX recruitment at ES cell telomeres was likely to be mediated via interaction with H3.3 up to 12 d of induction of cellular differentiation.
Figure 5.
Colocalization of ATRX and MYC-H3.3. (A) ATRX (iii, green) clearly colocalized with MYC-H3.3 (ii, red) in undifferentiated (0 day) interphase ES129.1 cells. (B) The colocalization of ATRX (iii, green) with MYC-H3.3 (ii, red) remained prominent after 6 d of differentiation. (C) However, after 12 d of differentiation, the colocalization of ATRX (iii, green) with MYC-H3.3 (ii, red) was greatly reduced. Occasionally, strong ATRX signals were detected in the 6- and 12-d cells, but these signals were not colocalized with the telomeres (Biii,Ciii, green; arrowheads).
As we previously described, H3.3S31P staining at the telomeres in ES cells diminished 3 to 6 d (instead of after 12 d) following the induction of differentiation (Wong et al. 2009). The reason for the faster rate of H3.3S31P dissociation than of MYC-H3.3 and ATRX (Figs. 4, 5) from the telomeres following the induction of differentiation is unknown. Nevertheless, our study shows that the more rapid loss in the level of H3.3S31P at the telomeres in metaphase cells (Wong et al. 2009) was not due to an overt loss of H3.3 association at the telomeres but likely to be the consequence of a reduction in the phosphorylation of H3.3, the regulatory mechanism of which remains the subject of future studies.
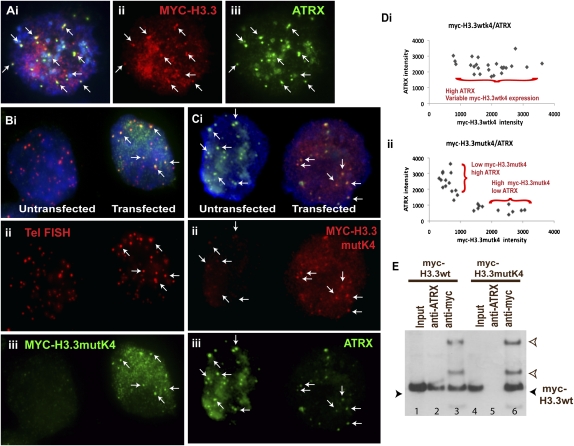
ATRX directly interacts with H3.3 through its K4 residue
Based on the known association of PHD domain with K4 residue on histone H3, we further hypothesized that the interaction between ATRX and H3.3 at the telomeres occurred through the direct association of ATRX with the K4 residue of H3.3. As a positive control, we showed clear localization of MYC-H3.3wtK4 (wild type) with ATRX in ES cells (Fig. 6A). To investigate how the loss of K4 in MYC-H3.3 might affect ATRX and H3.3 interaction, we generated a MYC-H3.3 construct carrying a mutation in K4. The mutation was introduced using PCR-based site-directed mutagenesis, followed by transient transfection into ES129.1 cells. The overexpression of MYC-H3.3mutK4 did not affect its localization to telomeres in ES129.1 (Fig. 6B), but it resulted in a significant reduction in the level of ATRX association with MYC-H3.3mutK4 at telomeres compared to nontransfected ES129.1 cells (Fig. 6C), with the extent of this association being inversely proportional to the expression level of MYC-H3.3mutK4 (Fig. 6D; Supplemental Table S3). As for the control cells, ATRX signal intensity remained prominent and was not affected by the expression levels of MYC-H3.3wtK4.
Figure 6.
The effects of H3.3 lysine-4 mutation on its interaction with ATRX by immunofluorescence and immunoprecipitation/Western blot analysis. (A) As a positive control, we showed a clear colocalization of MYC-H3.3 (ii, red) with ATRX (iii, green) in ES129.1 cells. (B,C) The mutation of K4 (lysine to alanine) residue was introduced by site-directed PCR mutagenesis. The MYC-H3.3mutK4 plasmid DNA was transiently transfected into mouse ES129.1 cells. The expression of MYC-H3.3mutK4 (Biii, green; indicated by arrows) in ES129.1 cells (cell on the right) did not affect its telomeric localization (as indicated by telomere FISH signals; Bii, red) but resulted in a significant reduction in the association of ATRX (Ciii, green) with MYC-H3.3 (Cii, red; indicated by arrows) at telomeres compared with cells not expressing MYC-H3.3mutK4 (cell on the left). (D) In ES129.1 cells expressing either MYC-H3.3wtK4 or MYC-H3.3mutK4, the signal intensities of MYC-H3.3wtK4/ATRX and MYC-H3.3mutK4/ATRX were quantitated. For each cell, the intensities for five sets of colocalizing MYC-H3.3wtK4/ATRX and MYC-H3.3mutK4/ATRX signals were measured, and the average intensities were determined. In total, 25 cells were assessed (Supplemental Table S3). As shown in the scattered plot, in ES cells expressing MYC-H3.3wtK4, ATRX signal intensity remained high regardless of MYC-H3.3wtK4 signal intensity, whereas, an inverse relationship between the expression level of MYC-H3.3mutK4 and ATRX signal intensity was observed, suggesting that MYC-H3.3mutK4 expression affects ATRX association. (E) Immunoprecipitation with anti-ATRX antiserum pulled down MYC-H3.3 in ES129.1 cells expressing MYC-H3.3 (lane 2) but failed to pull down MYC-H3.3mutK4 in ES129.1 cells expressing MYC-H3.3mutK4 (lane 5). As a positive control, immunoprecipitation with anti-MYC antiserum also pulled down MYC-H3.3 (lane 3) and MYC-H3.3mutK4 (lane 6). The two bands above MYC-H3.3 in lanes 3 and 6 are heavy and light chains of the anti-MYC antiserum (clear arrowheads).
To further determine the interaction between ATRX and H3.3, we transiently transfected ES129.1 cells with either MYC-H3.3 or MYC-H3.3mutK4 and performed immunoprecipitation and Western blot analysis using anti-ATRX and anti-MYC antisera (since anti-H3.3 antiserum is not available). As shown in Figure 6E, immunoprecipitation with anti-ATRX antiserum successfully “pulled down” MYC-H3.3 in ES129.1 cells expressing MYC-H3.3 but did not pull down MYC-H3.3mutK4 in ES129.1 cells expressing MYC-H3.3mutK4, providing further evidence for the interaction between the two proteins.
ATRX RNAi-knockdown affects telomere integrity in mouse ES cells
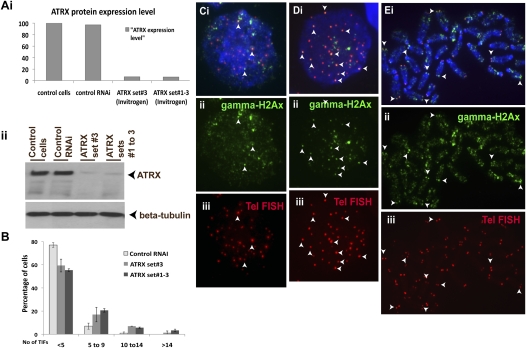
To study the normal functional role of ATRX at the telomeres in ES cells, we used two sources of RNAi-oligonucleotides system (Ambion and Invitrogen) to deplete ATRX (Fig. 7; Supplemental Fig. S7B). Initial experiments by real-time reverse transcription PCR assay confirmed the specificity and efficiency of three sets of oligonucleotides for the knockdown in mouse ES129.1 cells, with these siRNA oligonucleotide duplexes giving up to about 70%–80% depletion of ATRX expression (data not shown). Importantly, these siRNA duplexes also resulted in a robust reduction in ATRX at the protein level as shown in Western blot analysis (Fig. 7; Supplemental Fig. S7A). ATRX RNAi-depletion had no immediate effect on cell-growth during the initial 24 h, although it led to a slight reduction in cell-growth 48 h post RNAi-knockdown (data not shown; and in agreement with a similar observation previously reported [Garrick et al. 2006]). Changes in telomere length in mouse ES cells subjected to ATRX RNAi-knockdown for 48 h were determined using Southern blot analysis; however, no significant change was observed (Supplemental Fig. S7C).
Figure 7.
Effect of ATRX RNAi-knockdown on telomere integrity in mouse ES cells. (A) Western blot analysis of ES129.1 cells transfected with ATRX-specific RNAi oligonucleotide sets (set 3 and sets 1–3 from Invitrogen) using anti-ATRX and anti-beta-tubulin antisera. (i) Data are presented in histograms by normalization of the intensity of ATRX levels against the intensity of beta-tubulin levels. (ii) ATRX RNAi-depleted cells showed a significant reduction of ∼90% in ATRX protein level (arrows; lanes 3,4) 48 h after the transfection, compared with nontransfected cells (lane 1) and cells transfected with scrambled control RNAi oligonucleotides (lane 2). The equal loading of protein was achieved by normalization against the beta-tubulin level. (B) Induction of TIFs by ATRX-inhibition using ATRX RNAi oligonucleotide (set 3 and sets 1–3 from Invitrogen and set 2 from Ambion, respectively) for 48 h. Data are presented in histograms by subgrouping the cells according to the number of TIFs per cell (less than five 5 TIFs, five to nine TIFs, 10 to 14 TIFs, and more than 14 TIFs). A normal cell can contain one to two TIFs on average; thus, a threshold of four or more TIFs was used, as described in other studies (Hockemeyer et al. 2005). When transfected with ATRX-RNAi oligonucleotides, the number of cells with five or more TIFs (85 cells were counted for each experiment) by three- to fourfold (increased from 9.41% to as high as 29.8%–34.90%, with an average increase of 20.39%–25.49%). In this study, we only counted the number of TIFs in interphase ES129.1 cells although in some metaphase cells, TIFs were also present at the telomeres following RNAi-depletion of ATRX. We also performed RNAi-knockdown using ATRX-specific RNAi oligonucleotide sets purchased from Ambion, showing a significant reduction of ∼80% in ATRX protein level and a fivefold increase in the number of cells with five or more TIFs 48 h after the transfection (see Fig. S7). (C–E) Immunofluorescence analysis of ES129.1 cells subjected to 48-h knockdown with either control (C) or ATRX-specific (D,E) RNAi-duplex oligonuclotides using anti-gamma-H2AFX (Cii,Eii; green) antiserum. Increased number of TIFs was detected at telomeres (indicated by telomere FISH analysis; Diii,Eiii) in cells depleted of endogenous ATRX (arrowheads show some examples of TIFs).
We have previously shown that RNAi-depletion of H3.3 in mouse ES cells induced telomere-dysfunction phenotypes (Wong et al. 2009), as indicated by an increase in the number of cells with telomere induced dysfunctional foci (TIF; a marker for DNA damage at the telomeres identified by gamma-H2AFX [formerly known as gamma-H2AX] staining) (Hockemeyer et al. 2005). To assess the effects of ATRX-depletion on telomere structural integrity, we quantitated the level of TIFs in cells subjected to RNAi-depletion of ATRX (Fig. 7C–E, Supplemental Fig. S7B; Supplemental Table S4). In ES129.1 cells transfected with control RNAi oligonucleotides, only about 5.88%–9.41% of cells showed five or more TIFs per cell. Importantly, ATRX-specific RNAi-knockdown caused three- to fivefold (9.41% increased to 29.80%–34.90%, and 5.88% increased to 29.41%, depending on the source of RNAi-oligonucleotides system) increase in the population of cells showing five or more TIFs per cell (Supplemental Table S4). Although TIFs were mainly seen at telomeres in interphase cells, they were occasionally seen at the telomeres in metaphase cells (Fig. 7E). In contrast, ATRX RNAi-knockdown in mouse NIH3T3 cells did not lead to a significant increase in the number of cells containing five or more TIFs (data not shown). These results indicated a direct and specific contribution of ATRX to the functional integrity of telomere chromatin in ES cells.
ATRX RNAi-knockdown affects CBX5 binding at the telomeres in mouse ES cells
HP1 was first discovered in Drosophila melanogaster as a non-histone chromosomal protein associated with heterochromatin (James and Elgin 1986). It is phylogenetically conserved in many eukaryotes. Three mammalian HP1 proteins have been identified: HP1 alpha (CBX5), HP1 beta (CBX1), and HP1 gamma (CBX3). A variety of functions have been described for HP1, including heterochromatin organization, transcription regulation, DNA replication and repair, and chromatin remodeling (Maison and Almouzni 2004; Fanti and Pimpinelli 2008). In Drosophila, HP1 is a protein essential for telomere stability; mutations in HP1 cause severe telomere fusions and chromosome breakage. Although there is evidence in mammalian cells for HP1 involvement in telomere metabolism (Minc et al. 1999; Sharma et al. 2003), the function of HP1 in telomere homeostasis is less well defined.
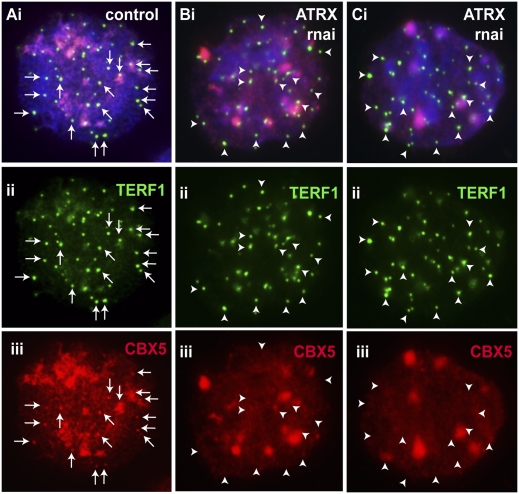
Interestingly, our immunofluorescence analysis showed prominent CBX5 colocalization with ATRX at the telomeres, in addition to enrichment at pericentric heterochromatin, in undifferentiated mouse ES cells (Fig. 8A). The signal intensity of CBX5 telomeric association in mouse ES cells was much higher compared with those in somatic human and mouse cells (for example, see Fig. 8D). Furthermore, as was seen for ATRX and MYC-H3.3, the induction of differentiation in ES cells by LIF withdrawal and RA treatment led to a decrease in the association of CBX5 with the telomeres (Fig. 8B,C), whereas CBX5 signal at the pericentric heterochromatin remained prominently high. Given the strong association of ATRX with CBX5, immunofluorescence analysis was performed in ES129.1 cells to examine the effect of ATRX-depletion on CBX5 recruitment at the telomeres. As shown in Figure 9, RNAi-mediated knockdown of ATRX caused an extreme reduction in the binding of CBX5 at the telomeres in ES cells, suggesting a direct function of ATRX as chromatin remodeler in recruiting CBX5 to the telomere chromatin in ES cells. It is also interesting to note that CBX5 signals remained high at the pericentric heterochromatin following ATRX-depletion. This is not surprising, given that CBX5 could be recruited by an alternative mechanism such as via binding to the H3K9me3 on pericentric heterochromatin.
Figure 8.
Colocalization of CBX5 with ATRX at the telomeres in ES cells. (A) A triple staining/immunofluorescence analysis showing colocalization of CBX5 (iv, blue) with ATRX (ii, green) at the pericentric heterochromatin (arrowheads) and telomeres (iii, red; indicated by telomere FISH). (B,C) CBX5 (Biii, red) colocalized with TERF1 (Bii, green) at the telomeres (examples shown by the arrows) in undifferentiated (0 day) interphase ES 129.1 cells. CBX5 signal remained apparent at the telomeres after 6 and 9 d of induction (data not shown). However, CBX5 localization (Ciii, red) at the telomeres (Cii, green) was greatly decreased 12 d following the induction of differentiation. (D) In somatic cells, e.g., NIH3T3, CBX5 (iii, red) localized mainly at pericentric heterochromation, but its association with the telomeres (arrowheads; ii, green) was below detection.
Figure 9.
Effect of ATRX RNAi-knockdown on telomeric association of CBX5 in mouse ES cells. (A–C) ES129.1 cells transfected with scramble RNAi oligonucleotide (A) and ATRX-specific RNAi oligonucleotide sets (B–C; set 3 and sets 1–3 [Invitrogen]) for a period of 48 h. Transfection of ES129.1 cells with scrambled RNAi oligonucleotide did not affect localization of CBX5 (Aiii, red) at the telomeres (arrows), as indicated by costaining with TERF1 (Aii, green). However, RNAi depletion of ATRX with RNAi oligonucleotide sets, set 3 (B) and sets 1–3 (C) led to a significant dissociation of CBX5 from the telomeres (arrowheads) in ES129.1 cells.
Discussion
The localization of ATRX to the pericentric heterochromatin (recruited by CBX5 and MECP2) (Kourmouli et al. 2005; Nan et al. 2007) and the PML nuclear bodies (recruited by DAXX) (Xue et al. 2003; Ishov et al. 2004) has been well documented. Here, we have demonstrated the localization of ATRX at the interphase telomeres in two different pluripotent cell types (ES and EG cells), in addition to its binding at the pericentric heterochromatin as previously reported (Kourmouli et al. 2005; Nan et al. 2007) for nonpluripotent cell types such as NIH3T3, L cells, and HT1080 cells. This observation, in conjunction with our previous report of a distinct enrichment of H3.3 in the ES cells (Wong et al. 2009), suggests the presence of a unique chromatin at the telomere that is modulated in a significant way by ATRX and H3.3. Interestingly, the prominent presence of ATRX at the telomeres up to 12 d of induction of differentiation in these cells indicates that ATRX does not dissociate from the telomeres immediately after the loss of cellular pluripotency and may continue to play an as yet undefined role in the early stages of the undifferentiated cellular state.
We have shown that ATRX binding at the telomeres in ES cells occurs during late S/G2 phase and is coupled with the events of H3.3 loading and telomere replication timing at S phase of the cell cycle. By immunofluorescence and immunoprecipitation analysis in ES cells, we have further demonstrated a direct interaction between ATRX and H3.3. The reduction in the colocalization of these two proteins at the telomeres following the mutation of the K4 residue on H3.3 clearly indicates that H3.3 acts as a docking site for ATRX recruitment. Our observation of a direct interaction between ATRX and H3.3 is the first to be reported in mammalian cells.
Although ATRX depletion did not induce an extremely severe telomere dysfunction phenotype such as telomere fusions, we observed a robust increase in DNA damage/TIF phenotype following ATRX depletion, indicating a novel role of ATRX for the proper maintenance of telomere chromatin integrity in ES cells. A similar DNA damage/TIF phenotype was previously observed in H3.3 knockdown in these cells (Wong et al. 2009). The lack of a more severe telomere dysfunction phenotype could be due to the relatively short knockdown-time permissible in these experiments. Another possibility is that compromise in the telomere chromatin integrity following ATRX depletion may take many cell divisions to take effect. Consistent with this, an earlier study demonstrated that ES cells lacking ATRX, despite having a slower rate of cell division, did not exhibit any serious change in mitotic index (Garrick et al. 2006). This indicates that loss of ATRX does not immediately cause a severe cell cycle defect and is consistent with the absence of an observable severe telomere dysfunction phenotype and cell-growth defect in our ATRX-depleted ES cells.
The role of ATRX as a chromatin modifier is well documented. It is possible that ATRX, through interaction with H3.3 (Wong et al. 2009), acts to maintain a unique telomere chromatin state. Specifically, we propose that such an interaction, coupled to the telomere replication machinery (Verdun and Karlseder 2006, 2007), provides a primary epigenetic determinant that directly controls the assembly of a functionally essential and protected telomere structure in ES cells. Our observation of the direct function of ATRX in recruiting CBX5 to the telomeres further supports its role in the maintenance of telomere chromatin integrity in ES cells. Although it may seem conflicting that H3.3, being a marker for active chromatin, takes part in the recruitment of ATRX and CBX5 (given their common association with repressive chromatin), these proteins may act to strike a balance in telomere chromatin state to allow continual telomere-renewal but yet maintain an essential compacted structure. The phenomenon of having such a dynamic “open and closed chromatin state” is not new. Indeed, it has been well documented that a dynamic balance between open chromatin and closed heterochromatin is essential for vertebrate centromere activity (Wong et al. 2007; Nakano et al. 2008; Chueh et al. 2009). Future studies looking at the detailed mechanisms of action between these chromatin regulators and their interaction with histone methyltransferases SUV39H1/2 and SUV420H1/2 and with DNA methyltransferases DNMT3A/B and DNMT1 should elucidate novel pathways on the regulation of telomeric chromatin and telomere length homeostasis.
Although HP1 is known to play a direct role in regulating telomere length, capping, and transcription activity of retroelements in Drosophila (Fanti et al. 1998), less is known of its direct function at mammalian telomeres. Interestingly, a recent study described the association of HP1 proteins (CBX1, CBX3, and CBX5) with ALT (alternative lengthening of telomeres)-associated PML nuclear bodies and its role in compacting telomeric DNA in human ALT cancer cells (Jiang et al. 2007, 2009). It remains to be investigated if ALT cancer cells or other tumors have adopted a similar mechanism as ES cells in the regulation of telomere chromatin. Furthermore, it will be interesting to examine if H3.3, ATRX, and CBX5 also play a role in the establishment of telomeric chromatin during the differentiation and reprogramming of early mouse embryos, given that an alternative ALT-like recombination-based mechanism involving telomere sister-chromatid exchange has been proposed to be the driving force for the resetting of telomere length at early cleavage embryos (Liu et al. 2007).
Methods
Cell cultures
Cell lines including human HT1080 (fibrosarcoma), HeLa (cervical cancer cells), mouse NIH3T3, and L cells (transformed fibroblasts) were cultured in DMEM with 10% FCS. Mouse ES129.1 and ES-W9.5 in DMEM with 15% heat-inactivated FCS and 1 × 103 units/mL of leukemic inhibiting factor (LIF) and 0.1 mM β-mercaptoethanol. EG cell lines EGRA2 and EGRA3 were derived from primordial germ cells of strain 129 mice (J McWhir, The Roslin Institute, Scotland). They were cultured on LIF-expressing STO fibroblast feeder cells.
Antibodies
Primary antibodies used were as follows: rabbit polyclonal antisera against TERF1 (Iwano et al. 2004) and ATRX (H300 and D19; Santa Cruz Biotechnology); mouse monoclonal antiserum against MYC tag (Invitrogen), OCT4 (also known as POU5F1; Santa Cruz Biotechnology); gamma-H2AFX phosphorylated at Ser 139 (Upstate Biotechnology), CBX5 (Chemicon), and BrdU (Roche; conjugated with FITC); and human autoimmune anti-centromere antiserum CREST6 (predominantly anti-CENPA and -CENPB). Secondary antisera were donkey anti-human Texas Red (Jackson Laboratory), and donkey anti-mouse or anti-rabbit Alexa Fluoro 488/594/647 (Molecular Probes, Invitrogen).
Immunofluorescence analysis, FISH, and BrdU incorporation
Cells were treated with microtubule-depolymerizing agent Colcemid for 1 h at 37°C, harvested for hypotonic treatment in 0.075 M KCl, cytospun on slides, and incubated in KCM buffer (120 mM KCl, 20 mM NaCl, 10 mM Tris.HCl at pH 7.2, 0.5 mM EDTA, 0.1% [v/v] Triton X-100, and protease inhibitor) (Uren et al. 2000; Wong et al. 2005, 2007). Slides were blocked in KCM buffer containing 1% BSA and incubated with the relevant primary and secondary antibodies for 1 h at 37°C. After each round of antibody incubation, slides were washed three times in KCM buffer. Slides were then fixed in KCM with 4% formaldehyde and mounted in mounting medium (Vetashield). Images were collected using a fluorescence microscope linked to a CCD camera system.
For telomere-FISH, slides were fixed in cold methanol followed by dehydration in an ethanol series prior to denaturation and hybridization with Cy3-conjugated telomere DNA probe (Dako). For BrdU staining, cells were incubated with 10 μM BrdU (Sigma) for 1 h. BrdU incorporation was assessed by a denaturation protocol using 0.5 N hydrochloric acid, followed by immunofluorescence analysis with anti-BrdU FITC antibody (Roche).
Cell-cycle synchronization and FACS analysis
Cells were treated with 2 mM thymidine for 12 h, released from cell cycle blockage by washing with warm medium, and incubated in medium at 37°C for various time points. For FACS analysis, cells were harvested, washed in 1× PBS, fixed with ice-cold ethanol, stained with propidium iodide, and analyzed on a LSR II Becton Dickinson Flow Cytometry Analyzer.
Generation and transfection of MYC-tagged H3.3 and MYC-tagged H3.3mutK4 constructs
The generation of pcDNA4/TO/MYC-H3.3 (tetracycline-inducible expression vector expressing MYC-H3.3) has been described (Wong et al. 2009). The mutation on K4 residue was performed by PCR/site-directed mutagenesis. Using pcDNA4/TO/MYC-H3.3 as DNA template, the 5′ and 3′ fragments of pcDNA4/TO/MYC-H3.3K4 carrying a mutation in K4 residue were generated using the following primers: H3.3K4Aforward, 5′-GCTCGTACAGCGCAGACTGCC-3′ (carrying a mutated site AAG to GCG, underlined); H3.3K4Areverse, 5′-GGCAGTCTGCGCTGTACGAGC-3′ (carrying a mutated site CGT to CGC, underlined); MYCforward, 5′-CGCAAATGGGCGGTAGGCGTG-3′; H3.3MYCreverse, 5′-GAAGGGCCCTTAAGCACGTTCTCCACGTAT-3′ (carrying an ApaI site, underlined).
The two fragments generated were purified, annealed, and subjected to PCR extension. Subsequently, overlapping PCR amplification was performed using MYCforward primer and H3.3cMYCreverse primer corresponding to 5′ and 3′ fragments of MYC-H3.3, respectively. The resulting MYC-H3.3mutK4 fragment was digested with BamHI and ApaI and cloned into pcDNA4/TO/MYC-HisTA at BamHI and ApaI.
RNAi-depletion of ATRX and real-time PCR
Transfections of ATRX–RNAi-duplex oligonucleotides were performed using Lipofectamine 2000 (Invitrogen). The RNAi-duplex oligonucleotides (three sets of oligonucleotides) corresponding to the ATRX gene were purchased from Invitrogen and Ambion. These oligonucleotides have been prevalidated to confirm their targeting specificity. A set of control scrambled RNAi-duplex oligonucleotides was also used. Cells were harvested 24–48 h post-RNAi-knockdown for the assessment of the levels of ATRX by real-time PCR analysis using the SYBR system (Applied Biosystems). Four different sets of primers against ATRX cDNA were used for real-time PCR analysis, and average ATRX expression levels post-RNAi-knockdown were calculated for each experiment. As internal controls, specific primers corresponding to the housekeeping gene actin were used in real-time PCR analysis. Changes in expression levels were calculated according to the manufacturer's instruction. The ATRX and actin primer sets used for real-time PCR analysis were as follows:
ATRX exon 1–2: 5′ primer, 5′-GTCCGAGCCAAAAACATGAC-3′; 3′ primer, 5′-GTCATGAAGCTTCTGCACCA-3′
ATRX exon 18–19: 5′ primer, 5′-GAGAAGTGGCAGGAGGGATT-3′; 3′ primer, 5′-TGAGGACGTTTCACAGTTGC-3′
ATRX exon 27–28: 5′ primer, 5′-AGCCAGTCGCTCATATCTCTG-3′; 5′ primer, 5′-TCGGATCCACTTTCCTTCAC-3′
ATRX exon 34–35: 5′ primer, 5′-CGTGAAACGAAGAGAAGCAA-3′; 3′ primer, 5′-TGCTGGAGCCGTCTATTCAT-3′
Actin: 5′ primer, 5′-TCCCTGGAGAAGAGCTACGA-3′; 3′ primer, 5′-AGCACTGTGTTGGCGTACAG-3′.
Western blot analysis and protein immunoprecipitation
Cells were washed with ice cold 1× PBS lysed with ice-cold 1× modified RIPA buffer (150 mM NaCl, 50 mM Tris-HCl at pH 7.5, 0.25% sodium deoxycholate, 0.1% NP40, 1 mM NaF, 1 mM sodium orthovanadate, and protease inhibitor), followed by a 10-sec pulse sonication. The lysate was collected after a 10-min centrifugation at 12,000 rpm and boiled in SDS/PAGE sample buffer prior to protein gel electrophoresis and Western blot analysis.
For immunoprecipitation, cells were lysed in ice-cold TBS buffer (10 mM Tris at pH 7.5, 3 mM CaCl2, 2 mM MgCl2, and protease inhibitor) containing 0.5% Tween 40 for 1 h and homogenized using a Dounce Homogeniser “Tight/A” pestle. Nuclei were pelleted and incubated in digestion buffer (50 mM Tris at pH 7.5, 320 mM sucrose, 1 mM CaCl2, 4 mM MgCl2, and protease inhibitor) with MNase (4–16 units/mg DNA) at 37°C for various time-courses. Reaction was stopped with the addition of 1/100 of 0.5 M EDTA. The nuclei were pelleted, and the supernatant was collected. The pelleted nuclei were then lysed further in 1× lysis buffer (1 mM Tris at pH 7.5, 0.1 mM EDTA, and protease inhibitor) and centrifugated, and the supernatant was collected. The lysates were then pooled and mixed (1:1 ratio) with incubation buffer (50 mM NaCl, 20 mM Tris at pH 7.2, 5 mM EDTA, and protease inhibitor).
Cell lysates were subjected to immunoprecipitation with 1–5 μg of antibodies overnight at 4°C with constant agitation. Fifty microliters of 50% protein G–Dynabead (Invitrogen) was added to the mixture for capturing the protein/antibody complexes. The immunoprecipitated complexes were washed with wash buffer three times (first wash with 50 mM Tris at pH 7.2, 10 mM EDTA, and 50 mM NaCl; second wash with 50 mM Tris at pH 7.2, 10 mM EDTA, and 100 mM NaCl; third wash with 50 mM Tris at pH 7.2, 10 mM EDTA, and 150 mM NaCl). The proteins were eluted by the addition of 80 μL of 2× SDS-PAGE sample buffer followed by boiling for 5 min. The beads were pelleted by centrifugation in a microfuge, and the supernatant containing proteins were subjected to SDS-PAGE analysis.
Pulse field gel electrophoresis and Southern blot analysis
Total genomic DNA was digested with RsaI and HinfI and subjected to either standard or pulse-field (6 V/cm for 20 h at 14°C; switch times ramped linearly from 0.5 to 6 sec using CHEF PFGE Bioradsystem) gel electrophoresis, followed by Southern blot analysis with a [γ-32P]ATP-end-labeled (TTAGGG)4 telomere repeat probe. Signal intensities were analyzed with Typhoon PhosphoImagerSystem and ImageQuant software.
Dot blot analysis
DNA (50 ng) was diluted with 0.6 M NaCl, denatured (by heating DNA for 5 min at 100°C and cooling on ice for 5 min), transferred onto Hybond N+ nitrocellulose membrane, and rinsed in 0.5 M NH2SO4. The membrane was incubated in 0.4 N NaOH at room temperature for 5 min, followed by neutralization with 2× SSC twice. Subsequently, the membrane was subjected to Southern blot analysis with a [γ-32P]ATP-end-labeled (TTAGGG)4 telomere probe. Signal intensities were analyzed with Typhoon PhosphoImager System and ImageQuant software.
Acknowledgments
We thank Jim McWhir for EG cell lines. This work was supported by the National Health and Medical Research Council of Australia.
Footnotes
[Supplemental material is available online at http://www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.101477.109.
References
- Argentaro A, Yang JC, Chapman L, Kowalczyk MS, Gibbons RJ, Higgs DR, Neuhaus D, Rhodes D. Structural consequences of disease-causing mutations in the ATRX-DNMT3-DNMT3L (ADD) domain of the chromatin-associated protein ATRX. Proc Natl Acad Sci. 2007;104:11939–11944. doi: 10.1073/pnas.0704057104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berube NG, Jagla M, Smeenk C, De Repentigny Y, Kothary R, Picketts DJ. Neurodevelopmental defects resulting from ATRX overexpression in transgenic mice. Hum Mol Genet. 2002;11:253–261. doi: 10.1093/hmg/11.3.253. [DOI] [PubMed] [Google Scholar]
- Berube NG, Mangelsdorf M, Jagla M, Vanderluit J, Garrick D, Gibbons RJ, Higgs DR, Slack RS, Picketts DJ. The chromatin-remodeling protein ATRX is critical for neuronal survival during corticogenesis. J Clin Invest. 2005;115:258–267. doi: 10.1172/JCI22329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienz M. The PHD finger, a nuclear protein-interaction domain. Trends Biochem Sci. 2006;31:35–40. doi: 10.1016/j.tibs.2005.11.001. [DOI] [PubMed] [Google Scholar]
- Cardoso C, Lutz Y, Mignon C, Compe E, Depetris D, Mattei MG, Fontes M, Colleaux L. ATR-X mutations cause impaired nuclear location and altered DNA binding properties of the XNP/ATR-X protein. J Med Genet. 2000;37:746–751. doi: 10.1136/jmg.37.10.746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chueh AC, Northrop EL, Brettingham-Moore KH, Choo KH, Wong LH. LINE retrotransposon RNA is an essential structural and functional epigenetic component of a core neocentromeric chromatin. PLoS Genet. 2009;5:e1000354. doi: 10.1371/journal.pgen.1000354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanti L, Pimpinelli S. HP1: A functionally multifaceted protein. Curr Opin Genet Dev. 2008;18:169–174. doi: 10.1016/j.gde.2008.01.009. [DOI] [PubMed] [Google Scholar]
- Fanti L, Giovinazzo G, Berloco M, Pimpinelli S. The heterochromatin protein 1 prevents telomere fusions in Drosophila. Mol Cell. 1998;2:527–538. doi: 10.1016/s1097-2765(00)80152-5. [DOI] [PubMed] [Google Scholar]
- Garcia-Cao M, O'Sullivan R, Peters AH, Jenuwein T, Blasco MA. Epigenetic regulation of telomere length in mammalian cells by the Suv39h1 and Suv39h2 histone methyltransferases. Nat Genet. 2004;36:94–99. doi: 10.1038/ng1278. [DOI] [PubMed] [Google Scholar]
- Garrick D, Sharpe JA, Arkell R, Dobbie L, Smith AJ, Wood WG, Higgs DR, Gibbons RJ. Loss of Atrx affects trophoblast development and the pattern of X-inactivation in extraembryonic tissues. PLoS Genet. 2006;2:e58. doi: 10.1371/journal.pgen.0020058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalo S, Garcia-Cao M, Fraga MF, Schotta G, Peters AH, Cotter SE, Eguia R, Dean DC, Esteller M, Jenuwein T, et al. Role of the RB1 family in stabilizing histone methylation at constitutive heterochromatin. Nat Cell Biol. 2005;7:420–428. doi: 10.1038/ncb1235. [DOI] [PubMed] [Google Scholar]
- Gonzalo S, Jaco I, Fraga MF, Chen T, Li E, Esteller M, Blasco MA. DNA methyltransferases control telomere length and telomere recombination in mammalian cells. Nat Cell Biol. 2006;8:416–424. doi: 10.1038/ncb1386. [DOI] [PubMed] [Google Scholar]
- Guerrini R, Shanahan JL, Carrozzo R, Bonanni P, Higgs DR, Gibbons RJ. A nonsense mutation of the ATRX gene causing mild mental retardation and epilepsy. Ann Neurol. 2000;47:117–121. [PubMed] [Google Scholar]
- Hake SB, Garcia BA, Kauer M, Baker SP, Shabanowitz J, Hunt DF, Allis CD. Serine 31 phosphorylation of histone variant H3.3 is specific to regions bordering centromeres in metaphase chromosomes. Proc Natl Acad Sci. 2005;102:6344–6349. doi: 10.1073/pnas.0502413102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D, Sfeir AJ, Shay JW, Wright WE, de Lange T. POT1 protects telomeres from a transient DNA damage response and determines how human chromosomes end. EMBO J. 2005;24:2667–2678. doi: 10.1038/sj.emboj.7600733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishov AM, Vladimirova OV, Maul GG. Heterochromatin and ND10 are cell-cycle regulated and phosphorylation-dependent alternate nuclear sites of the transcription repressor Daxx and SWI/SNF protein ATRX. J Cell Sci. 2004;117:3807–3820. doi: 10.1242/jcs.01230. [DOI] [PubMed] [Google Scholar]
- Iwano T, Tachibana M, Reth M, Shinkai Y. Importance of TRF1 for functional telomere structure. J Biol Chem. 2004;279:1442–1448. doi: 10.1074/jbc.M309138200. [DOI] [PubMed] [Google Scholar]
- James TC, Elgin SC. Identification of a nonhistone chromosomal protein associated with heterochromatin in Drosophila melanogaster and its gene. Mol Cell Biol. 1986;6:3862–3872. doi: 10.1128/mcb.6.11.3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang WQ, Zhong ZH, Henson JD, Reddel RR. Identification of candidate alternative lengthening of telomeres genes by methionine restriction and RNA interference. Oncogene. 2007;26:4635–4647. doi: 10.1038/sj.onc.1210260. [DOI] [PubMed] [Google Scholar]
- Jiang WQ, Zhong ZH, Nguyen A, Henson JD, Toouli CD, Braithwaite AW, Reddel RR. Induction of alternative lengthening of telomeres-associated PML bodies by p53/p21 requires HP1 proteins. J Cell Biol. 2009;185:797–810. doi: 10.1083/jcb.200810084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourmouli N, Sun YM, van der Sar S, Singh PB, Brown JP. Epigenetic regulation of mammalian pericentric heterochromatin in vivo by HP1. Biochem Biophys Res Commun. 2005;337:901–907. doi: 10.1016/j.bbrc.2005.09.132. [DOI] [PubMed] [Google Scholar]
- Liu L, Bailey SM, Okuka M, Munoz P, Li C, Zhou L, Wu C, Czerwiec E, Sandler L, Seyfang A, et al. Telomere lengthening early in development. Nat Cell Biol. 2007;9:1436–1441. doi: 10.1038/ncb1664. [DOI] [PubMed] [Google Scholar]
- Loyola A, Bonaldi T, Roche D, Imhof A, Almouzni G. PTMs on H3 variants before chromatin assembly potentiate their final epigenetic state. Mol Cell. 2006;24:309–316. doi: 10.1016/j.molcel.2006.08.019. [DOI] [PubMed] [Google Scholar]
- Lu X, Meng X, Morris CA, Keating MT. A novel human gene, WSTF, is deleted in Williams syndrome. Genomics. 1998;54:241–249. doi: 10.1006/geno.1998.5578. [DOI] [PubMed] [Google Scholar]
- Maison C, Almouzni G. HP1 and the dynamics of heterochromatin maintenance. Nat Rev Mol Cell Biol. 2004;5:296–304. doi: 10.1038/nrm1355. [DOI] [PubMed] [Google Scholar]
- McDowell TL, Gibbons RJ, Sutherland H, O'Rourke DM, Bickmore WA, Pombo A, Turley H, Gatter K, Picketts DJ, Buckle VJ, et al. Localization of a putative transcriptional regulator (ATRX) at pericentromeric heterochromatin and the short arms of acrocentric chromosomes. Proc Natl Acad Sci. 1999;96:13983–13988. doi: 10.1073/pnas.96.24.13983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minc E, Allory Y, Worman HJ, Courvalin JC, Buendia B. Localization and phosphorylation of HP1 proteins during the cell cycle in mammalian cells. Chromosoma. 1999;108:220–234. doi: 10.1007/s004120050372. [DOI] [PubMed] [Google Scholar]
- Nagamine K, Peterson P, Scott HS, Kudoh J, Minoshima S, Heino M, Krohn KJ, Lalioti MD, Mullis PE, Antonarakis SE, et al. Positional cloning of the APECED gene. Nat Genet. 1997;17:393–398. doi: 10.1038/ng1297-393. [DOI] [PubMed] [Google Scholar]
- Nakano M, Cardinale S, Noskov VN, Gassmann R, Vagnarelli P, Kandels-Lewis S, Larionov V, Earnshaw WC, Masumoto H. Inactivation of a human kinetochore by specific targeting of chromatin modifiers. Dev Cell. 2008;14:507–522. doi: 10.1016/j.devcel.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan X, Hou J, Maclean A, Nasir J, Lafuente MJ, Shu X, Kriaucionis S, Bird A. Interaction between chromatin proteins MECP2 and ATRX is disrupted by mutations that cause inherited mental retardation. Proc Natl Acad Sci. 2007;104:2709–2714. doi: 10.1073/pnas.0608056104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picketts DJ, Higgs DR, Bachoo S, Blake DJ, Quarrell OW, Gibbons RJ. ATRX encodes a novel member of the SNF2 family of proteins: Mutations point to a common mechanism underlying the ATR-X syndrome. Hum Mol Genet. 1996;5:1899–1907. doi: 10.1093/hmg/5.12.1899. [DOI] [PubMed] [Google Scholar]
- Ritchie K, Seah C, Moulin J, Isaac C, Dick F, Berube NG. Loss of ATRX leads to chromosome cohesion and congression defects. J Cell Biol. 2008;180:315–324. doi: 10.1083/jcb.200706083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma GG, Hwang KK, Pandita RK, Gupta A, Dhar S, Parenteau J, Agarwal M, Worman HJ, Wellinger RJ, Pandita TK. Human heterochromatin protein 1 isoforms HP1Hsα and HP1Hsβ interfere with hTERT-telomere interactions and correlate with changes in cell growth and response to ionizing radiation. Mol Cell Biol. 2003;23:8363–8376. doi: 10.1128/MCB.23.22.8363-8376.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uren AG, Wong L, Pakusch M, Fowler KJ, Burrows FJ, Vaux DL, Choo KH. Survivin and the inner centromere protein INCENP show similar cell-cycle localization and gene knockout phenotype. Curr Biol. 2000;10:1319–1328. doi: 10.1016/s0960-9822(00)00769-7. [DOI] [PubMed] [Google Scholar]
- Verdun RE, Karlseder J. The DNA damage machinery and homologous recombination pathway act consecutively to protect human telomeres. Cell. 2006;127:709–720. doi: 10.1016/j.cell.2006.09.034. [DOI] [PubMed] [Google Scholar]
- Verdun RE, Karlseder J. Replication and protection of telomeres. Nature. 2007;447:924–931. doi: 10.1038/nature05976. [DOI] [PubMed] [Google Scholar]
- Wong LH, Saffery R, Anderson MA, Earle E, Quach JM, Stafford AJ, Fowler KJ, Choo KH. Analysis of mitotic and expression properties of human neocentromere-based transchromosomes in mice. J Biol Chem. 2005;280:3954–3962. doi: 10.1074/jbc.M410047200. [DOI] [PubMed] [Google Scholar]
- Wong LH, Brettingham-Moore KH, Chan L, Quach JM, Anderson MA, Northrop EL, Hannan R, Saffery R, Shaw ML, Williams E, et al. Centromere RNA is a key component for the assembly of nucleoproteins at the nucleolus and centromere. Genome Res. 2007;17:1146–1160. doi: 10.1101/gr.6022807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong LH, Ren H, Williams E, McGhie J, Ahn S, Sim M, Tam A, Earle E, Anderson MA, Mann J, et al. Histone H3.3 incorporation provides a unique and functionally essential telomeric chromatin in embryonic stem cells. Genome Res. 2009;19:404–414. doi: 10.1101/gr.084947.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Gibbons R, Yan Z, Yang D, McDowell TL, Sechi S, Qin J, Zhou S, Higgs D, Wang W. The ATRX syndrome protein forms a chromatin-remodeling complex with Daxx and localizes in promyelocytic leukemia nuclear bodies. Proc Natl Acad Sci. 2003;100:10635–10640. doi: 10.1073/pnas.1937626100. [DOI] [PMC free article] [PubMed] [Google Scholar]