Abstract
Here, we demonstrate how comparative sequence analysis facilitates genome-wide base-pair-level interpretation of individual genetic variation and address two questions of importance for human personal genomics: first, whether an individual's functional variation comes mostly from noncoding or coding polymorphisms; and, second, whether population-specific or globally-present polymorphisms contribute more to functional variation in any given individual. Neither has been definitively answered by analyses of existing variation data because of a focus on coding polymorphisms, ascertainment biases in favor of common variation, and a lack of base-pair-level resolution for identifying functional variants. We resequenced 575 amplicons within 432 individuals at genomic sites enriched for evolutionary constraint and also analyzed variation within three published human genomes. We find that single-site measures of evolutionary constraint derived from mammalian multiple sequence alignments are strongly predictive of reductions in modern-day genetic diversity across a range of annotation categories and across the allele frequency spectrum from rare (<1%) to high frequency (>10% minor allele frequency). Furthermore, we show that putatively functional variation in an individual genome is dominated by polymorphisms that do not change protein sequence and that originate from our shared ancestral population and commonly segregate in human populations. These observations show that common, noncoding alleles contribute substantially to human phenotypes and that constraint-based analyses will be of value to identify phenotypically relevant variants in individual genomes.
As the sequencing of human genomes becomes routine, a growing challenge is how to assess the functional consequences of the genetic variation carried by a given individual. Of the >3 million variants present in any given human genome (Levy et al. 2007; Bentley et al. 2008; Wang et al. 2008; Kim et al. 2009; Mardis et al. 2009; McKernan et al. 2009), only a small fraction are expected to be phenotypically relevant (Kimura 1983; Ng et al. 2008; Mardis et al. 2009); among those that are, there is a great range in the degree to which they contribute to phenotype (Boyko et al. 2008). Therefore, to fully leverage the benefits of genome-wide variation data, methods for detecting and evaluating functional variants across the entire genome are required, as is an improved understanding of the nature of functional human genetic variation.
One strategy for identifying potentially important genetic variants is to focus on polymorphisms that fall into regions that have well-defined molecular functions, such as protein-coding exons. Several methods exist to estimate the impact of nonsynonymous changes on protein function, using data on the physicochemical properties of amino acids, three-dimensional structures and enzymatic functions of proteins, as well as sequence conservation (Sunyaev et al. 2001; Ng and Henikoff 2003; Stone and Sidow 2005; Chun and Fay 2009). However, many critically important functions are found outside protein-coding exons, as hypothesized more than 30 yr ago (King and Wilson 1975) and proven to be true in many functional and computational studies of the human and related genomes (Lander et al. 2001; Venter et al. 2001; Mouse Genome Sequencing Consortium 2002; The ENCODE Project Consortium 2007). In particular, elements involved in the regulation of transcription and in the processing and control of RNA transcripts play important cellular and developmental roles, and in some cases mutations within them contribute to disease (e.g., Orkin et al. 1982; Hata et al. 2007; Hirose et al. 2008; Verlaan et al. 2009). Indeed, it is likely that restricting analysis of human variation to coding sequences alone will overlook the majority of functional variants present in the human genome. In comparison with coding exons, however, comprehensive experimental identification of the majority of human noncoding functional elements remains largely elusive, despite large-scale efforts such as ENCODE (The ENCODE Project Consortium 2007). Consequently, computational and experimental interpretations of the functional impact of noncoding variation have been weaker than that of coding variation, and alternative approaches for assessing noncoding functional variation need to be devised.
Motivated by the relative lack of a comprehensive annotation of noncoding elements, many genomes have recently been sequenced with the goal of generating comparative annotations for human or model organisms (e.g., Mouse Genome Sequencing Consortium 2002; Kellis et al. 2003; Drosophila 12 Genomes Consortium 2007; Margulies et al. 2007). Concomitant with the increases in comparative data, methodologies have been developed that identify evolutionarily constrained noncoding sequences in the target genomes (Cooper et al. 2005; Siepel et al. 2005; Asthana et al. 2007a; Margulies et al. 2007). Three major lines of evidence suggest that these strategies are successful in identifying functionally important regions, despite an inability to comprehensively and precisely define their molecular functions. First, large fractions of noncoding constrained elements have been shown to be functional in vivo, driving expression of reporter genes in zebrafish (Woolfe et al. 2005) and mouse embryos (Pennacchio et al. 2006; Visel et al. 2008), as well as human cell lines (Cooper et al. 2006). Second, a wide variety of functional categories of sequence, including transcripts, protein–DNA binding sites, and DNase hypersensitive sites, among others (The ENCODE Project Consortium 2007), harbor strong signatures of evolutionary constraint. Third, polymorphisms are depleted from human conserved noncoding sequences and are present at reduced allele frequencies, suggesting that negative selection inferred during mammalian evolution has persisted during recent human demographic history (Drake et al. 2006; Asthana et al. 2007b; Chen et al. 2007; Katzman et al. 2007). The present study addresses the relative importance of coding versus noncoding variation in humans.
Also important to personal genomics is the extent to which an individual's functional variation comes from common versus rare alleles. Much functional variation is expected to be deleterious, and therefore purifying selection is likely to reduce the frequency of alleles in functional sequences relative to alleles in neutrally evolving regions (Drake et al. 2006; Asthana et al. 2007b; Katzman et al. 2007; Lohmueller et al. 2008). On the other hand, functional polymorphisms may be able to reach high frequencies in modern human populations, either through weakened negative selection caused by demography (Marth et al. 2004; Lohmueller et al. 2008) or positive selection (Akey et al. 2004; Bustamante et al. 2005; Pickrell et al. 2009). To date, the majority of variants identified in association studies are common alleles predicted to have very modest effects (Bodmer and Bonilla 2008; McCarthy et al. 2008), although medical resequencing studies make a case for the strong functional impact of rare alleles (e.g., Ahituv et al. 2007; Romeo et al. 2007). Thus, there is debate over whether the functional variation carried by an individual is likely to be comprised of a large number of common variants interacting with each other and the environment or a smaller number of rare variants with more dramatic effects (Gibbs 2005; Bodmer and Bonilla 2008; McCarthy et al. 2008).
Here, we show that high-resolution comparative sequence analyses shed light on genome-wide personal genetic variation without requiring prior annotation by functional assays and without biasing against noncoding variation. We use evolutionary constraint as a metric to first stratify each base in the human genome according to its potential functional significance, and then use this as a framework to interpret single nucleotide variation present in the human population and in recently sequenced genomes. We employed two relevant data sets. To obtain data enriched for variation at constrained sites, we resequenced 575 short regions containing constrained elements in a sample of 432 individuals from five populations, from both coding and noncoding sequence. The depth of this sample allowed us to ascertain rare polymorphisms with high accuracy and also permitted us to conduct an in-depth comparison of functional variation between individuals from multiple geographically distinct populations. We then applied our analysis genome-wide to three published individual sequences (a Yoruba [Bentley et al. 2008], a Chinese [Wang et al. 2008], and a Caucasian American [Levy et al. 2007]), to obtain a more comprehensive view of the functional variation in individual human genomes and to understand the overlap in putatively functional coding and noncoding variation among individuals of diverse ancestry.
Results
Ascertainment of variation in regions harboring constrained elements
To obtain a high-quality data set enriched for potentially functional variation, we identified 575 short regions (74–1427 bp, median 463 bp) in which there is a deficiency of evolutionary variation, using genomic evolutionary rate profiling (GERP) (Cooper et al. 2005) on mammalian multiple sequence alignments (Margulies et al. 2007). These include 100 regions with demonstrated promoter activity in a luciferase reporter assay (Cooper et al. 2006). We then sequenced these putatively functional, constrained elements (CEs) in 432 individuals from five distinct geographic locations: two African populations (the Yoruba from Nigeria and the Luhya from Kenya), two Asian populations (Han Chinese and Japanese), and the Centre d'Etude du Polymorphisime Humain (CEPH) Utah population representing the European population (Table 1, see Methods). Since most CEs are smaller than the PCR amplicons used to capture them, most sequenced regions also contain flanking neutral DNA. In aggregate, the sequenced sample comprises approximately equal proportions of neutral and constrained sites.
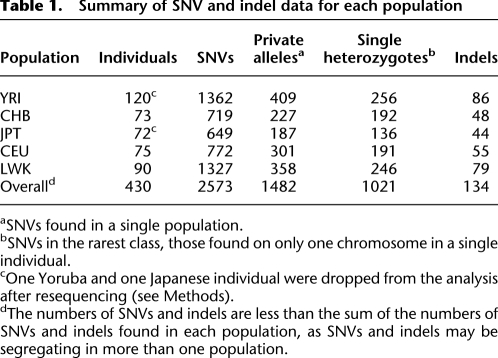
Table 1.
Summary of SNV and indel data for each population
aSNVs found in a single population.
bSNVs in the rarest class, those found on only one chromosome in a single individual.
cOne Yoruba and one Japanese individual were dropped from the analysis after resequencing (see Methods).
dThe numbers of SNVs and indels are less than the sum of the numbers of SNVs and indels found in each population, as SNVs and indels may be segregating in more than one population.
To quantify the importance of rare functional polymorphisms compared with common ones, we needed to ascertain rare polymorphisms with high accuracy. To this end, candidate single nucleotide variants (SNVs) were confirmed through visual inspection of sequence reads, yielding a total of 2573 sites with SNVs (Table 1). After quality control and filtering, 516 regions encompassing 227,252 bp were analyzed. For 2214 variants in these regions we inferred the derived allele by comparison with chimp and baboon and obtained a derived allele frequency (DAF) spectrum (Fig. 1A). The DAF spectrum is strongly skewed toward rare alleles, with 1537 SNVs occurring at a DAF of 1% or less (Fig. 1A), providing evidence that rare variation was more successfully captured than in previous studies of functional variation on fewer individuals, in which the skew is much less pronounced (Drake et al. 2006; Katzman et al. 2007; Bhangale et al. 2008; Lohmueller et al. 2008).
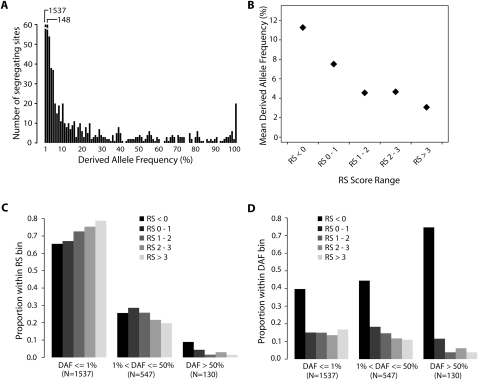
Figure 1.
Derived allele frequency (DAF) compared with evolutionary constraint. (A) DAF spectrum of all SNVs. Each category is one percentile DAF. Note the much higher number of SNVs with 0%–1% and 1%–2% DAF compared with the rest of the data. (B) Mean DAF as a function of RS score. Sites with RS > 0 are binned by increasing level of constraint. (C) Proportion of sites of the indicated DAF within each of five RS bins. Bars of each RS bin add up to 1 but are organized by DAF to facilitate visual comparison between the RS bins. The greater the RS of a site, the rarer is its derived allele. (D) Proportion of sites within the indicated RS bins at sites of rare, intermediate to common, and very common DAF. The greater the DAF, the more SNVs avoid constrained sites.
Selection against variation at constrained sites
For each site in an alignment, GERP calculates a “rejected substitution” (RS) score, so called because it is the estimated amount of evolutionary variation that was “rejected” by past negative selection (see Methods). These site-specific scores are a quantitative measure of evolutionary constraint, with higher scores corresponding to greater constraint. For each site in each of the ENCODE Pilot regions, we calculated an RS score based on alignments containing 24–26 mammalian sequences (Margulies et al. 2007). We find a strong negative trend between the RS score at a given position and the DAF for polymorphisms at that site (Fig. 1B), illustrating the impact of negative selection on derived alleles at evolutionarily constrained sites. This trend largely disappears when comparing DAF and the RS of sites adjacent with SNVs (Supplemental Fig. S1), highlighting the ability of GERP to enrich at the nucleotide level for sites under selection. The same effect is also seen in the normalized DAF spectrum, in which the proportion of SNVs that have low allele frequencies (e.g., <1%) is higher for sites with high RS, and SNVs that have higher allele frequencies (e.g., between 1% and 50%) are increasingly underrepresented as RS increases (Fig. 1C). Similarly, the RS spectrum shifts toward neutral sites as allele frequency increases (Fig. 1D; g-test, P = 2.76 × 10−12; degrees of freedom = 10), and the number of SNVs at constrained sites in all DAF bins is less than expected by chance, including in the rarest DAF class (<0.125%; 1 SNV in >800 chromosomes) (Supplemental Fig. S2). These results, which are consistent across all five study populations (Supplemental Figs. S3, S4), demonstrate that site-specific estimates of evolutionary constraint can be used to enrich for functionally deleterious variation and that SNVs across the entire derived allele frequency spectrum have been subject to recent purifying selection.
In addition to our resequencing data from the ENCODE Pilot regions, we also obtained SNVs from the genomes of three individuals: J. Craig Venter, a Caucasian American male (Levy et al. 2007); an anonymous Chinese male (Wang et al. 2008); and an anonymous Yoruba male (Bentley et al. 2008). To account for the fact that the human reference sequence contains many polymorphisms found in the wider human population, we compared each individual genome with the others and with the human (hg18) and chimpanzee (panTro2) reference sequences to identify additional sites in which each individual harbored a derived allele. This provided us with a data set of 3.2–3.6 million sites in each individual (Supplemental Table 1) that are polymorphic in this sample (the three individuals plus hg18) and carry a derived allele.
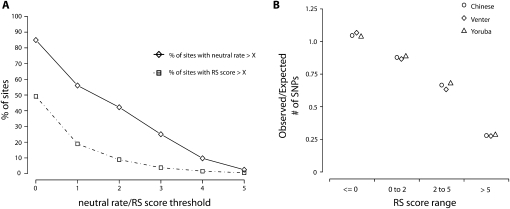
We obtained site-specific constraint scores (RS) for each site in the human genome using the mammalian sequences from 44-way vertebrate MULTIZ/TBA alignments for each human chromosome (http://hgdownload.cse.ucsc.edu/goldenPath/hg18/multiz44way/). These data are the deepest chromosome-level mammalian species alignments available to date, with ∼42% of the human genome aligned to a depth of at least two substitutions per site (Fig. 2A). We classified sites with RS > 2 as evolutionarily constrained, which captures ∼8.9% of the human genome, a value similar to recent estimates of the portion of the human genome that is selectively conserved among mammals (Siepel et al. 2005; Asthana et al. 2007a; Margulies et al. 2007). In all three individuals, variation at constrained sites is significantly suppressed: The most evolutionarily constrained bases harbor over 35% fewer SNVs than expected (Fig. 2B; Supplemental Table S1), with the depletion of variation correlating with the level of constraint. This demonstrates that negative selection has acted against variation at constrained sites throughout the human genome and throughout human population history, leaving behind a strong signature that can be detected in any given human genome.
Figure 2.
Variation and evolutionary constraint in three personal genomes. (A) Coverage of the human genome by mammalian alignment depth (solid line) and level of constraint (broken line). (B) Ratio of the number of SNVs observed in the three individual genomes at sites within the given RS score range to the number expected, given the distribution of RS scores across the human genome.
Relative contribution of common and rare alleles to functional variation
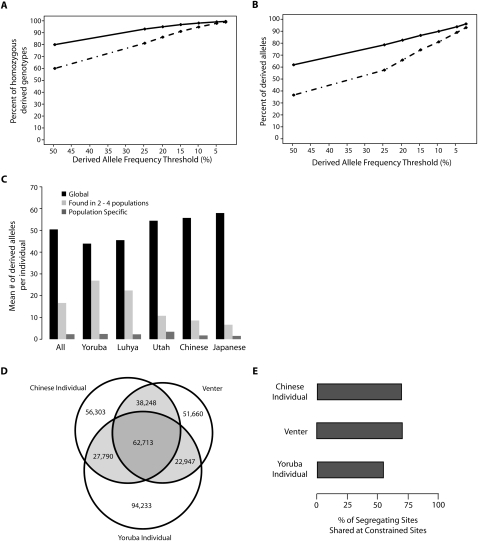
While the majority of the total variation in any given individual is known to be common (The International HapMap Consortium 2005; Frazer et al. 2007; Li et al. 2008), we find that polymorphisms at constrained positions are more likely to be rare (Fig. 1). This prompted us to ask whether common alleles also dominate putatively functional variation, as the large number of rare alleles in the resequencing data from the 432 individuals provides the power to compare the contributions of rare and common variants with the functional variation carried by a given individual. In the average individual at constrained sites, high-frequency alleles (DAF > 25%) contribute more than 80% of all homozygous derived genotypes (Fig. 3A) and more than 50% of total derived alleles (Fig. 3B).
Figure 3.
Properties of derived alleles at constrained sites. (A) Percentage of sites homozygous for derived alleles, where the derived alleles are of a frequency above the threshold indicated along the x-axis, per individual, at all sites (solid line) or at constrained sites only (broken lines). (B) Percentage of derived alleles of a frequency above the threshold indicated along the x-axis, per individual, at all sites (solid line) or at constrained sites only (broken lines). (C) Mean number of sites per individual that bear derived alleles found in all five populations (global SNVs), are found only in the indicated population (population specific), or are shared between two to four populations. (D) Number of segregating sites in each individual at highly constrained (RS > 2) sites that are shared between individuals or are private to each individual. (E) Percentage of segregating sites at constrained (RS > 2) sites in each individual that are shared with at least one of the other individuals.
The sizeable proportion of currently segregating functional variation originating from common derived alleles suggests that a large portion of this variation may be shared among populations. To test this hypothesis, we quantified the amount of variation at constrained sites per individual coming from global (found in all five sampled populations), somewhat restricted (found in two to four populations), or population-specific (found in one population only) variants. Within a given genome, global SNVs contribute over 20 times more derived alleles at constrained sites (Fig. 3C) than do population-specific variants. Thus, while alleles at neutral sites are more likely to be shared between populations than alleles at constrained positions (Fig. 3A,B), shared variation nevertheless contributes to the majority of observed functional variation as well.
A possible consequence of the fact that a large amount of an individual's functional variation comes from alleles that are shared between populations is that individuals in different populations may carry similar amounts of functional variation. Indeed, we find that the average number of derived alleles affecting evolutionarily constrained sites carried by African individuals is virtually indistinguishable from that carried by European or Asian individuals (Supplemental Fig. S5). Consistent with the effects of demographic history and analyses of nonsynonymous SNVs (Lohmueller et al. 2008), Europeans and Asians have a greater proportion of derived alleles at constrained sites in the homozygous state (Supplemental Fig. S6). If one assumes all derived alleles are recessive, variants at constrained sites would be expected to have a greater functional impact on European and Asian individuals than on African individuals. Since the ascertained functional variation is enriched for deleterious alleles, this would imply that Europeans and Asians also carry a higher genetic load (Lohmueller et al. 2008). However, if the degrees of dominance of the derived alleles in our data set range from completely recessive to completely additive, the implication would instead be that geographically disparate humans with considerably different demographic histories would harbor similar amounts of functional variation, and thus carry a similar genetic load.
For a genome-wide view of the degree to which putatively functional variation is shared between individuals with different geographic origins, we returned to the three previously described genome-wide SNV data sets. We detected a considerable amount of overlap in SNVs at constrained sites between individuals, ranging from 41% (SNVs in the Yoruba individual found in Venter) to 57% (SNVs in Venter found in the Chinese individual) (Fig. 3D). As expected, the Chinese individual and Venter shared more variation with each other than with the Yoruba individual (Marth et al. 2004; The International HapMap Consortium 2005; Frazer et al. 2007). However, even in the Yoruba individual, over 50% of SNVs at constrained sites are shared with one or both of the other two individuals, with this percentage rising to ∼70% in the Caucasian and the Chinese individuals (Fig. 3D,E). Thus, we conclude that unrelated individuals from geographically and genetically distant populations share a substantial amount of putatively functional variation.
Relative functional importance of variation at coding and noncoding sites
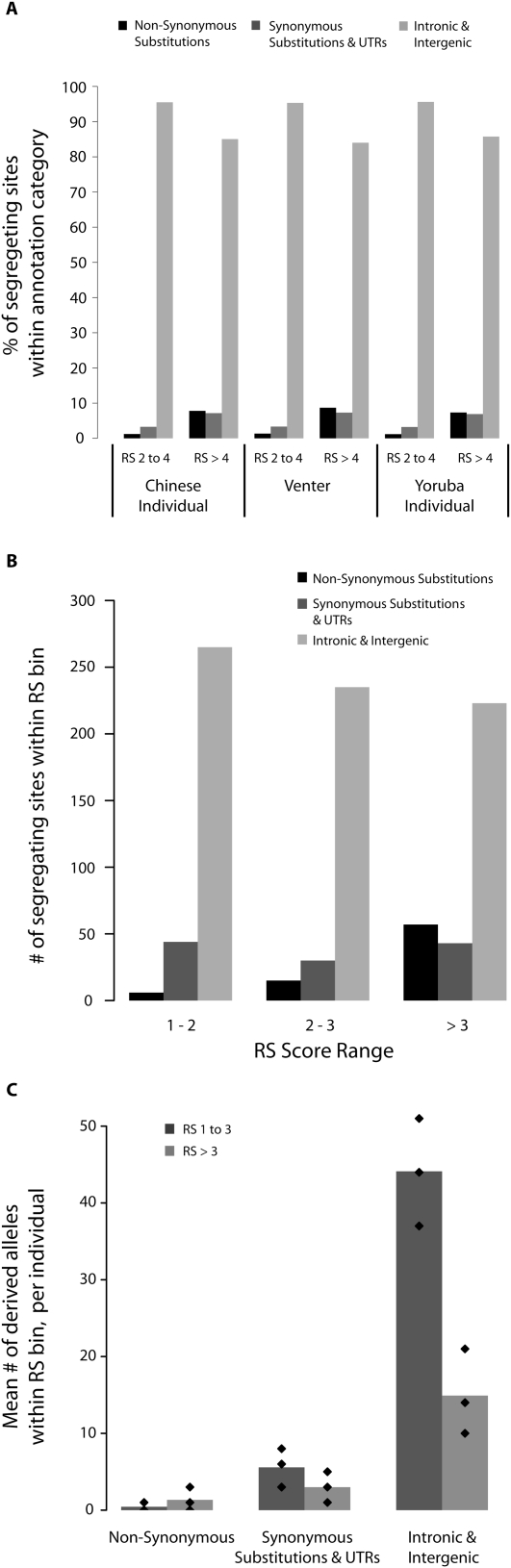
To examine how SNVs at constrained sites could be affecting function, we divided SNVs into three categories: those that result in nonsynonymous substitutions; variants within exons that do not alter the primary protein sequence but that may affect transcript stability or translation (i.e., synonymous substitutions and untranslated regions [UTRs]); and those that are outside of exons (intronic and intergenic). We find that noncoding sites dominate putatively functional variation in all three individuals. In the human genome as a whole, 82% of sites with RS > 2 are intronic or intergenic, but in each individual, >90% of SNVs affecting sites with RS > 2 are intronic or intergenic (Fig. 4A). Of the ∼6% of SNVs in each individual that affect sites within mature transcripts that have RS > 2, roughly a third cause amino acid replacements (Fig. 4A). While the proportion of nonsynonymous SNVs increases as the constraint threshold moves from RS > 2 to RS > 4, >90% of variation is still accounted for by SNVs that do not change protein sequences (Fig. 4A). Although the importance of nonsynonymous SNVs is well known (Bustamante et al. 2005; Boyko et al. 2008; Ng et al. 2008), our data show that the vast majority of an individual's functional variants do not result in amino acid replacements.
Figure 4.
Relative abundance of coding and noncoding functional variation. SNVs were divided into three categories: those that cause nonsynonymous substitutions; those that cause synonymous substitutions or changes in the UTRs; and those that do not occur in exons (intronic and intergenic). (A) Contribution of SNVs in each category to total variation at constrained sites in each resequenced individual genome at sites with RS 2 to 4 and at sites with RS > 4. (B) Total number of segregating sites in our ENCODE resequencing sample that occur at constrained sites in our three annotation categories. Constrained sites are divided into bins of increasing constraint. (C) Mean number of segregating sites carried by the individuals in our ENCODE resequencing sample in all three categories, at moderately (RS 1 to 3) and highly (RS > 3) constrained positions. Diamonds correspond to the 10%, 50%, and 90% quantiles.
The sites we identify as constrained are a mixture of truly constrained sites and neutrally evolving sites that appear constrained by chance (Cooper et al. 2005; Eddy 2005). If the false discovery rate (FDR) is dramatically higher for noncoding sequence than it is for coding sequence, this could explain the preponderance of noncoding SNVs we observe at constrained sites in an individual's genome. To investigate this possibility, we employed a Poisson model to predict the distribution of RS scores at neutrally evolving nonexonic sites, which gave an estimated FDR of 65% at sites with RS > 2 (Supplemental material). However, an FDR of >90% at sites with RS > 2 for noncoding sequence coupled with a FDR of zero for coding sequence would be required for an individual to have an equal number of functional coding and noncoding variants. Furthermore, two observations suggest that false positives are unlikely to account for the observation that most functional variants are noncoding. First, we note that many studies using a variety of neutral models have concluded that at least 5% (Mouse Genome Sequencing Consortium 2002; Rat Genome Sequencing Project Consortium 2004), and more likely 7%–10% (Siepel et al. 2005; Asthana et al. 2007a; Margulies et al. 2007), of the genome is under constraint, and an RS > 2 only captures 8.9% of the genome. Second, at more strongly constrained sites, those with RS > 4 (1.6% of the genome), ∼92% of the SNVs in an individual genome are noncoding (Fig. 4A), and we estimate an FDR of at most 13% at this threshold (Supplemental material). We conclude that a decisive majority of variants at functionally constrained sites in human genomes is noncoding.
Noncoding SNVs also predominate our deep ENCODE resequencing data, despite this data set being enriched for exons and for rare alleles (Fig. 1A). Of the 918 SNVs affecting mildly to strongly constrained positions (RS > 1), 78 are nonsynonymous substitutions and 840 are synonymous or noncoding (Fig. 4B; Supplemental Table S2). Noncoding variants are particularly prevalent among sites with weaker constraint, but even among the most constrained positions (RS > 3, N = 323), 82.3% of variants are synonymous or noncoding (Fig. 4B; Supplemental Table S2). We note that 258 of these 323 variants have a DAF of <1% and that selection against variation at constrained sites is consistent across all annotation categories (Supplemental Fig. S7). At the individual level, >90% of such putatively functional variation comes from noncoding sites, with most individuals harboring only a very small number of nonsynonymous changes (Fig. 4C). Thus, noncoding variation consistently outnumbers coding variation at comparable levels of constraint, even in a sample dominated by rare variants and even among the most constrained sites.
Discussion
This work provides an in-depth analysis of functional human variation at the individual level, encompassing both coding and noncoding sites and rare and common alleles. The results support two conclusions about the nature of human functional variation. First, an individual's phenotype is heavily influenced by SNVs that do not change protein sequence. Second, functional variation in a given genome is dominated by polymorphisms inherited from our shared ancestral population and remaining at significant frequencies in many modern-day human populations. By focusing on variation at the individual level, our findings provide two key insights that are directly relevant to future strategies for analysis of normal phenotypic variation and disease.
First, while the average deleteriousness of a nonsynonymous SNV may be greater than that of a noncoding or silent functional SNV (Boyko et al. 2008; Lohmueller et al. 2008; Ng et al. 2008), the sheer number of noncoding SNVs even among the most constrained sites indicates that they are likely to make a major contribution to the phenotype of a generally healthy individual, even when a large FDR for constrained noncoding sites is considered. It is worth noting that an analysis of the results of published genome-wide association studies revealed that 88% of trait or disease-associated SNVs are found outside of coding exons (Hindorff et al. 2009), a figure that is consistent with the proportion of SNVs at constrained sites within an individual that are noncoding (Fig. 4). This suggests that explicit and implicit reliance on the discovery of coding SNVs will overlook many important human variants.
Second, functional variation is dominated by common alleles that are shared between populations. As association studies have the highest power when causative alleles are also at high frequencies and present in multiple cohorts for the replication of associations (Bodmer and Bonilla 2008; McCarthy et al. 2008), genome-wide approaches for detecting statistical associations between alleles and phenotypes may yet be able to identify many functionally important alleles, particularly as resequencing efforts expand to more comprehensively define common but lower-frequency variants (i.e., 1%–10%) (McCarthy et al. 2008; Siva 2008; http://www.1000genomes.org). Our observation that well over 90% of potentially functional derived alleles within an individual come from SNVs with a DAF > 5% (Fig. 3B) indicates common variants shared among populations may collectively explain a large amount of functional variation between individuals. This is consistent with the intuition that, despite some phenotypic differentiation that correlates with geography (such as skin color), within-population variance of multiple morphological and physiological traits is similar among different populations.
Given that noncoding function is particularly difficult to investigate experimentally with base-pair-level resolution, and given that most functional variation in an individual is noncoding, comparative analyses of the sort we describe will be important when the sequencing of individual genomes becomes routine. They will facilitate comprehensive assessment of the potentially functional variation independent of annotation or allele frequency. Evolutionary constraint may also be of use in the fine-mapping of causative alleles after an association has been found for a particular genomic region or tagging SNV. Likewise, in parts of the genome with evidence for strong and recent selective pressure (Akey et al. 2004; Bustamante et al. 2005; Pickrell et al. 2009), measures of constraint could help to identify the selected variant, as such regions tend to be depleted of both functional and neutral variation (Cai et al. 2009).
However, there are some caveats to consider. SNVs with greater functional impact are more likely to be rare than neutral variants. This implies that the ascertainment of rare variation and of lesions other than SNVs, which are fewer in number but affect more bases (Kidd et al. 2008), remains important, particularly for extreme phenotypes and severe diseases (Romeo et al. 2007; Bodmer and Bonilla 2008; McCarthy et al. 2008; Mardis et al. 2009; Ng et al. 2009). On the other hand, given that SNVs with lesser functional impact are more likely to be common, many of the functional alleles carried by an individual may alone have small effects that are difficult to detect through an association study. Additionally, not all sites we identified as constrained are functional. As mentioned previously, some neutral sites may appear to be constrained simply by chance (Cooper et al. 2005; Eddy 2005), while other sites may have been functionally constrained during mammalian evolution but have more recently lost function in the human or ape lineages. Conversely, there are some functional sites in the genome that cannot be detected through comparative means, either because of limited alignment depth at these sites or because the function they perform is limited to a subset of the mammalian tree containing human (Cooper and Brown 2008). Nevertheless, by focusing on constrained sites one can obtain a set of SNVs that is highly enriched for functional variants.
The clear and strong signal of negative selection on variation affecting evolutionarily constrained sites suggests that, with additional comparative sequence and the increased power it would bring over what was available for our study, a large fraction of putatively functional variation can be identified in sequenced personal genomes. But until genome-wide resequencing is more economical, it would be logical to construct the next generation of genotyping tools to include alleles of common and intermediate frequency at sites that are more likely to be functional, i.e., those that are evolutionarily constrained. We estimate that the pool of human variation harbors about 1,000,000 polymorphisms that affect strongly constrained sites and have a global DAF > 1%, a number well within the capacity of modern genotyping platforms. Because most functional variants predate the geographic diversification of humans, such tools would be generally applicable and help uncover the genetic basis of normal phenotypic variation regardless of ethnicity.
Methods
DNA resequencing in the ENCODE Pilot regions
Five-hundred-seventy-five regions (74–1427 bp, median length 463 bp) containing CEs along with flanking neutral DNA were PCR amplified from a panel of 432 individuals from five populations: Yoruba from Ibadan, Nigeria (Yoruba; 121 individuals); Luhya from Webuye, Kenya (Luhya; 90 individuals); CEPH Utah (Utah; 75 individuals); Han Chinese from Beijing (Chinese; 73 individuals); and Japanese from Tokyo (Japanese; 73 individuals) (see Supplemental Methods). All individuals were unrelated. Equal numbers of males and females were resequenced. A complete list of samples, including the Coriell catalog ID for each sample, is given in Supplemental Table S6. A complete list of the Coriell plates from which our samples were obtained is given in Supplemental Table S7.
In total, 247,748 sites were resequenced. All sequencing reads were visually inspected for quality and acceptable reads were assembled for each resequenced region using phrap v.990319 (Ewing and Green 1998; Ewing et al. 1998). SNVs were identified using PolyPhred 5 (Stephens et al. 2006), and all SNV and indel calls were confirmed by visual inspection of the sequencing traces using consed (Gordon et al. 1998). Sequencing was performed at the Stanford Human Genome Center. SNVs and indels that could not be genotyped in at least 80% of the individuals in every one of the five sample populations were discarded. Two individuals were removed from the study, as genotyping of these individuals failed at >20% of loci.
The derived allele for each SNV was identified by comparison to the aligned position in baboon and chimp. SNVs at positions where the sequence differed between baboon and chimp, or where data were missing for one of the species, were removed from further analysis. Likewise, regions without at least one SNV for which the derived allele could be ascertained were also excluded. See Supplemental Methods for the details of our SNV filtering process. After filtering, 2214 SNVs and 134 indels in 516 regions totaling 227,252 bp in length remained. The DAF spectrum of the 2214 SNVs is strongly skewed toward rare alleles with 1537 SNVs occurring at a DAF of 1% or less (Fig. 1A).
Whole-genome variation data
SNV data for Craig Venter's genome were downloaded from the J. Craig Venter Institute (JCVI) public FTP site (ftp://ftp.jcvi.org/pub/data/huref/), and for the anonymous Chinese individual from the Beijing Genome Institute (BJI) website for the YanHuang Project (http://yh.genomics.org.cn/download.jsp). SNV data from the anonymous Yoruba individual were obtained from Illumina. Within each individual, we considered only the sites where that individual carried at least one derived allele. Derived and ancestral alleles were identified via comparison to the chimpanzee genome.
About 1% of the sites in the human reference sequence (hg18) differ from the chimpanzee reference sequence (panTro2) (The Chimpanzee Sequencing and Analysis Consortium 2005). Many of these sites are due to substitutions that have fixed in the human or chimpanzee lineages (The Chimpanzee Sequencing and Analysis Consortium 2005), but some of the apparent differences between hg18 and panTro2 are due to sites that are polymorphic in humans and at which hg18 happens to contain the derived allele. The SNVs available for each of the individual genomes were obtained through comparison to hg18 (Levy et al. 2007; Bentley et al. 2008; Wang et al. 2008), so if an individual happens to be homozygous for the derived allele at a site that is polymorphic in the human population but where hg18 also carries the derived allele, that site will not be detected as polymorphism in that individual. To uncover such “hidden” SNVs, we compared all sites with SNVs from all three individual genomes to each other and to hg18 and panTro2 to uncover additional sites in each individual that were polymorphic in this sample (the three individuals plus hg18) and that carried a derived allele. This comparison revealed an additional 200,000 to 500,000 polymorphic sites harboring derived alleles in each individual, creating a final data set of 3.2–3.6 million SNVs per individual (Supplemental Table S1).
Measurement of evolutionary constraint
In the ENCODE Pilot regions, CEs were initially identified in multiple alignments of ENCODE Pilot regions using GERP 1.0 (Cooper et al. 2005) with default parameters. During the course of sequencing these regions, deeper alignments, containing 24–26 mammalian sequences, became available, affording greater resolution, and an improved version of GERP (2.1) (http://mendel.stanford.edu/SidowLab/downloads/gerp/index.html) was used to obtain the site-specific RS scores. Multisequence alignments for each of the 44 ENCODE Pilot regions constructed using the Threaded Blockset Aligner (TBA) program (Blanchette et al. 2004), and the September 2007 ENCODE sequence data freeze were used as input for GERP. In this set of alignments, the nonhuman sequences were reordered to be syntenic with the human sequence, such that all human bases are present in each alignment and have at most one aligned nucleotide from each other species. Further details about the alignments can be found in Margulies et al. (2007). We processed these alignments by removing all nonmammalian species and any species that had extensive gaps or was missing sequence for >50% of the ENCODE Pilot regions. To make the human sequence continuous, we removed from the alignment any positions that were gapped in human. The expected RS score of a neutral site is zero. In our alignments the maximum possible RS score is 4.22, reflecting the depth of the neutral tree relating the aligned sequences. The branch lengths of the neutral tree were calculated using fourfold degenerate sites.
To obtain site-specific constraint (RS) scores for as much of the human genome as possible, we used GERP 2.2 with 44-way MULTIZ/TBA alignments, downloaded from the University of California Santa Cruz (UCSC) Genome Browser FTP site (ftp://hgdownload.cse.ucsc.edu/goldenPath/hg18/multiz44way/). All nonmammalian sequences, as well as the human sequence, were removed from the alignment, and only sites where at least three mammalian species remained were used to calculate RS scores. Approximately 42% of the human genome was aligned with a depth sufficient for detection of sites having a RS score > 2 (Fig. 2A). All sites that were not aligned to any other mammals or were aligned at an insufficient depth (<0.5 substitutions/site) were given an RS score of zero. The maximum RS score possible from the 44-way alignments is 5.82.
Genome annotations
Annotations from the “knownGenes” track were downloaded from the from the UCSC ENCODE browser (http://genome.ucsc.edu/ENCODE/downloads.html) for version hg18 of the human genome. Each base was annotated as belonging to a coding exon, an intron, or a UTR, or as intergenic if it did not fall into any of these three categories. All SNVs in coding exons were further divided into nonsynonymous and synonymous.
All statistical analyses were conducted using R (http://www.r-project.org/).
Acknowledgments
We thank E. Margulies and G. McEwen at the NHGRI, and the NISC Comparative Sequencing Program, for alignments of the ENCODE Pilot regions and phylogenetic trees; N. Vo and C. Eastman at the Stanford Human Genome Center for assistance with verification of SNV calls; and D. Petrov for helpful comments and discussion. We thank M. Ross and G. Schroth at Illumina for providing us with SNV data from the Yoruba individual. Our paper was greatly improved by the insightful and constructive comments of our anonymous reviewers. D.L.G. is a Lucille P. Markey Biomedical Research Stanford Graduate Fellow. G.M.C. is supported by a Merck, Jane Coffin Childs Memorial Fund Fellowship. This work was funded by an ENCODE Pilot Project Grant (NIH/NHGRI) to R.M.M., S.B., and A.S.
Footnotes
[Supplemental material is available online at http://www.genome.org. All sequences can be retrieved from the NCBI Trace Archive (http://www.ncbi.nlm.nih.gov/Traces/trace.cgi) using the search string CENTER_NAME = “SHGC” and SPECIES_CODE = “HOMO SAPIENS”.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.102210.109.
References
- Ahituv N, Kavaslar N, Schackwitz W, Ustaszewska A, Martin J, Hebert S, Doelle H, Ersoy B, Kryukov G, Schmidt S, et al. Medical sequencing at the extremes of human body mass. Am J Hum Genet. 2007;80:779–791. doi: 10.1086/513471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akey JM, Eberle MA, Rieder MJ, Carlson CS, Shriver MD, Nickerson DA, Kruglyak L. Population history and natural selection shape patterns of genetic variation in 132 genes. PLoS Biol. 2004;2:e286. doi: 10.1371/journal.pbio.0020286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asthana S, Roytberg M, Stamatoyannopoulos J, Sunyaev S. Analysis of sequence conservation at nucleotide resolution. PLoS Comput Biol. 2007a;3:e254. doi: 10.1371/journal.pcbi.0030254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asthana S, Noble WS, Kryukov G, Grant CE, Sunyaev S, Stamatoyannopoulos JA. Widely distributed noncoding purifying selection in the human genome. Proc Natl Acad Sci. 2007b;104:12410–12415. doi: 10.1073/pnas.0705140104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhangale TR, Rieder MJ, Nickerson DA. Estimating coverage and power for genetic association studies using near-complete variation data. Nat Genet. 2008;40:841–843. doi: 10.1038/ng.180. [DOI] [PubMed] [Google Scholar]
- Blanchette M, Kent WJ, Riemer C, Elnitski L, Smit AF, Roskin KM, Baertsch R, Rosenbloom K, Clawson H, Green ED, et al. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 2004;14:708–715. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodmer W, Bonilla C. Common and rare variants in multifactorial susceptibility to common diseases. Nat Genet. 2008;40:695–701. doi: 10.1038/ng.f.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyko AR, Williamson SH, Indap AR, Degenhardt JD, Hernandez RD, Lohmueller KE, Adams MD, Schmidt S, Sninsky JJ, Sunyaev SR, et al. Assessing the evolutionary impact of amino acid mutations in the human genome. PLoS Genet. 2008;4:e1000083. doi: 10.1371/journal.pgen.1000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante CD, Fledel-Alon A, Williamson S, Nielsen R, Hubisz MT, Glanowski S, Tanenbaum DM, White TJ, Sninsky JJ, Hernandez RD, et al. Natural selection on protein-coding genes in the human genome. Nature. 2005;43:1153–1157. doi: 10.1038/nature04240. [DOI] [PubMed] [Google Scholar]
- Cai JJ, Macpherson JM, Sella G, Petrov DA. Pervasive hitchhiking at coding and regulatory sites in humans. PLoS Genet. 2009;5:e1000336. doi: 10.1371/journal.pgen.1000336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CT, Wang JC, Cohen BA. The strength of selection on ultraconserved elements in the human genome. Am J Hum Genet. 2007;80:692–704. doi: 10.1086/513149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Chimpanzee Sequencing and Analysis Consortium. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- Chun S, Fay JC. Identification of deleterious mutations within three human genomes. Genome Res. 2009;19:1553–1561. doi: 10.1101/gr.092619.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper GM, Brown CD. Qualifying the relationship between sequence conservation and molecular function. Genome Res. 2008;18:201–205. doi: 10.1101/gr.7205808. [DOI] [PubMed] [Google Scholar]
- Cooper GM, Stone EA, Asimenos G, Green ED, Batzoglou S, Sidow A. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 2005;15:901–913. doi: 10.1101/gr.3577405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper SJ, Trinklein ND, Anton ED, Nguyen L, Myers RM. Comprehensive analysis of transcriptional promoter structure and function in 1% of the human genome. Genome Res. 2006;16:1–10. doi: 10.1101/gr.4222606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake JA, Bird C, Nemesh J, Thomas DJ, Newton-Cheh C, Reymond A, Excoffier L, Attar H, Antonarakis SE, Dermitzakis ET, et al. Conserved noncoding sequences are selectively constrained and not mutation cold spots. Nat Genet. 2006;38:223–227. doi: 10.1038/ng1710. [DOI] [PubMed] [Google Scholar]
- Drosophila 12 Genomes Consortium. Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- Eddy SR. A model of the statistical power of comparative genome sequence analysis. PLoS Biol. 2005;3:e10. doi: 10.1371/journal.pbio.0030010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, Belmont JW, Boudreau A, Hardenbol P, Leal SM, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs R. Deeper into the genome. Nature. 2005;437:1233–1234. doi: 10.1038/4371233a. [DOI] [PubMed] [Google Scholar]
- Gordon D, Abajian C, Green P. Consed: A graphical tool for sequence finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- Hata J, Matsuda K, Ninomiya T, Yonemoto K, Matsushita T, Ohnishi Y, Saito S, Kitazono T, Ibayashi S, Iida M, et al. Functional SNP in an Sp1-binding site of AGTRL1 gene is associated with susceptibility to brain infarction. Hum Mol Genet. 2007;16:630–639. doi: 10.1093/hmg/ddm005. [DOI] [PubMed] [Google Scholar]
- Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose Y, Chiba K, Karasugi T, Nakajima M, Kawaguchi Y, Mikami Y, Furuichi T, Mio F, Miyake A, Miyamoto T, et al. A functional polymorphism in THBS2 that affects alternative splicing and MMP binding is associated with lumbar-disc herniation. Am J Hum Genet. 2008;82:1122–1129. doi: 10.1016/j.ajhg.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzman S, Kern AD, Bejerano G, Fewell G, Fulton L, Wilson RK, Salama SR, Haussler D. Human genome ultraconserved elements are ultraselected. Science. 2007;317:915. doi: 10.1126/science.1142430. [DOI] [PubMed] [Google Scholar]
- Kellis M, Patterson N, Endrizzi M, Birren B, Lander ES. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423:241–254. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- Kidd JM, Cooper GM, Donahue WF, Hayden HS, Sampas N, Graves T, Hansen N, Teague B, Alkan C, Antonacci F, et al. Systematic assessment of copy number variant detection via genome-wide SNP genotyping. Nature. 2008;453:56–64. doi: 10.1038/ng.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JI, Ju YS, Park H, Kim S, Lee S, Yi JH, Mudge J, Miller NA, Hong D, Bell CJ, et al. A highly annotated whole-genome sequence of a Korean individual. Nature. 2009;460:1011–1015. doi: 10.1038/nature08211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. The neutral theory of molecular evolution. Cambridge University Press; Cambridge, UK: 1983. [Google Scholar]
- King MC, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Levy S, Sutton G, Ng PC, Feuk L, Halpern AL, Walenz BP, Axelrod N, Huang J, Kirkness EF, Denisov G, et al. The diploid genome sequence of an individual human. PLoS Biol. 2007;5:e254. doi: 10.1371/journal.pbio. 0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JZ, Absher DM, Tang H, Southwick AM, Casto AM, Ramachandran S, Cann HM, Barsh GS, Feldman M, Cavalli-Sforza LL, et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- Lohmueller KE, Indap AR, Schmidt S, Boyko AR, Hernandez RD, Hubisz MJ, Sninsky JJ, White TJ, Sunyaev SR, Nielsen R, et al. Proportionally more deleterious genetic variation in European than in African populations. Nature. 2008;451:994–997. doi: 10.1038/nature06611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, Koboldt DC, Fulton RS, Delehaunty KD, McGrath SD, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361:1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies EH, Cooper GM, Asimenos G, Thomas DJ, Dewey CN, Thomas DJ, Dewey CN, Siepel A, Birney E, Keefe D, et al. Analyses of deep mammalian sequence alignments and constraint predictions for 1% of the human genome. Genome Res. 2007;17:760–774. doi: 10.1101/gr.6034307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marth GT, Czabarka E, Murvai J, Sherry ST. The allele frequency spectrum in genome-wide human variation data reveals signals of differential demographic history in three large world populations. Genetics. 2004;166:351–372. doi: 10.1534/genetics.166.1.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, Ioannidis JP, Hirschhorn JN. Genome-wide association studies for complex traits: Consensus, uncertainty and challenges. Nat Rev Genet. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- McKernan KJ, Peckham HE, Costa GL, McLaughlin SF, Fu Y, Tsung EF, Clouser CR, Duncan C, Ichikawa JK, Lee CC, et al. Sequence and structural variation in a human genome uncovered by short-read, massively parallel ligation sequencing using two-base encoding. Genome Res. 2009;19:1527–1541. doi: 10.1101/gr.091868.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouse Genome Sequencing Consortium. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Ng P, Henikoff S. Sift: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng PC, Levy S, Huang J, Stockwell TB, Walenz BP, Li K, Axelrod N, Busam DA, Strausberg RL, Venter JC. Genetic variation in an individual human exome. PLoS Genet. 2008;4:e1000160. doi: 10.1371/journal.pgen.1000160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SB, Turner EH, Robertson PD, Flygare SD, Bigham AW, Lee C, Shaffer T, Wong M, Bhattacharjee A, Eichler EE, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–276. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin SH, Kazazian HH, Jr, Antonarakis SE, Goff SC, Boehm CD, Sexton JP, Waber PG, Giardina PJ. Linkage of β-thalassaemia mutations and β-globin gene polymorphisms with DNA polymorphisms in human β-globin gene cluster. Nature. 1982;296:627–631. doi: 10.1038/296627a0. [DOI] [PubMed] [Google Scholar]
- Pennacchio LA, Ahituv N, Moses AM, Prabhakar S, Nobrega MA, Shoukry M, Minovitsky S, Dubchak I, Holt A, Lewis KD, et al. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444:499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- Pickrell JK, Coop G, Novembre J, Kudaravalli S, Li JZ, Absher D, Srinivasan BS, Barsh GS, Myers RM, Feldman MW, et al. Signals of recent positive selection in a worldwide sample of human populations. Genome Res. 2009;19:826–837. doi: 10.1101/gr.087577.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rat Genome Sequencing Project Consortium. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature. 2004;428:493–521. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- Romeo S, Pennacchio LA, Fu Y, Boerwinkle E, Tybjaerg-Hansen A, Hobbs HH, Cohen JC. Population-based resequencing of ANGPTL4 uncovers variations that reduce triglycerides and increase HDL. Nat Genet. 2007;39:513–516. doi: 10.1038/ng1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siva N. 1000 Genomes project. Nat Biotechnol. 2008;26:256. doi: 10.1038/nbt0308-256b. [DOI] [PubMed] [Google Scholar]
- Stephens M, Sloan JS, Robertson PD, Scheet P, Nickerson DA. Automating sequence-based detection and genotyping of SNPs from diploid samples. Nat Genet. 2006;38:375–381. doi: 10.1038/ng1746. [DOI] [PubMed] [Google Scholar]
- Stone EA, Sidow A. Physicochemical constraint violation by missense substitutions mediates impairment of protein function and disease severity. Genome Res. 2005;15:978–986. doi: 10.1101/gr.3804205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunyaev S, Ramensky V, Koch I, Lathe W, Kondrashov A, Bork P. Prediction of deleterious human alleles. Hum Mol Genet. 2001;10:591–597. doi: 10.1093/hmg/10.6.591. [DOI] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Verlaan DJ, Berlivet S, Hunninghake GM, Madore AM, Larivière M, Moussette S, Grundberg E, Kwan T, Ouimet M, Ge B, et al. Allele-specific chromatin remodeling in the ZPBP2/GSDMB/ORMDL3 locus associated with the risk of asthma and autoimmune disease. Am J Hum Genet. 2009;85:377–393. doi: 10.1016/j.ajhg.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Prabhakar S, Akiyama JA, Shoukry M, Lewis KD, Holt A, Plajzer-Frick I, Afzal V, Rubin EM, Pennacchio LA. Ultraconservation identifies a small subset of extremely constrained developmental enhancers. Nat Genet. 2008;40:158–160. doi: 10.1038/ng.2007.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Wang W, Li R, Li Y, Tian G, Goodman L, Fan W, Zhang J, Li J, Zhang J, et al. The diploid genome sequence of an Asian individual. Nature. 2008;456:60–65. doi: 10.1038/nature07484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolfe A, Goodson M, Goode DK, Snell P, McEwen GK, Vavouri T, Smith SF, North P, Callaway H, Kelly K, et al. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol. 2005;3:e7. doi: 10.1371/journal.pbio.0030007. [DOI] [PMC free article] [PubMed] [Google Scholar]