Abstract
Antimicrobial peptides (AMPs) are a crucial component of the natural immune system in insects. Five types of AMPs have been identified in the tobacco hornworm Manduca sexta, including attacin, cecropin, moricin, gloverin, and lebocin. Here we report the isolation of lebocin-related cDNA clones and antibacterial activity of their processed protein products. The seventeen cDNA sequences are composed of a constant 5′ end and a variable 3′ region containing 3∼16 copies of an 81-nucleotide repeat. The sequence of the corresponding gene isolated from a M. sexta genomic library and Southern blotting results indicated that the gene lacks introns and exists as a single copy in the genome. The genomic sequence contained 13 complete and one partial copy of the 81-nucleotide repeat. Northern blot analysis revealed multiple transcripts with major size differences. The mRNA level of M. sexta lebocin increased substantially in fat body after larvae had been injected with bacteria. The RXXR motifs in the protein sequences led us to postulate that the precursors are processed by an intracellular convertase to form four bioactive peptides. To test this hypothesis, we chemically synthesized the peptides and examined their antibacterial activity. Peptide 1 killed Gram-positive and Gram-negative bacteria. Peptide 2, similar in sequence to a Galleria mellonella AMP, did not affect the bacterial growth. Peptide 3 was inactive but peptide 3 with an extra Arg at the carboxyl terminus was active against E. coli at a high minimum inhibitory concentration. Peptide 4, encoded by the 81-bp repeat, was inactive in the antibacterial tests. The hypothesis that posttranslational processing of the precursor proteins produces multiple bioactive peptides for defense purposes was validated by identification of peptides 1, 2, and 3 from larval hemolymph via liquid chromatography and tandem mass spectrometry. Comparison with the orthologs from other lepidopteran insects indicates that the same mechanism may be used to generate several functional products from a single precursor.
1. Introduction
Antimicrobial peptides (AMPs) participate in innate immunity of various organisms ranging from prokaryotes to plants, invertebrates, and vertebrates [1,2]. Most of them are less than 5 kDa, hydrophobic, membrane-active, and carry positive net charge at physiological pH. These peptides are either absent or present at low constitutive levels in naïve insects. Upon microbial infection, association of host recognition molecules and microorganisms triggers extracellular serine proteases to activate a spätzle precursor via limited proteolysis [3]. Spätzle then binds to the Toll receptor to initiate an intracellular pathway that relays the signal into nucleus, where transcription factors of the Rel family induce the AMP gene expression [4]. Identification of orthologous genes in the Anopheles gambiae, Aedes aegypti, Apis mellifera, Tribolium castaneum, and Bombyx mori genomes suggests that similar signaling pathways exist in other insects to induce the production of defense proteins by fat body and hemocytes [5-9]. Fat body, a tissue analogous to liver, synthesizes AMPs and secretes these heat-stable compounds into the plasma to kill the invading microbes.
Insect AMPs can be categorized into the following groups: α-helical peptides (e.g. cecropin, moricin), disulfide-stabilized peptides (e.g. defensin, drosomycin), proline-rich peptides (e.g. lebocin, drosocin), glycine-rich peptides (e.g. gloverin, diptericin), and others [2]. In the tobacco hornworm Manduca sexta, at least five types of AMPs have been identified, including attacin, cecropin, moricin, gloverin, and lebocin [10-13]. As a step towards understanding the role of AMPs in humoral immune responses of M. sexta, we have characterized a lebocin-like protein, its processing products, and corresponding gene in this work.
Lebocins are proline-rich AMPs first purified from the silkworm, Bombyx mori [14]. These 32-residue peptides are glycosylated at Thr15 and this modification is important for the antimicrobial activity against Gram-negative bacteria. cDNA and gene cloning indicates that the active peptide is located near the carboxyl terminus, after a signal peptide and a 102-residue pro-segment [15,16]. Lebocins increase the permeability of liposomes at a low ionic strength and have weak antibacterial activities under physiological conditions. They seem to function as synergists by reducing the minimum inhibitory concentration of cecropin D [17]. Lebocin cDNAs have been isolated from Trichoplusia ni [18] and Pseudoplusia includens [19]. In lebocin homologs of Samia cynthia [20], the pro-segment aligns well with the Bombyx mori sequences, but the part corresponding to the mature lebocin differs significantly. Two peptides, purified from hemolymph of Helicoverpa armigera and Galleria mellonella, are similar to a region in the pro-segment of B. mori lebocins [21,22]. The 42-residue anionic peptide-1 from the greater wax moth G. mellonella is active against Micrococcus luteus, Listeria monocytogenes and filamentous fungi, but neither of these peptides inhibits the growth of Gram-negative bacteria (e.g. Escherichia coli). It is unclear how these peptides are derived from their protein precursors (pro-lebocins). Here we report the cDNA and genomic cloning of lebocin-related proteins from M. sexta, which suggests a conserved mechanism in Lepidoptera to generate structural/functional diversity in products derived from pro-lebocins. This mechanism is validated by identification in hemolymph of three processing products by mass spectrometry. For simplicity, we use “lebocin” in parts of the paper to describe the entire gene, cDNA, and protein that are related but not the same as the mature lebocin peptide.
2. Materials and methods
2.1. Insect rearing and bacterial challenge
M. sexta (eggs purchased from Carolina Biological Supply) were reared on an artificial diet [23]. Day 2, fifth instar larvae were injected with formalin-killed E. coli (2×108 cells/larva). Alternatively, day 2, fifth instar larvae or day 1, male adults were injected with 100 μg M. luteus (Sigma) or with sterile water as a control. At various time points after injection, hemocytes and fat body were collected for RNA preparation. Muscle tissues or gut-removed carcasses were used for genomic DNA isolation.
2.2. Screening for full-length lebocin cDNA clones
A 280 bp M. sexta lebocin cDNA fragment [11] was labeled with [α-32P]-dCTP using Multiprime DNA Labeling System (GE Healthcare Life Science). A M. sexta induced fat body cDNA library [24] was screened according to Sambrook and Russell [25]. Positive plaques were purified to homogeneity via secondary and tertiary screening. Plasmids, in vivo excised from the positive bacteriophages, were sequenced using a BigDye Terminator Cycle Sequencing Ready Reaction Kit (PE Applied Biosystems). Sequences were assembled using MacVector Sequence Analysis Software (Oxford Molecular Ltd).
2.3. Multiple sequence alignment and phylogenetic analysis
Based on BLAST search of GenBank (http://www.ncbi.nlm.nih.gov/) and ButterflyBase (http://butterflybase.ice.mpg.de/) using M. sexta lebocin as query, homologous protein sequences were retrieved and compared using ClustalX 1.83 (ftp://ftp-igbmc.u-strasbg.fr/pub/ClustalX/) [26]. A Blosum 30 matrix [27], with a gap penalty of 10 and an extension gap penalty of 0.1 were selected for multiple sequence alignment and unrooted phylogenetic tree was constructed based on neighbor-joining algorithm. Treeview (http://taxonomy.zoology.gla.ac.uk/rod/treeview.html) [28] was used to display Phylogram.
2.4. Genomic library screening, subcloning, and DNA sequencing
The insert from a full-length lebocin cDNA clone (NC2), obtained after digestion with EcoRI and XhoI, was labeled with [α-32P]-dCTP and used as a probe to screen a M. sexta genomic library in λGEM11, kindly provided by Dr. Yucheng Zhu at the Southern Insect Management Research Unit (USDA ARS). Following plaque purification and amplification, phage DNA was isolated using Wizard Lambda Preps DNA Purification System (Promega). To determine its restriction map, the DNA was digested with one, two or three of the enzymes (XhoI, ApaI, EcoRI, HindIII, KpnI, SacI, SalI and XbaI) and separated by 0.8% agarose gel electrophoresis. After transferring onto a GenScreen Plus membrane (NEN Life Science Products), the DNA fragments were hybridized with the full-length lebocin cDNA, labeled by DIG-High Prime DNA Labeling Detection Kit (Roche Applied Science). Fragments of the lebocin gene were subcloned, sequenced, and assembled as described above.
2.5. Genomic Southern blot analysis
M. sexta genomic DNA was extracted from muscles of a single fifth instar larva using a DNeasy Blood and Tissue Kit (Qiagen). The genomic DNA was also isolated from two larval carcasses according to Bradfield and Wyatt [29]. About 10 μg of DNA was digested with XhoI and ScaI at 37 °C overnight and separated by 1% agarose gel electrophoresis. After capillary transfer onto a nitrocellulose membrane, hybridization was carried out using 32P-labeled XhoI-ScaI fragment of the lebocin cDNA.
2.6. Northern blot and RT-PCR analysis
Fat body RNA samples were prepared from the individual insects using Micro-to-Midi Total RNA Purification System (Invitrogen) or the method described by Chinzei et al [30]. Denatured total RNA samples were separated on a 1% agarose gel containing 2.2 M formaldehyde. After electrophoresis, RNA was transferred to a membrane and hybridized with 32P-labeled XhoI-ScaI fragment of the cDNA. Similarly, the time course of lebocin expression in larvae and adults were examined. For RT-PCR analysis, hemocyte and fat body total RNA samples were prepared from naïve and injected larvae. In each reaction, RNA (2∼4 μg), oligo(dT) (0.5 μg) and dNTPs (1 μl, 10 mM each) were mixed with DEPC-treated H2O in a final volume of 12 μl and denatured at 65 °C for 5 min. M-MLV reverse transcriptase (1 μl, 200 U, Invitrogen), 5×buffer (4 μl), 0.1 M dithiothreitol (2 μl), and RNase OUT (1 μl, 40 U) were added to the RNA for cDNA synthesis at 37 °C for 50 min. The M. sexta ribosomal protein S3 mRNA was used as an internal control to normalize the cDNA samples in a PCR using primers j501 (5′-GCCGTTCTTGCCCTGTT-3′) and j504 (5′-CGCGAGTTGACTTCGGT-3′). The lebocin cDNA fragment was amplified using forward (5′-CTGATTTTGGGCGTTGCGCTG-3′) and reverse (5′-GCGCGTATCTTCTATCTGGA-3′) primers under conditions empirically chosen to avoid saturation: 30 cycles of 94 °C, 30 s; 50 °C, 30 s; 72 °C, 30 s. The relative levels of lebocin mRNA in the normalized samples were determined by 1.5% agarose gel electrophoresis.
2.7. Peptide synthesis
Eight lebocin-related peptides (∼10 mg each) (LP1, QRFSQPTFKLPQGRLTLSRKFR; LP1A, QRFSQPTFKLPQGRLTLSRKF; LP2, ESGNEPLWLYQGDNIPKAPSTAEHPFLPSIIDDVKFNPDRRYAR; LP2A, ESGNEPLWLYQGDNIPKAPSTAEHPFLPSIIDDVKFNPDRRYA; LP3, SLGTPDHYHGGRHSISRGSQSTGPTHPGYNRRNAR; LP3A, SLGTPDHYHGGRHSISRGSQSTGPTHPGYNRRNA; LP4, SVETLASQEHLSSLPMDSQETLLRGTR; LP4A, SVETLASQEHLSSLPMDSQETLLRGT) were prepared by stepwise solid-phase synthesis using Fmoc-amino acid derivatives (Bio-Synthesis Inc). Following deprotection and cleavage, the peptides were purified by reversed-phase HPLC to >95% purity and analyzed by mass spectrometry.
2.8. Antibacterial activity assay
The synthetic peptides were separately tested against pathogenic strains of Salmonella typhimurium, E. coli O157:H7, Klebsiella pneumoniae, S. typhimurium DT104, L. monocytogenes, Staphylococcus aureus, and two strains of methicillin-resistant S. aureus, kindly provided by Dr. Guolong Zhang in the Department of Animal Science at Oklahoma State University. The minimum inhibitory concentrations (MICs) were determined in a broth micro-dilution assay [31]. Briefly, overnight bacterial cultures were subcultured into 4 ml of Trypticase Soy Broth for 3-5 h until the bacteria reached mid-log phase. After centrifugation at 1000 × g at 4 °C and washing with 10 mM Tris-HCl (pH 7.4), the cells were suspended in 5% Trypticase Soy Broth (5×105 cfu/ml). Aliquots of the diluted cultures (90 μl) were mixed with 10 μl of the synthetic peptide at 1000, 500, 250, 125, 62.5 μg/ml. All bacteria were cultured at 37 °C for overnight in a 96-well cell culture plate, and the lowest concentration of peptide that caused no visible growth was recorded. The experiment was performed at least three times for each strain to obtain MICs of the M. sexta lebocin-related peptides against the bacterial strains.
2.9. Identification of lebocin-related peptides in plasma by mass spectrometry
Synthetic peptides and biological samples were characterized using a hybrid LTQ-Orbitrap mass spectrometer (Thermo Fisher Scientific) coupled to a New Objectives PV-550 nanoelectrospray ion source and an Eksigent NanoLC-2D chromatography system. MS and MS/MS spectra of LP1A through LP4A were first collected by infusion using 1-micron infusion tips (New Objective). These peptides were further analyzed by trapping on a 2.5 cm ProteoPrepII pre-column (New Objective), followed by analytical separation on a 75 μm ID fused silica column packed in house with10 cm of Magic C18 AQ, terminated with an integral fused silica emitter pulled in house. The peptides were eluted using a 3-70% acetonitrile (AcCN)/0.1% formic acid gradient performed over 33 min at a flow rate of 300 nL/min. During each one-second full-range FT-MS scan (nominal resolution of 60,000 FWHM, 300 to 2000 m/z), the three most intense ions were analyzed via MS/MS in the linear ion trap. MS/MS spectra were collected using a trigger threshold of 1000 counts and monoisotopic precursor selection. Parent ions were rejected for MS/MS if their charge states were unassignable, if they were previously identified as contaminants on blank gradient runs, or if they had already been twice selected for MS/MS (data dependent acquisition using a dynamic exclusion for 150% of the typical chromatographic peak width). Column performance was monitored using the peptide standards, and via blank injections between samples to assay for contamination. A 100 μl aliquot of pooled hemolymph from bacteria-injected M. sexta larvae was heated at 95 °C for 5 min and centrifuged at 4 °C for 10 min at 12,000g. Polypeptides in the supernatant (40 μl) was cleaned up on a PepClean™ C-18 Spin Column (Pierce) according to the manufacturer's instructions. Peptide samples eluted with 5-40% AcCN and 40-70% AcCN were lyophilized, reconstituted in 0.1% formic acid, and combined for LC-MS/MS analysis as described above. The resultant LC-MS/MS files were inspected manually, using the synthetic peptide's retention time, m/z, charge states, isotopic distributions, and characteristic MS/MS decay spectra as identifying criteria. For use as a standard to show that the natural LPA1 has a pyroGlu at the amino terminus, the first Gln in the synthetic peptide was partly converted to pyroGlu by incubating LPA1 (1 μl, 1 mg/ml) with 50 μl, 100 mM NH4HCO3 (pH 8.2) overnight at 37 °C [32]. The peptide mixture was enriched on a ZipTipC18 (Millipore) and analyzed by LC and tandem mass spectrometry as described above.
3. Results
During the cDNA cloning of M. sexta hemolin [33], we isolated a false positive clone (pP4-7) which encodes a region 48% identical to B. mori lebocin in the amino-terminal pro-segment followed by twelve copies of a 27-residue repeated amino acid sequence. In a separate project to identify bacteria-induced genes by suppression subtractive hybridization and molecular cloning, Zhu et al. [11] identified 5 cDNA fragments (BI262584, BI262626, BI262686, BI262688, and BI262708) encoding sequences similar to pP4-7 and the proregion of silkworm lebocins. Intrigued by the unique structure of pP4-7 and similarity of the PCR-derived clones to lebocin, we screened an induced fat body cDNA library using the 5′ unrepeated region as a probe and isolated fifteen positive clones.
Sequencing from both ends of the positive clones indicated that all of the cDNAs contained a 5′ region identical to that found in pP4-7, which includes a 5′ untranslated region (5′-UTR) and a 363-nucleotide sequence encoding a 20-residue signal peptide and a 101-residue segment similar to the proregion of B. mori lebocin (Fig. 1A). Following this region, an 81-nucleotide sequence was repeated 3∼16 times in different clones. Four clones (NC2, NC3, NC10 and NC11) contain thirteen repeats and the other twelve (NC7, G6, G1, NC6, NC9, NC13, NC1, NC5, G2, NC8, pP4-7 and C5) have 3, 3, 5, 7, 8, 8, 9, 10, 11, 12, 12 and 16 repeats, respectively. We have identified in DQ115324 an open reading frame of the pro-lebocin with the constant region and 6.3 repeats, out-of-frame fused with M. sexta HP23 cDNA as an artifact of library construction [34]. The repeats in these seventeen clones are nearly identical at the nucleotide level and they encode peptide sequences that are highly conserved. Following these complete repeats is a constant region consisting of a 33 bp partial repeat, a stop codon, and a 3′-UTR.
Fig. 1. Nucleotide and deduced amino acid sequences of M. sexta lebocin cDNA (A) and gene (B).
Amino acid sequence is listed below the coding region using one letter code under the middle nucleotide of each codon. The signal peptide is underlined. The processing enzyme cleavage sites (RXXR) are double underlined. The Ser, His, and Val residues, which deviate from the consensus of the 27-residue repeats, and the PstI, XhoI and ScaI sites flanking the DNA repeats are in bold and underlined. Synonymous and nonsynonymous substitutions, which differ from the consensus of the 81-nuclotide repeats, are shaded grey and green, respectively. The polyadenylation signal (AATAAA) is double underlined. (A) NC6 cDNA (FJ530954). The intact and partial repeats are shaded and unshaded alternatively. (B) Gene (FJ530955). Three GATA motifs are boxed, two NF-κB sites double underlined, one ISRE site shaded, and mismatched nucleotides in lower case. The TATA box and CCAGT sequence for transcription initiation are underlined.
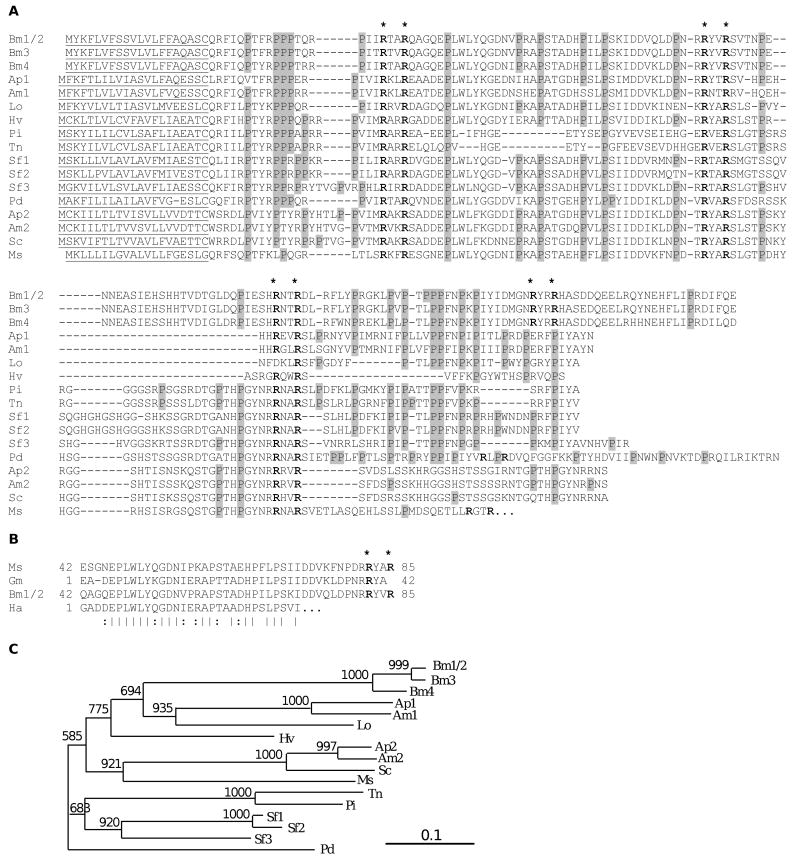
Amino acid sequence comparison shows that the 101-residue pro-segment in M. sexta lebocin is similar to its counterpart in T. ni (45%), P. includens (49%), Papilio dardanus (68%), Spodoptera frugiperda (66% for 1, 64% for 2, 70% for 3), B. mori (64%), Heliothis virescens (72%), S. cynthia (78%), Antheraea mylitta (67% for 1 and 75% for 2), Antheraea pernyi (70% for 1 and 73% for 2), Lonomia obliqua (81%) (Fig. 2). In the remaining carboxyl-terminal region, the T. ni and P. includens sequences are 70% identical to each other, the identity between A. mylitta-1 and A. pernyi-1 is 76%, and the S. cynthia, A. mylitta-2 and A. pernyi-2 sequences are ∼80% identical. However, this high similarity in the carboxyl terminus is not conserved in lepidopterans from different families. It appears that rapid evolution has given rise to major variations in length and sequence of the M. sexta repeat, B. mori mature lebocin, and their counterparts in the above three groups. In fact, this region in S. cynthia, A. mylitta-2 and A. pernyi-2 are Ser/Thr, Gly, His-rich (instead of Pro-rich), whereas the 27-residue repeat in M. sexta lebocin contains eight Ser or Thr, five Leu, and only one Pro.
Fig. 2. Multiple sequence alignment of lebocin-related proteins from lepidopteran insects.
(A) M. sexta pro-lebocin is compared with its homologs from B. mori [15, 16], P. includens [19], T. ni [18], S. cynthia [20], A. mylitta (EU557311a, EU557312a), A. pernyi (DG666499a, DG666500a), S. frugiperda (SFC03896b, SFC05898b, SFC03407b), P. dardanus (PDC00100b), H. virescens (K. Shelby, personal communication), and L. obliqua (LOC00221b) [a and b: GenBank (http://www.ncbi.nlm.nih.gov/) and ButterflyBase (http://butterflybase.ice.mpg.de/) accession numbers, respectively]. Putative signal peptide for secretion is underlined in each sequence. The conserved Arg residues in RXXR motif are marked *, and Pro residues are shaded gray. (B) Residues 42-85 (LP2) of M. sexta lebocin and B. mori lebocin-1/2 aligned with G. mellonella anionic peptide 1 [22] and H. armigera P1 peptide [21]. Identical and similar residues are indicated with “|” and “:”, respectively. (C) Phylogenetic tree based on the alignment shown in panel A. Bootstrap values from 1000 trials are indicated at nodes.
Three mechanisms that might account for variable numbers of the 81-nucleotide repeats in M. sexta sequences are: 1) multiple lebocin genes in the M. sexta genome, 2) allelic differences in the colony of insects used for cDNA library construction, or 3) alternative splicing of exons coding for the repeats. To explore these possibilities, we screened a M. sexta genomic library and obtained one positive clone, L45. Southern blot analysis of L45 using the full-length cDNA probe identified two hybridizing bands (Fig. 3A). The 2.9 kb XhoI-HindIII and 2.5 kb XhoI-XhoI fragments were subcloned and completely sequenced. Comparison of the cDNA and genomic sequences indicated that the M. sexta lebocin gene did not contain any intron or alternative splicing site flanked by GT and AG. The B. mori lebocin gene is also intron-free [16].
Fig. 3. Subcloning strategy (A), genomic Southern blot (B), and structure (C) of the M. sexta lebocin gene.
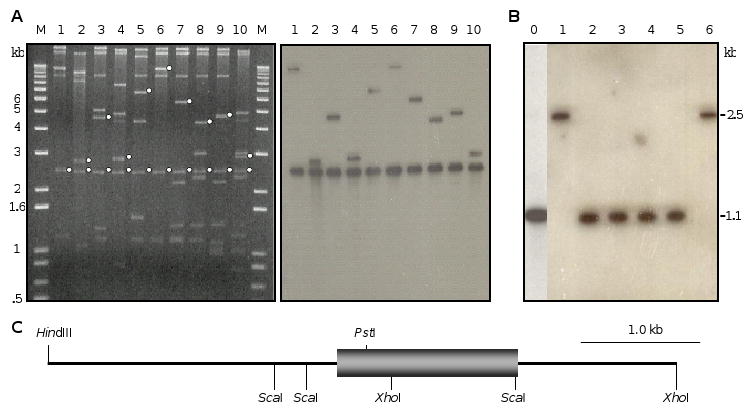
(A) The bacteriophage DNA isolated from the positive λ clone was digested by XhoI (lane 1), XhoI and ApaI (lane 2), XhoI and EcoRI (lane 3), XhoI and HindIII (lane 4), XhoI and KpnI (lane 5), XhoI and SacI (lane 6), XhoI and SalI (lane 7), XhoI and XbaI (lane 8), XhoI, EcoRI and SalI (lane 9), or XhoI, HindIII and XbaI (lane 10). The reaction mixtures and DNA size markers were subjected to gel electrophoresis, capillary transfer, and hybridization using the full-length lebocin cDNA as a probe. Left panel, stained gel with hybridizing fragments marked with white dots; right panel, chemioluminescence detection of the hybridization signals; (B) Following XhoI-ScaI double digestion, genomic DNA (10 μg) from multiple (lane 0) or single (lanes 1∼6: A1 to A6) insects was separated by gel electrophoresis, transferred onto a membrane, and hybridized with 32P-labeled XhoI-ScaI fragment of lebocin cDNA corresponding to the repetitive region. Positions and sizes of the DNA markers are indicated. (C) Restriction map of the subcloned HindIII-XhoI and XhoI-XhoI fragments with the single exon of M. sexta lebocin gene shown as a box.
The M. sexta lebocin gene includes a 1,452 bp coding sequence ranging from nucleotides 2581-4032. The open reading frame encodes a 483-residue polypeptide, which includes the putative signal peptide, pro-segment, and 13.4 repeats (Fig. 1B). Repeats 1-4, 6-10, 12 and 13 contain 1-8 synonymous substitutions while repeats 7, 8 and 10 also contain 3 nonsynonymous changes (ProCCG to SerTCG; GlnCAA to HisCAC). In the partial repeat, four nucleotide substitutions do not alter the amino acid residues, while five other substitutions change HisCAT to ValGTA and SerAGC to a stop codon (TGA). A polyadenylation signal (AATAAA) is located at 89 bp after the TGA. Except for the repeat numbers, there is no major difference between the gene and cDNA sequences. For instance, clone NC6 contains 7.4 (instead of 13.4) repeats (Fig. 1A).
Computer analysis of the 5′ flanking sequence allows us to identify potential regulatory elements in the lebocin gene. Two imperfect NF-κB motifs (9/10 match) [35] are present at nucleotides 634 (minus strand) and 764 (plus strand). One GATA box is located on the plus strand at position 1308 and two are on the minus strand at positions 1523 and 2305. There is an interferon-stimulated response element at nucleotide 115, matching twelve of the thirteen positions in the consensus motif. The CCAGT sequence at position 1406 closely resembles the consensus (TCAGT) which typically resides within ten nucleotides before or after the transcription initiation site in arthropod genes [36]. At 38 bp upstream of CCAGT, there is a TATAAT sequence, reminiscent of the TATA or Goldberg-Hogness box.
In Southern blot analysis of the genomic DNA isolated from multiple insects (Fig. 3B, lane 0), digestion with XhoI-ScaI released a 1.1 kb fragment hybridizing with a cDNA probe that only recognized the repeats. Since no other hybridization signal was found, this result suggests a single lebocin gene in the genome, which contains an XhoI-ScaI fragment identical in size to the genomic clone (Fig. 3C) and to the cDNAs (NC2, NC3, NC10, and NC11) with 13.4 repeats. We repeated the experiment using DNA samples isolated from six different insects randomly chosen from the colony. Four of them (A2-A5) showed the 1.1 kb band that corresponded to 13.4 repeats, but the other two (A1 and A6) exhibited a single band at 2.5 kb (Fig. 3B, lanes 1-6). Further analyses indicated that the 2.5 kb band resulted from XhoI cleavage (Fig. 3C), rather than XhoI-ScaI double digestion. Sequencing of the PCR products of genomic DNA from A1 and A6 revealed 13.4 repeats and a point mutation that abolished the ScaI site (data not shown).
Northern blot analysis of fat body RNA from insects A1, A4, A5, and A6 showed three bands at 1.9, 2.5 and 4.2 kb positions (Fig. 4A). The 1.9 kb band represented the major transcript, consistent in size with the cDNAs containing 13.4 repeats. The other two bands, due to their large sizes, may result from transcription at an alternative initiation or termination site rather than containing more than twenty repeats. There was no significant variation in the transcript sizes among these four insects. Neither was there a ladder of mRNA with different numbers of repeats, although we cannot rule out that smaller transcripts were present at a low level. We carried out RT-PCR to test whether lebocin transcription occurred in fat body and hemocytes in response to the bacterial injection. The lebocin mRNA in hemocytes or fat body of naïve larvae was below the detection limit, and a PCR product at the expected size was amplified from fat body but not hemocytes of the insects after injection of E. coli (Fig. 4B). This is consistent with the results from Northern analysis of fat body RNA from M. luteus-injected larvae and adults, indicating that lebocin gene expression is highly induced upon immune challenges (Fig. 4C). The mRNA level and its increase in larvae seem to be higher than those in adults.
Fig. 4. Detection of lebocin mRNA in fat body of M. sexta injected with bacteria.
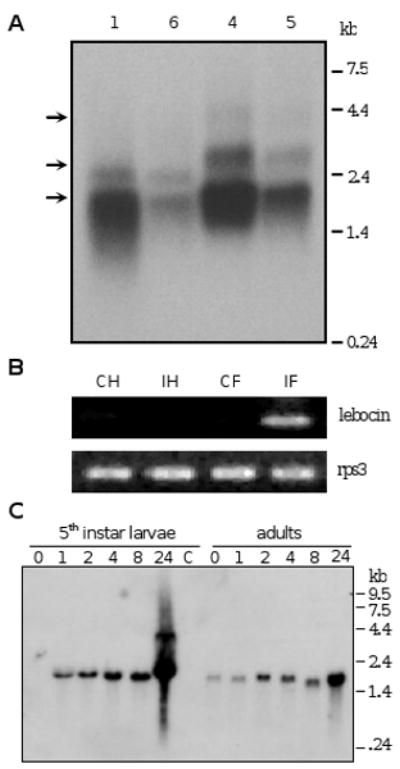
(A) Transcript sizes. Fat body total RNA samples (6 μg) from the four E. coli-injected larvae (A1, A4, A5, A6 in Fig. 3B) were individually separated by agarose gel electrophoresis, transferred onto a membrane, and hybridized with the 32P-labeled repetitive region in the M. sexta lebocin cDNA. Positions of the 1.9, 2.5, and 4.2 kb bands are marked with arrows. (B) Reverse transcription-PCR. CH and CF: hemocytes and fat body from naïve larvae (control); IH and IF: hemocytes and fat body collected from larvae at 24 h after E. coli injection. The cDNA samples were normalized with M. sexta ribosomal protein S3 (rpS3) transcripts. (C) Time course. Fat body total RNA (10 μg), isolated from M. sexta larvae or adults at different time after injection with M. luteus, were subjected to Northern blot analysis using the full-length cDNA (pP4-7) as the probe. Lane labels indicate hours after injection. The positions and sizes of the RNA markers are indicated.
Although its transcription pattern suggests a role in defense, we do not know the exact function of M. sexta lebocin, especially those repeats which correspond to the silkworm mature lebocins in position but not sequence. Do these repeats function in a whole or separately as a processed 27-residue peptide? If processing does occur, where is the scissile bond located? Does the conserved pro-segment carry any function? To address these questions, we synthesized the peptide (RSVETLASQEHLSSLPMDSQETLLRGT). The synthetic peptide did not kill any bacteria (data not shown). Recombinant expression of the pro-lebocin was unsuccessful using E. coli or insect cells. These negative results stimulated us to closely examine the precursor sequences and propose that processing of the repeats yields 27-residue peptides ending with RGTR. This prediction is based on an RXXR motif recognized by intracellular processing enzymes [37,38], often followed by a hydrophilic residue (e.g. Ser). We have identified three RXXR motifs in the pro-segment of M. sexta lebocin: R19KFR22*E, R63YAR66*S and R98NAR101*S (Fig. 1). These Arg residues at the -4 and -1 positions are 100% conserved in the lebocin-related proteins from other insects (Fig. 2A). A convertase in the secretory pathway may cleave the lebocin precursors to form five peptides: LP1, Q1-R22 (22 mer); LP2, E23-R66 (44 mer); LP3, S67-R101 (35 mer); LP4, S102-R128 (27 mer); LP5, SVETLASQEVL (11-residue partial repeat). LP4 (SVETLASQEHLSSLPMDSQETLLRGTR) would be produced in multiple copies since each polypeptide contains 3∼16 of the 27-residue repeats.
To test their biological functions, we chemically synthesized and purified four of the peptides. Their experimental masses were 2,691.95 (LP1), 5,056.39 (LP2), 3,814.12 (LP3) and 2,988.89 (LP4), nearly identical to the theoretical values (2,691.19, 5,054.64, 3,814.09 and 2,985.33 Da). LP1 is highly cationic with a calculated isoelectric point of 12.9. LP2 (pI = 4.8) is similar in sequence to the anionic AMP isolated from G. mellonella [22] (Fig. 2B). LP3 has a calculated pI of 11.8, whereas LP4, the repetitive sequence of 27 residues (SVET…RGTR), is anionic (pI = 4.6). We tested these peptides for antibacterial activity against Gram-negative and Gram-positive bacterial strains using a broth micro-dilution assay [31]. LP1 was active against both Gram-positive and Gram-negative bacteria. Its MIC was 25 μg/ml against all the bacteria except for S. aureus, whose growth was completely blocked by LP1 at 100 μg/ml (Table 1). LP2 did not exhibit antimicrobial activity against the bacterial strains tested, differing from its homolog in G. mellonella [22]. LP3 had a low activity against E. coli O157:H7 with a high MIC (200 μg/ml). LP4 showed no activity against the bacteria. After realizing that the carboxyl-terminal Arg in these four peptides is likely removed by a carboxyl peptidase in vivo, we synthesized another set of peptides LP1A, LP2A, LP3A, and LP4A, one residue shorter than the corresponding LPs, and tested antimicrobial activities against the same panel of bacteria strains. The only difference we observed was between LP3A and LP3: missing the terminal Arg rendered LP3A inactive against the E. coli strain (Table 1). The extra positively charged residue in LP3 may have facilitated the interaction between the peptide and negatively charged bacterial cell surface.
Table 1.
MICs of M. sexta lebocin-related peptidesa
| ATCC | MIC of Peptides (μg/ml) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| LP1 | LP1A | LP2 | LP2A | LP3 | LP3A | LP4 | LP4A | ||
| Gram-negative | |||||||||
| E. coli O157:H7 | 25922 | 25 | 25 | - | - | 200 | - | - | - |
| S. typhimurium | 14028 | 25 | 25 | - | - | - | - | - | - |
| K. pneumoniae | 13883 | 25 | 25 | - | - | - | - | - | - |
| S. typhimurium DT104 | 700408 | 25 | 25 | - | - | - | - | - | - |
| Gram-positive | |||||||||
| L. monocytogenes | 19115 | 25 | 25 | - | - | - | - | - | - |
| S. aureus | 25923 | 100 | 100 | - | - | - | - | - | - |
| S. aureus (MRSA) b | 43300 | 100 | 100 | - | - | - | - | - | - |
| S. aureus (MRSA) b | BAA-39 | 100 | 100 | - | - | - | - | - | - |
MIC is defined as the lowest peptide concentration that gives no visible growth after overnight incubation in 100% Muller-Hinton broth. Conditions for the broth microdilution assay are specified by Clinical Laboratory Standard Institute. The experiments were repeated at least three times for each bacterial strain.
Methicillin-resistant S. aureus.
To identify lebocin-related peptides in larval plasma, we raised a polyclonal antiserum against a mixture of the four synthetic peptides, which only detected one immunoreactive band with mobility similar to that of synthetic LP2 (data not shown). Concrete evidence for the existence of LP2 and other peptides came from analysis liquid chromatography (LC) and tandem mass spectrometry. We first assayed synthetic LP1A through LP4A to obtain chromatographic retention times, peptide m/z's, and MS/MS fragmentation fingerprints (Table 2, left columns). For instance, LP2A eluted at 22 min and the peak was composed primarily of +5 and +6 ions with monoisotopic m/z of 980.489 and 817.242, respectively. These ions corresponded to a neutral molecular mass of 4897.45 Da, a value close to the calculated LP2A mass of 4897.42 Da. Upon MS/MS fragmentation, each ion of synthetic LP2A ion yielded a characteristic fingerprint identical to that of a peptide in the induced plasma (Table 2, right columns). This peptide, with the same retention time of 22 min, represented a major peptide constituent of the hemolymph (data not shown). While LP3A was found in the same way (Table 2), we did not detect LP4A in the induced plasma. The identification of LP1A in the induced plasma was complicated by a posttranslational modification. Synthetic LP1A eluted at 19 min and yielded +4 and +5 ions that correspond to a neutral mass of 2534.43 Da (Table 2). When the biological sample was analyzed, the 19-min major peak was dominated by ions corresponding to a neutral mass of 2517.42 Da. The mass decrease of 17.01 Da suggested that the amino-terminal Gln of LP1A was cyclized in vivo to form pyroGlu. Consistent with this interpretation, the predominant MS/MS fragments generated by the biological ions were nearly identical with those observed for synthetic LP1A (Table 2). We converted the first Gln of synthetic LP1A to pyroGln and analyzed the peptide mixture by NanoLC-MS/MS, which yielded ions corresponding to 2534.43 Da (original) and 2517.42 Da (converted) peptides (data not shown). Fragmented +4 and +5 ions (630.35 and 504.47 Da) from the modified LP1A were identical in mass fingerprints to those from the 19-min fraction of the biological sample.
Table 2.
LC-MS/MS analysis of lebocin-related peptides from chemical synthesis or M. sexta hemolympha
| peptide name (calculated mass) | Synthetic peptide | Hemolymph | ||||
|---|---|---|---|---|---|---|
| Intact peptide ion (m/z) | Elution time (min) | Predominant MS/MS fragments | Intact peptide ion (m/z) | Elution time (min) | Predominant MS/MS fragments | |
| LP1A (2534.41) | 507.887* | 19 | 508→473 | 504.484* | 19 | 505→473 |
| 508→498 | 505→498 | |||||
| 508→625 | 505→625 | |||||
| 508→630 | 505→630 | |||||
| 508→631 | 505→631 | |||||
| 634.608** | 19 | 635→594 | 630.353** | 19 | (630→589) | |
| (635→626) | ||||||
| 635→652 | 630→652 | |||||
| 635→709 | 630→709 | |||||
| 635→773 | 630→773 | |||||
| 635→846 | 630→846 | |||||
| LP2A (4897.42) | 817.242*** | 22 | 817→776 | 817.241*** | 22 | 817→776 |
| 817→798 | 817→798 | |||||
| 817→877 | 817→877 | |||||
| 817→969 | 817→969 | |||||
| 980.489* | 22 | 980→969 | 980.486* | 22 | 980→969 | |
| 980→1044 | 980→1044 | |||||
| 980→1096 | 980→1096 | |||||
| LP3A (3656.73) | 610.459*** | 15-18 | 611→693 | 610.462*** | 14-17 | 611→693 |
| 611→689 | 611→689 | |||||
| 611→661 | 611→661 | |||||
| 732.353* | 15-18 | 733→661 | 732.350* | 14-17 | 733→661 | |
| 733→689 | 733→689 | |||||
| 733→773 | 733→773 | |||||
| 733→826 | 733→826 | |||||
| 733→866 | 733→866 | |||||
Peptides were analyzed by LC-MS/MS as described in Section 2. Characteristic precursor and fragment ions are listed. m/z = (M+n)/n, wherein m/z is ratio of ion mass to ion charge, M is neutral molecular mass, z is charge (*: +5; **: +4; ***: +6).
4. Discussion
Insects and other multicellular organisms rely on a network of host defense mechanisms to survive and prosper in microbe-rich environments [4]. One such mechanism involves the synthesis and secretion into hemolymph of AMPs that are effective against a broad spectrum of pathogens including bacteria, fungi, and protozoa [1,39]. Gene duplication and sequence divergence has given rise to a sizable repertoire of AMPs in various insect groups. For instance, analysis of the silkworm genome has revealed over thirty AMP genes in the cecropin, moricin, gloverin, attacin, lebocin, and enbocin families [40]. X-tox genes in lepidopteran species encode proteins with multiple defensin-like domains that are not processed or active as AMPs [41-44].
M. sexta lebocin was first reported as a differentially expressed gene in response to bacterial injection in a subtractive suppression hybridization experiment [11]. In this study, we confirmed its inducible transcription and discovered an interesting feature of the cDNAs: 3∼16 copies of an 81 bp repeat between the 5′ and 3′ constant regions (Fig. 1). Gene sequencing and Southern blotting (Fig. 3 and Fig. 4) ruled out three potential mechanisms (i.e. multiple genes, allelic variations, and alternative splicing) for generating these variations. The negative results led us to suspect that the variation in copy numbers was an artifact caused by the tandem repeats when these cDNAs were first synthesized. No change was observed during DNA subcloning or propagation.
Regardless of the number of repeats in the transcript, five different peptides can be derived from the protein precursors via proteolytic processing after RXXR (Fig. 2). LP1 and LP3 were both active against bacteria, although the latter had a high MIC. These two peptides, as well as probably also G. mellonella anionic peptide 1, result via proteolytic processing from the amino-terminal region previously thought to be a pro-segment in lebocin-related proteins. In the initial biochemical study, B. mori lebocin-1/2 and -3 were names for 32-residue polypeptides active against Gram-negative bacteria [14], which are now known to be processed from the carboxyl-terminal end of a larger precursor. However, the carboxyl-terminal region in their counterparts in the M. sexta, T. ni, P. includens, S. cynthia, A. mylitta, A. pernyi, S. frugiperda, P. dardanus, H. virescens, and L. obliqua homologs differ significantly from the B. mori lebocins in amino acid sequence (Fig. 2). We propose to expand the original definition of lebocin to include the entire precursor proteins. Then, the processed products can be sequentially named from amino to carboxyl terminus as lebocin peptide 1 (LP1) through LPn. For instance, the M. sexta peptides are named LP1 through LP5: LP1, LP2 and LP3 come from the “pro-segment”; LP4 (27 mer) and LP5 (11 mer) correspond to the complete and partial repeats (Fig. 1A). B. mori LP4 is the same as lebocin by the original definition and LP5 represents the 25-residue fragment at the carboxyl terminus. While B. mori LP5 results from processing at R151YRR154*H, there is no LP5 counterpart in T. ni, P. includens, S. cynthia, A. mylitta, A. pernyi, S. frugiperda, H. virescens, or L. obliqua lebocins.
B. mori LP4 variants were thought to be encoded by members of a gene family in the genome [15,16]. Since the genome analysis only revealed a single gene [40], the minor differences in the cDNA and gene sequences may reflect allelic variations in the silkworms used for protein purification and library construction. No evidence was available for the existence of such multigene family in T. ni [18], H. virescens, L. obliqua, or P. dardanus. On the other hand, A. pernyi and A. mylitta have two genes: lebocin-2's are most similar to the S. cynthia lebocin [20] and lebocin-1's to the B. mori variants (Fig. 2C). Lebocin gene duplication must have occurred in the evolution of Saturniidae and, in another species from the same family (S. cynthia), a separate lebocin gene may exist to encode a precursor protein more similar to A. pernal-1, A. mylitta-1 and B. mori lebocins. In one lineage of the superfamily Noctuoidea, gene duplication has generated two to three lebocins in S. frugiperda, which form a clade with their homologs of P. includens and T. ni (Fig. 2C). In the pyrosequence analysis of immunity-related cDNAs from M. sexta [44], we identified four contigs (#5813, #6639, #6760 and #6851) coding for polypeptides similar to a part of the prosegment (LP1-LP2 region). Sequence comparison suggested two additional lebocin-like genes in the M. sexta genome (data not shown). One or both of them may be more similar to silkworm lebocins than the one reported herein. Taken together, the sequence-based phylogenetic relationships are consistent with those derived from morphological characteristics.
B. mori, T. ni, P. includens and S. cynthia lebocin genes were mainly expressed in fat body of immune challenged insects [15,18-20]. Their mRNA levels were much lower in hemocytes even after bacterial injection. Consistent with the presence of immune regulatory elements in its gene (Fig. 1B), M. sexta lebocin expression was up-regulated during antimicrobial responses in larvae and adults (Fig. 4C). Similarly, cecropin B of M. sexta was mainly produced in fat body [10]. These results are consistent with the observation that lebocin and cecropin expression was up-regulated by injecting B. mori spätzle into M. sexta larvae [45].
Although direct evidence is limited, it seems to be a common mechanism in lepidopteran insects to generate several active peptides from a lebocin polypeptide precursor. This is supported by the purification of LP2 from G. mellonella and H. armigera hemolymph [21,22] and by the detection of antibacterial activities of M. sexta LP1, LP3, and G. mellonella LP2 (i.e. anionic peptide 1) (Fig. 2B). The purification and functional characterization of B. mori LP4 variants (i.e. lebocin-1/2 and 3) further supported this hypothesis [14]. In this study, our antiserum prepared toward a mixture of the four synthetic lebocin peptides recognized synthetic LP2 efficiently and labeled a polypeptide in the induced plasma, which is similar to synthetic LP2 in apparent Mr (data not shown). Direct evidence for the existence of LP2A (as well as LP1A and LP3A) at a high level was obtained by LC-MS/MS analysis of the larval hemolymph (Table 2): 1) two plasma polypeptides were identical to synthetic LP2A and LP3A in all molecular properties; 2) a peptide with MS/MS fragments identical to those of synthetic LP1A had its amino-terminal Gln converted to a pyroGlu in vivo. Surprisingly, we did not detect any LP4A, which should be approximately 13 times as abundant as the other peptides. There are three possible reasons for this negative result: 1) the repeated region is resistant to processing, 2) LP4A attaches to hemocytes or proteins that precipitate during sample preparation; 3) LP4A is rapidly degraded in the plasma. The existence of the perfect repeats in multiple copies, which is not found in the other lepidopteran species, suggests that LP4A plays a unique role in the M. sexta defense system. On the other hand, the high sequence similarity of the LP1, LP2 and LP3 regions in other species suggest that these predicted processing products are functionally conserved. A very recent peptidomics study of AMPs in hemolymph of G. mellonella [46] has revealed the same mechanism discovered in this investigation.
Processing of large precursors is a strategy commonly used by neuroendocrine systems of vertebrates and invertebrates to generate biologically active peptides. Rules have been proposed to predict the processing sites in insect neuropeptide precursors [37,38]. This study provides initial evidence that the insect immune system may use the same tactic to produce functional products by processing protein precursors. This hypothesis would support the notion that parts of the nervous, hormonal, and immune systems have a common evolutionary origin.
Acknowledgments
We thank Dr. Kent Shelby at USDA ARS for sharing the unpublished sequence of H. virescens lebocin and Drs. Steffi Gebauer-Jung, Hendrik Tilger, and Alexie Papanicolaou at Max Planck Institute for Chemical Ecology in Germany for developing and maintaining ButterflyBase. This work was supported by National Institutes of Health Grants GM58634 (to H.J.) and AI31084 (to M.K), as well as National Science Foundation Award 0722494 (to S.H.) for the LTQ mass spectrometer. This article was approved for publication by the Director of Oklahoma Agricultural Experimental Station and supported in part under project OKLO2450.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bulet P, Stocklin R, Menin L. Antimicrobial peptides: from invertebrates to vertebrates. Immunol Rev. 2004;198:169–84. doi: 10.1111/j.0105-2896.2004.0124.x. [DOI] [PubMed] [Google Scholar]
- 2.Reddy KVR, Yedery RD, Aranha C. Antimicrobial peptides: premises and promises. Int J Antimicrobial Agents. 2004;24:536–47. doi: 10.1016/j.ijantimicag.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 3.Jiang H, Kanost MR. The clip-domain family of serine proteinases in arthropods. Insect Biochem Mol Biol. 2000;30:95–105. doi: 10.1016/s0965-1748(99)00113-7. [DOI] [PubMed] [Google Scholar]
- 4.Lemaitre B, Hoffmann J. The host defense of Drosophila melanogaster. Ann Rev Immunol. 2007;25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615. [DOI] [PubMed] [Google Scholar]
- 5.Christophides GK, Zdobnov E, Barillas-Mury C, Birney E, Blandin S, et al. Comparative genome and proteome analysis of Anopheles gambiae and Drosophila melanogaster. Science. 2002;298:159–65. doi: 10.1126/science.1077061. [DOI] [PubMed] [Google Scholar]
- 6.Evans JD, Aronstein K, Chen YP, Hetru C, Imler JL, Jiang H, Kanost M, Thompson GJ, Zou Z, Hultmark D. Immune pathways and defense mechanisms in honey bees Apis mellifera. Insect Mol Biol. 2006;15:645–56. doi: 10.1111/j.1365-2583.2006.00682.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zou Z, Evans JD, Lu Z, Zhao P, Williams M, Sumathipala N, Hetru C, Hultmark D, Jiang H. Comparative genomic analysis of the Tribolium immune system. Genome Biol. 2007;8:R177. doi: 10.1186/gb-2007-8-8-r177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Waterhouse RM, Kriventseva EV, Meister S, Xi Z, et al. Evolutionary dynamics of immune-related genes and pathways in disease-vector mosquitoes. Science. 2007;316:1738–43. doi: 10.1126/science.1139862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tanaka H, Ishibashi J, Fujita K, Nakajima Y, et al. A genome-wide analysis of genes and gene families involved in innate immunity of Bombyx mori. Insect Biochem Mol Biol. 2008;38:1087–110. doi: 10.1016/j.ibmb.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 10.Dickinson L, Russell V, Dunn PE. A family of bacteria-regulated, cecropin D-like peptides from Manduca sexta. J Biol Chem. 1988;263:19424–9. [PubMed] [Google Scholar]
- 11.Zhu Y, Johnson TJ, Myers AA, Kanost MR. Identification by subtractive suppression hybridization of bacteria-induced genes expressed in Manduca sexta fat body. Insect Biochem Mol Biol. 2003;33:541–59. doi: 10.1016/s0965-1748(03)00028-6. [DOI] [PubMed] [Google Scholar]
- 12.Gorman MJ, Kankanala P, Kanost MR. Bacterial challenge stimulates innate immune responses in extra-embryonic tissues of tobacco hornworm eggs. Insect Mol Biol. 2004;13:19–24. doi: 10.1111/j.1365-2583.2004.00454.x. [DOI] [PubMed] [Google Scholar]
- 13.Dai H, Rayaprolu S, Gong Y, Huang R, Prakash O, Jiang H. Solution structure antibacterial activity and expression profile of Manduca sexta moricin. J Peptide Sci. 2008;14:855–63. doi: 10.1002/psc.1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hara S, Yamakawa M. A novel antibacterial peptide family isolated from the silkworm, Bombyx mori. Biochem J. 1995;310:651–6. doi: 10.1042/bj3100651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chowdhury S, Taniai K, Hara S, Okuda KK, et al. cDNA cloning and gene expression of lebocin, a novel member of antibacterial peptides from the silkworm, Bombyx mori. Biochem Biophy Res Comm. 1995;214:271–8. doi: 10.1006/bbrc.1995.2284. [DOI] [PubMed] [Google Scholar]
- 16.Furukawa S, Taniai K, Ishibashi J, Hara S, Shono T, Yamakawa M. A novel member of lebocin gene family from the silkworm, Bombyx mori. Biochem Biophy Res Comm. 1997;238:769–74. doi: 10.1006/bbrc.1997.7386. [DOI] [PubMed] [Google Scholar]
- 17.Hara S, Yamakawa M. Cooperative antibacterial relationship between lebocin and cecropin D, antibacterial peptides isolated from the silkworm, Bombyx mori. Appl Entomol Zool. 1995;30:606–8. [Google Scholar]
- 18.Liu G, Kang D, Steiner H. Trichoplusia ni lebocin, an inducible immune gene with a downstream insertion element. Biochem Biophy Res Comm. 2001;269:803–7. doi: 10.1006/bbrc.2000.2366. [DOI] [PubMed] [Google Scholar]
- 19.Lavine MD, Chen G, Strand MR. Immune challenge differentially affects transcript abundance of three antimicrobial peptides in hemocytes from the moth Pseudoplusia includens. Insect Biochem Mol Biol. 2005;35:1335–46. doi: 10.1016/j.ibmb.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 20.Bao Y, Yamano Y, Morishima I. A novel lebocin-like gene from eri-silkworm, Samia cynthia ricini, that does not encode the antibacterial peptide lebocin. Comp Biochem Physiol. 2005;140:127–31. doi: 10.1016/j.cbpc.2004.09.022. [DOI] [PubMed] [Google Scholar]
- 21.Mackintosh JA, Gooley AA, Karuso PH, Beattie AJ, Jardine DR, Veal DA. A gloverin-like antibacterial protein is synthesized in Helicoverpa armigera following bacterial challenge. Dev Comp Immunol. 1998;22:387–99. doi: 10.1016/s0145-305x(98)00025-1. [DOI] [PubMed] [Google Scholar]
- 22.Cytrynska M, Mak P, Barabas AZ, Suder P, Jakubowicz T. Purification and characterization of eight peptides from Galleria mellonella immune hemolymph. Peptides. 2007;28:533–46. doi: 10.1016/j.peptides.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 23.Dunn P, Drake D. Fate of bacteria injected into naïve and immunized larvae of the tobacco hornworm, Manduca sexta. J Invert Pathol. 1983;41:77–85. [Google Scholar]
- 24.Jiang H, Wang Y, Gu Y, Guo X, Zou Z, Scholz F, Trenczek TE, Kanost MR. Molecular identification of a bevy of serine proteinases in Manduca sexta hemolymph. Insect Biochem Mol Biol. 2005;35:931–43. doi: 10.1016/j.ibmb.2005.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambrook J, Russell DW. Molecular cloning, a laboratory manual. 3rd. Cold Spring Harbor Laboratory; New York: 2001. pp. 2.90–2.100. [Google Scholar]
- 26.Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;35:3497–600. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Henikoff S, Henikoff JG. Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci USA. 1992;89:10915–9. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Page RDM. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–8. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 29.Bradfield JY, Wyatt GR. X-linkage of a vitellogenin gene in Locusta migratoria. Chromosoma. 1983;88:190–3. [Google Scholar]
- 30.Chinzei Y, White BN, Wyatt GR. Vitellogenin mRNA in locust fat body: identification, isolation and quantitative changes induced by juvenile hormone. Can J Biochem. 1982;60:243–51. doi: 10.1139/o82-029. [DOI] [PubMed] [Google Scholar]
- 31.National Committee for Clinical Laboratory Standards. Approved Standard M7-A6. 6th. Wayne, PA: 2003. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. [Google Scholar]
- 32.Khandke KM, Fairwell T, Chait BT, Manjula BN. Influence of ions on cyclization of the amino terminal glutamine residues of tryptic peptides of Streptococcal PepM49: resolution of cyclized peptides by HPLC and characterization by mass spectrometry. Int J Peptide Protein Res. 1989;34:118–23. doi: 10.1111/j.1399-3011.1989.tb01499.x. [DOI] [PubMed] [Google Scholar]
- 33.Ladendorff NE, Kanost MR. Bacteria-induced protein P4 (hemolin) from Manduca sexta: a member of the immunoglobulin superfamily which can inhibit hemocyte aggregation. Arch Insect Biochem Physiol. 1991;18:285–300. doi: 10.1002/arch.940180410. [DOI] [PubMed] [Google Scholar]
- 34.Wang Y, Zou Z, Jiang H. An expansion of the dual clip-domain serine proteinase family in Manduca sexta: gene organization, expression, and evolution of prophenoloxidase-activating proteinase-2, hemolymph proteinase 12, and other related proteinases. Genomics. 2006;87:399–409. doi: 10.1016/j.ygeno.2005.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hultmark D. Immune reactions in Drosophila and other insects, a model for innate immunity. Trends Genetics. 1993;9:178–83. doi: 10.1016/0168-9525(93)90165-e. [DOI] [PubMed] [Google Scholar]
- 36.Cherbas L, Cherbas P. The arthropod initiator: the capsite consensus plays an important role in transcription. Insect Biochem Mol Biol. 1993;23:81–90. doi: 10.1016/0965-1748(93)90085-7. [DOI] [PubMed] [Google Scholar]
- 37.Devi L. Consensus sequence for processing of peptide precursors at monbasic sites. FEBS Lett. 1991;280:189–94. doi: 10.1016/0014-5793(91)80290-j. [DOI] [PubMed] [Google Scholar]
- 38.Veenstra JA. Mono and dibasic proteolytic cleavage sites in insect neuroendocrine peptide precursors. Arch Insect Biochem Physiol. 2000;43:49–63. doi: 10.1002/(SICI)1520-6327(200002)43:2<49::AID-ARCH1>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 39.Engstrom Y. Induction and regulation of antimicrobial peptides in Drosophila. Dev Comp Immunol. 1999;23:345–58. doi: 10.1016/s0145-305x(99)00016-6. [DOI] [PubMed] [Google Scholar]
- 40.Cheng T, Zhao P, Liu C, Xu P, Gao Z, Xia Q, Xiang Z. Structures, regulatory regions, and inductive expression patterns of antimicrobial peptide genes in the silkworm Bombyx mori. Genomics. 2006;87:356–65. doi: 10.1016/j.ygeno.2005.11.018. [DOI] [PubMed] [Google Scholar]
- 41.Destoumieux-Garzón D, Brehelin M, Bulet P, Boublik Y, Girard PA, Baghdiguian S, Zumbihl R, Escoubas JM. Spodoptera frugiperda X-tox protein, an immune related defensin rosary, has lost the function of ancestral defensins. PLoS One. 2009;4:e6795. doi: 10.1371/journal.pone.0006795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seitz V, Clermont A, Wedde M, Hummel M, Vilcinskas A, Schlatterer K, Podsiadlowski L. Identification of immunorelevant genes from greater wax moth (Galleria mellonella) by a subtractive hybridization approach. Dev Comp Immunol. 2003;27:207–15. doi: 10.1016/s0145-305x(02)00097-6. [DOI] [PubMed] [Google Scholar]
- 43.Taniai K, Lee JH, Lee IH. Bombyx mori cell line as a model of immune-system organs. Insect Mol Biol. 2006;15:269–79. doi: 10.1111/j.1365-2583.2006.00639.x. [DOI] [PubMed] [Google Scholar]
- 44.Zou Z, Najar F, Wang Y, Roe B, Jiang H. Pyrosequence analysis of expressed sequence tags for Manduca sexta hemolymph proteins involved in immune responses. Insect Biochem Mol Biol. 2008;38:677–82. doi: 10.1016/j.ibmb.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Y, Cheng T, Rayaprolu S, Zou Z, Xia Q, Xiang Z, Jiang H. Proteolytic activation of pro-spätzle is required for the induced transcription of antimicrobial peptide genes in lepidopteran insects. Dev Comp Immunol. 2007;31:1002–12. doi: 10.1016/j.dci.2007.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brown SE, Howard A, Kasprzak AB, Gordon KH, East PD. A peptidomics study reveals the impressive antimicrobial peptide arsenal of the wax moth Galleria mellonella. Insect Biochem Mol Biol. 2009;39:792–800. doi: 10.1016/j.ibmb.2009.09.004. [DOI] [PubMed] [Google Scholar]