Summary
Cellular senescence is a permanent cell cycle arrest and a potent tumor suppression mechanism. The p53 tumor suppressor is a sequence-specific transcription factor and acts as a central hub sensing various stress signals and activating an array of target genes to induce cell cycle arrest, apoptosis, and senescence. Recent reports showed that restoration of p53 induces premature senescence and tumor regression in mice with hepatocarcinomas or sarcomas. Thus, p53-mediated senescence is capable of eliminating cancer cells in vivo. p63 and p73, two homologues of p53, have similar function in cell cycle arrest and apoptosis. However, the role of p63 and p73 in cellular senescence is elusive. In this review, we will discuss how p53 regulates senescence and future studies about p53 family members in senescence.
Keywords: p53, p21, cellular senescence, tumor suppression
Introduction
Cellular senescence is an irreversible cell cycle arrest in response to stress signals, including telomere dysfunction [1], oncogene activation [2], and DNA damage [3]. Cellular senescence is a safeguard mechanism that may prevent aged cells or cells bearing mutations from expansion by inducing a permanent cell cycle arrest [4]. Indeed, cellular senescence was shown to be activated at the initial steps of premalignant transformation [5–8]. Conditional activation of K-rasV12 in mice leads to the development of both adenomas (premalignant) and adeno-carcinomas (malignant). Interestingly, senescent cells are only present in premalignant lesions, but not in malignant tumors. Thus, it was hypothesized that oncogene-induced senescence may help to restrict tumor progression and loss of senescence in malignant tumor cells may be partly due to loss of the p53 tumor suppressor. However, whether induction of senescence is sufficient to repress tumor in vivo is controversial. Recent reports showed that conditional restoration of p53 in mice with hepatocarcinomas, sarcoma, or lymphoma is able to promote tumor regression [9–11]. Although regression of lymphoma was associated with apoptosis, regression of hepatocarcinomas and sarcomas was associated with senescence. Thus, in addition to p53-mediated apoptosis, p53-mediated cellular senescence is a potent pathway to eliminate cancer cells in vivo.
The p53 tumor suppressor is the most commonly mutated gene in human cancers [12]. Loss of p53 in both mutant mice and Li-Fraumeni syndrome patients correlates with early onset of multiple tumors [13–15]. p53 functions as a sequence-specific transcription factor at the crossroads of cellular stress response pathways [16]. In response to various stress signals such as DNA damage, hypoxia, or activated oncogenes, the p53 protein is activated in a specific manner by posttranslational modifications and leads to DNA repair, cell cycle arrest, apoptosis, or cellular senescence [17–19]. It has been shown that the level and/or activity of p53 were increased in senescent cells [2] and overexpression of p53 was sufficient to induce premature senescence in p53-null cells [20, 21]. In contrast, inactivation of p53 using viral proteins (SV40 large T antigen or HPV-16 E6 protein) or gene targeting results in an extension of cell lifespan [22]. Similarly, loss of p53 heterozygosity or expression of dominant negative p53 also extends cell lifespan [23, 24]. Thus, p53 is a pivotal mediator of cellular senescence. However, although loss of p53 alone is sufficient for mouse cells to bypass senescence, additional inhibition of p16 is required for human cells to bypass senescence [25]. In addition, lack of p53 significantly diminishes but does not abrogate DNA damage-induced premature senescence [26]. These suggest that senescence can occur through a p53-independent mechanism.
What is cellular senescence?
In the early 1960s, Hayflick and Moorhead observed that normal human diploid cells have a limited number of passages in vitro after which they enter a metabolic active but non-proliferative phase defined as “replicative senescence” or “Haylick’s limit” [27, 28]. Senescent cells are characterized by enlarged cell size, flattened morphology, inability to synthesize DNA, and expression of the biomarker, senescence-associated β-galactosidase (SA-β-gal) [29]. Senescent cells express a high level of lysosomal β-gal which can be detected at suboptimal pH 6.0, whereas overexpression of lysosomal β-gal itself is not able to initiate senescence [30–32]. Thus, the SA-β-gal activity is the outcome of the highly elevated expression of lysosomal β-gal. Years later, progressive telomere attrition during cell divisions was identified as the underlying mechanism for replicative senescence [1, 33]. The telomeres are a sequence of repetitive bases, TTAGGG in humans, at the ends of linear chromosomes followed by a single-strand overhang [34]. Binding of telomere-specific proteins to the single and double strand regions forms a nucleoprotein complex which caps the chromosome ends and protects chromosome termini from degradation, recombination, and end-fusion. Uncapping of the chromosome ends due to progressive telomere loss during each population doubling in somatic cells leads to cellular senescence by activating the DNA damage pathway [35]. In addition, disruption of normal telomere status also leads to rapid induction of growth arrest [36]. For example, TRF2, a key component of shelterin, the telomere-specific protein complex, binds to the duplex TTAGGG repeat array of mammalian chromosome ends [37]. Loss of TRF2 or expression of mutant forms of TRF2 induces end-to-end chromosome fusions leading to a cell cycle arrest with the characteristics of cellular senescence [36].
In addition to stress signals originated from telomere dysfunction, aberrant oncogenic activities are also able to induce cellular senescence. Among oncogenes, Ras is the most frequently mutated gene in human cancers [38]. The Ras oncogene family encodes small GTP-binding proteins that transduce mitogenic signals from G-protein coupled receptors in response to extracellular stimuli [39]. Combined activation of Ras with a cooperating oncogene, such as Myc, E1A, and SV40 large T antigen [40, 41], or with loss of a tumor suppressor, such as p53 and p16, leads to transformation of primary cells and tumor formation in animals [42, 43]. Interestingly, prolongd expression of an active Ras protein, H-RasV12, provokes an acute permanent cell cycle arrest, termed as “premature senescence”, which is indistinguishable from replicative senescence [2]. There are several underlying mechanisms for oncogene-induced senescence (OIS). First, Ras-induced senescence partly depends on the activation of the Raf-MEK-ERK MAPK pathway. Since Ras induces cellular transformation via the same pathway, it is likely that the Ras-induced senescence is a fail-safe mechanism to limit the transformation potential of excessive Ras mitogenic signaling. Besides the Raf-MEK-ERK cascade, oncogenic Ras also activates the JNK-p38 MAPK pathway [44, 45]. Consistent with this, many but not all Ras effectors, including activated RAF, MEK, p38, and BRAF, were shown to induce senescence [46, 47]. Second, Ras proteins induce senescence by altering the intracellular levels of reactive oxygen species (ROS) [48]. It has been shown that expression of RasV12 results in an increase of intracellular and in particular, mitochondrial reactive oxygen species. In contrast, the extent of RasV12-induced senescence is shown to be decreased by repressing the production of ROS by culturing cells in a low oxygen environment (1% oxygen) compared to normoxia (20% oxygen). Third, OIS is an outcome of a DNA damage response [49]. Cells undergoing senescence induced by oncogenic Ras contain clearly detectable DNA damage foci, whereas inactivation of DNA damage response pathway abrogates OIS and promotes cellular transformation. In addition, this robust DNA damage response in cells undergoing OIS is demonstrated to be triggered by oncogene-induced DNA hyper-replication.
Given the importance of DNA damage response in cell cycle arrest and senescence, it is not surprising that DNA damage agents, such as radiation, chemotherapeutic drugs, and oxidative stresses, induce premature senescence. Typically, stress-induced senescence (SIS) and OIS do not lead to significant telomere shortening [50, 51]. For example, hydrogen peroxide treatment ceases DNA replication and induces a permanent cell cycle arrest in human diploid fibroblasts [52, 53]. In addition, oxidative stress originated from UVB irradiation promotes premature senescence in human skin diploid fibroblasts [54]. Moreover, appropriate levels of doxorubicin or etoposide, inhibitors of topoisomerase II that can induce DNA double-strand breaks [55], induce a permanent cell cycle arrest in normal proliferating fibroblasts [56]. Contrary to normal somatic cells, most cancer cells have extended or infinite life spans. Thus, tumor cells were thought to have escaped from senescence. However, recent studies showed that cancer chemotherapeutic drugs, including doxorubicin, camptothecin, and cisplatin, are able to arrest tumor cells by initiating premature senescence in vitro and in vivo [3, 57–61]. Camptothecin is an inhibitor of topoisomerase I and can induce DNA double strand breaks [62]. Cisplatin creates DNA lesions by binding to DNA and in particular the formation of intrastrand cross-links between adjacent guanines [63]. In a summary, cellular senescence is a common stress response in response to genomic instability originated from telomere dysfunction, oncogene activation, and DNA damage.
Here, we would like to note that cellular senescence is defined as an irreversible cell cycle arrest, but is not necessarily irreversible depending on the expression of pRb and p53. Inactivation of p53 was found to stimulate the re-entrance into the cell cycle of senescent cells with low levels of p16, but not those with high levels of p16 [64]. In addition, several reports showed that upon inactivation of pRb, the pRb-induced senescent cells resumed to synthesize DNA, but most died in the process rather than proliferated [65–67]. Moreover, a small number of p16-induced senescent cells appeared to enter S phase upon removal of p16-expression [68].
p53 is an effector of stress signals leading to senescence
The role of p53 in replicative senescence
Several studies have revealed that DNA damage foci appear at the telomeres of senescent cells [35, 69]. These foci contain multiple DNA damage-response proteins, such as γ-H2AX, 53BP1, MDC1, NBS1, MRE11, and RAD17. Moreover, senescent cells express activated forms of Ataxia-telangiectasia mutated (ATM) and its downstream target, Chk2. Phosphorylation of p53 at Ser-15 by ATM and at Ser-20 by Chk2 inhibits p53 degradation by MDM2, a RING finger E3 ligase which ubiquitinates and degrades p53 through ubiquitin-mediated proteolysis. However, loss of ATM does not attenuate senescence as expected. It has been shown that human diploid fibroblasts from AT patients with mutation in the ATM gene are associated with accelerated telomere shortening and undergo senescence [69]. Indeed, other than functioning as a DNA damage response mediator for telomere attrition, ATM also plays a role in maintaining the integrity of telomeres [70]. In addition, localization of ATM- and Rad3-related (ATR) and ATR-interacting protein (ATRIP) to telomeres was observed in the ATM−/− cells [71]. ATR activates its substrate ChK1 and then phosphorylates p53 at Ser 15 and regulates p53 activity. It has been shown that ATM plays a major role transmitting the telomere DNA damage signal in normal cells but ATR becomes more prominent in ATM−/− cells [71]. Moreover, ectopic activation of ATR is sufficient to trigger a cell cycle arrest in ATM−/− cells suggesting that ATR has complementary role for ATM deficiency [71].
The role of p53 in OIS
While the adenovirus E1A and c-Myc activate p53 to promote apoptosis, oncogenic Ras induces p53 to initiate premature senescence. It has been shown that oncogenic Ras arrests cells at G1 along with a significant increase in the abundance of p53 [2]. Upregulation of p53 in oncogene-induced senescence results from two major signaling pathways. First, activation of ARF via the MAPK pathway has been shown to be required for oncogenes signaling to p53. Indeed, it has been shown that ARF interacts with MDM2 and prevents MDM2-mediated degradation of p53 [72]. Mice lacking ARF alone are highly prone to spontaneous tumor formation, which is similar to p53-deficient mice [73, 74]. In addition, ectopic expression of ARF induces cell cycle arrest in cells containing wild-type p53 but not mutant p53, whereas ARF-null MEFs do not undergo replicative senescence and can be transformed by oncogenic H-Ras alone [73]. Moreover, in the absence of ARF, coexpression of oncogenic Ras and p53 was unable to promote premature senescence in MEF [75]. However, ARF is not required for OIS in human cells [76]. These data suggest that ARF may function as a mediator of Ras signaling to p53 in mouse cells. Second, DNA damage response is required for the activation of p53 in response to oncogenes. It has been reported that OIS is accompanied by DNA replicative stress, including prematurely terminated DNA replication forks and DNA double-strand breaks caused by hyper-DNA replication [49, 77]. Consistent with this, oncogenic Ras-induced senescence is accompanied with activation of DNA damage response effectors, such as ATM/ATR and Chk2/Chk1, and inactivation of these DNA damage response effectors by RNA interference attenuates OIS [78–81]. Moreover, increased production of reactive oxygen species by oncogenic Ras also acts through the p53 pathway to induce senescence [82].
The role of p53 in SIS
In response to DNA damage stress signals, including oxidative stress, ionizing radiation, and chemotherapies, the ATM/ATR-p53 pathway is activated leading to premature senescence in both normal and tumor cells [26]. Upon treatment with doxorubicin, camptothecin, and cisplatin, SA-β-gal was present in multiple human cancer cells lines carrying wild-type p53, including MCF7, HCT116, LS174T, HCA-7, A2780, and HT1080, but not in DLD1, MDA-MB-231, and PC3 cells with mutant p53 [60]. In addition, knockdown of p53 in MCF7 and HT1080 cells, or knockout of p53 in HCT116 cells significantly decreases, but not abolishes doxorubicin-induced premature senescence [83, 84]. Moreover, in response to doxorubicin treatment, SA-β-gal positive cells were still observed in p53 null Saos-2 cells, in SW480 and U251 cells carrying mutant p53, and in Hela and Hep-2 cells in which p53 is inactivated by E6 protein [60]. These data suggest that p53 plays an important role in SIS, but some other genes may also play a role in SIS in cancer cells. Interestingly, cell cycle arrest is the initial step of cellular senescence, and it has been well established that DNA damage also activates p53 to induce cell cycle arrest and cell death [85, 86]. The outcome of p53 activation may depend on the cell type, amount of DNA damage, levels of p53 induced, different p53 post-translational modifications, recruitment of different co-factors to p53, and transcriptional activation of different sets of p53 target genes [87, 88]. Some cell cycle regulatory genes, such as CIP/KIP family of cyclin-dependent kinase inhibitors (p21/p27/p57) [71, 89–92] and Rb family of pocket proteins (pRb/p130/p107) [8, 93–95], are shown to play a role in cellular senescence. Therefore, it is likely that cells make a decision between undergoing premature senescence and apoptosis in response to DNA damage via a similar mechanism to that by which cells make a decision between cell cycle arrest and apoptosis. However, this needs to be further explored.
p53 target genes involved in senescence
The tumor suppression functions of p53 are manifested through the activation of its downstream genes resulting in cell cycle arrest, senescence, and apoptosis [96]. Although many target genes have been identified, those involved in p53-dependent cellular senescence are still poorly understood [97]. Several molecular markers, such as p21, PML, PAI-1, and DEC1, are shown to be sufficient to mediate senescence downstream of p53.
p21, a pleiotropic inhibitor of cyclin/cyclin-dependent kinases, is a classic p53 target [16]. p21 initiates growth arrest by preventing pRb phosphorylation by cyclin-dependent kinases [98], or by binding to and inactivating E2F [99] and PCNA [100, 101]. Thus, p21 plays a major role in inducing p53-dependent G1 arrest following DNA damage [16, 102]. In addition, in senescent cells, increased p53 activity is accompanied by increased binding of p53 to the p21 promoter [103]. Thus, it has been hypothesized that p21 is an important effector of p53-dependent cellular senescence. p21 was first identified as an overexpressed gene in senescent cells [104] and later was found to be capable of inducing premature senescence in p53-null H1299 cells [105]. However, disruption of p21 in human diploid fibroblasts results in extended cellular lifespan but not immortalization [92]. In agreement with this, MEFs derived from p21-null mice eventually enter senescence [106]. Moreover, lack of p21 diminishes but does not abrogate DNA damage-induced premature senescence in tumor cells [26]. Furthermore, p53 and p21 double-null cells undergo senescence induced by coexpression of oncogenic Ras and p53 [107]. These data suggest that p21 is not essential for p53-mediated senescence, although is sufficient to trigger senescence in the absence of p53.
Promyelocytic leukemia PML, a RING finger nuclear phosphoprotein, is a tumor suppressor and originally identified in acute promyelocytic leukemia (APL) patients [108]. PML is an essential component of the PML nuclear bodies (PML-NBs), which function as a docking area recruiting regulatory proteins including p53 to organize nuclear processes [109]. The level of PML was found to be increased in both Ras-induced premature senescence and replicative senescence [110, 111]. Overexpression of PML is capable of inducing premature senescence by stabilizing p53 via promoting p53 acetylation on Lys-382 and phosphorylation on Ser-15 and -46 [110–112]. In contrast, deacetylation of p53 antagonizes PML-induced premature senescence [113]. PML is expressed as seven isoforms. However, only PML-IV is able to regulate p53 activity and cause senescence [112]. Moreover, PML is capable of enhancing p53 stability through sequestering MDM2 [114] or inhibiting MDM2-mediated p53 degradation via prolonging the phosphorylation of p53 on serine-20 by Chk2 [115]. Interestingly, it has been shown that PML is a direct p53 target and functions as an effector for p53 activities [116]. Thus, in response to oncogene activation and DNA damage, induction of PML is p53-dependent and knockdown of PML attenuates p53-induced apoptosis and senescence. Together, PML and p53 form a positive regulatory feedback loop during cellular senescence.
PAI-1, plasminogen activator inhibitor-1, is upregulated in aging fibroblasts in vivo and in vitro and is considered a marker of replicative senescence [117]. PAI-1 inhibits cell proliferation by physically association with uPA and inhibits its activity. uPA, a secreted protease, promotes G1/S transition through activating a mitogenic signaling cascade by increasing the bioavailability of growth factors [118]. PAI-1 is transcriptionally regulated by p53 [119, 120]. Inhibition of PAI-1 by RNA interference leads to escape from replicative senescence and ectopic expression of PAI-1 induces premature senescence in proliferating p53-deficient mouse or human fibroblasts [121]. Therefore, PAI-1 is a critical downstream target of p53 in the senescence response of both mouse and human diploid fibroblasts.
DEC1, a basic helix-loop-helix transcription factor, was firstly identified as a gene expressed in differentiated embryo-chrondrocyte and induced by retinoic acid in mouse [122, 123]. DEC1 functions as a transcription repressor by directly binding to class B E-boxes [124, 125], by interacting with components of the basal transcription machinery [123, 126, 127], or by recruiting an HDAC co-repressor complex [128]. DEC1 is implicated in cell cycle regulation, differentiation, circadian rhythm, and apoptosis in response to various extracellular stimuli [123, 129–132]. In addition, DEC1 is a novel senescence marker which is upregulated in oncogene K-rasV12-induced senescence [7]. We recently showed that DEC1 is a target gene of the p53 family members and mediates p53-dependent cellular senescence. In addition, overexpression of DEC1 alone is sufficient to initiate G1 arrest and cellular senescence, whereas knockdown of DEC1 attenuates DNA damage-induced senescence in MCF7 cells [84]. Moreover, DEC1 inhibits the expression of ID1 [133], a member of the ID (inhibitor of differentiation or DNA binding) subfamily of the bHLH transcription factors [134]. ID1 plays an important role in senescence. It has been shown that ID1 is an oncogene and down-regulated in arrested or senescent cells [135]. In contrast, ectopic expression of ID1 extends the life-span of human keratinocytes [136, 137]. We found that ID1 expression is repressed upon DNA damage, whereas knockdown of p53 or DEC1 releases this inhibition. In addition, DEC1 represses ID1 expression through binding to the E-box elements in the proximal promoter region of the ID1 gene. Thus, we hypothesized that p53 trans-repressional activity on ID1 expression is mediated by its own target DEC1. However, knockdown of DEC1 or overexpression of ID1 partially attenuates DNA damage-induced senescence, which indicates the involvement of other unknown targets.
Over 100 target genes of p53 have been identified [96, 138–140]. For example, Bax, Puma, Noxa, Fas and Killer/DR5 are involved in apoptosis. However, genetic studies have shown that p53 is still able to induce apoptosis in the absence of each individual one. This suggests that p53 activates a redundant set of genes to fulfill its functions: some genes yet to be found may play an indispensable role in response to a specific senescence signal, or different combinations may be required in different tissues. Given the importance of the p53 transcriptional activities in regulating cellular responses, it is likely that other target genes are involved in p53-dependent cellular senescence.
p53 family members, p63 and p73, in cellular senescence
p53 was believed to be expressed predominantly as a single isoform. Recently, it was found that multiple isoforms of p53 are produced by using alternative promoters and undergoing alternative splicing [141]. One of the isoforms, named ΔNp53 in human and p44 in mouse, is generated from an alternative translation start site located in exon 4 at codon 40 in human and condon 41 in mouse. In the absence of the N-terminal transactivation domain, ΔNp53 is not able to transactivate p21 and MDM2 in p53-null background [142]. Consistent with this, ΔNp53 is tumorigenic in cells deficient in p53 [143], but is growth suppressive in cells with wild-type p53 [144]. Moreover, p44-knockin mice exhibit a phenotype of growth suppression and premature aging, which may be due to the altered expression ratio between p53 and p44 [145]. Therefore, the ratio of expression levels of p53 isoforms is important for p53 activity in growth suppression and senescence.
In addition to p53, p63 and p73 were identified to be members of the p53 family [146, 147]. p63 and p73 share similar structure with p53, especially in the DNA binding and activation domains [148]. Both p63 and p73 are expressed as multiple isoforms due to utilization of two separate promoters, the upstream P1 promoter and the P2 promoter in intron 3 [96, 149]. The P1 promoter produces transcriptional active (TA) isoforms, whereas the P2 promoter produces N-terminally deleted (ΔN) isoforms. Both TA and ΔN p63 and p73 transcripts can undergo C-terminal splicing resulting in three p63 isoforms (α–γ) and seven p73 isoforms (α-η). In addition, several p73 isoforms are produced through alternative N-terminal splicings. The presence of these different isoforms indicates the functional diversity of p63 and p73.
The sequence homology among p53, p63 and p73 indicates the possibility of similar functions for this gene family. Since p53 is a crucial tumor suppressor, it is not unreasonable to suspect that p63 and p73 may be tumor suppressors. Surprisingly, inactivating mutations of p63 and p73 are rarely observed in human cancers [150]. In addition, p63 and p73 knockout mice exhibit severe developmental abnormalities but not increased cancer susceptibility [151–153]. p63−/− mice are defected in limb and skin formation and lack ectodermal derivatives, including mammary glands, teeth, and hair [151, 153]. p73−/− mice are born with severe neurological defects, including hippocampal dysgenesis and hydrocephalus, chronic infection and inflammation, abnormalities in pheromone sensory pathways [152]. These suggest that p63 and p73 play a key role in development. However, p63−/− mice are born alive, but they die hours later due to dehydration. p73−/− mice live up to 4 to 6 weeks and die due to chronic infections. These put an obstacle to observing the role of p63 and p73 in cancer development. To overcome this, mouse models with knockout of specific isoforms of p63 and p73 were generated, and suggested that p63 and p73 are also involved in tumor formation and regulating cellular senescence. It has been shown that issue-specific p63 conditional knockout mice have a shortened lifespan and display features of accelerated aging correlated with induction of cellular senescence by conditional p63 ablation in skin [154]. Similar early-aging associated phenotypes were observed in mice with a deletion mutation of the first six exons in the p53 gene [155]. In addition, p63+/−;p73+/− mice develop spontaneous tumors and further loss of p63 or p73 in p53+/− mice exhibit higher tumor burden and metastasis compared to p53+/− mice [156]. Moreover, mice deficient in TAp73 are tumor prone and show accelerated aging [157]. These observations suggest that as a member of the p53 family, p63 and p73 play a role in tumorigenesis and aging.
Because of highly conserved sequence identity in the DNA binding domains among the p53 family, both p63 and p73 are capable of binding to the consensus p53 response element and activate some p53 targets, including p21, DEC1, and MDM2 [84, 96, 158], and transient expression of p63 and p73 induces cell cycle arrest and apoptosis [158, 159]. In addition, p63 and p73 are found to participate in DNA damage-induced apoptosis [160, 161]. Interestingly, it has been shown that p63 and p73 are required for p53-dependent apoptosis in MEFs upon DNA damage [162], but not in mature T cells upon treatment with radiation or glucocorticoid [163]. Thus, the role of p63 and p73 in the p53-pathway may vary under different conditions. However, whether p63 and p73 are also capable of initiating senescence is not clear. It has been shown that like p53, p73 can regulate the expression of human telomerase reverse transcriptase (hTERT), which adds TTAGGG repeats to chromosome ends [164, 165]. Telomere elongation by enforced expression of hTERT extends the lifespan of normal cells [166]. Interestingly, p53 is able to repress hTERT mRNA expression via blocking the accessibility of Sp1 to the Sp1 sites within the hTERT core promoter [167, 168] or indirectly through p21 by forming a repressive pRb/E2F complex on an atypical E2F site of the hTERT promoter [169]. Like p53, TAp73 isoforms can also inhibit the hTERT gene expression via Sp1 binding sites within the hTERT core promoter [164, 165]. Although ectopic expression of TAp73 isoforms (α, β, γ, σ) was found to downregulate the hTERT promoter activity in H1299 cells [164], other report showed that ectopic expression of TAp73β does not lead to suppression of hTERT transcription in the same cell system [170]. Moreover, both TAp73α and TAp73β can release p53-mediated suppression on the hTERT promoter by reducing p53 level through upregulation of MDM2 [170]. Furthermore, ΔNp73 antagonizes p53 and TAp73-mediated inhibition of hTERT via interfering with the activity of p53, TAp73 and, in addition, induces hTERT expression by interfering with E2F-RB-mediated repression of the hTERT core promoter [165]. These data indicate that p73 regulates hTERT through p53-like or unique mechanisms. However, whether regulation of hTERT, a regulator of replicative senescence, is a mechanism by which p73 modulates cellular senescence remain to be characterized. In addition, the common targets of p53 and p73 or the unique p73 targets involved in cellular senescence need to be identified in the future.
For p63, induced ablation of p63 in primary keratinocytes causes an arrest with enhanced expression of senescence markers, SA-β-gal and PML [154]. Reports showed that both p53 and p16 pathways are involved in senescence induced by p63 deficiency [171]. In addition, several regulators of cell cycle and senescence, such as PML, p21, 14-3-3σ, and IGFBP3, are shown to be positively or negatively regulated by TAp63/ΔNp63 [171]. However, how p63 fits into the senescence-regulatory pathway needs to be further explored. Taken together, the story of p53 family members in cellular senescence is far more complicated than we expected. Development of efficient antibodies specifically against each isoform and generation of cell lines or mice models with single isoform expression, knockdown, or knockout are needed to further elucidate the role of p53 family proteins in cellular senescence.
Conclusions
It is clear that cellular senescence is an important anti-cancer barrier and the p53 tumor suppressor plays a pivotal role in this process. How p53 family members are involved in the senescence pathway was summarized in Fig. 1. However, the complexity of the p53 family proteins makes this story even more complicated. For example, what is the function of each individual p53 isoform in senescence? How p63 and p73 are implicated in cellular senescence? Since p63 and p73 are rarely mutated in tumors, what is the significance of p63 and p73 in mediating senescence? Do p53 family proteins affect each other in this process? Therefore, future studies uncovering these questions will further our understanding of the integrated functions of the p53 family members in normal tissue development, cancer formation, and aging. This will fulfill our knowledge about the p53 family members in tumorigenesis and may provide valuable clues to solve the current clinical problems.
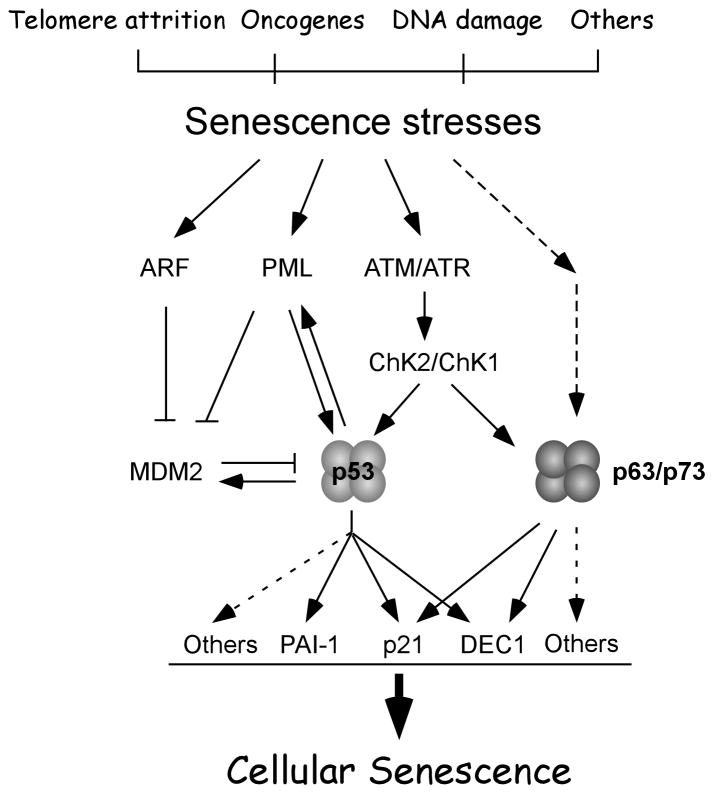
Fig 1.
p53 pathways in cellular senescence. Solid lines, known pathways. Dashed lines, undefined pathways.
References
- 1.Harley CB, Futcher AB, Greider CW. Telomeres shorten during ageing of human fibroblasts. Nature. 1990;345(6274):458–60. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- 2.Serrano M, et al. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88(5):593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 3.te Poele RH, et al. DNA damage is able to induce senescence in tumor cells in vitro and in vivo. Cancer Res. 2002;62(6):1876–83. [PubMed] [Google Scholar]
- 4.Sherwood SW, et al. Defining cellular senescence in IMR-90 cells: a flow cytometric analysis. Proc Natl Acad Sci U S A. 1988;85(23):9086–90. doi: 10.1073/pnas.85.23.9086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dimri GP. What has senescence got to do with cancer? Cancer Cell. 2005;7(6):505–12. doi: 10.1016/j.ccr.2005.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campisi J. Suppressing cancer: the importance of being senescent. Science. 2005;309(5736):886–7. doi: 10.1126/science.1116801. [DOI] [PubMed] [Google Scholar]
- 7.Collado M, et al. Tumour biology: senescence in premalignant tumours. Nature. 2005;436(7051):642. doi: 10.1038/436642a. [DOI] [PubMed] [Google Scholar]
- 8.Braig M, et al. Oncogene-induced senescence as an initial barrier in lymphoma development. Nature. 2005;436(7051):660–5. doi: 10.1038/nature03841. [DOI] [PubMed] [Google Scholar]
- 9.Xue W, et al. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature. 2007;445(7128):656–60. doi: 10.1038/nature05529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ventura A, et al. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007;445(7128):661–5. doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- 11.Martins CP, Brown-Swigart L, Evan GI. Modeling the therapeutic efficacy of p53 restoration in tumors. Cell. 2006;127(7):1323–34. doi: 10.1016/j.cell.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 12.Olivier M, et al. The IARC TP53 database: new online mutation analysis and recommendations to users. Hum Mutat. 2002:607–14. doi: 10.1002/humu.10081. [DOI] [PubMed] [Google Scholar]
- 13.Li FP, Fraumeni JF., Jr Soft-tissue sarcomas, breast cancer, and other neoplasms. A familial syndrome? Ann Intern Med. 1969;71(4):747–52. doi: 10.7326/0003-4819-71-4-747. [DOI] [PubMed] [Google Scholar]
- 14.Gasco M, I, Yulug G, Crook T. TP53 mutations in familial breast cancer: functional aspects. Hum Mutat. 2003;21(3):301–6. doi: 10.1002/humu.10173. [DOI] [PubMed] [Google Scholar]
- 15.Birch JM, et al. Relative frequency and morphology of cancers in carriers of germline TP53 mutations. Oncogene. 2001;20(34):4621–8. doi: 10.1038/sj.onc.1204621. [DOI] [PubMed] [Google Scholar]
- 16.el-Deiry WS, et al. Definition of a consensus binding site for p53. Nat Genet. 1992;1(1):45–9. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- 17.Hofseth LJ, Hussain SP, Harris CC. p53: 25 years after its discovery. Trends Pharmacol Sci. 2004;25(4):177–81. doi: 10.1016/j.tips.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 18.Prives C, Hall PA. The p53 pathway. J Pathol. 1999;187(1):112–26. doi: 10.1002/(SICI)1096-9896(199901)187:1<112::AID-PATH250>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 19.Ko LJ, Prives C. p53: puzzle and paradigm. Genes Dev. 1996;10(9):1054–72. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 20.Wang Y, et al. Induced p53 expression in lung cancer cell line promotes cell senescence and differentially modifies the cytotoxicity of anti-cancer drugs. Oncogene. 1998;17(15):1923–30. doi: 10.1038/sj.onc.1202113. [DOI] [PubMed] [Google Scholar]
- 21.Sugrue MM, et al. Wild-type p53 triggers a rapid senescence program in human tumor cells lacking functional p53. Proc Natl Acad Sci U S A. 1997;94(18):9648–53. doi: 10.1073/pnas.94.18.9648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Itahana K, Dimri G, Campisi J. Regulation of cellular senescence by p53. Eur J Biochem. 2001;268(10):2784–91. doi: 10.1046/j.1432-1327.2001.02228.x. [DOI] [PubMed] [Google Scholar]
- 23.Rovinski B, Benchimol S. Immortalization of rat embryo fibroblasts by the cellular p53 oncogene. Oncogene. 1988;2(5):445–52. [PubMed] [Google Scholar]
- 24.Bond JA, Wyllie FS, Wynford-Thomas D. Escape from senescence in human diploid fibroblasts induced directly by mutant p53. Oncogene. 1994;9(7):1885–9. [PubMed] [Google Scholar]
- 25.Smogorzewska A, de Lange T. Different telomere damage signaling pathways in human and mouse cells. EMBO J. 2002;21(16):4338–48. doi: 10.1093/emboj/cdf433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmitt CA. Cellular senescence and cancer treatment. Biochim Biophys Acta. 2007;1775(1):5–20. doi: 10.1016/j.bbcan.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 27.Hayflick L, Moorhead PS. The serial cultivation of human diploid cell strains. Exp Cell Res. 1961;25:585–621. doi: 10.1016/0014-4827(61)90192-6. [DOI] [PubMed] [Google Scholar]
- 28.Hayflick L. The Limited In Vitro Lifetime Of Human Diploid Cell Strains. Exp Cell Res. 1965;37:614–36. doi: 10.1016/0014-4827(65)90211-9. [DOI] [PubMed] [Google Scholar]
- 29.Dimri GP, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci U S A. 1995;92(20):9363–7. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee BY, et al. Senescence-associated beta-galactosidase is lysosomal beta-galactosidase. Aging Cell. 2006;5(2):187–95. doi: 10.1111/j.1474-9726.2006.00199.x. [DOI] [PubMed] [Google Scholar]
- 31.Kurz DJ, et al. Senescence-associated (beta)-galactosidase reflects an increase in lysosomal mass during replicative ageing of human endothelial cells. J Cell Sci. 2000;113(Pt 20):3613–22. doi: 10.1242/jcs.113.20.3613. [DOI] [PubMed] [Google Scholar]
- 32.Le Gall JY, et al. Lysosomal enzyme activities during ageing of adult human liver cell lines. Mech Ageing Dev. 1979;11(4):287–93. doi: 10.1016/0047-6374(79)90008-3. [DOI] [PubMed] [Google Scholar]
- 33.Lundblad V, Szostak JW. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell. 1989;57(4):633–43. doi: 10.1016/0092-8674(89)90132-3. [DOI] [PubMed] [Google Scholar]
- 34.Blackburn EH. The molecular structure of centromeres and telomeres. Annu Rev Biochem. 1984;53:163–94. doi: 10.1146/annurev.bi.53.070184.001115. [DOI] [PubMed] [Google Scholar]
- 35.d’Adda di Fagagna F, et al. A DNA damage checkpoint response in telomere-initiated senescence. Nature. 2003;426(6963):194–8. doi: 10.1038/nature02118. [DOI] [PubMed] [Google Scholar]
- 36.Karlseder J, Smogorzewska A, de Lange T. Senescence induced by altered telomere state, not telomere loss. Science. 2002;295(5564):2446–9. doi: 10.1126/science.1069523. [DOI] [PubMed] [Google Scholar]
- 37.van Steensel B, Smogorzewska A, de Lange T. TRF2 protects human telomeres from end-to-end fusions. Cell. 1998;92(3):401–13. doi: 10.1016/s0092-8674(00)80932-0. [DOI] [PubMed] [Google Scholar]
- 38.Bos JL. ras oncogenes in human cancer: a review. Cancer Res. 1989;49(17):4682–9. [PubMed] [Google Scholar]
- 39.Karnoub AE, Weinberg RA. Ras oncogenes: split personalities. Nat Rev Mol Cell Biol. 2008;9(7):517–31. doi: 10.1038/nrm2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Land H, Parada LF, Weinberg RA. Tumorigenic conversion of primary embryo fibroblasts requires at least two cooperating oncogenes. Nature. 1983;304(5927):596–602. doi: 10.1038/304596a0. [DOI] [PubMed] [Google Scholar]
- 41.Ruley HE. Adenovirus early region 1A enables viral and cellular transforming genes to transform primary cells in culture. Nature. 1983;304(5927):602–6. doi: 10.1038/304602a0. [DOI] [PubMed] [Google Scholar]
- 42.Chintala SK, et al. Adenovirus-mediated p16/CDKN2 gene transfer suppresses glioma invasion in vitro. Oncogene. 1997;15(17):2049–57. doi: 10.1038/sj.onc.1201382. [DOI] [PubMed] [Google Scholar]
- 43.Kemp CJ, et al. Reduction of p53 gene dosage does not increase initiation or promotion but enhances malignant progression of chemically induced skin tumors. Cell. 1993;74(5):813–22. doi: 10.1016/0092-8674(93)90461-x. [DOI] [PubMed] [Google Scholar]
- 44.Minden A, et al. Differential activation of ERK and JNK mitogen-activated protein kinases by Raf-1 and MEKK. Science. 1994;266(5191):1719–23. doi: 10.1126/science.7992057. [DOI] [PubMed] [Google Scholar]
- 45.Whitmarsh AJ, et al. Role of p38 and JNK mitogen-activated protein kinases in the activation of ternary complex factors. Mol Cell Biol. 1997;17(5):2360–71. doi: 10.1128/mcb.17.5.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang W, et al. Sequential activation of the MEK-extracellular signal-regulated kinase and MKK3/6-p38 mitogen-activated protein kinase pathways mediates oncogenic ras-induced premature senescence. Mol Cell Biol. 2002;22(10):3389–403. doi: 10.1128/MCB.22.10.3389-3403.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun P, et al. PRAK is essential for ras-induced senescence and tumor suppression. Cell. 2007;128(2):295–308. doi: 10.1016/j.cell.2006.11.050. [DOI] [PubMed] [Google Scholar]
- 48.Lee AC, et al. Ras proteins induce senescence by altering the intracellular levels of reactive oxygen species. J Biol Chem. 1999;274(12):7936–40. doi: 10.1074/jbc.274.12.7936. [DOI] [PubMed] [Google Scholar]
- 49.Di Micco R, et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 2006;444(7119):638–42. doi: 10.1038/nature05327. [DOI] [PubMed] [Google Scholar]
- 50.Wei S, Sedivy JM. Expression of catalytically active telomerase does not prevent premature senescence caused by overexpression of oncogenic Ha-Ras in normal human fibroblasts. Cancer Res. 1999;59(7):1539–43. [PubMed] [Google Scholar]
- 51.Gorbunova V, Seluanov A, Pereira-Smith OM. Expression of human telomerase (hTERT) does not prevent stress-induced senescence in normal human fibroblasts but protects the cells from stress-induced apoptosis and necrosis. J Biol Chem. 2002;277(41):38540–9. doi: 10.1074/jbc.M202671200. [DOI] [PubMed] [Google Scholar]
- 52.Chen Q, Ames BN. Senescence-like growth arrest induced by hydrogen peroxide in human diploid fibroblast F65 cells. Proc Natl Acad Sci U S A. 1994;91(10):4130–4. doi: 10.1073/pnas.91.10.4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bladier C, et al. Response of a primary human fibroblast cell line to H2O2: senescence-like growth arrest or apoptosis? Cell Growth Differ. 1997;8(5):589–98. [PubMed] [Google Scholar]
- 54.Borlon C, et al. Transient increased extracellular release of H2O2 during establishment of UVB-induced premature senescence. Ann N Y Acad Sci. 2007;1119:72–7. doi: 10.1196/annals.1404.002. [DOI] [PubMed] [Google Scholar]
- 55.Nelson WG, Kastan MB. DNA strand breaks: the DNA template alterations that trigger p53-dependent DNA damage response pathways. Mol Cell Biol. 1994;14(3):1815–23. doi: 10.1128/mcb.14.3.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Robles SJ, et al. Permanent cell cycle arrest in asynchronously proliferating normal human fibroblasts treated with doxorubicin or etoposide but not camptothecin. Biochem Pharmacol. 1999;58(4):675–85. doi: 10.1016/s0006-2952(99)00127-6. [DOI] [PubMed] [Google Scholar]
- 57.Elmore LW, et al. Adriamycin-induced senescence in breast tumor cells involves functional p53 and telomere dysfunction. J Biol Chem. 2002;277(38):35509–15. doi: 10.1074/jbc.M205477200. [DOI] [PubMed] [Google Scholar]
- 58.Han Z, et al. Role of p21 in apoptosis and senescence of human colon cancer cells treated with camptothecin. J Biol Chem. 2002;277(19):17154–60. doi: 10.1074/jbc.M112401200. [DOI] [PubMed] [Google Scholar]
- 59.Roninson IB. Tumor cell senescence in cancer treatment. Cancer Res. 2003;63(11):2705–15. [PubMed] [Google Scholar]
- 60.Chang BD, et al. A senescence-like phenotype distinguishes tumor cells that undergo terminal proliferation arrest after exposure to anticancer agents. Cancer Res. 1999;59(15):3761–7. [PubMed] [Google Scholar]
- 61.Farrell N, et al. Cytotoxicity, DNA strand breakage and DNA-protein crosslinking by a novel transplatinum compound in human A2780 ovarian and MCF-7 breast carcinoma cells. Biochem Pharmacol. 2004;68(5):857–66. doi: 10.1016/j.bcp.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 62.Liu LF, et al. Mechanism of action of camptothecin. Ann N Y Acad Sci. 2000;922:1–10. doi: 10.1111/j.1749-6632.2000.tb07020.x. [DOI] [PubMed] [Google Scholar]
- 63.Reedijk J, Lohman PH. Cisplatin: synthesis, antitumour activity and mechanism of action. Pharm Weekbl Sci. 1985;7(5):173–80. doi: 10.1007/BF02307573. [DOI] [PubMed] [Google Scholar]
- 64.Beausejour CM, et al. Reversal of human cellular senescence: roles of the p53 and p16 pathways. EMBO J. 2003;22(16):4212–22. doi: 10.1093/emboj/cdg417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tiemann F, Hinds PW. Induction of DNA synthesis and apoptosis by regulated inactivation of a temperature-sensitive retinoblastoma protein. EMBO J. 1998;17(4):1040–52. doi: 10.1093/emboj/17.4.1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Alexander K, Yang HS, Hinds PW. pRb inactivation in senescent cells leads to an E2F-dependent apoptosis requiring p73. Mol Cancer Res. 2003;1(10):716–28. [PubMed] [Google Scholar]
- 67.Xu HJ, et al. Reexpression of the retinoblastoma protein in tumor cells induces senescence and telomerase inhibition. Oncogene. 1997;15(21):2589–96. doi: 10.1038/sj.onc.1201446. [DOI] [PubMed] [Google Scholar]
- 68.Dai CY, Enders GH. p16 INK4a can initiate an autonomous senescence program. Oncogene. 2000;19(13):1613–22. doi: 10.1038/sj.onc.1203438. [DOI] [PubMed] [Google Scholar]
- 69.Takai H, Smogorzewska A, de Lange T. DNA damage foci at dysfunctional telomeres. Curr Biol. 2003;13(17):1549–56. doi: 10.1016/s0960-9822(03)00542-6. [DOI] [PubMed] [Google Scholar]
- 70.Pandita TK. ATM function and telomere stability. Oncogene. 2002;21(4):611–8. doi: 10.1038/sj.onc.1205060. [DOI] [PubMed] [Google Scholar]
- 71.Herbig U, et al. Telomere shortening triggers senescence of human cells through a pathway involving ATM, p53, and p21(CIP1), but not p16(INK4a) Mol Cell. 2004;14(4):501–13. doi: 10.1016/s1097-2765(04)00256-4. [DOI] [PubMed] [Google Scholar]
- 72.Zhang Y, Xiong Y. Control of p53 ubiquitination and nuclear export by MDM2 and ARF. Cell Growth Differ. 2001;12(4):175–86. [PubMed] [Google Scholar]
- 73.Kamijo T, et al. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell. 1997;91(5):649–59. doi: 10.1016/s0092-8674(00)80452-3. [DOI] [PubMed] [Google Scholar]
- 74.Donehower LA, et al. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356(6366):215–21. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 75.Ferbeyre G, et al. Oncogenic ras and p53 cooperate to induce cellular senescence. Mol Cell Biol. 2002;22(10):3497–508. doi: 10.1128/MCB.22.10.3497-3508.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wei W, Hemmer RM, Sedivy JM. Role of p14(ARF) in replicative and induced senescence of human fibroblasts. Mol Cell Biol. 2001;21(20):6748–57. doi: 10.1128/MCB.21.20.6748-6757.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bartkova J, et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444(7119):633–7. doi: 10.1038/nature05268. [DOI] [PubMed] [Google Scholar]
- 78.Halazonetis TD, V, Gorgoulis G, Bartek J. An oncogene-induced DNA damage model for cancer development. Science. 2008;319(5868):1352–5. doi: 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- 79.Mallette FA, Gaumont-Leclerc MF, Ferbeyre G. The DNA damage signaling pathway is a critical mediator of oncogene-induced senescence. Genes Dev. 2007;21(1):43–8. doi: 10.1101/gad.1487307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mallette FA, Ferbeyre G. The DNA damage signaling pathway connects oncogenic stress to cellular senescence. Cell Cycle. 2007;6(15):1831–6. doi: 10.4161/cc.6.15.4516. [DOI] [PubMed] [Google Scholar]
- 81.Toledo LI, et al. ATR signaling can drive cells into senescence in the absence of DNA breaks. Genes Dev. 2008;22(3):297–302. doi: 10.1101/gad.452308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Itahana K, et al. Control of the replicative life span of human fibroblasts by p16 and the polycomb protein Bmi-1. Mol Cell Biol. 2003;23(1):389–401. doi: 10.1128/MCB.23.1.389-401.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chang BD, et al. Role of p53 and p21waf1/cip1 in senescence-like terminal proliferation arrest induced in human tumor cells by chemotherapeutic drugs. Oncogene. 1999;18(34):4808–18. doi: 10.1038/sj.onc.1203078. [DOI] [PubMed] [Google Scholar]
- 84.Qian Y, et al. DEC1, a basic helix-loop-helix transcription factor and a novel target gene of the p53 family, mediates p53-dependent premature senescence. J Biol Chem. 2008;283(5):2896–905. doi: 10.1074/jbc.M708624200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Caelles C, Helmberg A, Karin M. p53-dependent apoptosis in the absence of transcriptional activation of p53-target genes. Nature. 1994;370(6486):220–3. doi: 10.1038/370220a0. [DOI] [PubMed] [Google Scholar]
- 86.Sabbatini P, et al. Essential role for p53-mediated transcription in E1A-induced apoptosis. Genes Dev. 1995;9(17):2184–92. doi: 10.1101/gad.9.17.2184. [DOI] [PubMed] [Google Scholar]
- 87.Wahl GM, Carr AM. The evolution of diverse biological responses to DNA damage: insights from yeast and p53. Nat Cell Biol. 2001;3(12):E277–86. doi: 10.1038/ncb1201-e277. [DOI] [PubMed] [Google Scholar]
- 88.Hansen R, Oren M. p53; from inductive signal to cellular effect. Curr Opin Genet Dev. 1997;7(1):46–51. doi: 10.1016/s0959-437x(97)80108-6. [DOI] [PubMed] [Google Scholar]
- 89.Alexander K, Hinds PW. Requirement for p27(KIP1) in retinoblastoma protein-mediated senescence. Mol Cell Biol. 2001;21(11):3616–31. doi: 10.1128/MCB.21.11.3616-3631.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Collado M, et al. Inhibition of the phosphoinositide 3-kinase pathway induces a senescence-like arrest mediated by p27Kip1. J Biol Chem. 2000;275(29):21960–8. doi: 10.1074/jbc.M000759200. [DOI] [PubMed] [Google Scholar]
- 91.Nijjar T, et al. p57KIP2 expression and loss of heterozygosity during immortal conversion of cultured human mammary epithelial cells. Cancer Res. 1999;59(20):5112–8. [PubMed] [Google Scholar]
- 92.Brown JP, Wei W, Sedivy JM. Bypass of senescence after disruption of p21CIP1/WAF1 gene in normal diploid human fibroblasts. Science. 1997;277(5327):831–4. doi: 10.1126/science.277.5327.831. [DOI] [PubMed] [Google Scholar]
- 93.Kiyono T, et al. Both Rb/p16INK4a inactivation and telomerase activity are required to immortalize human epithelial cells. Nature. 1998;396(6706):84–8. doi: 10.1038/23962. [DOI] [PubMed] [Google Scholar]
- 94.Kapic A, et al. Cooperation between p53 and p130(Rb2) in induction of cellular senescence. Cell Death Differ. 2006;13(2):324–34. doi: 10.1038/sj.cdd.4401756. [DOI] [PubMed] [Google Scholar]
- 95.Lehmann BD, et al. Distinct roles for p107 and p130 in Rb-independent cellular senescence. Cell Cycle. 2008;7(9):1262–8. doi: 10.4161/cc.7.9.5945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Harms K, Nozell S, Chen X. The common and distinct target genes of the p53 family transcription factors. Cell Mol Life Sci. 2004;61(7–8):822–42. doi: 10.1007/s00018-003-3304-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Levine AJ, Hu W, Feng Z. The P53 pathway: what questions remain to be explored? Cell Death Differ. 2006;13(6):1027–36. doi: 10.1038/sj.cdd.4401910. [DOI] [PubMed] [Google Scholar]
- 98.el-Deiry WS, et al. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75(4):817–25. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 99.Afshari CA, et al. A role for a p21-E2F interaction during senescence arrest of normal human fibroblasts. Cell Growth Differ. 1996;7(8):979–88. [PubMed] [Google Scholar]
- 100.Li R, et al. Differential effects by the p21 CDK inhibitor on PCNA-dependent DNA replication and repair. Nature. 1994;371(6497):534–7. doi: 10.1038/371534a0. [DOI] [PubMed] [Google Scholar]
- 101.Waga S, et al. The p21 inhibitor of cyclin-dependent kinases controls DNA replication by interaction with PCNA. Nature. 1994;369(6481):574–8. doi: 10.1038/369574a0. [DOI] [PubMed] [Google Scholar]
- 102.el-Deiry WS, et al. WAF1/CIP1 is induced in p53-mediated G1 arrest and apoptosis. Cancer Res. 1994;54(5):1169–74. [PubMed] [Google Scholar]
- 103.Jackson JG, Pereira-Smith OM. p53 is preferentially recruited to the promoters of growth arrest genes p21 and GADD45 during replicative senescence of normal human fibroblasts. Cancer Res. 2006;66(17):8356–60. doi: 10.1158/0008-5472.CAN-06-1752. [DOI] [PubMed] [Google Scholar]
- 104.Noda A, et al. Cloning of senescent cell-derived inhibitors of DNA synthesis using an expression screen. Exp Cell Res. 1994;211(1):90–8. doi: 10.1006/excr.1994.1063. [DOI] [PubMed] [Google Scholar]
- 105.Wang Y, Blandino G, Givol D. Induced p21waf expression in H1299 cell line promotes cell senescence and protects against cytotoxic effect of radiation and doxorubicin. Oncogene. 1999;18(16):2643–9. doi: 10.1038/sj.onc.1202632. [DOI] [PubMed] [Google Scholar]
- 106.Pantoja C, Serrano M. Murine fibroblasts lacking p21 undergo senescence and are resistant to transformation by oncogenic Ras. Oncogene. 1999;18(35):4974–82. doi: 10.1038/sj.onc.1202880. [DOI] [PubMed] [Google Scholar]
- 107.Castro ME, et al. Cellular senescence induced by p53-ras cooperation is independent of p21waf1 in murine embryo fibroblasts. J Cell Biochem. 2004;92(3):514–24. doi: 10.1002/jcb.20079. [DOI] [PubMed] [Google Scholar]
- 108.Reymond A, et al. The tripartite motif family identifies cell compartments. EMBO J. 2001;20(9):2140–51. doi: 10.1093/emboj/20.9.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Borden KL. Pondering the promyelocytic leukemia protein (PML) puzzle: possible functions for PML nuclear bodies. Mol Cell Biol. 2002;22(15):5259–69. doi: 10.1128/MCB.22.15.5259-5269.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Pearson M, et al. PML regulates p53 acetylation and premature senescence induced by oncogenic Ras. Nature. 2000;406(6792):207–10. doi: 10.1038/35018127. [DOI] [PubMed] [Google Scholar]
- 111.Ferbeyre G, et al. PML is induced by oncogenic ras and promotes premature senescence. Genes Dev. 2000;14(16):2015–27. [PMC free article] [PubMed] [Google Scholar]
- 112.Bischof O, et al. Deconstructing PML-induced premature senescence. EMBO J. 2002;21(13):3358–69. doi: 10.1093/emboj/cdf341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Langley E, et al. Human SIR2 deacetylates p53 and antagonizes PML/p53-induced cellular senescence. EMBO J. 2002;21(10):2383–96. doi: 10.1093/emboj/21.10.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Bernardi R, et al. PML regulates p53 stability by sequestering Mdm2 to the nucleolus. Nat Cell Biol. 2004;6(7):665–72. doi: 10.1038/ncb1147. [DOI] [PubMed] [Google Scholar]
- 115.Louria-Hayon I, et al. The promyelocytic leukemia protein protects p53 from Mdm2-mediated inhibition and degradation. J Biol Chem. 2003;278(35):33134–41. doi: 10.1074/jbc.M301264200. [DOI] [PubMed] [Google Scholar]
- 116.de Stanchina E, et al. PML is a direct p53 target that modulates p53 effector functions. Mol Cell. 2004;13(4):523–35. doi: 10.1016/s1097-2765(04)00062-0. [DOI] [PubMed] [Google Scholar]
- 117.Mu XC, Higgins PJ. Differential growth state-dependent regulation of plasminogen activator inhibitor type-1 expression in senescent IMR-90 human diploid fibroblasts. J Cell Physiol. 1995;165(3):647–57. doi: 10.1002/jcp.1041650324. [DOI] [PubMed] [Google Scholar]
- 118.Andreasen PA, Egelund R, Petersen HH. The plasminogen activation system in tumor growth, invasion, and metastasis. Cell Mol Life Sci. 2000;57(1):25–40. doi: 10.1007/s000180050497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kunz C, et al. Differential regulation of plasminogen activator and inhibitor gene transcription by the tumor suppressor p53. Nucleic Acids Res. 1995;23(18):3710–7. doi: 10.1093/nar/23.18.3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhao R, et al. Analysis of p53-regulated gene expression patterns using oligonucleotide arrays. Genes Dev. 2000;14(8):981–93. [PMC free article] [PubMed] [Google Scholar]
- 121.Kortlever RM, Higgins PJ, Bernards R. Plasminogen activator inhibitor-1 is a critical downstream target of p53 in the induction of replicative senescence. Nat Cell Biol. 2006;8(8):877–84. doi: 10.1038/ncb1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Shen M, et al. Molecular characterization of the novel basic helix-loop-helix protein DEC1 expressed in differentiated human embryo chondrocytes. Biochem Biophys Res Commun. 1997;236(2):294–8. doi: 10.1006/bbrc.1997.6960. [DOI] [PubMed] [Google Scholar]
- 123.Boudjelal M, et al. Overexpression of Stra13, a novel retinoic acid-inducible gene of the basic helix-loop-helix family, inhibits mesodermal and promotes neuronal differentiation of P19 cells. Genes Dev. 1997;11(16):2052–65. doi: 10.1101/gad.11.16.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Li Y, et al. DEC1 negatively regulates the expression of DEC2 through binding to the E-box in the proximal promoter. J Biol Chem. 2003;278(19):16899–907. doi: 10.1074/jbc.M300596200. [DOI] [PubMed] [Google Scholar]
- 125.St-Pierre B, et al. Stra13 homodimers repress transcription through class B E-box elements. J Biol Chem. 2002;277(48):46544–51. doi: 10.1074/jbc.M111652200. [DOI] [PubMed] [Google Scholar]
- 126.Zawel L, et al. DEC1 is a downstream target of TGF-beta with sequence-specific transcriptional repressor activities. Proc Natl Acad Sci U S A. 2002;99(5):2848–53. doi: 10.1073/pnas.261714999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Shen M, et al. Basic helix-loop-helix protein DEC1 promotes chondrocyte differentiation at the early and terminal stages. J Biol Chem. 2002;277(51):50112–20. doi: 10.1074/jbc.M206771200. [DOI] [PubMed] [Google Scholar]
- 128.Sun H, Taneja R. Stra13 expression is associated with growth arrest and represses transcription through histone deacetylase (HDAC)-dependent and HDAC-independent mechanisms. Proc Natl Acad Sci U S A. 2000;97(8):4058–63. doi: 10.1073/pnas.070526297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yoon DY, et al. Identification of genes differentially induced by hypoxia in pancreatic cancer cells. Biochem Biophys Res Commun. 2001;288(4):882–6. doi: 10.1006/bbrc.2001.5867. [DOI] [PubMed] [Google Scholar]
- 130.Miyazaki K, et al. Identification of functional hypoxia response elements in the promoter region of the DEC1 and DEC2 genes. J Biol Chem. 2002;277(49):47014–21. doi: 10.1074/jbc.M204938200. [DOI] [PubMed] [Google Scholar]
- 131.Li Y, et al. Abundant expression of Dec1/stra13/sharp2 in colon carcinoma: its antagonizing role in serum deprivation-induced apoptosis and selective inhibition of procaspase activation. Biochem J. 2002;367(Pt 2):413–22. doi: 10.1042/BJ20020514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Honma S, et al. Dec1 and Dec2 are regulators of the mammalian molecular clock. Nature. 2002;419(6909):841–4. doi: 10.1038/nature01123. [DOI] [PubMed] [Google Scholar]
- 133.Qian Y, Chen X. ID1, inhibitor of differentiation/DNA binding, is an effector of the p53-dependent DNA damage response pathway. J Biol Chem. 2008;283(33):22410–6. doi: 10.1074/jbc.M800643200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Norton JD. ID helix-loop-helix proteins in cell growth, differentiation and tumorigenesis. J Cell Sci. 2000;113(Pt 22):3897–905. doi: 10.1242/jcs.113.22.3897. [DOI] [PubMed] [Google Scholar]
- 135.Hara E, et al. Id-related genes encoding helix-loop-helix proteins are required for G1 progression and are repressed in senescent human fibroblasts. J Biol Chem. 1994;269(3):2139–45. [PubMed] [Google Scholar]
- 136.Alani RM, et al. Immortalization of primary human keratinocytes by the helix-loop-helix protein, Id-1. Proc Natl Acad Sci U S A. 1999;96(17):9637–41. doi: 10.1073/pnas.96.17.9637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Nickoloff BJ, et al. Id-1 delays senescence but does not immortalize keratinocytes. J Biol Chem. 2000;275(36):27501–4. doi: 10.1074/jbc.C000311200. [DOI] [PubMed] [Google Scholar]
- 138.Nakamura Y. Isolation of p53-target genes and their functional analysis. Cancer Sci. 2004;95(1):7–11. doi: 10.1111/j.1349-7006.2004.tb03163.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Harris SL, Levine AJ. The p53 pathway: positive and negative feedback loops. Oncogene. 2005;24(17):2899–908. doi: 10.1038/sj.onc.1208615. [DOI] [PubMed] [Google Scholar]
- 140.Agarwal ML, et al. The p53 network. J Biol Chem. 1998;273(1):1–4. doi: 10.1074/jbc.273.1.1. [DOI] [PubMed] [Google Scholar]
- 141.Bourdon JC, et al. p53 isoforms can regulate p53 transcriptional activity. Genes Dev. 2005;19(18):2122–37. doi: 10.1101/gad.1339905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Courtois S, et al. DeltaN-p53, a natural isoform of p53 lacking the first transactivation domain, counteracts growth suppression by wild-type p53. Oncogene. 2002;21(44):6722–8. doi: 10.1038/sj.onc.1205874. [DOI] [PubMed] [Google Scholar]
- 143.Mowat M, et al. Rearrangements of the cellular p53 gene in erythroleukaemic cells transformed by Friend virus. Nature. 1985;314(6012):633–6. doi: 10.1038/314633a0. [DOI] [PubMed] [Google Scholar]
- 144.Rovinski B, et al. Deletion of 5′-coding sequences of the cellular p53 gene in mouse erythroleukemia: a novel mechanism of oncogene regulation. Mol Cell Biol. 1987;7(2):847–53. doi: 10.1128/mcb.7.2.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Maier B, et al. Modulation of mammalian life span by the short isoform of p53. Genes Dev. 2004;18(3):306–19. doi: 10.1101/gad.1162404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kaghad M, et al. Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell. 1997;90(4):809–19. doi: 10.1016/s0092-8674(00)80540-1. [DOI] [PubMed] [Google Scholar]
- 147.Schmale H, Bamberger C. A novel protein with strong homology to the tumor suppressor p53. Oncogene. 1997;15(11):1363–7. doi: 10.1038/sj.onc.1201500. [DOI] [PubMed] [Google Scholar]
- 148.Courtois S, de Fromentel CC, Hainaut P. p53 protein variants: structural and functional similarities with p63 and p73 isoforms. Oncogene. 2004;23(3):631–8. doi: 10.1038/sj.onc.1206929. [DOI] [PubMed] [Google Scholar]
- 149.Prives C, Manfredi JJ. The continuing saga of p53--more sleepless nights ahead. Mol Cell. 2005;19(6):719–21. doi: 10.1016/j.molcel.2005.08.024. [DOI] [PubMed] [Google Scholar]
- 150.Moll UM, Slade N. p63 and p73: roles in development and tumor formation. Mol Cancer Res. 2004;2(7):371–86. [PubMed] [Google Scholar]
- 151.Mills AA, et al. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999;398(6729):708–13. doi: 10.1038/19531. [DOI] [PubMed] [Google Scholar]
- 152.Yang A, et al. p73-deficient mice have neurological, pheromonal and inflammatory defects but lack spontaneous tumours. Nature. 2000;404(6773):99–103. doi: 10.1038/35003607. [DOI] [PubMed] [Google Scholar]
- 153.Yang A, et al. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398(6729):714–8. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
- 154.Keyes WM, et al. p63 deficiency activates a program of cellular senescence and leads to accelerated aging. Genes Dev. 2005;19(17):1986–99. doi: 10.1101/gad.342305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Tyner SD, et al. p53 mutant mice that display early ageing-associated phenotypes. Nature. 2002;415(6867):45–53. doi: 10.1038/415045a. [DOI] [PubMed] [Google Scholar]
- 156.Flores ER, et al. Tumor predisposition in mice mutant for p63 and p73: evidence for broader tumor suppressor functions for the p53 family. Cancer Cell. 2005;7(4):363–73. doi: 10.1016/j.ccr.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 157.Tomasini R, et al. TAp73 knockout shows genomic instability with infertility and tumor suppressor functions. Genes Dev. 2008;22(19):2677–91. doi: 10.1101/gad.1695308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Zhu J, et al. The potential tumor suppressor p73 differentially regulates cellular p53 target genes. Cancer Res. 1998;58(22):5061–5. [PubMed] [Google Scholar]
- 159.Dohn M, Zhang S, Chen X. p63alpha and DeltaNp63alpha can induce cell cycle arrest and apoptosis and differentially regulate p53 target genes. Oncogene. 2001;20(25):3193–205. doi: 10.1038/sj.onc.1204427. [DOI] [PubMed] [Google Scholar]
- 160.Stiewe T, Putzer BM. Role of the p53-homologue p73 in E2F1-induced apoptosis. Nat Genet. 2000;26(4):464–9. doi: 10.1038/82617. [DOI] [PubMed] [Google Scholar]
- 161.Yang A, et al. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol Cell. 1998;2(3):305–16. doi: 10.1016/s1097-2765(00)80275-0. [DOI] [PubMed] [Google Scholar]
- 162.Flores ER, et al. p63 and p73 are required for p53-dependent apoptosis in response to DNA damage. Nature. 2002;416(6880):560–4. doi: 10.1038/416560a. [DOI] [PubMed] [Google Scholar]
- 163.Senoo M, et al. p63 and p73 are not required for the development and p53-dependent apoptosis of T cells. Cancer Cell. 2004;6(1):85–9. doi: 10.1016/j.ccr.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 164.Racek T, et al. C-terminal p73 isoforms repress transcriptional activity of the human telomerase reverse transcriptase (hTERT) promoter. J Biol Chem. 2005;280(49):40402–5. doi: 10.1074/jbc.C500193200. [DOI] [PubMed] [Google Scholar]
- 165.Beitzinger M, et al. Regulation of telomerase activity by the p53 family member p73. Oncogene. 2006;25(6):813–26. doi: 10.1038/sj.onc.1209125. [DOI] [PubMed] [Google Scholar]
- 166.Bodnar AG, et al. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279(5349):349–52. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- 167.Xu D, et al. Downregulation of telomerase reverse transcriptase mRNA expression by wild type p53 in human tumor cells. Oncogene. 2000;19(45):5123–33. doi: 10.1038/sj.onc.1203890. [DOI] [PubMed] [Google Scholar]
- 168.Kanaya T, et al. Adenoviral expression of p53 represses telomerase activity through down-regulation of human telomerase reverse transcriptase transcription. Clin Cancer Res. 2000;6(4):1239–47. [PubMed] [Google Scholar]
- 169.Shats I, et al. p53-dependent down-regulation of telomerase is mediated by p21waf1. J Biol Chem. 2004;279(49):50976–85. doi: 10.1074/jbc.M402502200. [DOI] [PubMed] [Google Scholar]
- 170.Toh WH, Kyo S, Sabapathy K. Relief of p53-mediated telomerase suppression by p73. J Biol Chem. 2005;280(17):17329–38. doi: 10.1074/jbc.M500044200. [DOI] [PubMed] [Google Scholar]
- 171.Keyes WM, Mills AA. p63: a new link between senescence and aging. Cell Cycle. 2006;5(3):260–5. doi: 10.4161/cc.5.3.2415. [DOI] [PubMed] [Google Scholar]