Abstract
Background
Decreased epithelial expression of mRNA for S100A7 (Psoriasin) and S100A8/A9 (Calprotectin) have been reported in chronic rhinosinusitis (CRS).
Objectives
To assess whether the expression of S100 proteins is also altered in the sinonasal cavity of patients with CRS.
Methods
We determined levels of S100 proteins in nasal lavage and sinonasal tissue extracts from CRS patients using ELISA and immunohistochemical analysis of nasal polyp tissue from CRSwNP patients and uncinate tissue from all three groups.
Results
Expression levels of S100 proteins were decreased compared to control subjects in nasal lavage fluids from both CRS groups (p < 0.05). Similarly, tissue expression of these proteins assessed by immunohistochemistry demonstrated clear reductions primarily in the epithelial lining. Interestingly, levels of Calprotectin were increased in nasal polyp tissue despite lower levels in lavage fluid. Levels of Calprotectin in nasal tissues were correlated with levels of neutrophils as assessed by quantification of neutrophil elastase.
Conclusions
Several S100 proteins are in the epidermal differentiation complex of genes and have been demonstrated to play a role in maintenance of barrier function and formation of an antimicrobial shield. We demonstrate significantly decreased levels of expression of S100 proteins in epithelium of CRS patients, which may lead to diminished innate immune response and barrier function. Increased levels of Calprotectin in nasal polyp tissue may reflect neutrophil recruitment and a compensatory mechanism. Future studies will be important to determine whether reduced levels of S100 proteins lead to decreased antimicrobial responses in the upper airways and sinuses and whether this reduction plays an etiologic role in CRS pathogenesis and susceptibility to infectious disease.
Keywords: Chronic Rhinosinusitis, S100A7, S100A8/A9, Psoriasin, Calprotectin, Epithelium, Barrier, Epidermal Differentiation Complex
INTRODUCTION
Chronic Rhinosinusitis (CRS) is characterized by persistent symptomatic inflammation of the nasal mucosa and is one of the most frequently reported chronic diseases in the United States, with an estimated 22 million annual physician visits. Despite various hypotheses in regard to its cause, CRS, defined as a chronic inflammatory disease of the nasal and paranasal sinus mucosa, remains poorly understood. CRS is commonly divided into two subtypes: CRS with nasal polyps (CRSwNP) and CRS without nasal polyps (CRSsNP).1 Sinonasal tissue from patients with CRSsNP displays a predominant infiltration of neutrophils and T-helper type 1 (Th1) cytokines whereas CRSwNP tissue is characterized by more intense eosinophilic infiltration and a Th2-biased cytokine profile.2 Although complex host genetic factors are widely believed to influence CRS pathogenesis, recent investigations have highlighted environmental factors such as allergens, fungal or bacterial colonization, biofilms, and superantigens.1 While it is unlikely that any one environmental factor accounts for all the manifestations of CRS, comparatively little interest has focused on potential host defects or susceptibilities in this disorder.
There is increasing evidence of the importance of epithelial cell activation and function in disease. Recent studies of allergic diseases in the skin and lungs such as atopic dermatitis (AD) and asthma have uncovered evidence that there may be deficiencies in the barrier function of skin and airway mucosal epithelium in these diseases.3 The respiratory epithelium provides a barrier to entry of pathogens through mucociliary function and tight junctions, and epithelial cells lining the airway have been shown to respond to the presence of microorganisms by producing natural antimicrobial factors and mounting an inflammatory response.4 Because of the importance of the epithelium as a mediator of immune defense, defects in a broad set of epithelial-related genes, in theory, could contribute to a dysfunctional innate immune response to environmental agents in the upper airways as they appear to do in the skin and lungs.
The S100 proteins comprise a multigene family of low molecular weight proteins that serve diverse functions in a wide variety of cell types and tissues. A majority of the S100 protein genes are encoded in the epidermal differentiation complex (EDC) located on chromosome 1q21. This region is of heightened interest since it encodes many genes that are expressed in epidermal keratinocytes. Thus, the finding that many S100 genes are clustered within the EDC has increased interest in their role in the epithelium. Of particular interest in barrier function are the proteins S100A7 and S100A8/S100A9, initially found to be overexpressed in psoriasis.5
Using epithelial cell scrapings from three groups of patients: normals, CRSsNP, and CRSwNP, a recent study from our laboratories demonstrated reduced expression of mRNA for selected barrier and S100 genes in the upper airways.6 S100 proteins have also been shown to be involved in epithelial defense and repair and in the skin their expression is regulated by cytokines of the IL-10/IL-22 family.7 Recent studies from Glaser, Meyer and colleagues have demonstrated that S100A7 is the principal antimicrobial compound responsible for killing of E. Coli in both the skin and tongue of humans.8,9 Similarly, Calprotectin has been shown to be a potent antimicrobial capable of killing both bacteria and fungi. The objective of the present study was to determine if the levels of expression of S100 proteins are altered in the upper airways and sinuses of patients with CRS (both with and without polyps) when compared with a control population. Decreased expression levels of these proteins in epithelial cells from patients with CRS could increase susceptibility to colonization of the upper airways and sinuses by both bacteria and fungi. We found reduced expression of S100 proteins in epithelium and mucosal lavage fluids from CRS patients, an observation that suggests that dysfunction of epithelial innate immune mechanisms may be present in CRS.
METHODS
Patients and Tissue samples
Patients with CRS were recruited from the allergy and otolaryngology clinics at Northwestern University Feinberg School of Medicine. Sinonasal and polyp tissues were obtained from routine functional endoscopic sinus surgery and nasal lavages were obtained as previously described.10 Specimens from patients without CRS were obtained during the performance of skull base tumor excisions, facial fracture repair, lacrimal duct surgery and orbital decompression surgery. Any patients that have been using oral (or intranasal) steroids within 2 weeks of surgery or had prior surgery were excluded from sample collection. Details of subjects’ characteristics are described in Table I.
Table I. Subject Characteristics.
| Normal | CRSsNP | CRSwNP | AR | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total no. of subjects | n=48 (27M/21F) | n=46 (18M/28F) | n=69 (41M/28F) | n=6 (2M/4F) | ||||||||
| Age (y), median (range) | 41 (16–77) 16U | 43 (23–63) 15U | 44 (22–74) 27 U | 6U | ||||||||
| Y | N | U | Y | N | U | Y | N | U | Y | N | U | |
| Atopy | 1 | 35 | 12 | 22 | 15 | 9 | 28 | 20 | 21 | 4 | 0 | 2 |
| Asthma | 1 | 34 | 13 | 8 | 33 | 5 | 37 | 21 | 11 | 3 | 1 | 2 |
| Methodology used | ||||||||||||
| P | ||||||||||||
| Tissue extract | n=19 (13M/6F) | n=19 (7M/12F) | n=16 (7M/9F) | n=5 (2M/3F) | - | |||||||
| Age (y), median (range) | 44 (16–49) | 37 (23–63) | 43 (22–58) | 41 (29–65) | ||||||||
| Immunohistochemistry | n=6 (3M/3F) | n=11 (5M/6F) | n=10 (7M/3F) | n=10 (9M/1F) | - | |||||||
| Age (y), median (range) | 48 (25–77) | 36 (23–62) | 39 (27–74) | 51 (27–74) | ||||||||
| Nasal lavage | n=14 (7M/7F) | n=16 (6M/10F) | n=13 (11M/2F) | n=6 (2M/4F) | ||||||||
| Age (y), median (range) | 35 (26–48) 6U | 44 (28–60) 6U | 42 (27–63) 7U | 6U | ||||||||
| Serum | n=11 (5M/6F) | n=11 (4M/7F) | n=26 (13M/13F) | - | ||||||||
| Age (y), median (range) | 30 (10U) | 55 (9U) | 45 (35–58) 20U | - | ||||||||
M=Male, F=Female, U=Unknown, P=Polyp, Y=Yes, N=No
Preparation of Polyp/sinus tissue extracts and nasal lavages
Detergent extracts of sinonasal surgical tissue samples were prepared as described previously.10 Briefly, 100 mg of polyp, uncinate or inferior turbinate tissue was suspended in 1ml of PBS (Phosphate Buffered Saline)-Tween in the presence of a cocktail of protease inhibitors (Sigma Chemical Co., St. Louis, MO) added at a 1:100 dilution. The samples were then homogenized for approximately 1–5 minutes on ice at 24,000 rpm with an IKA Ultra-Turrax T8 Homogenizer. The tissue suspension was allowed to settle for 30 minutes on ice. The suspensions were then centrifuged at 4000 rpm for 20 minutes at 4°C. The supernatants were collected and stored at −80 °C until analysis. Nasal lavage was performed using PBS without Calcium and Magnesium. Nasal lavage fluids were centrifuged in 15mL conical tubes for 10 min at 3000 rpm. The resulting supernatants were then transferred and concentrated using an Amicon Ultra-4 Centrifugal Filter Device (10k mw cutoff) and centrifuged at 3,000 rpm for 5–10 minutes at 4°C or until the sample was concentrated 2 fold. The concentrated supernatants were aliquoted into tubes and stored at −20°C until use. The protein concentrations for tissue extracts and nasal lavage fluids were determined by the BCA Protein Assay Kit (Pierce/Thermo Scientific, Rockford, IL).
ELISA
Prior to analysis, samples were thawed at room temperature and vortexed to ensure a well-mixed sample. S100A7 and S100A8/A9 were assayed using commercially available assay kits (MBL Internatl, Woburn, MA and Cell Sciences, Canton, MA). The color intensity was measured using a Bio-Rad Spectrophotometer Model 680 Microplate Reader with associated software applied to the sandwich enzyme immunoassay technique. Concentrations of S100 proteins in the tissue homogenate and nasal lavages were normalized to the concentration of total protein.
Immunohistochemistry (IHC)
Analysis of S100 protein expression was performed using specific monoclonal antibodies against S100A7 and S100A8/A9 (Imgenex and Abcam). Data from the manufacturers indicate that specificities for human S100A7 and S100A8/A9 have been confirmed by Western blot. Formalin-fixed, 3μm sections of paraffin-embedded tissue were incubated overnight with S100A7 or S100A8/A9 antibodies at a dilution of 1:400 overnight. Specimens included polyp tissue from CRSwNP patients (n = 10) as well as uncinate tissue from control (n = 7), CRSsNP (n = 10), and CRSwNP (n = 11). As a negative control, sections were incubated overnight without the primary antibody. After rinsing, biotinylated horse anti-mouse secondary antibody (Vector Laboratories, Burlingame, CA) was used for 1 hour at a dilution of 1:500. After several washings, sections were incubated with avidin-peroxidase complex (Vectastain Elite ABC Kit; Vector Laboratories) for 1 hour. After several rinsings, sections were incubated with diaminobenzidine chromagen (Invitrogen) for approximately 10 minutes. Counterstaining with Gill modified hematoxylin (EMD, San Diego, CA) was performed for 5–10 seconds. Sections were then dehydrated, cleared, and mounted.
Semiquantitative analysis was performed using an Olympus IX71 inverted research microscope and a MicroFire AR digital microscope camera (Optronics, Goleta, CA). Slides were blinded and then rated by two independent observers on a scale of zero to three in three different locations of tissue structure (epithelium, glands, and subepithelial stroma). A rating of zero indicated no staining, one was mild staining, two was moderate staining, and three was intense staining. Photographs were taken at 400x objective magnification using PictureFrame software (Optronics).
Statistical Analysis
All ELISA data were analyzed using the Mann-Whitney U test and for immunohistochemical staining grades, the Kruskal-Wallis ANOVA on ranks with Dunnett’s post hoc testing was used. Statistical analysis was performed using Prism GraphPad version 5 software. A p value of less than 0.05 was considered statistically significant.
RESULTS
S100A7 in sinonasal tissue extracts
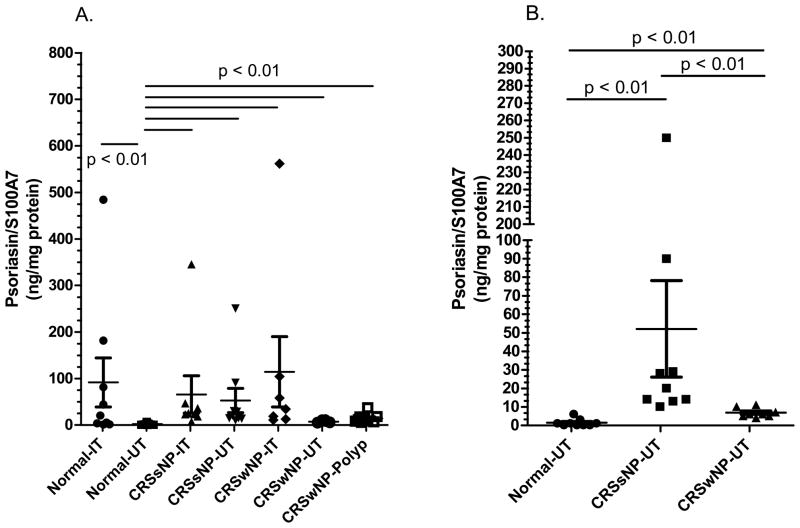
In previous studies, we found reduced mRNA for S100A7 in epithelial scrapings from CRS patients.6 We first tested the levels of this protein in whole tissue extracts using ELISA on a total of 61 sinonasal tissue extracts (Figure 1). Expression of S100A7 protein in normal subjects was significantly higher in inferior turbinate tissue (IT) extracts when compared to extracts of uncinate tissue (UT - Figure 1A, p < 0.01). Levels of S100A7 in all CRS tissue extracts were elevated when compared to normal uncinate tissue extracts. Of note, an outlier was eliminated in the CRSwNP-IT group in Figure 1A which did not alter the statistical analysis. Examining only uncinate tissue, S100A7 was significantly higher in CRSsNP extracts than in extracts from normal and CRSwNP (Figure 1B, p < 0.01).
Figure 1. Evaluation of expression of S100A7 using ELISA in sinonasal tissue extracts.
A) Expression of S100A7 protein is increased in inferior turbinate tissue (IT) as compared to uncinate tissue (UT) from normal subjects. The statistical bars indicate a significant difference among the corresponding groups. B) Levels of S100A7 were increased in all CRS tissue extracts when compared to extracting uncinate tissue from controls. The statistical bars indicate a significant difference among the corresponding groups. p values <0.05 are considered statistically significant.
S100A8/A9 and Human Neutrophil Elastase in sinonasal tissue extracts
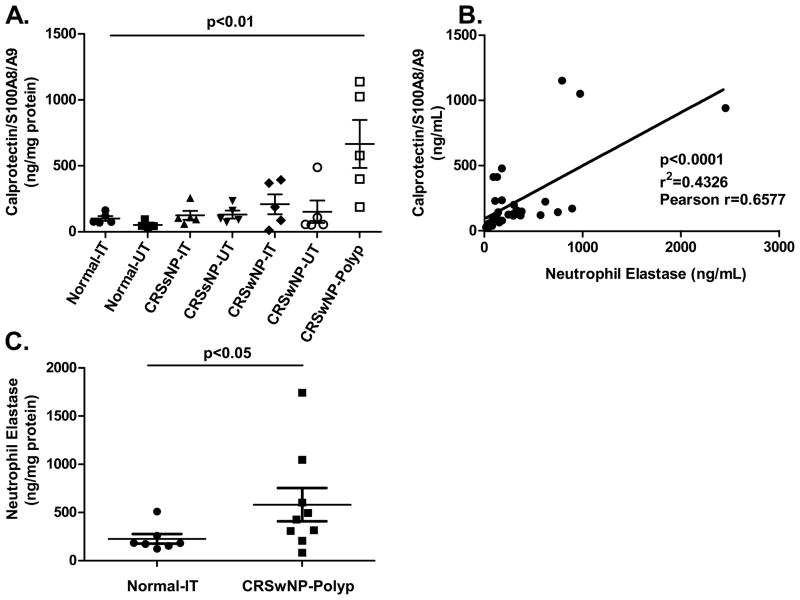
We next analyzed S100A8/A9 and neutrophil elastase protein expression from a total of 35 and 59 sinonasal tissue extracts, respectively (Figure 2). Expression of S100A8/A9 was significantly increased in polyp tissue compared to turbinate and uncinate tissue from CRSwNP, CRsSNP, and controls (Figure 2A, p < 0.01). Given that S100A8/A9 expression is typically found in neutrophils, we determined whether there was a correlation between S100A8/A9 and the presence of neutrophils using elastase as a marker for neutrophils. Elastase was highly expressed and increased in polyp tissue when compared with normal turbinate and uncinate tissue (Figure 2B and C, p < 0.05). Using linear regression, we observed a clear correlation between the expression of S100A8/A9 and neutrophil elastase, suggesting that S100A8/A9 may be produced by the infiltrating neutrophils (Figure 2B, r = 0.66, p < 0.0001). Only tissue extracts tested for both neutrophil elastase and S100A8/A9 were used for this correlation analysis.
Figure 2. Evaluation of expression of human calprotectin (S100A8/A9) and human neutrophil elastase using ELISA in sinonasal tissue extracts.
A) Human S100A8/A9 protein levels in tissue extracts normalized to total protein. Polyp tissue from CRSwNP patients demonstrate a significant increase in human calprotectin when compared to normal tissue extracts (p <0.01). B) Linear regression analysis of matching S100A8/A9 and human neutrophil elastase samples demonstrate a significant correlation between the two proteins (p < 0.0001, r = 0.6577). C) Human neutrophil elastase protein levels in polyp tissue from CRSwNP patients are significantly increased when compared to normal tissue extracts. (p < 0.05).
S100A7 and S100A8/A9 in nasal lavages
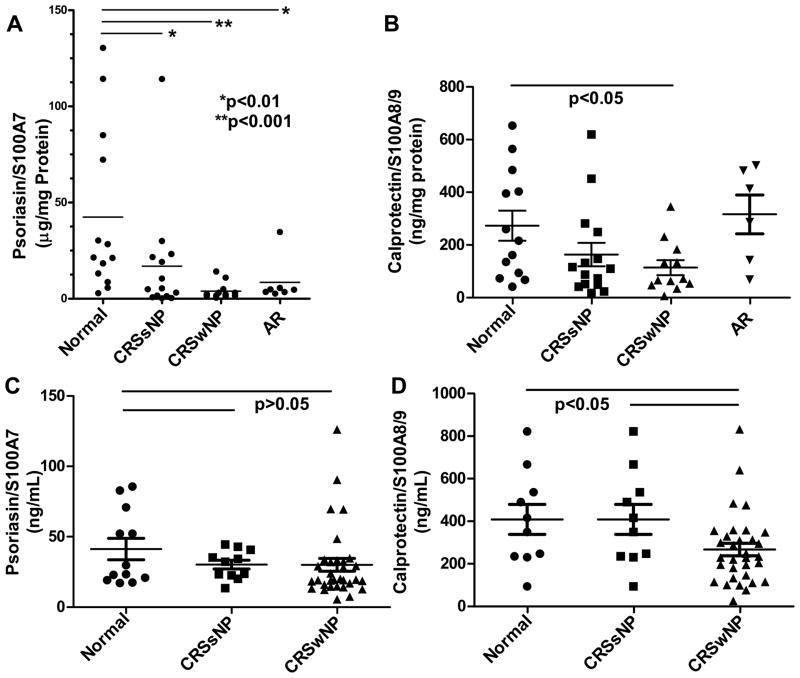
We examined the expression of S100 proteins using ELISA in nasal lavage samples from 40 control, CRSsNP, and CRSwNP patients (Figure 3A and B). Substantial levels of S100A7 were detected using this methodology and levels were dramatically decreased in lavage samples from both groups of CRS patients when compared to normals (p < 0.01). S100A7 expression was also reduced in nasal lavage fluids taken from allergic rhinitis (AR) patients when compared to controls (Figure 3A, p < 0.01). Given that some of the patients in the AR group are atopic, we analyzed the data further separating non-atopic vs. atopic subsets. There continues to be a statistically significant difference in S100A7 nasal lavage from normal vs. atopic CRSwNP and no statistical difference between the atopic and non-atopic S100A7 lavages (data not shown). As with S100A7, S100A8/A9 protein levels were also substantial and clearly decreased in CRSwNP nasal lavage fluids when compared to normals (Figure 3B, p < 0.05). In contrast to S100A7, which was reduced in AR, there was no statistically significant difference in the levels of S100A8/A9 in lavage fluids from AR patients when compared to normal controls (Figure 3B).
Figure 3. Expression of S100A7 and S100A8/A9 in nasal lavage fluids and serum samples using ELISA.
A). Human S100A7 (* p < 0.01, ** p < 0.001) and B) Human S100A8/A9 protein levels in nasal lavage fluids. Human S100A7 protein levels are significantly decreased in nasal lavages of CRS and AR patients when compared to normals (p < 0.05). Human S100A8/A9 protein levels are significantly decreased in CRSwNP nasal lavages when compared to normals. C) Human S100A7 (p > 0.05) and D) Human S100A8/A9 (p < 0.05) protein levels in serum. Only human S100A8/A9 protein levels are significantly decreased in CRSwNP when compared to normals. p values < 0.05 are considered statistically significant.
S100A7 and S100A8/A9 in serum
Because S100 proteins are known to be detected in the circulation, we determined whether there were any differences in S100 protein levels in sera from normal subjects and CRS patients (Figure 3C and D). S100A7 protein levels were decreased in CRS sera compared to normal sera, but the results did not reach statistical significance (Figure 3C, p > 0.05). However, S100A8/A9 levels in sera of CRSwNP patients were significantly reduced when compared to normal subjects (Figure 3D, p < 0.05)
Immunohistochemical analysis of S100A7 and S100A8/A9 in sinonasal tissue
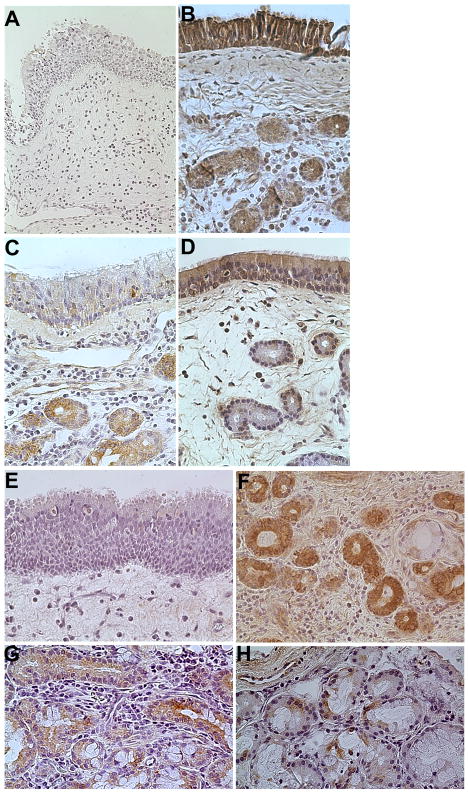
To further characterize the epithelial expression of S100 proteins in CRS, we performed immunohistochemical analysis of surgical samples from normal subjects and CRS patients to determine whether S100A7 and S100A8/A9 expression could be detected. Consistent with the ELISA data from nasal lavage fluids, S100A7 protein was present in the nasal respiratory epithelium. As seen in Figure 4, we detected S100A7 staining in both glandular and mucosal epithelial cells in tissue samples as well as in the subepithelial stroma. Cellular staining was graded by two blinded observers for intensity as described in methods. As shown in Table II, a semiquantitative analysis showed significantly more intense S100A7 staining in the epithelium of control tissue when compared with either polyp tissue in CRSwNP or with uncinate tissue from CRS subjects in both groups. A composite S100A7 staining score was also significantly greater in control specimens, with the lowest score observed in polyp tissue. A similar analysis was also performed for S100A8/A9 immunostaining in nasal respiratory tissue using an antibody that recognizes the heterodimer (Table III and Figure 5). Although epithelial staining for S100A8/A9 was more modest than staining for S100A7, S100A8/A9 expression was detected in the epithelium of normal and CRS uncinate tissue. There was a statistically significant decrease in expression of S100A8/A9 in epithelium, glands and stroma of polyp tissue when compared with normal and CRS uncinate tissue (Table III). In both S100A7 and S100A8/A9, cells that stained positively were primarily the basal epithelial cells of the sinonasal mucosa as well as the glandular epithelial cells. In addition, the S100A8/A9 IHC results show large numbers of intensely staining leukocytes that we believe to be neutrophils (data not shown).
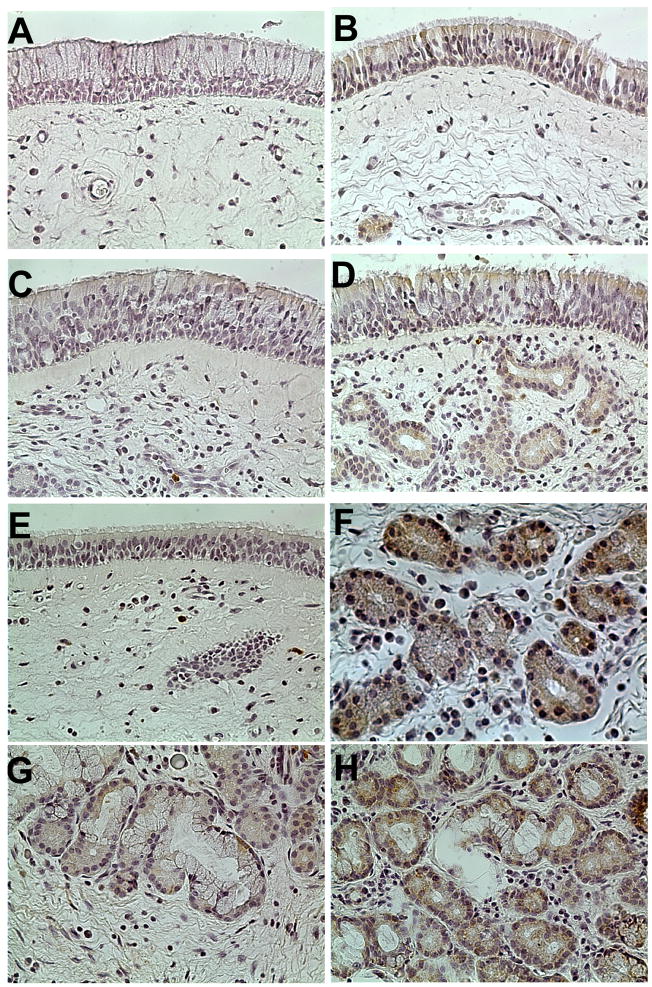
Figure 4. Immunohistochemical staining for S100A7 in representative tissue samples from the uncinate process and nasal polyps (400x original magnification).
(A) Negative control of uncinate from representative normal subject did not stain. (B) S100A7 staining of uncinate from a control subject showed intense staining in the epithelial and glandular tissue, whereas light to moderate staining of S100A7 was seen in uncinate samples from (C) CRSsNP and (D) CRSwNP. (E) Minimal S100A7 staining in epithelial cells was seen in polyp tissue. (F) Intense glandular staining for S100A7 in the uncinate of a control subject. Less staining was seen in the uncinate from (G) CRSsNP and (H) CRSwNP. Original magnification was 400x.
Table II. Semi-quantitative analysis of S100A7 Immunostaining.
Mean semiquantitative scores of S100A7 staining in epithelium, glands, and subepithelial stroma
The difference between the groups is displayed in the Kruskal-Wallis ANOVA on ranks. The p values in italics reflect post hoc comparison with normal samples, with statistically significant values in bold. S100A7 staining was significantly decreased in epithelium, glands, and subepithelial stroma in CRSwNP, CRSsNP, and polyp tissue compared with normal samples. Among the groups, polyp tissue showed the least staining in all areas.
| CRSwNP (n = 11) | CRSsNP (n = 10) | Polyp (n = 10) | Normal (n = 7) | Kruskal-Wallis ANOVA p Value | |
|---|---|---|---|---|---|
| Epithelium (scale = 0–3) |
1.55 p < 0.01 |
1.8 p < 0.01 |
0.7 p < 0.001 |
2.57 | < 0.0001 |
| Glands (scale = 0–3) |
1.60 p < 0.01 |
2.15 p = 0.0539 |
1.39 p < 0.01 |
2.86 | < 0.01 |
| Subepithelial stroma (scale = 0–3) |
1.10 p < 0.01 |
1.45 p < 0.05 |
0.85 p < 0.01 |
2 | < 0.001 |
| Total (scale = 0–9) |
4.25 p < 0.01 |
5.4 p < 0.01 |
2.94 p < 0.01 |
7.43 | < 0.0001 |
Table III. Semi-quantitative analysis of S100A8/A9 Immunostaining.
Mean semiquantitative scores of S100A8/A9 staining in epithelium, glands, and subepithelial stroma
The difference between the groups is displayed in the Kruskal-Wallis ANOVA on ranks. The p values in italics reflect post hoc comparison with normal samples, with statistically significant values in bold. S100A8/A9 staining was significantly decreased in epithelium, glands, and subepithelial stroma in polyp tissue compared with normal samples. Among the groups, polyp tissue showed the least staining in all areas.
| CRSwNP (n = 11) | CRSsNP (n = 10) | Polyp (n = 10) | Normal (n = 6) | Kruskal-Wallis ANOVA p Value | |
|---|---|---|---|---|---|
| Epithelium (scale = 0–3) |
0.95 p = 0.7573 |
0.89 p = 0.8412 |
0.05 p < 0.05 |
0.83 | < 0.01 |
| Glands (scale = 0–3) |
0.68 | 0.7 | 0.11 | 0.92 | 0.135 |
| Subepithelial stroma (scale = 0–3) |
0.045 | 0.1 | 0.05 | 0.17 | 0.8654 |
| Total (scale = 0–9) |
1.68 p = 1.0000 |
1.69 p = 0.9527 |
0.21 p < 0.05 |
1.92 | < 0.01 |
Figure 5. Immunohistochemical staining for S100A8/A9 in representative tissue samples from the uncinate process and nasal polyps (400x original magnification).
(A) Negative control of uncinate from normal subject did not stain. (B) S100A8/A9 staining of uncinate from a control subject showed moderate staining in the epithelial and glandular tissue. Also note several intensely staining leukocytes that we believe to be neutrophils in (D). Some light staining of S100A8/A9 was seen in uncinate samples from (C) CRSsNP and (D) CRSwNP. (E) Minimal S100A8/A9 staining in epithelial cells was seen in polyp tissue. (F) Glandular staining in the uncinate of a control subject with moderate staining for S100A8/A9. Diffuse staining was seen in the uncinate from (G) CRSsNP and (H) CRSwNP. Original magnification was 400x.
DISCUSSION
The S100 family of peptides is now appreciated to play a myriad of roles in inflammation, barrier function, cancer and innate immunity. In the present study we examined the expression of S100A7 and S100A8/A9 in nasal tissue from normal control subjects and patients with CRS. These members of the S100 family of proteins were found at decreased levels of expression in the epithelium of uncinate tissue and nasal polyps from CRS patients when compared to controls. S100A7, also called Psoriasin, has calcium binding-properties like other members of this family. It was originally identified in the skin of psoriasis patients where it was found to be highly elevated.11 In normal human epidermis, S100A7 is present at the cell periphery in terminally differentiated keratinocytes.12 S100A7 has been shown to function as a potent cytokine and as an attractant for CD4+ lymphocytes and neutrophils.13 In addition to chemotactic properties, S100A7 has also been shown to directly kill bacteria and plays a key antimicrobial role in human keratinocytes, particularly in the defense against E. Coli.14 Expression of S100A7 is increased in atopic dermatitis, wound exudates, and following skin barrier disruption.15,16 In the present study, we found a clear reduction in S100A7 protein levels in nasal lavage fluid of CRS individuals and reduced expression of S100A7 at the sinonasal epithelium was confirmed using immunohistochemical analysis. This agrees with earlier findings showing reduced mRNA for S100A7 in epithelial scrapings from CRS patients.6 In extracts of uncinate tissue, S100A7 levels were relatively low and were slightly increased in tissue from CRS patients compared to normal tissue. Interestingly, S100A7 appeared to be increased in normal turbinate tissue when compared to normal uncinate tissue. This tissue specific difference in normal S100A7 expression could be attributed to the proximal location of the inferior turbinate, which results in higher exposure to material in inhaled air when compared to the uncinate. Along similar lines, Meyer et al found higher levels of S100A7 at the tip of the tongue than at the base or margin of the tongue.9 In the present study, S100A7 appeared to be expressed in the subepithelial stroma and glands of nasal respiratory tissue as well as mucosal epithelium; all locations showed significantly decreased expression in CRS. Interestingly, S100A7 was recently identified in nasal lavage fluid using 2-dimensional gel electrophoresis and mass spectrometry and its expression was lower in lavage fluids from allergic subjects than from non-allergic individuals.17 Our results confirm these findings of Bryborn et al, as we found that S100A7 levels were reduced in nasal lavage fluids from allergic rhinitis patients compared with non-allergic individuals. This may suggest a possible role of reduced Psoriasin in allergic airway inflammation or may reflect an inhibitory effect of allergic inflammation (e.g. Th2 cytokines) on expression of S100A7.
A recent report has demonstrated a similarity of the three-dimensional structures of S100A7 and amoebapore A, an ancient antimicrobial, pore-forming peptide from Entamoeba histolytica, and shown that S100A7 exerts it antimicrobial activity via disruption of microbial membranes at low pH.18 This raises the possibility that reduced levels of S100A7 may reduce the antimicrobial activity of the epithelium in patients with CRS. Our findings indicate that the level of S100A7 normalized to total protein was 10–20 fold greater in nasal lavage fluids when compared to tissue extracts, and immunohistochemical analysis showed intense staining primarily in the epithelium, suggesting that it is primarily located at the mucosal surface. Moreover, S100A7 was detectable in the supernatant of nasal epithelial cells cultured for 24 hours and appears to be inducible in stimulated human bronchial epithelial cells (Tieu et al, unpublished observations). Taken together, these findings suggest a role for secreted S100A7 as an antimicrobial substance in the sinonasal cavity and suggest that reduced levels should be considered as a possible contributing factor in CRS pathogenesis.
Recent studies in psoriasis have examined the possible role of S100A7 as a circulating marker for inflammatory disease and demonstrated decreased levels in psoriatic patients.19 We examined the levels of S100A7 in sera of CRS individuals and observed decreased levels when compared to controls. It is doubtful that S100A7 would be useful as a marker for CRS disease states, but this finding suggests either that the apparent deficiency in S100A7 production in the sinonasal cavity may reflect similar reductions at other locations that contribute to the circulating pool or that respiratory epithelium is a primary source of this peptide in the circulation.
We examined the levels and tissue expression of S100A8/A9 (Calprotectin) in CRS. S100A8/A9 is a heterodimer of two small, calcium-binding proteins of the S100 family, and is thought to be an important component of the innate immune response.20 S100A8/A9 is produced by neutrophils, monocytes, and epithelial cells and is found in inflammatory exudates and saliva. It is a potent proinflammatory chemoattractant for neutrophils and monocytes and is involved in transendothelial migration of leukocytes by inducing neutrophil adhesion to fibrinogen by activation of the β2 integrin Mac-1.21, 22 Calprotectin has also been shown to induce the release of neutrophils from bone marrow and direct their migration to inflammatory sites in response to LPS and monosodium urate crystals.21,23 Like S100A7, Calprotectin has been shown to directly kill microorganisms and it has a somewhat different specificity than S100A7, targeting both bacteria (e.g. Prophyromonas gingivalis) and fungi (e.g. Candida albicans).24,25 We found that S100A8/A9 was dramatically increased in polyp tissue of CRSwNP individuals when compared with control tissues. Because S100A8/A9 is known to be derived from neutrophils, it was important to determine if the increase in S100A8/A9 in polyp tissue could be attributed to the infiltrating neutrophils. We observed a significant increase in neutrophil elastase in CRSwNP polyp and regression analysis demonstrated a strong correlation between the levels of S100A8/A9 and the levels of neutrophil elastase. We do not know the exact origin of S100A8/A9 production in the tissue, but this correlation analysis suggests that neutrophils may be the main source in the tissue. In contrast to the finding in CRS tissue, S100A8/A9 was significantly decreased in nasal lavage fluids from CRS patients when compared to controls. Unlike S100A7, levels of S100A8/A9 in lavage fluids were not different between controls and AR individuals. Our previous study had shown reduced mRNA for S100A8/A9 in epithelial scrapings from CRS patients and in the present study we found that the levels of S100A8/A9 protein were dramatically reduced in sinonasal epithelium by immunohistochemical staining.6 Our immunohistochemical studies indicate that S100A8/A9 is present at moderate levels in the sinonasal epithelium and diffusely expressed in the subepithelial stroma and glandular tissue. Epithelial expression of S100A8/A9 was diminished in polyp tissue. Diminished levels of S100A8/A9 in nasal lavage may reflect lower epithelial production of the peptides in CRS. Since levels of S100A8/A9 were higher in CRS tissues, this finding suggests that the content of lavage fluids primarily reflects epithelial production rather than tissue production. The observation of low levels in lavage despite high levels in polyp tissue suggests that the peptides are associated with cells or other tissue structures and are not readily transported from the stroma to the lumen of the airways. Recent studies in other inflammatory disease states such as chronic inflammatory bowel disease (IBD) have uncovered a potential utility of S100A8/A9 as a marker for inflammation.26 Serum levels of S100A8/A9 have been helpful in differentiating between active and non-active IBD and possibly in monitoring disease activity. We examined the S100A8/A9 levels in sera of normal and CRS patients. The level of S100A8/A9 was significantly diminished in sera of CRSwNP patients when compared to controls. Given the potential role for S100A8/A9 at the epithelial interface and in the lamina propria, further studies are needed to determine whether decreased circulating S100A8/A9 reflects events that are occurring exclusively in the airway and can be used as a marker of CRS disease state.
In addition to its chemoattractant properties, S100A8/A9 possesses a host of antimicrobial and antifungal properties. Squamous mucosal epithelial cells in the oral cavity have been shown to constitutively express S100A8/A9 and this expression confers epithelial resistance to invading bacteria (Listeria monocytogenes, Porphyromonas gigivalis, and Salmonella enterica).24 Mechanistic studies indicate that calcium-binding loops I and II of S100A8/A9 are essential for bacterial resistance in keratinocytes.27 More recently, S100A8/A9 was found to be critical for inflammatory cell transmigration into epithelial tissues during pancreatitis and directly dissociated epithelial cell-cell contacts in a highly calcium dependent manner.28 We detected inducible S100A8/A9 expression in primary bronchial epithelial cell cultures (Tieu et al, unpublished observations), suggesting that expression may be varied based on exposure to pathogens and cytokines. Published studies suggest that S100A8/A9 is involved in dermal inflammation and wound repair29 and critical in pancreatic epidermal cell-cell contacts.28 The role of this protein in airway epithelium remains elusive and further studies are needed to define its function in both antimicrobial responses and inflammation in CRS.
Based on our studies and those of other laboratories, it is reasonable to hypothesize that both S100A7 and S100A8/A9 play important roles in innate immune defense of the airways. While both of these molecules are produced by epithelial cells, S100A8/A9 appears to have other important cellular sources in sinonasal tissue from CRS patients. Accumulating evidence indicates that epithelial cells play important roles in the initiation, maintenance, and regulation of innate and adaptive immune responses in the airways. With the recent focus on epithelial genes and the discovery of the EDC and S100 proteins, there has been a heightened emphasis on barrier function and dysfunction in the pathogenesis of inflammatory airway diseases such as AD, psoriasis, inflammatory bowel disease, asthma, COPD and CRS. Here, we demonstrate the presence of S100A7 and S100A8/A9 at the epithelial interface and show that levels of these S100 proteins are significantly diminished in CRS. Taken together, these findings suggest a potential deficiency in CRS patients of the immune barrier, a barrier comprised of both molecules involved in epithelial integrity as well as antimicrobial defense proteins.30 Defects in immune barrier function could serve as a primary etiologic mechanism in CRS pathogenesis by rendering the patient susceptible to microbial colonization and/or invasion leading to a heightened inflammatory response. Future studies will be required to elucidate the role of S100 proteins and other genes in the EDC in immune barrier function and disease.
Acknowledgments
Grant Support: NIH NHLBI RO1 HL78860
NIH NIAID RO1 AI072570
Ernest S. Bazley grant to Northwestern Memorial Hospital and Northwestern University
Abbreviations used
- AD
Atopic Dermatitis
- AR
Allergic Rhinitis
- COPD
Chronic Obstructive Pulmonary Disease
- CRS
Chronic Rhinosinusitis
- CRSsNP
Chronic Rhinosinusitis without Polyps
- CRSwNP
Chronic Rhinosinusitis with Polyps
- EDC
Epidermal Differentiation Complex
- IBD
Inflammatory Bowel Disease
- IT
Inferior Turbinate
- PBS
Phosphate Buffered Saline
- Th1/2
T-helper type 1 and 2
- UT
Uncinate Tissue
Footnotes
Clinical Implications: Reduced immune barrier function in CRS patients may underlie their increased susceptibility to infections and/or bacterial and fungal colonization in the upper airways and sinuses.
Capsule Summary: Epithelial expression of S100 host defense proteins is reduced in the nose and sinuses of patients with Chronic Rhinosinusitis.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Meltzer EO, Hamilos DL, Hadley JA, Lanza DC, Marple BF, Nicklas RA, et al. Rhinosinusitis: establishing definitions for clinical research and patient care. J Allergy Clin Immunol. 2004;114:155–212. doi: 10.1016/j.jaci.2004.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van Zele T, Claeys S, Gevaert P, Van Maele G, Holtappels G, Van Cauwenberge P, et al. Differentiation of chronic sinus diseases by measurement of inflammatory mediators. Allergy. 2006;61:1280–9. doi: 10.1111/j.1398-9995.2006.01225.x. [DOI] [PubMed] [Google Scholar]
- 3.Cookson W. The immunogenetics of asthma and eczema: a new focus on the epithelium. Nat Rev Immunol. 2004;4:978–88. doi: 10.1038/nri1500. [DOI] [PubMed] [Google Scholar]
- 4.Avila PC, Schleimer RP. Allergy and Allergic Disease. 2. chapter 16. 2008. Airway Epithelium; pp. 366–97. [Google Scholar]
- 5.Madsen P, Rasmussen HH, Leffers H, et al. Molecular cloning and expression of a novel keratinocyte protein (psoriasis-associated fatty acid-binding protein [PA-FABP]) that is highly up-regulated in psoriatic skin and that shares similarity to fatty acid-binding proteins. J Invest Dermatol. 1991;97 (4):701–712. doi: 10.1111/1523-1747.ep12616641. [DOI] [PubMed] [Google Scholar]
- 6.Richer S, Truong-Tran A, Conley D, et al. Epithelial genes in chronic rhinosinusitis with and without nasal polyps. AJR. 2008;22:228–234. doi: 10.2500/ajr.2008.22.3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wolk K, Witte E, Wallace E, et al. IL-22 regulates the expression of genes responsible for antimicrobial defense, cellular differentiation, and mobility in keratinocytes: a potential role in psoriasis. Eur J Immunol. 2006;36 (5):1309–1323. doi: 10.1002/eji.200535503. [DOI] [PubMed] [Google Scholar]
- 8.Glaser R, Harder J, Lange H, Bartels J, Christophers E, Schroder JM. Antimicrobial psoriasin (S100A7) protects human skin from Escherichia coli infection. Nat Immunol. 2005;6:57–64. doi: 10.1038/ni1142. [DOI] [PubMed] [Google Scholar]
- 9.Meyer JE, Harder J, Sipos B, Maune S, Kloppel G, Bartels J, et al. Psoriasin (S100A7) is a principal antimicrobial peptide of the human tongue. Mucosal Immunol. 2008;1:239–43. doi: 10.1038/mi.2008.3. [DOI] [PubMed] [Google Scholar]
- 10.Kato A, Peters A, Suh L, et al. Evidence of a role for B-cell activating factor of the TNF family in the pathogenesis of chronic rhinosinusitis with nasal polyps. J Allergy Clin Immunol. 2008;121 (6):1385–1392. doi: 10.1016/j.jaci.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Madsen P, Rasmussen HH, Leffers H, et al. Molecular cloning and expression of a novel keratinocyte protein (psoriasis-associated fatty acid-binding protein [PA-FABP]) that is highly up-regulated in psoriatic skin and that shares similarity to fatty acid-binding proteins. J Invest Dermatol. 1991;97 (4):701–712. doi: 10.1111/1523-1747.ep12616641. [DOI] [PubMed] [Google Scholar]
- 12.Broome AM, Ryan D, Eckert EL. S100 protein subcellular localization during epidermal differentiation and psoriasis. J Histochem Cytochem. 2003;51 (5):675–685. doi: 10.1177/002215540305100513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jinquan T, Vorum H, Larsen CG, et al. Psoriasin: a novel chemotactic protein. J Invest Dermatol. 1996;107 (1):5–10. doi: 10.1111/1523-1747.ep12294284. [DOI] [PubMed] [Google Scholar]
- 14.Glaser R, Harder J, Lange H, et al. Antimicrobial psoriasin (S100A7) protects human skin from E. coli infection. Nat Immunol. 2005;6:57–64. doi: 10.1038/ni1142. [DOI] [PubMed] [Google Scholar]
- 15.Lee KC, Eckert RL. S100A7 (Psoriasin)—mechanism of antibacterial action in wounds. J Invest Dermatol. 2006;127 (4):945–957. doi: 10.1038/sj.jid.5700663. [DOI] [PubMed] [Google Scholar]
- 16.Glaser R, Meyer-Hoffert U, Harder J, et al. The antimicrobial protein psoriasin (S100A7) is upregulated in atopic dermatitis and after experimental skin barrier disruption. J Invest Dermatol. 2008;129 (3):641–649. doi: 10.1038/jid.2008.268. [DOI] [PubMed] [Google Scholar]
- 17.Bryborn M, Adner M, Cardell LO. Psoriasin, one of several new proteins identified in nasal lavage fluid from allergic and non-allergic individuals using 2-dimensional gel electrophoresis and mass spectrometry. Respir Res. 2005;6:118. doi: 10.1186/1465-9921-6-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Michalek M, Gelhaus C, Hecht O, et al. The human antimicrobial protein psoriasin acts by permeabilization of bacterial membranes. Dev Comp Immunology. 2009;33 (6):74–746. doi: 10.1016/j.dci.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 19.Anderson KS, Wong J, Polyak K, et al. Detection of psoriasin/S100A7 in the sera of patient with psoriasis. Br J Dermatol. 2009 Feb;160(2):325–32. doi: 10.1111/j.1365-2133.2008.08904.x. [DOI] [PubMed] [Google Scholar]
- 20.Ross KF, Herzberg MC. Calprotectin expression by gingival epithelial cells. Infect Immun. 2001;69 (5):3248–3254. doi: 10.1128/IAI.69.5.3248-3254.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ryckman C, Vandal K, Rouleau P, et al. Proinflammatory activities of S100: proteins S100A8, S100A9, and S100A8/A9 induce neutrophil chemotaxis and adhesion. J Immunol. 2003;170 (6):3233–3242. doi: 10.4049/jimmunol.170.6.3233. [DOI] [PubMed] [Google Scholar]
- 22.Cornish CJ, Devery JM, Poronnik P, et al. S100 protein CP-10 stimulates myeloid cell chemotaxis without activation. J Cell Physiol. 1996;166 (2):437–47. doi: 10.1002/(SICI)1097-4652(199602)166:2<427::AID-JCP21>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 23.Vandal K, Rouleau P, Boivin A, et al. Blockade of S100A8 and S100A9 suppresses neutrophil migration in response to lipopolysaccharide. J Immunol. 2003;171 (5):2602–2609. doi: 10.4049/jimmunol.171.5.2602. [DOI] [PubMed] [Google Scholar]
- 24.Nisapakultorn K, Ross KF, Herzberg MC. Calprotectin in vitro by oral epithelial cells confers resistance to infection by Porphyromonas gingivalis. Infect Immun. 2001;69 (7):4242–4247. doi: 10.1128/IAI.69.7.4242-4247.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sohnle PG, Hahn BL, Santhanagolapan V. Inhibition of candida albicans growth by calprotectin in the absence of direct contact with the organisms. J Infect Dis. 1996;174 (6):1369–1372. doi: 10.1093/infdis/174.6.1369. [DOI] [PubMed] [Google Scholar]
- 26.Diamanti A, Colistro F, Basso MS, et al. Clinical role of calprotectin assay in determining histological relapses in childrn affected by inflammatory bowel diseases. Inflammatory Bowel Diseases. 2008;14:1229–1235. doi: 10.1002/ibd.20472. [DOI] [PubMed] [Google Scholar]
- 27.Champaiboon C, Sappington KJ, Guenther BD, et al. Calprotectin S100A9 calcium-binding loops I and II essential for keratinocyte resistance to bacterial invasion. J Biol Chem Article. 2009;284 (11):7078–7090. doi: 10.1074/jbc.M806605200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schneckenburger J, Schick V, Kruger B, et al. The calcium binding protein S100A9 is essential for pancreatic leukocyte infiltration and induces disruption of cell-cell contacts. J Cell Physiol. 2008;216 (2):558–567. doi: 10.1002/jcp.21433. [DOI] [PubMed] [Google Scholar]
- 29.Thorey IS, Roth J, Regenbogen J, et al. The Ca2+-binding proteins S100A8 and S100A9 are encoded by novel injury-regulated genes. J Biol Chem. 2001;276 (38):35818–25. doi: 10.1074/jbc.M104871200. [DOI] [PubMed] [Google Scholar]
- 30.Tieu DD, Kern RC, Schleimer RP. Perspectives: Alterations in epithelial barrier function and host defense responses in Chronic Rhinosinusitis. J Allergy Clin Immunol. 2009;124 (1):37–42. doi: 10.1016/j.jaci.2009.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]