Abstract
Background
Vascular endothelial growth factor A (VEGFA), a critical mediator of tumor angiogenesis, is a well-characterized target of hypoxia-inducible factor 1 (HIF-1). Murine arrest-defective protein 1A (mARD1A225) acetylates HIF-1α, triggering its degradation, and thus may play a role in decreased expression of VEGFA.
Methods
We generated ApcMin/+/mARD1A225 transgenic mice and quantified growth of intestinal polyps. Human gastric MKN74 and murine melanoma B16F10 cells overexpressing mARD1A225 were injected into mice, and tumor growth and metastasis were measured. VEGFA expression and microvessel density in tumors were assessed using immunohistochemistry. To evaluate the role of mARD1A225 acetylation of Lys532 in HIF-1α, we injected B16F10-mARD1A225 cell lines stably expressing mutant HIF-1α/K532R into mice and measured metastasis. All statistical tests were two-sided, and P values less than .05 were considered statistically significant.
Results
ApcMin/+/mARD1A225 transgenic mice (n = 25) had statistically significantly fewer intestinal polyps than ApcMin/+ mice (n = 21) (number of intestinal polyps per mouse: ApcMin/+ mice vs ApcMin/+/mARD1A225 transgenic mice, mean = 83.4 vs 38.0 polyps, difference = 45.4 polyps, 95% confidence interval [CI] = 41.8 to 48.6; P < .001). The growth and metastases of transplanted tumors were also statistically significantly reduced in mice injected with mARD1A225-overexpressing cells than in mice injected with control cells (P < .01). Moreover, overexpression of mARD1A225 decreased VEGFA expression and microvessel density in tumor xenografts (P < .04) and ApcMin/+ intestinal polyps (P = .001). Mutation of lysine 532 of HIF-1α in B16F10-mARD1A225 cells prevented HIF-1α degradation and inhibited the antimetastatic effect of mARD1A225 (P < .001).
Conclusion
mARD1A225 may be a novel upstream target that blocks VEGFA expression and tumor-related angiogenesis.
CONTEXTS AND CAVEATS
Prior knowledge
Angiogenesis plays a central role in tumor growth and is mediated by vascular endothelial growth factor A (VEGFA), which is expressed in many tumor types. VEGFA transcription is initiated by hypoxia-inducible factor 1α (HIF-1α). Arrest-defective protein 1A (ARD1A) induces the degradation of HIF-1α, resulting in lower VEGFA expression.
Study design
Growth and metastasis of intestinal tumors were quantified in transgenic mice expressing murine ARD1A. VEGFA expression and blood vessel proliferation in tumors were assessed in these mice relative to controls. The importance of HIF-1α degradation in suppression of VEGFA expression was tested by mutation of the HIF-1α acetylation site.
Contribution
Overexpression of ARD1A reduced the number and size of both primary tumors and metastases. In addition, ARD1A had an antiangiogenic effect by decreasing VEGFA expression and blood vessel density in tumors. This effect was inhibited by mutation of a single acetylation site in HIF-1α.
Implications
The ARD1A protein may act as a novel antitumorigenic agent by blocking tumor-related angiogenesis.
Limitations
The effects of the mutant HIF-1α did not fully override the increased expression of ARD1A, suggesting that additional targets of ARD1A may be involved in regulating tumor growth. The murine ARD1A isoform has not yet been identified in human cell lines.
From the Editors
Arrest-defective protein 1, homolog A (ARD1A) N-acetyltransferase, which was initially identified in yeast, is involved in the regulation of the cell cycle and sporulation (1–4). Murine ARD1A (mARD1A225) acetylates a lysine residue in the oxygen-dependent degradation domain of hypoxia-inducible factor 1, alpha subunit (HIF-1α), which enhances binding to the von Hippel–Lindau tumor suppressor protein (VHL) and promotes the subsequent ubiquitination and proteasomal degradation of HIF-1α (5,6).
HIF-1 is a key factor involved in cellular adaptation to hypoxia (7). Under normoxic conditions, HIF-1α is posttranslationally modified and degraded (5,8–10). However, during hypoxia, HIF-1α is stabilized and interacts with HIF-1β to form a functional HIF-1 complex (11). Transcriptional targets of HIF-1 include genes that promote angiogenesis and metastasis, most notably, the potent proangiogenic protein, vascular endothelial growth factor A (VEGFA) (7).
VEGFA modulates endothelial cell proliferation and vascular permeability (12) and is overexpressed in many tumor types (13–17), malignant carcinomas (18), and premalignant lesions (19). For example, a statistically significant positive association has been demonstrated between increased levels of HIF-1α and VEGFA in human colorectal carcinomas (18). In this study, we examined the role of mARD1A225 in the regulation of both primary tumor growth and metastatic disease. We generated mARD1A225-expressing transgenic mice to determine whether mARD1A225 plays a physiologically important role in regulating tumor growth via HIF-1α and VEGFA.
Methods
Construction of mARD1A225 Expression Vector for Generation of Transgenic Mice
Sequences encoding a 9His-HRV2-9Myc tag were fused to full-length mARD1A225 cDNA and cloned into the pCAGGS vector under the control of a chicken beta-actin promoter. To investigate the effect of mARD1A225-9His-9Myc fusion protein, 293T cells were transfected with pCAGGS-mARD1A225-9His-9Myc or mock vector (pCAGGS-9His-9Myc) using Lipofectamine 2000 Reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. Transfected 293T cells were treated with proteasome inhibitor 10 μM MG132 under hypoxia for 4 hours, after which the cells were harvested and lysed. Then, we performed an acetylation assay of HIF-1α using an anti–acetyl lysine antibody and western blot analysis with a rabbit polyclonal anti–HIF-1α antibody (1:1000) in whole-cell extract.
Mouse Models and Generation of mARD1A225 and ApcMin/+/mARD1A225 Transgenic Mice
All mouse experiments were approved by the Institutional Animal Care and Use Committees (IACUC) of Ewha Women's University and adhered to the National Research Council Guidelines. ApcMin/+ mice, a spontaneous mouse model of intestinal tumorigenesis with a C57BL/6 background, were purchased from the Jackson Laboratory (Bar Harbor, ME). Eight-week-old male C57BL/6 and BALB/cSlc-nude mice were purchased from the Jackson Laboratory and Japan Shizuoka Laboratory Center, Inc (Shizuoka, Japan), respectively. All mice were maintained in the Ewha Laboratory Animal Genomics Center under specific pathogen-free conditions.
Purified transgenic construct DNA was microinjected into the fertilized eggs collected from the superovulated C57BL/6 females. Genotyping was performed by polymerase chain reaction (PCR) and confirmed by Southern blot analysis of genomic DNA from the tail tissue of founder mice at 2 weeks of age. Eight founder mice were identified via PCR and Southern blot analysis (data not shown). Among these, expression of mARD1A225-His9-HRV2-Myc9 protein was observed through the germline in three lines (0041, 0043, and 0046). The expression of mARD1A225 protein was higher in major organs of the 0041 and 0043 lines than in those of the 0046 line (see Supplementary Figure 1, D, available online). To determine whether mARD1A225 is associated with tumorigenesis, we crossed ApcMin/+ males with mARD1A225 transgenic (0041 and 0043 lines) females to generate ApcMin/+/mARD1A225 transgenic mice. The PCR primers used were as follows: mARD1A225-forward, 5′_ GGA ATT CGC CGC CGC GAT GAA CAT_3′ and His9-HRV2-Myc9-reverse, 5′_ CAC CAC TAG TGT AAG CCG GG _3′; ApcMin/+-forward, 5′_ TTC CAC TTT GGC ATA AGG C_3′ and reverse, 5′_ GCC ATC CCT TCA CGT TAG_3′; Apcwild-forward, 5′_ TTC CAC TTT GGC ATA AGG C_3′ and reverse, 5′_ TTC TGA GAA AGA CAG AAG TTA_3′.
Reagents and Antibodies
MG132, a proteasome inhibitor, was purchased from Calbiochem (Darmstadt, Germany). Rabbit polyclonal anti–HIF-1α antibody was obtained from Cayman Chemical (Ann Arbor, MI). Rabbit polyclonal anti–von Willebrand Factor (vWF) antibody and mouse monoclonal anti-CD34 antibody were purchased from Dako (Glostrup, Denmark). Mouse monoclonal anti-Myc antibody and rabbit polyclonal anti–acetyl lysine antibody were obtained from Santa Cruz Biotechnology (Santa Cruz, CA) and Cell Signaling (Beverly, MA), respectively. Mouse monoclonal VEGFA antibody (for Western blot) and rabbit polyclonal VEGFA antibody (for immunohistochemistry) were obtained from Novus Biologicals (Littleton, CO) and Thermo (Fremont, CA), respectively. The rabbit polyclonal antibody against ARD1A was purchased from Dinona (Seoul, Korea) (6).
Cell Culture and Hypoxic Conditions
The murine melanoma cell lines B16F1 and B16F10 and human embryonic kidney 293T cells were purchased from the American Type Culture Collection (Manassas, VA). The human gastric cancer cell lines MKN74 and NUGC-3 were obtained from the Korea Cell Line Bank (Seoul, Korea). Those cell lines were maintained in Dulbecco's modified Eagle medium (DMEM) (Gibco/Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (Gibco/Invitrogen), 100 U/mL penicillin, and 100 μg/mL streptomycin (Gibco/Invitrogen). Human umbilical vein endothelial cells (HUVECs) were purchased from the American Type Culture Collection and were grown in Medium 199 supplemented with 20% fetal bovine serum, 100 U/mL penicillin, 100 μg/mL streptomycin, 3 ng/mL basic fibroblast growth factor, and 5 U/mL heparin. All cells were maintained at 37°C in a 5% carbon dioxide humidified incubator.
To generate hypoxic conditions, murine B16F10 melanoma and human gastric MKN74 tumor cells were incubated in 5% CO2 and 1% O2 balanced with N2 in a hypoxic chamber (Forma Scientific, Marietta, OH). Hypoxia was also induced in B16F10 and MKN74 cell lines by treatment with cobalt chloride (CoCl2; Sigma-Aldrich, St Louis, MO) at a final concentration of 200 μM. Cells treated with 200 μM CoCl2 exhibited induction of HIF-1α and VEGFA.
Construction of Plasmid and Stable Cell Lines
Sequences encoding a 9His-HRV2-9Myc tag were fused to full-length mARD1A225 cDNA and cloned into pcDNA3.1/His C (Invitrogen) expressing a neomycin resistance gene. We inserted the SV40 early promoter puromycin resistance gene cassette at the PvuII and ApaI sites of the pcDNA3.1-HIF-1α/K532R vector (5). This construct drives the expression of both puromycin resistance genes. Stable mARD1A225-expressing cell lines were generated with B16F1, B16F10, MKN74, and NUGC-3 cells. Those cell lines were transfected with pcDNA3.1/His C-mARD1A225-His9-HRV2-Myc9 vector or empty vector (pcDNA3.1/His C-His9-HRV2-Myc9), as a control, by using Lipofectamine 2000 Reagent (Invitrogen) according to the manufacturer's instructions. Transfected cells (B16F10, B16F1, and MKN74) were treated with 2 mg/mL geneticin (Roche, Nutley, NJ) for 3 weeks, after which single geneticin-resistant colonies were selected for expression of mARD1A225 in the B16F10 (clones A#1 and A#2), MKN74 (clones A#3 and A#4), and B16F1 (clones A#5 and A #6) cell lines and expression of pcDNA3.1/His C-9His-HRV2-9Myc in mock cell lines (B16F10-mock, MKN74-mock, and B16F1-mock) as the control. Transfected NUGC-3 cells were cultured for 2 weeks in medium containing 2 mg/mL geneticin, after which the geneticin-resistant cells were selected for expression of mARD1A225(NUGC-3-Ard) and pcDNA3.1/His C-His9-HRV2-Myc9 (NUGC-3-mock) in the NUGC-3 cells. These stable cells were not selected for a single clone and, as such, they formed a population with mixed geneticin-resistant colonies. Stable HIF-1α/K532R–expressing cell lines were generated with B16F10-mARD1A225 cells using Lipofectamine 2000. B16F10-mARD1A225 cells and B16F10-mock cells were transfected with pcDNA3.1-HIF-1α/K532R-puro vector and empty vector (pcDNA3.1-puro), respectively, by using Lipofectamine 2000 Reagent (Invitrogen) according to the manufacturer's instructions. Transfectants were treated with 1 μg/mL puromycin (Sigma-Aldrich) for 2 weeks, after which puromycin-resistant colonies were selected for expression of HIF-1α/K532R in B16F10-mARD1A225 (clone B16F10-mARD1A225-HIF-1α/K532R) cells and expression of pcDNA3.1-puro in B16F10-mock cells (B16F10-mock-mock), as the control. These stable cells were not selected for a single clone and, as such, they form a mixed population of puromycin-resistant colonies.
Western Blot Analysis for mARD1A225, HIF-1α, and VEGFA Protein
Murine melanoma B16F10 (B16F10, B16F10-mock, B16F10-mARD1A225 #1, and B16F10-mARD1A225 #2) and human gastric MKN74 (MKN74, MKN74-mock, MKN74-mARD1A225 #3, and MKN74-mARD1A225 #4) tumor cells were exposed to hypoxia for 4 and 6 hours, respectively, which resulted in the induction of HIF-1α. All cells were lysed in whole-cell extract buffer (10 mM HEPES, pH 7.9; 400 mM NaCl; 0.1 mM EDTA; 5% glycerol; 1 mM dithiothreitol (DTT); and protease inhibitors). Tumors (derived from mice injected with mARD1A225-overexpressing B16F10 and MKN74 cells) were lysed and homogenized using a MICCRA D-8 homogenizer (Art-moderne Labortechnik, Müllheim-Hügelheim, Germany) in protein extract buffer (20 mM HEPES, pH 7.9; 300 mM NaCl; 10 mM EDTA; 0.1% NP40; 100 mM KCl; and protease inhibitors). Total protein (40 μg) was fractionated on a 10% sodium dodecyl sulfate–polyacrylamide gel and transferred to nitrocellulose membranes (Amersham Biosciences, Piscataway, NJ). The membranes were probed with primary antibodies (rabbit polyclonal anti–HIF-1α, mouse monoclonal anti-Myc, and mouse monoclonal anti-VEGFA antibodies at 1:1000), followed by secondary antibodies (anti-rabbit and anti-mouse antibody, respectively, at 1:5000 [Zymed Laboratories, San Diego, CA]) conjugated with horseradish peroxidase. Protein–antibody complexes were detected with the ECL Plus system (Amersham Biosciences). Three independent experiments were performed.
Immunoprecipitation and In Vivo Acetylation Assay of HIF-1α
Murine melanoma B16F10 (B16F10, B16F10-mock, B16F10-mARD1A225 #1, and B16F10-mARD1A225 #2) and human gastric MKN74 (MKN74, MKN74-mock, MKN74-mARD1A225 #3, and MKN74-mARD1A225 #4) tumor cells were treated with 10 μM proteasome inhibitor MG132 under hypoxia for 4 and 6 hours, respectively.
All cells were lysed in whole-cell extract buffer (10 mM HEPES, pH 7.9; 400 mM NaCl; 0.1 mM EDTA; 5% glycerol; 1 mM DTT; and protease inhibitors). Anti-acetyl lysine antibody (1 μg) was added to the lysate, followed by the addition of Protein A agarose (Upstate Biotech, Lake Placid, NY) in TEG reaction buffer (20 mM Tris–HCl at pH 7.4, 1 mM EDTA, 10% glycerol, 1 mM DTT, and 150 mM NaCl), and the mixture was stirred overnight at 4°C. Immunoprecipitates were washed in TEG washing buffer (TEG reaction buffer containing 0.1% Triton-X 100) and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blot analysis with a rabbit polyclonal anti–HIF-1α antibody as previously described. Three independent experiments were performed in duplicates.
Reverse Transcription–PCR Analysis to Test for VEGFA RNA Expression
Total RNA was extracted from MKN74, MKN74-mock, and MKN74-mARD1A225 cells, which were under hypoxic conditions for 48 hours, using a RNA extraction kit (Invitrogen). Complementary DNA was synthesized from 4 μg of total RNA using an oligo-dT primer. The primers used for PCR were as follows: VEGFA forward, 5′ _GAGAATTCGGCCTCCGAAACCATGAACTTTCTGCT_ 3′ and reverse, 5′_ GAGCATGCCCTCCTGCCCGGCTCACCGC_ 3′; glyceraldehyde 3-phosphate dehydrogenase (GAPDH) forward, 5′_ ACCACAGTCCATGCCATCAC _3′ and reverse, 5′_ TCCACCACCCTGTTGCTGTA_ 3′. Thirty cycles of PCR were performed for amplification of VEGFA and GAPDH, and PCR products were detected with autoradiography. Three independent experiments were performed.
Quantification of VEGFA Expression by Enzyme-Linked Immunosorbent Assay
VEGFA protein levels were quantified in ApcMin/+, ApcMin/+/mARD1A225 transgenic mouse serum (12 mice per group, 110 days old), and in conditioned media (1% serum) from control and mARD1A225-expressing cells after treatment with CoCl2 at a final concentration of 200 μM to mimic hypoxia. Murine melanoma B16F10 (B16F10, B16F10-mock, B16F10-mARD1A225 #1, and B16F10-mARD1A225 #2) and human gastric cancer cells MKN74 (MKN74, MKN74-mock, MKN74-mARD1A225 #3, and MKN74-mARD1A225 #4) were treated with 200 μM CoCl2 for 4 and 48 hours, respectively. VEGFA levels were assayed with a commercially available mouse and human enzyme-linked immunosorbent assay kit (R&D Systems, Minneapolis, MN) according to the manufacturer's instructions. Three independent experiments were performed using conditioned media, each sample in duplicates. Of the three experiments, one experiment was performed for mouse serum with two replicates for each of 12 mice per group. Serial dilutions of a purified recombinant mouse or human VEGFA preparation with a known concentration were used to generate a standard curve. The total protein concentration in each conditioned medium was determined with Bio-Rad (Hercules, CA) protein assay reagent using bovine serum albumin to generate a standard curve.
Collection of Conditioned Media
mARD1A225-overexpressing MKN74 stable cell lines were grown in T75 flasks with 10% fetal bovine serum. After seeding, cells were incubated for 24 hours, washed gently with serum-free DMEM medium, and incubated with 4 mL of DMEM. MKN74, MKN74-mock, and MKN74-mARD1A225 (A#3 and A#4) cells and MKN74-mARD1A225 cells transfected with HIF-1α/K532R were exposed to hypoxia for 48 hours using hypoxic chamber. Also, MKN74-mARD1A225 cells were treated with 10 ng/mL recombinant human VEGFA protein (Sigma-Aldrich) after hypoxic condition. Conditioned media was collected, filtered, and stored at 4°C.
Tube Formation Assay
For the tube formation assay, 48-well plates were coated with cold Matrigel (BD Biosciences, Bedford, MA), which was allowed to polymerize at 37°C for 30 minutes. HUVECs were seeded (5 × 104 cells per well) onto Matrigel with 500 μL filtered conditioned medium from MKN74, MKN74-mock, and MKN74-mARD1A225 (A#3 and A#4) cells; MKN74-mARD1A225 cells treated with 10 ng/mL recombinant human VEGFA protein; and MKN74-mARD1A225 cells transfected with HIF-1α/K532R. Tube formation was assessed after 6 hours and quantified by determining the number of branching points. Each experiment was performed in three replicates.
Transwell Migration Assay
HUVEC migration was performed with the Transwell system (Corning, Inc, Corning, NY), which allows cells to pass through a polycarbonate membrane with a pore size of 8 μm in 24-well plates. The lower surface of the filter was coated with 20 μL of type I collagen (0.5 mg/mL). Cells were detached, counted, and plated at a concentration of 5 × 104 cells per well in the upper chamber with conditioned medium, and the lower chamber was filled with 600 μL of conditioned medium to serve as the chemoattractant. After 6 hours of incubation, migrating cells were counted under microscope. Each experiment was performed in three replicates.
Tumor Cell Invasion Assay into Matrigel
Invasion assays were performed using the Transwell system (Corning, Inc) with a polycarbonate membrane filter (6.5 mm diameter, 8 μm pore size). The upper surface of the filter was coated with 20 μL of Matrigel (0.3 mg/mL; BD Biosciences) and the lower side with 20 μL of type I collagen (0.5 mg/mL). HUVECs (5 × 104 cells per well) were detached from tissue culture plates, washed, resuspended in conditioned medium, and added to the upper compartment of the invasion chamber. Conditioned media were added to the lower compartment. Conditioned media of MKN74 cells served as a negative control. Matrigel invasion chambers were incubated at 37°C for 24 hours in 5% CO2. After incubation, the filter inserts were removed from wells, and cells on the upper side of the filter were removed using cotton swabs. Filters were fixed, mounted, and stained according to the manufacturer's instructions. Cells invading the Matrigel and located on the underside of the filter were counted. Three to five invasion chambers were used per condition. Values calculated were numbers of cells averaged over at least three filters.
Tumor Cell Growth and Morphology and Primary Tumor Growth and Metastasis in Mice
Growth curves were determined 3 days after seeding B16F1 (B16F1, B16F1-mock, B16F1-mARD1A225 #5, and B16F1-mARD1A225 #6; 5 × 104 cells per well), B16F10 (B16F10, B16F10-mock, B16F10-mARD1A225 #1, and B16F10-mARD1A225 #2; 5 × 104 cells per well), and MKN74 cell lines (MKN74, MKN74-mock, MKN74-mARD1A225 #3, and MKN74-mARD1A225 #4; 3 × 104 cells per well) in triplicate into 12-well plates. B16F1 and B16F10 cell line sizes were determined by fluorescence-activated cell sorter analysis. Cell morphology was examined by photography.
For tumor growth experiments, a total of 5 × 105 B16F10 cells (B16F10, B16F10-mock, B16F10-mARD1A225 #1, and B16F10-mARD1A225 #2) in 200 μL of 1× phosphate-buffered saline (PBS) (n = 8 mice per group) was injected subcutaneously into C57BL/6 mice. In addition, a total of 2 × 107 MKN74 cells (MKN74, MKN74-mock, MKN74-mARD1A225 #3, and MKN74-mARD1A225 #4; n = 10 mice per group) and 2 × 107 NUGC-3 cells (NUGC-3-mock and NUGC-3-mARD1A225-mix; n = 5 mice per group) in 200 μL of PBS were injected subcutaneously into BALB/cSlc-nude mice. When the major axis of the tumors reached approximately 10 mm in length, tumor measurements were initiated. Tumors were measured every 2–4 days with calipers to estimate the volume (0.5 × width2 × length). Mice with necrotic tumors were killed by CO2 asphyxiation, and tumors were isolated and weighed. For metastasis experiments, a total of 3 × 105 cells (B16F10, B16F10-mock, and B16F10-mARD1A225) in 100 μL of PBS/5% mouse serum (n = 8 mice per group) was injected into C57BL/6 mice via the tail vein. If any mouse in any group began to appear moribund, all mice in the entire experiment were killed together (day 19 after injection) by CO2 asphyxiation, and surface tumor nodules in the lung were counted under a dissecting microscope. In addition, a total of 1 × 106 cells of each of B16F10-mock (n = 40 mice), B16F10-mARD1A225 (n = 31 mice), B16F10-mARD1A225-HIF-1α/K532R (n = 11 mice), B16F10-mock-mock (n = 15 mice), and B16F10-mock-HIF-1α/K532R cell lines (n = 15 mice) in 100 μL of PBS/5% mouse serum were injected into C57BL/6 mice via the tail vein. Mice were killed 14 days after injection, and surface tumor nodules in the lung were counted under a dissecting microscope. Finally, a total of 5 × 105 cells of B16F10 cell lines (B16F10, B16F10-mock, B16F10-mARD1A225 #1, and B16F10-mARD1A225 #2) in 100 μL of PBS/5% mouse serum were injected into C57BL/6 mice via the tail vein (n = 9 mice per group), and overall survival was monitored.
Polyp Number Determination
For macroscopic assessment of adenoma formation, ApcMin/+ (n = 25) and ApcMin/+/mARD1A225 transgenic mice (0041 lines, n = 21; 0043 lines, n = 9) were killed by CO2 asphyxiation at 110 days old (when they exhibited sufficient tumor volume and numbers), and the small intestine and colon were isolated. The small intestine was subdivided into three equal segments (duodenum, jejunum, and ileum), dissected longitudinally, rinsed, spread flat on paper, and photographed with a digital camera. Tumor frequency and size were determined using the image analysis program with a semiautomated reading system, AxioVision (Carl Zeiss, Thornwood, NY).
Wound Healing Assay
B16F10 cell lines (B16F10, B16F10-mock, B16F10-mARD1A225 #1, B16F10-mARD1A225 #2, B16F10-mARD1A225-HIF-1α/K532R cells, B16F10-mock-mock, and B16F10-mock-HIF-1α/K532R cells) were cultured as 80% confluent monolayers and wounded by removing strips 300–500 μm wide across the well with a sterile pipette tip. Wounded monolayers were washed twice to remove nonadherent cells. Thereafter, cells were incubated in the presence of 200 μM CoCl2 for 24 hours. In addition, cells were treated with 3 μg/mL anti-mouse VEGFA antibody (B16F10, B16F10-mock, B16F10-mARD1A225-HIF-1α/K532R cells, B16F10-mock-mock, and B16F10-mock-HIF-1α/K532R cells; R&D Systems) and 25 ng/mL recombinant mouse VEGFA protein (B16F10-mARD1A225 cells; Sigma-Aldrich) for 24 hours, fixed, and photographed. Three independent experiments were performed, each in duplicates.
Histology
Intestinal polyps from ApcMin/+, ApcMin/+/mARD1A225 transgenic mice, and tumor tissues (derived from mice injected with mARD1A225-overexpressing B16F10 and MKN74 cells) were removed, immediately fixed with 10% formalin, embedded in paraffin, and 4-μm sections were stained with hematoxylin and eosin. Sections were deparaffinized in xylene and rehydrated in graded alcohol. Antigen retrieval was performed using sodium citrate buffer. Sections were stained with mouse monoclonal anti-CD34 (1:100; Dako), rabbit polyclonal anti-vWF (1:400; Dako), rabbit polyclonal anti-VEGFA (1:200; Thermo), and rabbit polyclonal anti-ARD1A (1:200; Dinona) antibodies. Following incubation with the respective biotinylated secondary antibodies and horseradish peroxidase avidin D (Vector Laboratories, Burlingame, CA), sections were visualized with the peroxidase substrate kit (DAB; Vector Laboratories), followed by counterstaining with hematoxylin.
Microvessel Determination
Microvessels were detected in intestinal polyps from ApcMin/+, ApcMin/+/mARD1A225 transgenic mice, or MKN74-mARD1A225 expressing tumor tissue using rabbit polyclonal anti-vWF (1:400; Dako) or mouse monoclonal anti-CD34 (1:100; Dako) antibodies, respectively. Vessels in four or six high-power fields from each of 4–8 mice per group were scored for quantification.
Statistical Analysis
Data are expressed as means with 95% confidence intervals (CIs). Statistical significance was determined with the two-sided Mann–Whitney U test or a two-way repeated measures analysis of variance (SPSS, version 12.0; Statistical Package for Social Sciences, Chicago, IL), where applicable. Survival of mice was assessed by the Kaplan–Meier method, and the difference in median time to death between groups was compared by log-rank tests (SPSS). All tests were two-sided. P values less than .05 were considered statistically significant.
Results
Effect of mARD1A225 Overexpression on the Number and Size of Intestinal Polyps and Survival in ApcMin/+ Mice
To determine the physiological relevance of mARD1A225, we generated a transgenic mouse containing a C-terminal 9His-9Myc epitope-tagged mARD1A225 transgene driven by the beta-actin promoter (see Supplementary Figure 1, A, available online). The effect of the mARD1A225-9His-9Myc fusion protein on stability and acetylation of HIF-1α was examined in 293T cells via acetylation assays. HIF-1α was acetylated in cells expressing mARD1A225-9His-9Myc fusion protein (lane 1) but not in mock cells (lane 2) (see Supplementary Figure 1, B, available online). Transgene expression in mARD1A225 transgenic mice was confirmed by PCR and Southern blot, and expression in the lung, liver, and colon were established by Western blot with an anti-Myc antibody (see Supplementary Figure 1, C and D, available online).
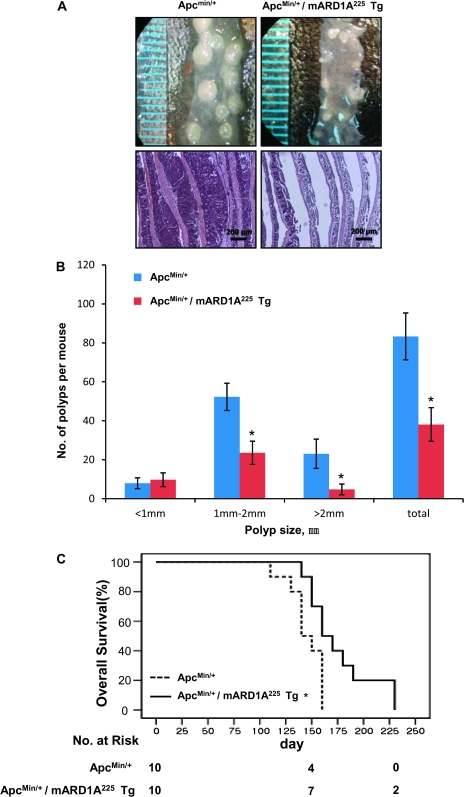
To verify whether an increase of mARD1A225 expression affects tumor initiation and/or progression, we crossbred mARD1A225 transgenic mice with a spontaneous mouse model of intestinal tumorigenesis (ApcMin/+ mice) to generate ApcMin/+/mARD1A225 transgenic mice (see Supplementary Figure 1, E, available online). At approximately 16 weeks of age, mice were killed and polyp numbers and sizes were measured. Gross inspection of polyps and examination of the small intestine with hematoxylin and eosin staining revealed a statistically significant decrease in both polyp size and number (Figure 1, A and B). The total number of intestinal polyps per mouse in ApcMin/+/mARD1A225 transgenic mice was less than 50% of that in ApcMin/+ mice (ApcMin/+ mice vs ApcMin/+/mARD1A225 mice, mean = 83.4 vs 38.0 polyps, difference = 45.4 polyps, 95% CI = 41.8 to 48.6, n = 25 and n = 21, respectively; P < .001) (Figure 1, B). Notably, overexpression of mARD1A225 was associated with 55% reduction in medium (1–2 mm) polyps (ApcMin/+: mean = 52.3, 95% CI = 45.3 to 59.3; ApcMin/+/mARD1A225: mean = 23.6 polyps, 95% CI = 17.6 to 29.6; P < .001) and 79% reduction in large (>2 mm) polyps (ApcMin/+: mean = 23.1 polyps, 95% CI = 15.6 to 30.6; ApcMin/+/mARD1A225: mean = 4.8 polyps, 95% CI = 2.1 to 6.9; P < .001) (Figure 1, B). Comparison of polyp distribution in different segments of the small intestine (duodenum, jejunum, and ileum) revealed similar reductions in each anatomic region (see Supplementary Table 1, available online). The inhibitory effect of mARD1A225 on intestinal tumorigenesis was confirmed in a second ApcMin/+/mARD1A225 transgenic line (n = 9, P = .03, see Supplementary Figure 2, available online). Moreover, ApcMin/+/mARD1A225 transgenic mice (n = 10) displayed a statistically significant survival advantage with a median life span of 171 days (95% CI = 150 to 191, P = .005, log-rank test) compared with ApcMin/+ mice (n = 10) with a median life span of 141 days (95% CI = 131 to 151) (Figure 1, C). These results suggest that mARD1A225 acts as a tumor suppressor in vivo.
Figure 1.
Effect of transgenic overexpression of mARD1A225 on intestinal tumorigenesis in ApcMin/+ mice. A) Representative images (top) and hematoxylin and eosin staining (bottom) of small intestinal polyps from 16-week-old ApcMin/+ mice and ApcMin/+/mARD1A225 transgenic mice (Tg). Magnification ×100. Scale bar = 200 μm. B) Tumor size distribution of polyps in the small intestine from ApcMin/+ mice (n = 21 per group) and ApcMin/+/mARD1A225 Tg (n = 25 per group). Polyps were classified according to size in millimeters. Results are means with 95% confidence intervals (error bars). *P < .001, compared with ApcMin/+ mice, determined with the two-sided Mann–Whitney U test. C) Kaplan–Meier survival plot of ApcMin/+ (mean = 141 days, 95% confidence interval = 131 to 151, n = 10 per group, solid line) and ApcMin/+/mARD1A225 Tg (mean = 170 days, 95% confidence interval = 150 to 191, n = 10 per group, dotted line). *P = .005, compared with ApcMin/+ mice, determined with log-rank tests. All statistical tests were two-sided.
Effect of Reduced VEGFA Expression on Tumor Angiogenesis in ApcMin/+/mARD1A225 Transgenic Mice
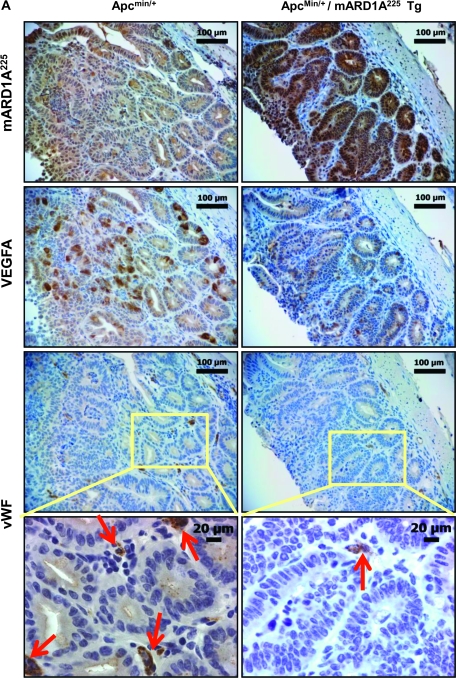
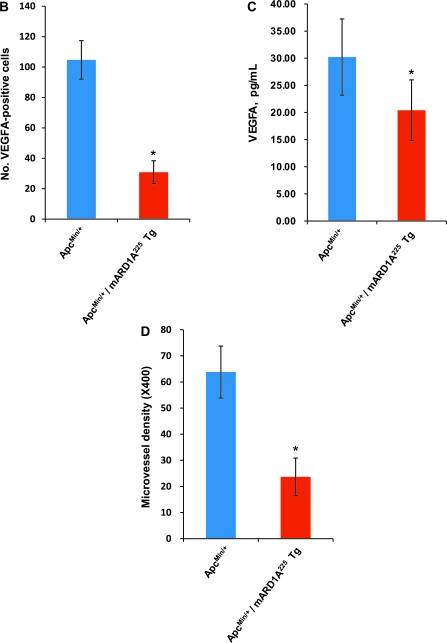
To examine the antiangiogenic effect of mARD1A225 via reduction of VEGFA expression, we immunostained intestinal polyps with the anti-mARD1A225 (Figure 2, A, top row panels, brown stain) and anti-VEGFA antibodies (Figure 2, A, second row panels, brown stain). VEGFA expression was statistically significantly decreased in intestinal polyps from ApcMin/+/mARD1A225 transgenic mice compared with those from ApcMin/+ mice (number of VEGFA-positive cells per field, ApcMin/+ mice: mean = 104.7, 95% CI = 92 to 117.4; ApcMin/+/mARD1A225: mean = 30.9, 95% CI = 23.4 to 38.4, n = 8; P = .001) (Figure 2, B) and in serum (ApcMin/+ mice: mean = 30.2 pg/mL, 95% CI = 23.2 to 37.2; ApcMin/+/mARD1A225: mean = 20.4 pg/mL, 95% CI = 14.8 to 26, n = 12; P = .023) (Figure 2, C). Microvessel density of polyps was quantified by immunostaining with an antibody specific for the endothelial marker, vWF (Figure 2, A, four lower panels). Notably, vWF-positive vessels of intestinal polyps from ApcMin/+/mARD1A225 transgenic mice were statistically significantly reduced compared with those of polyps from ApcMin/+ mice (number of vessels per field, ApcMin/+ mice: mean = 63.8, 95% CI = 53.9 to 73.8; ApcMin/+/mARD1A225: mean = 23.7, 95% CI = 16.5 to 7.2, n = 8; P = .001) (Figure 2, D). The data indicate that mARD1A225 inhibits tumor angiogenesis by reducing VEGFA expression in intestinal polyps of ApcMin/+ mice.
Figure 2.
Effect of transgenic overexpression of mARD1A225 on neovascularization and vascular endothelial growth factor A (VEGFA) expression in intestinal polyps in ApcMin/+/mARD1A225 transgenic mice. A) Immunohistochemical analysis of mARD1A225 (top row), VEGFA (second row), and von Willebrand Factor (vWF, third row) in intestinal polyps of ApcMin/+ mice and ApcMin/+/mARD1A225 transgenic mice at 16 weeks of age (n = 8 mice per group). Magnification ×200. Scale bars = 100 μm. Higher magnification of the selected area (third row, yellow boxes) on vWF with vessels indicated by red arrows within the polyps (bottom row). Magnification ×400. Scale bar = 20 μm. B) The numbers of VEGFA-positive cells in polyps are presented as means with 95% confidence intervals (error bars). VEGFA-positive cells in four high-power fields (×200) from each of the eight mice per group were scored for quantification. *P = .001, compared with the number of VEGFA-positive cells in ApcMin/+ mouse intestinal polyps, as determined with the two-sided Mann–Whitney U test. C) Concentrations of mouse VEGFA protein in serum from ApcMin/+ mice and ApcMin/+/mARD1A225 transgenic mice, respectively (n = 12 mice per group). VEGFA protein levels were quantified by enzyme-linked immunosorbent assay (ELISA). The experiment was performed in two replicate for each of 12 mice per group. The levels of VEGFA protein in serum are presented as means and 95% confidence intervals (error bars) that are derived from the means of the individual mice (n = 12). *P = .023, compared with the levels of VEGFA protein in ApcMin/+ mouse serum, as determined with the two-sided Mann–Whitney U test. D) The numbers of vWF-positive vessels in polyps are presented as means and 95% confidence intervals (error bars). Vessels from five high-power fields (×400) from each of the eight mice per group were scored for quantification. *P = .001, compared with the number of vWF-positive vessels in ApcMin/+ mouse intestinal polyps, as determined with the two-sided Mann–Whitney U test. Tg = transgenic mice.
Effect of mARD1A225 on Primary Tumor Growth
To determine the in vivo effects of mARD1A225 expression on primary tumor growth, we used a xenograft tumor model in which syngeneic or nude mice were injected subcutaneously with mARD1A225-expressing murine B16F10 melanoma and human gastric MKN74 tumor cells, respectively. These cell lines were established from single clones expressing different levels of mARD1A225 protein (see Supplementary Figure 3, A, available online). Clonal cell lines did not display statistically significant differences in proliferation (see Supplementary Figure 3, B, available online), cell size (see Supplementary Figure 3, C, available online), or morphological teratism under normoxic conditions (see Supplementary Figure 3, D, available online).
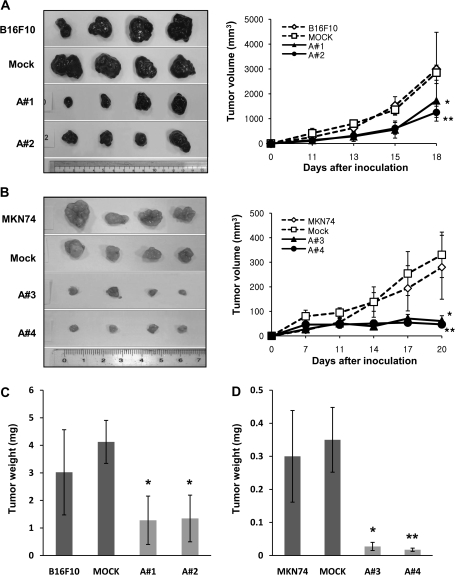
Tumors were isolated, weighed, and measured at day 18 (B16F10) or day 20 (MKN74). Tumor growth was statistically significantly suppressed in mice injected with B16F10-mARD1A225 and MKN74-mARD1A225 cells (Figure 3, A and B). At day 18, for B16F10-mARD1A225 cells, tumor volumes were as follows: B16F10, mean = 2989.5 mm2, 95% CI = 1502 to 4477; B16F10/mock, mean = 2851.9 mm2, 95% CI = 2547.1 to 3156.8; B16F10/mARD1A225 #1, mean = 1734.1 mm2, 95% CI = 1053.1 to 2415.2; B16F10/mARD1A225 #2, mean = 1255.4 mm2, 95% CI = 901 to 1609.7 (n = 8 per group, P = .003, clone A#1, P < .001, clone A#2) (Figure 3, A). For MKN74-mARD1A225 cells, at day 20, tumor volumes were as follows: MKN74, mean = 279.7 mm2, 95% CI = 149.4 to 410; MKN74/mock, mean = 330.4 mm2, 95% CI = 237.7 to 423; MKN74/mARD1A225 #3, mean = 60.5 mm2, 95% CI = 38.9 to 82.2; MKN74/mARD1A225 #4, mean = 46.7 mm2, 95% CI = 38 to 55.5 (n = 10 per group, P = .007, clone A#3, P < .001, clone A#4) (Figure 3, B). An inhibitory effect of mARD1A225 on primary tumor growth was additionally observed with the human gastric tumor cell line, NUGC-3 (see Supplementary Figure 4, available online).
Figure 3.
Effect of mARD1A225 on primary tumor growth in mice. A) Representative images of primary tumor masses (left panel) and growth curves (right panel) are shown. C57BL/6 mice were injected subcutaneously with B16F10 (open diamond), B16F10-mock (open square), or B16F10-mARD1A225 cells (two clones; closed symbols). Results are means and 95% confidence intervals (error bars). *P = .003, **P < .001, compared with mock control cells, as determined with two-way repeated measures analysis of variance (ANOVA). B) Representative images of primary tumor masses (left panel) and growth curves (right panel) are shown. Nude mice were injected subcutaneously with MKN74 (open diamond), MKN74-mock (open square), or MKN74-mARD1A225 cells (two clones; closed symbols). Results are means and 95% confidence intervals (error bars). *P = .007, **P < .001, compared with mock control cells, as determined with two-way repeated measures ANOVA. C) Tumor weights (n = 8 per group) were measured 3 weeks after inoculation with B16F10, B16F10-mock, or B16F10-mARD1A225 cells. Results are means and 95% confidence intervals (error bars). *P = .021, compared with mock control cells, as determined with the two-sided Mann–Whitney U test. D) Tumor weights (n = 10 per group) were measured 3 weeks after inoculation with MKN74, MKN74-mock, and MKN74-mARD1A225 cells. Results are means and 95% confidence intervals (error bars). *P = .017, **P = .015, compared with mock control cells, as determined with the two-sided Mann–Whitney U test. A#1= B16F10-mARD1A225 #1; A#2 = B16F10-mARD1A225 #2; A#3 = MKN74-mARD1A225 #3; A#4 = MKN74-mARD1A225 #4.
B16F10-mARD1A225 tumor weights were 36% of the weight of control tumors on average (B16F10, mean = 3.02 mg, 95% CI = 1.47 to 4.57; B16F10/mock, mean = 4.13 mg, 95% CI = 3.34 to 4.91; B16F10/mARD1A225 #1, mean = 1.28 mg, 95% CI = 0.4 to 2.16; B16F10/mARD1A225 #2, mean = 1.35 mg, 95% CI = 0.5 to 2.19; P = .021 for all comparisons) (Figure 3, C). Similarly, MKN74-mARD1A225 tumor weights were markedly reduced to less than 8% of control tumors (MKN74, mean = 0.3 mg, 95% CI = 0.161 to 0.439; MKN74/mock, mean = 0.35 mg, 95% CI = 0.252 to 0.448; MKN74/mARD1A225 #3, mean = 0.028 mg, 95% CI = 0.015 to 0.04; MKN74/mARD1A225 #4, mean = 0.018 mg, 95% CI = 0.013 to 0.022; P = .017, clone A#3, P = .015, clone A#4) (Figure 3, D). Taken together, the data suggest that mARD1A225 statistically significantly inhibited the growth of transplanted tumors in vivo.
Effect of mARD1A225 on Metastatic Lesions in the Lung
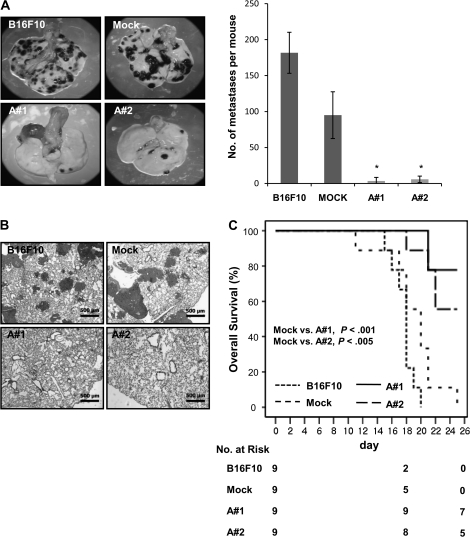
We determined the effect of mARD1A225 on the incidence of lung nodule formation using an experimental model of metastasis. We evaluated the number of lung nodules present at 3 weeks, following intravenous injection with equal numbers of B16F10 parental, B16F10-mock, and B16F10-mARD1A225 cells (Figure 4, A–C). Mice injected with B16F10-mARD1A225 cells contained few lung nodules, whereas numerous nodules were detected in groups injected with B16F10 parental or B16F10-mock cells. In mice injected with B16F10-mARD1A225 cells, the number of surface tumor nodules was markedly decreased to less than 4% of the number of nodules in mice injected with B16F10 or B16F10-mock cells (number of lung nodules per mouse, B16F10, mean = 181.75, 95% CI = 153.2 to 210.3; B16F10/mock, mean = 95, 95% CI = 62.5 to 127.5; B16F10/mARD1A225 #1, mean = 6.5, 95% CI = 0 to 16.0, P = .006; B16F10/mARD1A225 #2, mean = 5, 95% CI = 0 to 11.5, P = .006) (Figure 4, A). The suppression of lung metastases in mice injected with B16F10-mARD1A225 cells was further confirmed by hematoxylin and eosin staining of lung sections (Figure 4, B). Moreover, the overall survival of mice injected intravenously with B16F10-mARD1A225 cells was statistically significantly longer than those injected with B16F10 or B16F10-mock cells. Mice began to die 11 days after the injection of control cells (B16F10, B16F10-mock). By day 25, 100% mortality was observed in mice with control cells, in contrast to 22% and 44% mortality in mice injected with B16F10/mARD1A225 #1 (P < .001, log-rank test; n = 9 mice per group) and B16F10/mARD1A225 #2 (P = .002, log-rank test; n = 9 mice per group) cells, respectively (Figure 4, C). Thus, it appears that overexpression of mARD1A225 strongly suppresses the growth of metastases.
Figure 4.
Effect of mARD1A225 on lung nodule formation in mice. A and B) C57BL/6 mice were injected intravenously with B16F10, B16F10-mock, or B16F10-mARD1A225 cells. The incidence of lung nodules was determined 19 days after injection (n = 8 per group). A) Representative images of lungs (left). Numbers of surface tumor nodules (right). Results are means and 95% confidence intervals (CIs) (error bars) of two independent experiments performed with four mice per group. *P = .006, compared with the number of surface tumor nodules detected in B16F10-mock–injected mice, as determined with the two-sided Mann–Whitney U test. B) Hematoxylin and eosin staining of 4-μm lung tissue sections excised from mice described in (A). Magnification ×50. Scale bar = 500 μm. C) Kaplan–Meier survival plot of C57BL/6 mice injected with B16F10 (mean = 16.7 days, 95% CI = 15.7 to 17.7), B16F10-mock (mean = 18 days, 95% CI = 15.5 to 20.5), or B16F10-mARD1A225 cells (A#1, mean = 23.9 days, 95% CI = 22.5 to 25.3; A#2, mean = 22.7 days, 95% CI = 20.8 to 24.5) (n = 9 per group). Comparisons were made with log-rank tests. All statistical tests were two-sided.
Effect of mARD1A225 Overexpression on HIF-1α and VEGFA Expression In Vitro
To confirm the effects of mARD1A225 on the stability and acetylation of HIF-1α in cancer cell lines, we stably expressed epitope-tagged mARD1A225 cDNA. In B16F10 and MKN74 cells transfected with mARD1A225, a qualitative decrease in endogenous HIF-1α expression was evident during hypoxia (Figure 5, A). This inhibitory effect of mARD1A225 on HIF-1α expression was also observed in clones derived from B16F1 and NUGC-3 tumor cells (see Supplementary Figure 5, A and B, available online).
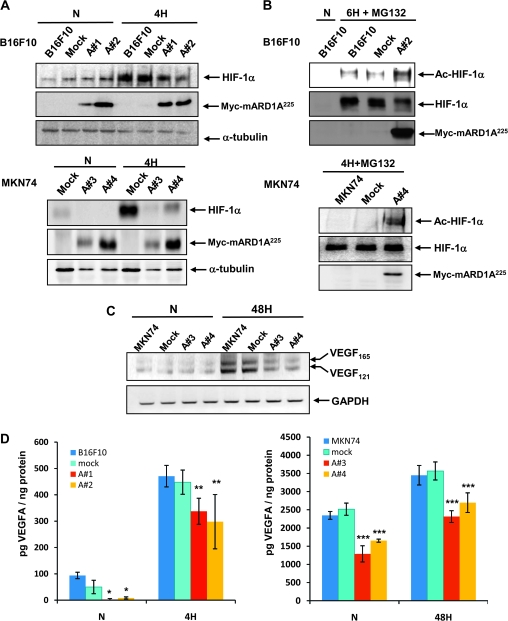
Figure 5.
Effect of mARD1A225 on VEGFA expression. A) B16F10, B16F10-mock, and B16F10-mARD1A225 cells were exposed to 1% O2 (4H) or 21% O2 (N) for 4 hours (top). MKN74-mock and MKN74-mARD1A225 cells were exposed to 1% O2 (4H) or 21% O2 (N) for 4 hours (bottom). HIF-1α and mARD1A225 protein levels were examined by Western blot analysis. Anti-Myc antibody was used for the detection of mARD1A225. B) B16F10, B16F10-mock, and B16F10-mARD1A225 cells were exposed to 21% O2 (N) or 10 μM MG132 and 1% O2 for 6 hours (6H + MG132) (top). MKN74, MKN74-mock, and MKN74-mARD1A225 cells were exposed to 21% O2 (N) (not shown) or 10 μM MG132 and 1% O2 for 4 hours (4H + MG132) (bottom). Cell lysates were immunoprecipitated with an anti–acetyl lysine antibody and subjected to Western blot analysis with an anti-HIF-1α antibody. The level of mARD1A225 was determined using an anti-Myc antibody. C) MKN74, MKN74-mock, and MKN74-mARD1A225 cells were exposed to 1% O2 (48H) or 21% O2 (N) for 48 hours. Reverse transcription–polymerase chain reaction analysis was performed to detect gene expression using specific primers for VEGFA and glyceraldehyde 3-phosphate dehydrogenase. D) VEGFA protein levels were quantified in conditioned media from control and mARD1A225-expressing cells treated with CoCl2 using enzyme-linked immunosorbent assay ELISA. B16F10, B16F10-mock, and B16F10-mARD1A225 cells were cultured in the absence (N) or presence of 200 μM CoCl2 for 4 hours (4H) (left). MKN74, MKN74-mock, and MKN74-mARD1A225 cells were cultured in the absence (N) or presence of 200 μM CoCl2 for 48 hours (48H) (right). Three independent experiments were performed, each in two replicates. Results are means and 95% confidence intervals (error bars) derived from the means of individual experiments (n = 3). *P = .046, **P = .05, ***P = .021, compared with mock control cells, determined with the two-sided Mann–Whitney U test.
To determine whether mARD1A225 acetylates HIF-1α in tumor cells, we performed an acetylation assay using an anti–acetyl lysine antibody after treatment with the proteasome inhibitor MG132 to equalize HIF-1α protein levels. Acetylation of HIF-1α was markedly increased in B16F10-mARD1A225 and MKN74-mARD1A225 cells (Figure 5, B), and in B16F1-mARD1A225 cells (see Supplementary Figure 5, C, available online), compared with parental or mock-infected tumor cells.
Effect of mARD1A225 on VEGFA Expression and Angiogenesis In Vitro
To determine the functional consequences of HIF-1α acetylation, we examined the expression of VEGFA, a known transcriptional target of HIF-1. The VEGFA mRNA level was appreciably decreased in MKN74-mARD1A225 cells during hypoxia compared with that in MKN74-mock cells (Figure 5, C). Moreover, VEGFA protein expression was decreased in mARD1A225-expressing tumor cell-conditioned medium, relative to control cells (Figure 5, D, P ≤ .05 for all comparisons). For B16F10 cells under normoxic condition, the concentration of VEGFA (in picograms) protein relative to total protein (in nanograms) was as follows: B16F10, mean = 93.75 pg/ng total protein, 95% CI = 81.5 to 106.25; B16F10/mock, mean = 50 pg/ng, 95% CI = 24.53 to 75.47; B16F10/mARD1A225 #1, mean = 2.08 pg/ng, 95% CI = 0 to 6.16; B16F10/mARD1A225 #2, mean = 8.3 pg/ng, 95% CI = 4.22 to 12.38; in the hypoxic condition: B16F10, mean = 470.83 pg/ng, 95% CI = 430 to 511.66; B16F10/mock, mean = 447.91 pg/ng, 95% CI = 401.9 to 493.92; B16F10/mARD1A225 #1, mean = 337.5 pg/ng, 95% CI = 288 to 387; B16F10/mARD1A225 #2, mean = 297.9 pg/ng, 95% CI = 194.85 to 400.95 (Figure 5, D, left panel). In MKN74 cells under normoxic conditions, the concentration of VEGFA (in picograms) relative to total protein (in nanograms) was as follows: MKN74, mean = 2346.6 pg/ng, 95% CI = 2239.2 to 2454.11; MKN74/mock, mean = 2517.7 pg/ng, 95% CI = 2350.1 to 2685.3; MKN74/mARD1A225 #3, mean = 1289.0 pg/ng, 95% CI = 1066.6 to 1311.6; MKN74/mARD1A225 #4, mean = 1652.0 pg/ng, 95% CI = 1608.9 to 1695.2; in the hypoxic condition: MKN74, mean = 3451.7 pg/ng, 95% CI = 3182.9 to 3720.5; MKN74/mock, mean = 3568.2 pg/ng, 95% CI = 3320.3 to 3816.1; MKN74/mARD1A225 #3, mean = 2314.9 pg/ng, 95% CI = 2153.9 to 2475.9; MKN74/mARD1A225 #4, mean = 2696.8 pg/ng, 95% CI = 2426.8 to 2966.7 (Figure 5, D, right panel). The data confirm that mARD1A225 overexpression leads to decreased expression of HIF-1α and VEGFA during hypoxia in both human and mouse cancer cell lines by promoting HIF-1α acetylation.
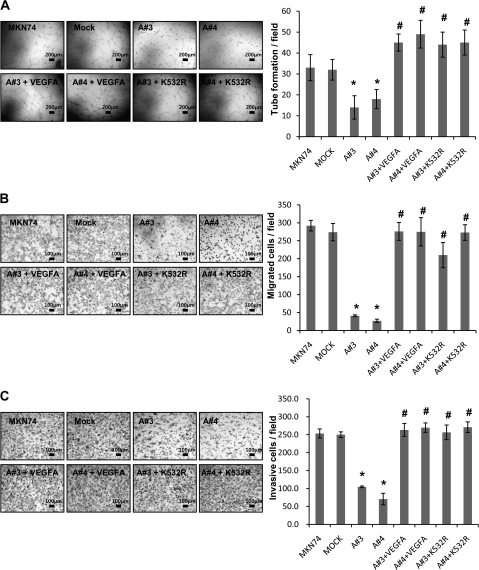
As an endothelial mitogen, VEGFA is a potent mediator of vascular permeability. To establish the antiangiogenic activity of mARD1A225, we examined endothelial cell activation in vitro. Capillary tube formation, migration, and invasion of HUVECs into Matrigel were all inhibited by hypoxic-conditioned media that were derived from mARD1A225-expressing MKN74 tumors cells (Figure 6, A). To confirm the specificity of the VEGFA-mediated effect, we added exogenous VEGFA protein to the conditioned media or used media derived from MKN74-mARD1A225 cells overexpressing HIF-1α/K532R (Figure 6, A), which enhanced tube formation, migration, and invasion of endothelial cells. The number of tubes formed per field was as follows: MKN74, mean = 35.7, 95% CI = 29.4 to 41.9; MKN74/mock, mean = 35, 95% CI = 30.1 to 39.9; MKN74/mARD1A225 #3, mean = 17.3, 95% CI = 11.8 to 22.9; MKN74/mARD1A225 #4, mean = 18.7, 95% CI = 14.1 to 23.2; MKN74-mARD1A225 #3+VEGFA, mean = 41.0, 95% CI = 36.9 to 45.1; MKN74-mARD1A225 #4+VEGFA, mean = 44.7, 95% CI = 38.0 to 51.3; MKN74-mARD1A225 #3+HIF-1α/K532R, mean = 42, 95% CI = 36 to 48; MKN74-mARD1A225 #4+HIF-1α/K532R, mean = 41, 95% CI = 35 to 47 (Figure 6, A). The number of migrated cells per field was as follows: MKN74, mean = 278, 95% CI = 263.2 to 292.8; MKN74/mock, mean = 289.7, 95% CI = 265.5 to 313.8; MKN74/mARD1A225 #3, mean = 39.7, 95% CI = 37.1 to 42.3; MKN74/mARD1A225 #4, mean = 31, 95% CI = 26.5 to 35.5; MKN74-mARD1A225 #3+VEGFA, mean = 271.7, 95% CI = 247 to 296.4; MKN74-mARD1A225 #4+VEGFA, mean = 237.7, 95% CI = 198.2 to 277.1; MKN74-mARD1A225 #3+HIF-1α/K532R, mean = 219.3, 95% CI = 184.2 to 254.5; MKN74-mARD1A225 #4+HIF-1α/K532R, mean = 276.3, 95% CI = 254.6 to 298.1 (Figure 6, B). The number of invasive cells per field was as follows: MKN74, mean = 253.3, 95% CI = 240.6 to 266.1; MKN74/mock, mean = 250.0, 95% CI = 242.1 to 257.9; MKN74/mARD1A225 #3, mean = 104.7, 95% CI = 102.3 to 107; MKN74/mARD1A225 #4, mean = 70.7, 95% CI = 54.7 to 86.7; MKN74-mARD1A225 #3+VEGFA, mean = 263.3, 95% CI = 245.3 to 281.4; MKN74-mARD1A225 #4+VEGFA, mean = 269.7, 95% CI = 256.6 to 282.8; MKN74-mARD1A225 #3+HIF-1α/K532R, mean = 256.7, 95% CI = 236.3 to 277; MKN74-mARD1A225 #4+HIF-1α/K532R, mean = 271, 95% CI = 256.2 to 285.8 (Figure 6, C). The results strongly suggest that mARD1A225 suppresses angiogenesis by reducing VEGFA expression and inhibits the activation of endothelial cells, preventing capillary tube formation, migration, and invasion (Figure 6, A–C, P = .05 for all comparisons).
Figure 6.
Effect of VEGFA treatment or HIF-1α/K532R overexpression on tube formation, migration, and invasion by mARD1A225. A) Human umbilical vein endothelial cells (HUVECs) were incubated for 6 hours on Matrigel with hypoxic-conditioned media from MKN74, MKN74-mock, and MKN74-mARD1A225 (A#3 and A#4) cells (top left) and MKN74-mARD1A225 cells treated with 10 ng/mL recombinant human VEGFA protein (A#3+VEGFA and A#4+VEGFA), and MKN74-mARD1A225 cells transfected with HIF-1α/K532R (A#3+K532R and A#4+K532R) (bottom left). Scale bar = 200 μm. Quantification of tube formation (right). Results are mean of three replicates from one experiment and their 95% confidence intervals (error bars) (n = 3). *P = .05, compared with the number of MKN74-mock cells; #P = .05, compared with the number of MKN74-mARD1A225 #3 or #4 cells, as determined with the two-sided Mann–Whitney U test. B) (Left) HUVECs and conditioned media were added to transwells, and migrating cells were counted. Data are averaged over three independent experiments. Scale bar = 100 μm. Quantification of migration (right). Results are mean of three replicates from one experiment and their 95% confidence intervals (error bars) (n = 3). *P = .05, compared with the number of MKN74-mock cells; #P = .05, compared with the number of MKN74-mARD1A225 #3 or #4 cells, as determined with the two-sided Mann–Whitney U test. C) Transwells were coated with Matrigel. Cells were stained with hematoxylin and eosin. Representative microscopic images illustrate HUVEC invasion (left). Scale bar = 100 μm. Quantification of invasion (right). Results are mean of three replicates from one experiment and their 95% confidence intervals (error bars) (n = 3). *P = .05, compared with the number of MKN74-mock cells; #P = .05, compared with the number of MKN74-mARD1A225 #3 or #4 cells, as determined with the two-sided Mann–Whitney U test. A#3 = MKN74-mARD1A225 #3; A#4 = MKN74-mARD1A225 #4; A#3+VEGFA = MKN74-mARD1A225 #3+ recombinant human VEGFA protein; A#4+VEGFA = MKN74-mARD1A225 #4+ recombinant human VEGFA protein; A#3+K532R = MKN74-mARD1A225 #3+HIF-1α/K532R; A#4+K532R = MKN74-mARD1A225 #4+HIF-1α/K532R.
Effect of mARD1A225 on HIF-1α Degradation and Neovascularization
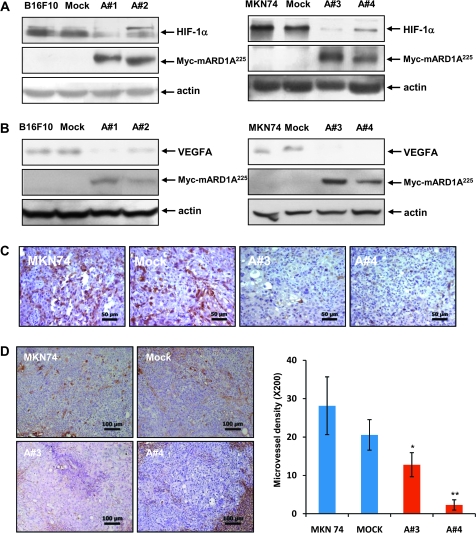
To determine whether HIF-1α and VEGFA levels are reduced in mARD1A225-expressing tumors, we analyzed expression of these proteins in tumors larger than 50 mm2 and thus likely to be hypoxic in the center (20). Tumors that were derived from mARD1A225-expressing B16F10 and MNK74 cells displayed a qualitative decrease in endogenous HIF-1α and VEGFA expression compared with parental and mock-infected cells (Figure 7, A and B). Similarly, immunohistochemical staining of MKN74-mARD1A225-expressing tumors displayed lower VEGFA expression than control tumors (MKN74, MKN74-mock) (Figure 7, C). To determine whether microvessel density was associated with VEGFA levels, we used an anti-CD34 antibody to identify endothelial cells and quantify the number of microvessels in these tumors. CD34 staining revealed that MKN74-mARD1A225–expressing tumors displayed lower microvessel density than those derived from mice injected with MKN74 or MKN74-mock cells (number of vessels per field: MKN74, mean = 28.2, 95% CI = 20.6 to 35.7; MKN74/mock, mean = 20.6, 95% CI = 16.6 to 24.5; MKN74/mARD1A225 #3, mean = 12.8, 95% CI = 9.6 to 15.9 P = .043; MKN74/mARD1A225 #4, mean = 2.3, 95% CI = 1.0 to 3.6, P = .021) (Figure 7, D). These data indicate that mARD1A225 inhibits tumor growth and angiogenesis by repressing VEGFA activity through degradation of its upstream regulator, HIF-1α.
Figure 7.
Effect of mARD1A225 on neovascularization. A) Western blot analysis of HIF-1α and mARD1A225 protein expression in transplanted B16F10 (left) and MNK74 (right) tumor cells. B) Western blot analysis of VEGFA protein expression in transplanted B16F10 (left) and MNK74 (right) tumor cells. C) BALB/cSlc-nude mice were injected subcutaneously with MKN74, MKN74-mock, or two MKN74-mARD1A225 clones, A#3 and A#4. After 3 weeks, formalin-fixed primary tumor sections from each mouse were prepared and stained with an anti-VEGFA antibody. The brown color signifies VEGFA in primary tumors. Magnification ×400. Scale bar = 50 μm. A#3 = MKN74-mARD1A225 #3; A#4 = MKN74-mARD1A225 #4. One representative image of five per cell line is shown. D) Anti-CD34 staining of xenograft tumors derived from MKN74, MKN74-mock, and MKN74-mARD1A225 cells, indicative of endothelial cells (left). Numbers of CD34-positive vessels in primary tumors (right) expressed as means and 95% confidence intervals (error bars). Vessels from four or five fields from each of the five tumors per group were scored for quantification. *P = .043, **P = .021, compared with the number of CD34-positive vessels in tumors derived from MKN74-mock cells, as determined with the two-sided Mann–Whitney U test. Magnification ×200. Scale bar = 100 μm.
Identification of the Critical Lysine Residue in HIF-1α for mARD1A225-Mediated Degradation
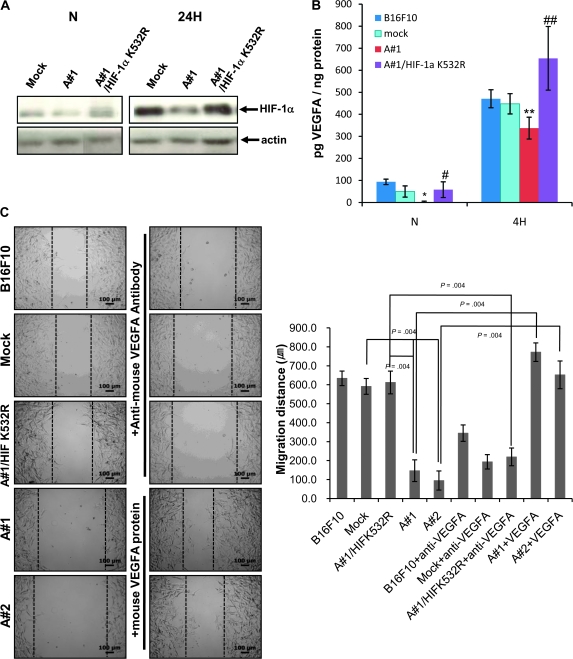
Previous studies reported that acetylation of lysine 532 of HIF-1α by mARD1A225 leads to HIF-1α ubiquitination and degradation (5). To determine whether lysine 532 is necessary and sufficient for HIF-1α degradation, we mutated this residue and generated cell lines stably expressing mutant HIF-1α/K532R in B16F10-mARD1A225 cells (B16F10-mARD1A225-HIF-1α/K532R). Examination of HIF-1α expression and acetylation after 24 hours of hypoxia revealed HIF-1α levels in mARD1A225-HIF-1α/K532R to be comparable to those in mock-infected tumor cell lines, suggesting that mARD1A225 is unable to acetylate and degrade HIF-1α in the absence of lysine 532 (Figure 8, A).
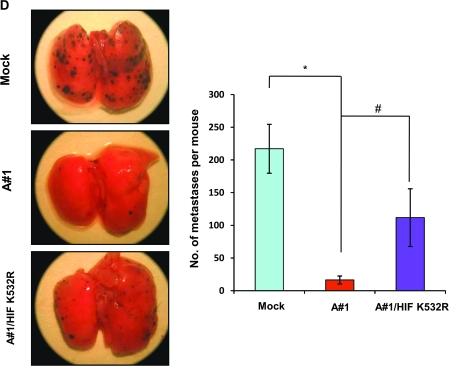
Figure 8.
Effects of lysine 532 mutation in HIF-1α. A) HIF-1α protein levels were determined by Western blot analysis. B16F10-mock, B16F10-mARD1A225, and B16F10-mARD1A225-HIF-1α/K532R cells were cultured in the absence (N) or presence of 200 μM CoCl2 for 24 hours (24H). Similar results were obtained with three independent experiments. B) VEGFA levels (in picograms per milliliter) were quantified using enzyme-linked immunosorbent assay. B16F10, B16F10-mock, B16F10-mARD1A225, and B16F10-mARD1A225-HIF-1α/K532R cells were cultured in the absence (N) or presence of 200 μM CoCl2 for 4 hours (4H). Three independent experiments were performed, each in two replicates. Results are means and 95% confidence intervals (error bars) derived from the means of the individual experiments (n = 3). *P = .05 for all comparisons as determined with the two-sided Mann–Whitney U test. C) B16F10, B16F10-mock, B16F10-mARD1A225, and B16F10-mARD1A225-HIF-1α/K532R cells were grown to 80% confluence. A linear injury line was created at the center of the cultured monolayer by scraping with a sterile pipette tip. Cells were incubated in the presence of 200 μM CoCl2 for 24 hours. B16F10, B16F10-mock, and B16F10-mARD1A225-HIF-1α/K532R cells were treated with 3 μg/mL anti-mouse VEGFA antibody. B16F10-mARD1A225 cells were treated with 25 ng/mL recombinant mouse VEGFA protein. Images of cell migration (left). Quantification of cell migration (right). Migration distance (in micrometers); P = .004, determined with the two-sided Mann–Whitney U test. Three independent experiments were performed, each in two replicates. Results are means and 95% confidence intervals (error bars) derived from the means of the individual experiments (n = 3). Magnification ×100. Scale bar = 100 μm. D) C57BL/6 mice were injected intravenously with B16F10-mock (n = 40 mice), B16F10-mARD1A225 (n = 31 mice) and B16F10-mARD1A225-HIF-1α/K532R cells (n = 11 mice). Lung nodule formation was determined 14 days after injection. Representative gross images of lungs are shown (left). Numbers of surface tumor nodules per mouse (right). Results are means and 95% confidence intervals (error bars). *P < .001, compared with the number of surface tumor nodules in B16F10-mock–injected mice, #P < .001, compared with the number of surface tumor nodules in B16F10-mARD1A225–injected mice, as determined with the two-sided Mann–Whitney U test. A#1 = B16F10-mARD1A225 #1; A#2 = B16F10-mARD1A225 #2; A#1/HIF K532R = B16F10-mARD1A225 #1-HIF-1α/K532R; B16F10 + anti-VEGFA = B16F10 + anti-mouse VEGFA antibody; Mock + anti-VEGFA = B16F10-Mock + anti-mouse VEGFA antibody; A#1/HIF K532R + anti-VEGFA = B16F10-mARD1A225 #1-HIF-1α/K532R + anti-mouse VEGFA antibody; A#1+VEGFA = B16F10-mARD1A225 #1 + recombinant mouse VEGFA protein; A#2+VEGFA = B16F10-mARD1A225 #2 + recombinant mouse VEGFA protein.
To confirm the functional consequences of HIF-1α overexpression, we measured VEGFA levels in the conditioned media of these cells (Figure 8, B). After 4 hours of hypoxia, VEGFA secretion by mARD1A225-HIF-1α/K532R–expressing cells was statistically significantly higher than that of mock-infected or parental cell lines, implying a critical role for lysine 532 in the regulation of HIF-1α stability (normoxic condition: B16F10/mock, mean = 50 pg/mg, 95% CI = 24.53 to 75.47; B16F10/mARD1A225 #1, mean = 2.08 pg/mg, 95% CI = 0 to 6.16; B16F10/mARD1A225/HIF-1α/K532R, mean = 58.33 pg/mg, 95% CI = 22.74 to 93.92; hypoxic condition: B16F10/mock, mean = 447.91 pg/mg, 95% CI = 401.9 to 493.92; B16F10/mARD1A225 #1, mean = 337.5 pg/mg, 95% CI = 288 to 387; B16F10/mARD1A225/HIF-1α/K532R, mean = 654.2 pg/mg, 95% CI = 509.9 to 798.5; P < .05 for all comparisons) (Figure 8, B).
VEGFA is an important endothelial mitogen but has also been reported to induce migration in a number of cell types, including tumor cells. Accordingly, we examined the ability of VEGFA generated by mARD1A225-HIF-1α/K532R–expressing tumor cells to potentiate cell migration in a scratch assay (Figure 8, C). A linear injury line was created at the center of a tissue culture dish, and tumor cell migration examined after 24 hours of incubation under hypoxic conditions. Regrowth and migration of B16F10 cells from the wound edge was inhibited by mARD1A225 (cell migration distances, B16F10/mock vs B16F10/mARD1A225 #1, mean = 591.4 vs 146.6 μm, difference = 444.8 μm, 95% CI = 429.4 to 460.3; B16F10/mock vs B16F10/mARD1A225 #2, mean = 591.4 vs 94.7 μm, difference = 497.7 μm, 95% CI = 488.1 to 505.4; P = .004 for all comparisons). As expected, increased expression of VEGFA in mutant HIF-1α/K532R cells led to enhanced migration compared with mARD1A225-expressing cells (B16F10/mARD1A225 #1-HIF-1α/K532R vs B16F10/mARD1A225 #1, mean = 611.9 vs 146.6 μm, difference = 465.3 μm, 95% CI = 462.5 to 468.2; P = .004). To confirm the specificity of VEGFA-mediated cell migration, we added a neutralizing anti-mouse VEGFA antibody or recombinant mouse VEGFA protein, which suppressed the migration of mARD1A225-HIF-1α/K532R–expressing tumor cells into the linear injury line (B16F10/mARD1A225 #1-HIF-1α/K532R vs B16F10-mARD1A225 #1-HIF-1α/K532R + anti-VEGFA, mean = 611.9 vs 219.5μm, difference = 392.4 μm, 95% CI = 379.2 to 405.7; P = .004) but enhanced migration of mARD1A225 cells (B16F10/mARD1A225 #1 vs B16F10-mARD1A225 #1 + VEGFA protein, mean = 146.6 vs 771.8 μm, difference = 625.2 μm, 95% CI = 616.8 to 633.6; B16F10/mARD1A225 #2 vs B16F10-mARD1A225 #2 + VEGFA protein, mean = 94.7 μm vs 652.2 μm, difference = 557.5 μm, 95% CI = 534.8 to 580.3; P = .004 for all comparisons; Figure 8, C). The data clearly suggest that reduction of VEGFA is a critical factor in mARD1A225-induced inhibition of B16F10 cell migration.
To determine the in vivo effects of the mARD1A225-HIF-1α/K532R mutants, we examined the formation of lung nodules after intravenous injection of B16F10-mock (n = 40), B16F10-mARD1A225 (n = 31), and B16F10-mARD1A225-HIF-1α/K532R (n = 11) cells into syngeneic C57BL/6 mice. The number of surface tumor nodules was higher in mice injected with B16F10-mARD1A225-HIF-1α/K532R cells compared with that in mice injected with B16F10-mARD1A225 cells (number of lung nodules per mouse: B16F10/mock vs B16F10/mARD1A225 vs B16F10/mARD1A225/HIF-1α/K532R, mean = 217.1 vs 16.38 vs 111.8; B16F10/mock vs B16F10/mARD1A225, difference = 200.72, 95% CI = 169.4 to 232.2; B16F10/mARD1A225 vs B16F10/mARD1A225/HIF-1α/K532R, difference = 95.42, 95% CI = 57.4 to 133.4; P < .001 for all comparisons; Figure 8, D). We tested the activity of a mutant HIF-1α/K532R, which was stably expressed under normoxic conditions (see Supplementary Figure 6, A, available online). Increased expression of mutant HIF-1α/K532R in B16F10-mock cells led to enhanced migration and metastasis compared with B16F10-mock-mock cells (see Supplementary Figure 6, B and C, available online). Therefore, stabilization of HIF-1α appears to reverse the tumor-suppressive effect of mARD1A225 on lung lesions. Based on these data, we propose that lysine 532 of HIF-1α is important for HIF-1α degradation, ultimately leading to the regulation of VEGFA expression.
Discussion
We demonstrate that the potent antitumor effects of mARD1A225 are mediated by HIF-1α degradation, resulting in reduced expression of its well-characterized target, VEGFA. Moreover, overexpression of mARD1A225 leads to inhibition of tumorigenesis in three different tumor models via reduction of VEGFA expression and its proangiogenic functions in both murine and human cancer cells.
The ApcMin/+ mice have been widely used as a model for studying early events in intestinal tumorigenesis. Recent reports indicate that VEGFA expression was increased and correlated with adenomatous polyposis coli (APC) gene mutations in primary human colon tumors (19). Also, this model showed that short-term treatment of ApcMin/+ mice with an anti-VEGFA monoclonal antibody results in reduced tumor burden and vessel density of intestinal polyps (21). Similarly, targeted deletion of VEGFA in intestinal epithelial cells of ApcMin/+ mice induced a statistically significant reduction in tumor growth (21), suggesting that VEGFA plays an important role in ApcMin/+ intestinal polyp growth. The offspring from a cross between the ApcMin/+ mouse and a mouse null for the endogenous anti–angiogenic protein thrombospondin 1 (THBS1−/−) showed a significant increase in polyp number and diameter (22), indicating a possible role for angiogenesis in polyp development. A study of another mouse model of intestinal adenomas, Apc716, which reported elevated VEGFA protein levels in polyps compared with normal intestinal epithelium control tissue, was also consistent with an association between VEGFA and polyp development (23). In our experiments, VEGFA expression was decreased in intestinal polyps of ApcMin/+/mARD1A225 transgenic mice, which, in turn, led to suppression of tumor growth and angiogenesis. Therefore, we propose that transgenic overexpression of mARD1A225 exerts an important tumor-suppressive effect via inhibition of VEGFA in ApcMin/+ mouse intestinal polyps.
Whereas the vast majority of tumors express VEGFA (24), it is clear that VEGFA regulation by HIF-1α plays a critical role in tumor angiogenesis and growth (25). Here, we show that overexpression of mARD1A225 in both human gastric cancer and mouse melanoma cells induces a statistically significant reduction of HIF-1α protein expression and consequent production of VEGFA. Furthermore, the growth of lung nodules in an experimental model of lung metastases was markedly inhibited by overexpression of mARD1A225 in cancer cells. Expression of HIF-1α and VEGFA triggered under conditions of intratumoral hypoxia was decreased by mARD1A225 in B16F10 and MKN74 tumors. Our in vitro studies demonstrate that mARD1A225 overexpression leads to suppression of VEGFA-mediated endothelial cell proliferation, migration, and capillary tube formation. The data collectively suggest that the decrease in VEGFA levels induced as a result of degradation of HIF-1α by mARD1A225 inhibits tumor growth and angiogenesis in vivo.
This study confirms the in vivo importance of a single lysine residue in regulating the stability of HIF-1α. To our knowledge, HIF-1α is the only known target of the acetyltransferase, mARD1A225. Our analyses of the mutant HIF-1α construct, HIF-1α/K532R, show that VEGFA overexpression is because of the stability of HIF-1α. However, the effects of the mutant HIF-1α in tumor cells did not fully override the increased expression of mARD1A225, such that the incidence of lung tumor lesions in our experimental model of lung metastases was higher than that in mARD1A225-overexpressing cells but not higher than in mock-infected tumor cells (Figure 8, D). This suggests that additional targets of mARD1A225 are involved in regulating tumor growth and/or progression. Other limitations of this study are that we only examined VEGFA expression as a measure of HIF-1α activity. We also could not exclude the possibility of other HIF-1α-targets involved in tumor growth and/or progression. In addition, the mARD1A225 isoform has not yet been identified in human cell lines. However, recent reports provide direct evidence for an inhibitory effect of human ARD1A on HIF-1α stability (26,27).
Our data collectively confirm the importance of mARD1A225 in regulating tumor growth and development and provide evidence of its antitumor and antiangiogenic activities. Notably, mARD1A225 regulates tumor growth via degradation of HIF-1α and consequent reduced expression of the proangiogenic protein, VEGFA. Therefore, we conclude that mARD1A225 presents a novel target in the regulation of HIF-1α stability and may have potent therapeutic effects in combination with currently approved anti-VEGFA treatments such as bevacizumab.
Funding
21st Century Frontier Functional Human Genome Project (FG06-2-9); National Research Foundation of Korea (NRF) grant funded by the Ministry of Education, Science & Technology (MEST) through the Creative Research Initiative Program (R16-2004-001010010 to K.-W.K., 2009); Research program for New Drug Target Discovery (M108KB010014-08K0201-01420) grant from MEST.
Supplementary Material
Footnotes
The funders did not have any involvement in the design of the study; the collection, analysis, and interpretation of the data; the writing of the manuscript; or the decision to submit the manuscript for publication.
We gratefully acknowledge the technical assistance of So-Young Kwon. We thank Dr Seongho Ryu in Cornell Medical School for advice on statistical analysis.
References
- 1.Mullen JR, Kayne PS, Moerschell RP, et al. Identification and characterization of genes and mutants for an N-terminal acetyltransferase from yeast. EMBO J. 1989;8(7):2067–2075. doi: 10.1002/j.1460-2075.1989.tb03615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee FJ, Lin LW, Smith JA. N alpha acetylation is required for normal growth and mating of Saccharomyces cerevisiae. J Bacteriol. 1989;171(11):5795–5802. doi: 10.1128/jb.171.11.5795-5802.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Whiteway M, Freedman R, Van Arsdell S, Szostak JW, Thorner J. The yeast ARD1 gene product is required for repression of cryptic mating-type information at the HML locus. Mol Cell Biol. 1987;7(10):3713–3722. doi: 10.1128/mcb.7.10.3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Whiteway M, Szostak JW. The ARD1 gene of yeast functions in the switch between the mitotic cell cycle and alternative developmental pathways. Cell. 1985;43(2, pt 1):483–492. doi: 10.1016/0092-8674(85)90178-3. [DOI] [PubMed] [Google Scholar]
- 5.Jeong JW, Bae MK, Ahn MY, et al. Regulation and destabilization of HIF-1alpha by ARD1-mediated acetylation. Cell. 2002;111(5):709–720. doi: 10.1016/s0092-8674(02)01085-1. [DOI] [PubMed] [Google Scholar]
- 6.Kim SH, Park JA, Kim JH, et al. Characterization of ARD1 variants in mammalian cells. Biochem Biophys Res Commun. 2006;340(2):422–427. doi: 10.1016/j.bbrc.2005.12.018. [DOI] [PubMed] [Google Scholar]
- 7.Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003;3(10):721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 8.Masson N, Willam C, Maxwell PH, Pugh CW, Ratcliffe PJ. Independent function of two destruction domains in hypoxia-inducible factor-alpha chains activated by prolyl hydroxylation. EMBO J. 2001;20(18):5197–5206. doi: 10.1093/emboj/20.18.5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jaakkola P, Mole DR, Tian YM, et al. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292(5516):468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- 10.Ivan M, Kondo K, Yang H, et al. HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science. 2001;292(5516):464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- 11.Kallio PJ, Wilson WJ, O'Brien S, Makino Y, Poellinger L. Regulation of the hypoxia-inducible transcription factor 1alpha by the ubiquitin-proteasome pathway. J Biol Chem. 1999;274(10):6519–6525. doi: 10.1074/jbc.274.10.6519. [DOI] [PubMed] [Google Scholar]
- 12.Folkman J. Role of angiogenesis in tumor growth and metastasis. Semin Oncol. 2002;29(6 suppl 16):15–18. doi: 10.1053/sonc.2002.37263. [DOI] [PubMed] [Google Scholar]
- 13.Carmeliet P, Jain RK. Angiogenesis in cancer and other diseases. Nature. 2000;407(6801):249–257. doi: 10.1038/35025220. [DOI] [PubMed] [Google Scholar]
- 14.Detmar M. Tumor angiogenesis. J Investig Dermatol Symp Proc. 2000;5(1):20–23. doi: 10.1046/j.1087-0024.2000.00003.x. [DOI] [PubMed] [Google Scholar]
- 15.Ellis LM, Fidler IJ. Angiogenesis and metastasis. Eur J Cancer. 1996;32A(14):2451–2460. doi: 10.1016/s0959-8049(96)00389-9. [DOI] [PubMed] [Google Scholar]
- 16.Ellis LM, Takahashi Y, Liu W, Shaheen RM. Vascular endothelial growth factor in human colon cancer: biology and therapeutic implications. Oncologist. 2000;5(suppl 1):11–15. doi: 10.1634/theoncologist.5-suppl_1-11. [DOI] [PubMed] [Google Scholar]
- 17.Hanahan D, Folkman J. Patterns and emerging mechanisms of the angiogenic switch during tumorigenesis. Cell. 1996;86(3):353–364. doi: 10.1016/s0092-8674(00)80108-7. [DOI] [PubMed] [Google Scholar]
- 18.Kuwai T, Kitadai Y, Tanaka S, et al. Expression of hypoxia-inducible factor-1alpha is associated with tumor vascularization in human colorectal carcinoma. Int J Cancer. 2003;105(2):176–181. doi: 10.1002/ijc.11068. [DOI] [PubMed] [Google Scholar]
- 19.Easwaran V, Lee SH, Inge L, et al. beta-Catenin regulates vascular endothelial growth factor expression in colon cancer. Cancer Res. 2003;63(12):3145–3153. [PubMed] [Google Scholar]
- 20.Folkman J. Tumor angiogenesis: therapeutic implications. N Engl J Med. 1971;285(21):1182–1186. doi: 10.1056/NEJM197111182852108. [DOI] [PubMed] [Google Scholar]
- 21.Korsisaari N, Kasman IM, Forrest WF, et al. Inhibition of VEGF-A prevents the angiogenic switch and results in increased survival of Apc+/min mice. Proc Natl Acad Sci U S A. 2007;104(25):10625–10630. doi: 10.1073/pnas.0704213104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gutierrez LS, Suckow M, Lawler J, Ploplis VA, Castellino FJ. Thrombospondin 1—a regulator of adenoma growth and carcinoma progression in the APC(Min/+) mouse model. Carcinogenesis. 2003;24(2):199–207. doi: 10.1093/carcin/24.2.199. [DOI] [PubMed] [Google Scholar]
- 23.Oshima M, Murai N, Kargman S, et al. Chemoprevention of intestinal polyposis in the Apcdelta716 mouse by rofecoxib, a specific cyclooxygenase-2 inhibitor. Cancer Res. 2001;61(4):1733–1740. [PubMed] [Google Scholar]
- 24.Senger DR, Van de Water L, Brown LF, et al. Vascular permeability factor (VPF, VEGF) in tumor biology. Cancer Metastasis Rev. 1993;12(3–4):303–324. doi: 10.1007/BF00665960. [DOI] [PubMed] [Google Scholar]
- 25.Roskoski R., Jr Vascular endothelial growth factor (VEGF) signaling in tumor progression. Crit Rev Oncol Hematol. 2007;62(3):179–213. doi: 10.1016/j.critrevonc.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 26.Yoo YG, Kong G, Lee MO. Metastasis-associated protein 1 enhances stability of hypoxia-inducible factor-1alpha protein by recruiting histone deacetylase 1. EMBO J. 2006;25(6):1231–1241. doi: 10.1038/sj.emboj.7601025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chang CC, Lin MT, Lin BR, et al. Effect of connective tissue growth factor on hypoxia-inducible factor 1alpha degradation and tumor angiogenesis. J Natl Cancer Inst. 2006;98(14):984–995. doi: 10.1093/jnci/djj242. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.