Summary
Adaptor protein 4 (AP-4) is the most recently discovered and least well-characterized member of the family of heterotetrameric adaptor protein (AP) complexes that mediate sorting of transmembrane cargo in post-Golgi compartments. Herein we report the interaction of an YKFFE sequence from the cytosolic tail of the Alzheimer’s Disease amyloid precursor protein (APP) with the μ4 subunit of AP-4. Biochemical and X-ray crystallographic analyses reveal that the properties of the APP sequence and the location of the binding site on μ4 are distinct from those of other signal-adaptor interactions. Disruption of the APP-AP-4 interaction decreases localization of APP to endosomes and enhances γ-secretase-catalyzed cleavage of APP to the pathogenic amyloid-β peptide. These findings demonstrate that APP and AP-4 engage in a distinct type of signal-adaptor interaction that mediates transport of APP from the trans-Golgi network (TGN) to endosomes, thereby reducing amyloidogenic processing of the protein.
Highlights.
A sorting signal in the cytosolic tail of APP interacts with the μ4 subunit of AP-4
X-ray crystallography reveals that the APP signal binds to a distinct site on μ4
Disruption of the APP-AP-4 interaction decreases APP localization to endosomes
Redistribution of APP enhances γ-secretase-mediated cleavage to amyloid-β peptide
Introduction
Sorting of transmembrane proteins to post-Golgi compartments of the endomembrane system such as endosomes, lysosomes, lysosome-related organelles (LROs) and the basolateral surface of polarized epithelial cells is mediated by interaction of (i) signals in the cytosolic domains of the proteins with (ii) adaptors that are components of protein coats (reviewed by Bonifacino and Traub, 2003). These signals are not strictly conserved sequences but degenerate arrays of amino acids fitting one of several consensus motifs, the most common of which are DXXLL, [DE]XXXL[LI], NPXY and YXXØ (where X is any amino acid, Ø an amino acid with a bulky hydrophobic side chain, and the rest specific amino acids denoted in single-letter code) (Bonifacino and Traub, 2003). A diverse set of monomeric or oligomeric adaptors recognize these signals with specificity that is, in most cases, dictated by the identity of the variable residues. DXXLL and NPXY signals are recognized by monomeric GGA and PTB-domain-containing clathrin adaptors, respectively (Bonifacino and Traub, 2003). [DE]XXXL[LI] and YXXØ signals, on the other hand, interact with the clathrin-associated, heterotetrameric adaptor protein (AP) complexes, AP-1 (γ-β1-μ1-σ1), AP-2 (α-β2-μ2-σ2) and AP-3 (δ-β3-μ3-σ3) (subunit composition in parenthesis) (Bonifacino and Traub, 2003). Biochemical analyses have shown that [DE]XXXL[LI] signals bind to a combination of two subunits (i.e., AP-1 γ-σ1, AP-2 α-σ2 and AP-3 δ-σ3) (Chaudhuri et al., 2007; Doray et al., 2007; Janvier et al., 2003), while YXXØ signals bind to the μ subunit of each AP complex (Ohno et al., 1996; Ohno et al., 1995). The structural bases for these interactions have been elucidated by X-ray crystallography of AP-2 (Kelly et al., 2008; Owen and Evans, 1998). These interactions produce different outcomes depending on the exact sequence and context of the signal, as well as the localization and function of each AP complex (Bonifacino and Traub, 2003). AP-1 has been implicated in bidirectional transport between the trans-Golgi network (TGN) and endosomes, and in sorting to the basolateral surface of polarized epithelial cells. AP-2 mediates rapid internalization of a subset of endocytic receptors from the plasma membrane. Finally, AP-3 directs proteins from endosomes to LROs such as melanosomes (Bonifacino and Traub, 2003; Dell’Angelica, 2009).
Despite these advances in the elucidation of the mechanisms that decode sorting signals in post-Golgi compartments, many aspects of this system remain poorly understood. Particularly tantalizing is the existence of a fourth AP complex, AP -4 (ε-β4-μ4-σ4) (Dell’Angelica et al., 1999; Hirst et al., 1999), w hose function as a signal-recognition adaptor is less well established. AP-4 is expressed in all mammalian cell types examined to date, wherein it localizes to the TGN in an ARF-regulated manner (Boehm et al., 2001; Dell’Angelica et al., 1999; Hirst et al., 1999). Unlike the other AP complexes, AP-4 does not interact with clathrin and is believed to be part of a non-clathrin coat (Dell’Angelica et al., 1999; Hirst et al., 1999). Yeast two- and three-hybrid (Y2H and Y3H, respectively) analyses have show n that AP-4 does not bind most canonical YXXØ and [DE]XXXL[LI] signals (Aguilar et al., 2001; Janvier and Bonifacino, 2005). The only naturally-occurring, canonical signals known to bind to the μ4 subunit of AP-4 are YXXØ-type sequences from the lysosomal membrane proteins, CD63 (Hirst et al., 1999), LAMP-1 (Stephens and Banting, 1998) and LAMP-2a (Aguilar et al., 2001). However, these interactions are exceedingly weak, and depletion of μ4 has no effect on the localization of any of these proteins to lysosomes (Janvier and Bonifacino, 2005; Simmen et al., 2002). Other studies have shown interaction of isolated μ4 or the whole AP-4 complex with several transmembrane proteins targeted to the basolateral surface of polarized epithelial cells (Simmen et al., 2002), to the δ2 orphan glutamate receptor (Yap et al., 2003) and to the transmembrane AMPA glutamate receptor regulatory proteins (TARPs) (Matsuda et al., 2008). However, the exact nature of the signals involved in these interactions and the structural basis for their recognition by AP-4 have not been defined.
We have investigated further the potential signal-recognition function of AP-4, aiming to achieve the same level of structural and functional understanding that is available for other signal-adaptor interactions. Herein we report the discovery of a specific and robust interaction of the sequence, YKFFE, from the cytosolic tail of the Alzheimer’s Disease (AD) amyloid precursor protein (APP) with the C-terminal domain of the AP-4 μ4 subunit. Although the YKFFE sequence fits the minimal consensus for YXXØ signals, mutational and binding analyses reveal unique features that identify it as a distinct type of signal. Moreover, X-ray crystallographic structure determination at near-atomic resolution shows that the YKFFE sequence binds to a site on μ4 that is different from the YXXØ-binding site on the homologous μ2 subunit of AP-2 (Owen and Evans, 1998). We also show that mutation of the YKFFE sequence or depletion of μ4 shifts the distribution of APP from endosomes to the trans-Golgi network (TGN), uncovering a role for AP-4 in TGN-to-endosome transport. Impaired transport of APP to endosomes enhances γ-secretase-catalyzed cleavage of APP to the pathogenic amyloid-β (Aβ) peptide, implying that this proteolytic event predominantly occurs at the TGN or in the late secretory pathway. These findings thus identify a novel type of signal-adaptor interaction that regulates APP trafficking in a manner that reduces amyloidogenic processing of the protein.
Results
An Unusual Sequence Motif Mediates Interaction of the APP Tail with the μ4 Subunit of AP-4
APP is an ubiquitously-expressed type-I transmembrane glycoprotein with a large N-terminal extracellular domain, a single membrane span, and a short C-terminal cytosolic tail (Fig. 1A). After synthesis in the endoplasmic reticulum (ER), APP traffics through the secretory pathway to attain a steady-state distribution that includes the Golgi complex, the plasma membrane and endosomes (Caporaso et al., 1994; Haass et al., 1992). During transport, APP undergoes proteolytic processing through two alternative pathways known as “non-amyloidogenic” and “amyloidogenic” (Small and Gandy, 2006). The amyloidogenic pathway produces the ~4-kDa Aβ peptide (Fig. 1A), which is secreted into the extracellular space and aggregates to form amyloid plaques, a pathogenic hallmark of AD (Small and Gandy, 2006). Because the endopeptidases (i.e., “secretases”) that participate in each pathway have different distributions within the cell, the itinerary followed by APP is a critical determinant of the amount of secreted Aβ. Understanding the mechanisms that control the intracellular trafficking of APP is thus crucial to the elucidation of AD pathogenesis.
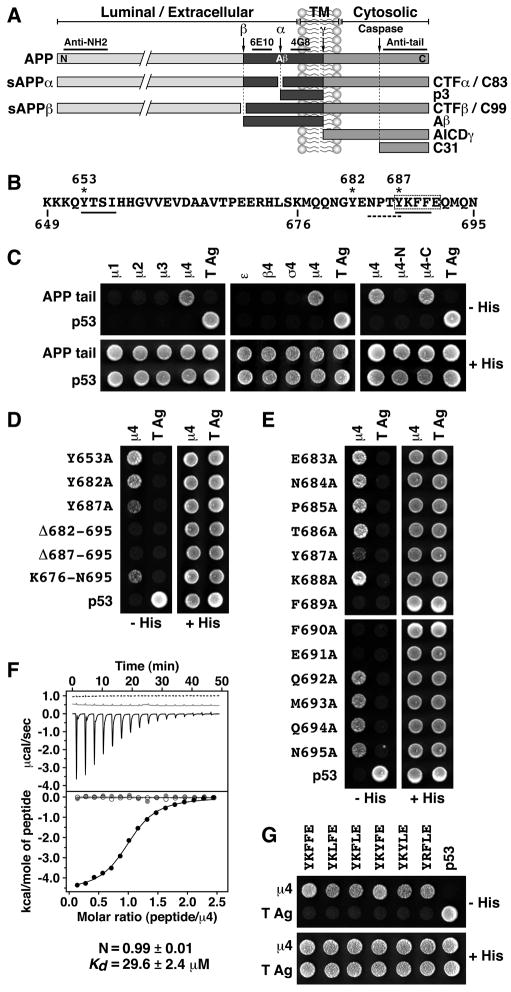
Fig. 1.
(A) Schematic representation of APP indicating its topological domains (TM: transmembrane; N: N -terminus; C: C-terminus), the position of the Aβ peptide, the α, β, γ and caspase cleavage sites, and the fragments produced. Underlined are regions in APP recognized by the antibodies used in this study. (B) Sequence of the APP cytosolic tail indicating residue numbers, two YXXØ motifs (solid underlines), Tyr residues (asterisks), an NPXY-type signal (dashed underline), and key residues for interaction with μ4 (dashed box). (C–E and G) Y2H analysis of the interaction of the APP cytosolic tail with μ4. Yeast were co-transformed with plasmids encoding Gal4bd fused to the wild-type or mutant APP constructs indicated on the left, and Gal4ad fused to the adaptor subunits indicated on top of each panel. Mouse p53 fused to Gal4bd and SV40 large T antigen (T Ag) fused to Gal4ad were used as controls. Co-transformed cells were spotted onto His-deficient (−His) or His-containing (+His) plates and incubated at 30°C. (F) ITC of ENPTYKFFEQ peptide (black line and solid circles) or ENPTAKAAEQ peptide (dashed line and open circles) with μ4, and of ENPTYKFFEQ peptide with μ4-R283D (grey line and circles). The Kd and stoichiometry (N) for the μ4-ENPTYKFFEQ interaction are expressed as the mean ± SEM (n=3).
In the course of a study on APP trafficking, we inspected the cytosolic tail of APP for potential sorting signals. We noticed two YXXØ motifs (YTSI, residues 653–656; YKFF, residues 687–690) (Fig. 1B, solid underlines) in addition to a previously reported (Chen et al., 1990; Perez et al., 1999) NPXY motif (NPTY, residues 684–687) (Fig. 1B, dashed underline). The presence of these YXXØ motifs prompted us to test for interaction of the APP tail with the μ subunits of the four AP complexes using the Y2H system. Strikingly, we found that the APP tail interacted exclusively with the μ4 subunit of AP-4 (Fig. 1C), a unique preference that had not been previously observed for any other cargo protein (Aguilar et al., 2001; Ohno et al., 1996; Ohno et al., 1995). The other three subunits of AP-4 (i.e., ε, β4 and σ4) did not bind to the APP tail in this system (Fig. 1C). Further analyses revealed that the binding site for the APP tail was on the C -terminal domain of μ4 (residues 156–453 of the human protein; Aguilar et al., 2001) (Fig. 1C, μ4-C).
Mutagenesis of the APP tail showed that Tyr-687 within the YKFF sequence was the only Tyr residue that contributed, albeit partially, to μ4 binding (Fig. 1D). Deletion of nine residues from the C-terminus of APP, including Tyr-687, completely abolished binding to μ4 (Fig. 1D, Δ687–695), suggesting a requirement for additional downstream residues. Moreover, the 18 C-terminal residues from the APP tail were sufficient for interaction (Fig. 1D, K676-N695). An Ala scan mutagenesis of this segment revealed an additional requirement of Phe-689, Phe-690 and Glu-691 for interaction (Fig. 1E). Therefore, the APP sequence recognized by μ4 is YKFFE (residues 687–691; dashed box in Fig. 1B), in which the Lys residue is unimportant. This interaction was confirmed in vitro by isotherm al titration calorimetry (ITC) using purified components. We found that a synthetic ENPTYKFFEQ peptide, but not a substituted ENPTAKAAEQ variant, bound to a single site on recombinant μ4 C-terminal domain with Kd = 29.6 ± 2.4 μM (Fig. 1F). Further Y2H analyses showed that Leu could substitute for either of the two Phe, and Tyr for the first Phe, in the YKFFE sequence, with only minor loss of interaction with μ4 (Fig. 1G). Although not exhaustive, these analyses defined a provisional motif for interaction with μ4 as YX[FYL][FL]E. This motif has unique features such as the [FYL] and E residues that distinguish it from other YXXØ-type signals and probably determine specific interaction with μ4. A search of protein sequence databases using this motif as query identified the sequences YKYLE and YRFLE from the cytosolic tails of two other type I transmembrane proteins, APLP1 and APLP2, respectively. We demonstrated experimentally that these two sequences indeed bind to μ4 (Fig. 1G). Notably, APLP1 and APLP2 are APP-related proteins that traffic and are proteolytically processed in a manner similar to APP (Anliker and Muller, 2006), suggesting that AP-4 might be a common adaptor for APP family members.
The YKFFE Sequence from APP Binds to a Novel Sit e on the Surface of μ4
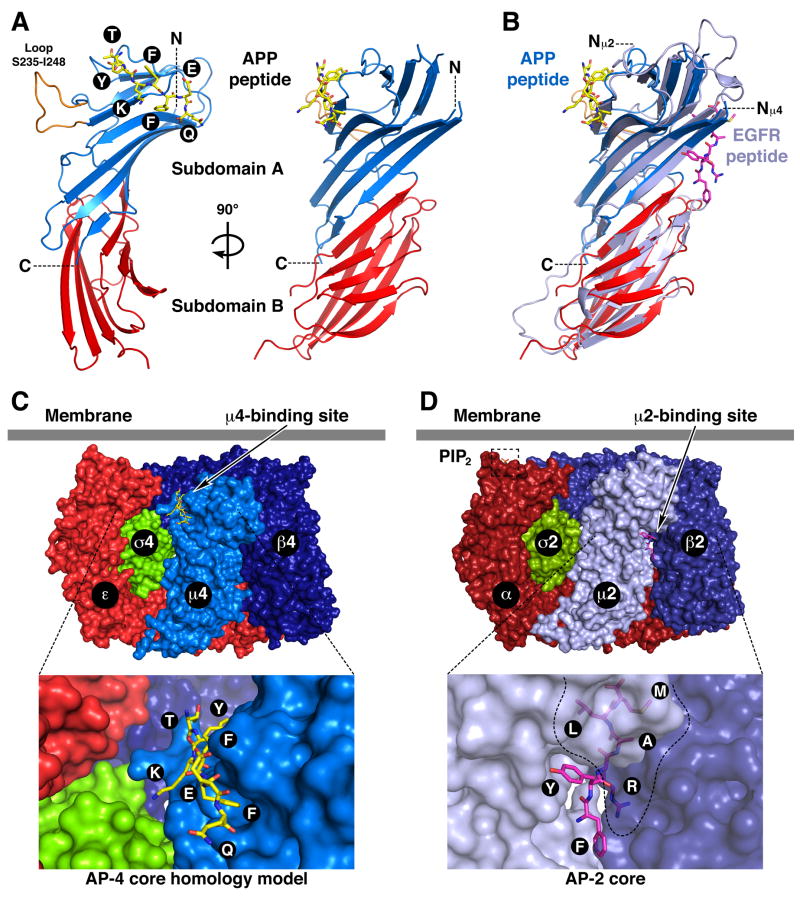
To elucidate the structural bases for the recognition of this unique subtype of YXXØ motif, w e solved the crystal structure of the C-terminal domain of μ4 (residues 185–453 of the human protein) in complex with an ENPTYKFFEQ peptide derived from the APP tail at 1.6 Å resolution (Figs. 2A, 3A, S1A; Table 1). The μ4 C-terminal domain has an immunoglobulin-like beta-sandwich fold consisting of 17 strands organized into two subdomains named A and B (Figs. 2A, S1A), similar to the structure of the C-terminal domain of μ2 (Figs. 2B, S1B) (Owen and Evans, 1998). The overall root mean square deviation for 222 superimposable Cα coordinates for the C-terminal domains of μ4 and μ2 is 1.83 Å. Of the ENPTYKFFEQ peptide, only the TYKFFEQ segment was visible in the density map (Figs. 2A, 3A, S1A). Unexpectedly, this segment was found to bind, in an extended conformation, to strands 4, 5 and 6 of μ4 (Figs. 2A, S1A), whereas YXXØ signals bind to strands 1 and 16 of μ2 (Figs. 2B, S1B) (Owen and Evans, 1998). The signal-binding sites on μ4 and μ2 are thus on opposite faces and separated by 30 Å on the surface of the proteins (Fig. 2B). Moreover, the signal-binding site on μ4 is predicted to be fully accessible for interactions in the context of the whole AP-4 complex core (Fig. 2C), in contrast to that on μ2, which is partially occluded by contacts with β2 in the AP-2 core complex (Fig. 2D) (Collins et al., 2002).
Fig. 2.
Crystal structure of the μ4 C-terminal domain in complex with a peptide signal from APP. (A) Ribbon representation of human μ4 C-terminal domain with subdomain A in blue, subdomain B in red, and the APP peptide (TYKFFEQ; stick model) in yellow. (B) Superposition of μ4 and rat μ2 (with the EGF receptor peptide in magenta; pdb entry 1BW8, Owen and Evans, 1998). In orange (A) is show n a loop that is disordered in μ2. The position of the N- (N, Nμ2 and Nμ4) and C-termini (C) are indicated. (C and D) Model of the AP-4 (C) and structure of the AP-2 core (D; Collins et al. 2002; pdb entry 1GW5) core complexes showing the position of the respective peptide-binding sites and the PIP2–binding site on the AP-2 α subunit. See also Fig. S1.
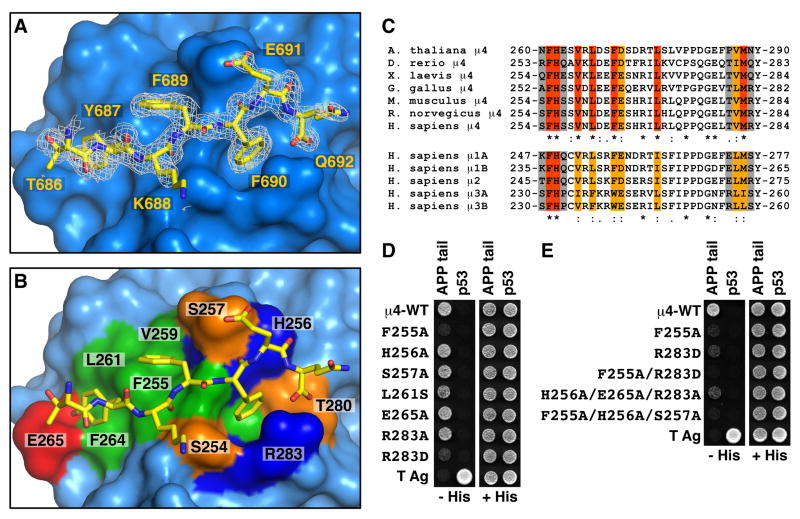
Fig. 3.
Details and verification of the signal-binding site on μ4. (A) Stick representation of the bound peptide TYKFFEQ (shown with carbon atoms colored yellow) superimposed on a 2Fo-Fc omit electron density map contoured at 0.8 σ. (B) Surface complementarity between the peptide and μ4. Surface colors for residues in contact with the TYKFFEQ peptide are green for hydrophobic, red for acidic, blue for basic, and orange for uncharged polar. (C) Alignments of the sequences containing the signal-binding site in μ4 from different species, and of the homologous μ1A, μ1B, μ2, μ3A and μ3B sequences from H. sapiens. Residues in the signal-binding site in human μ4 that are identical (red), conserved (orange), and non-conserved (gray) are indicated. (D, E) Y2H assays involving co-expression of Gal4ad fused to wild-type or mutant μ4, and Gal4bd fused to the APP tail, were performed as described in the legend to Fig. 1. See also Fig. S2.
Table 1.
Data collection, MAD phasing, and crystallographic refinement.
| Crystal | Native | Se-Met | ||
|---|---|---|---|---|
| Construct | H. sapiens μ 4 (160–453; C235S, C431S) | H. sapiens μ4 (160–453; C235S, L424M, C431S) | ||
| Space group | P21 | P21 | ||
| Cell dimension | a=46.7 Å, b=56.9 Å, c=60.7 Å, β =106.5 | a=46.7 Å, b=57.2 Å, c=60.7 Å, β =106.3 | ||
| X-ray source | SER-CAT 22-ID-D | GM/CA-CAT 23-ID-D | ||
| Wavelength (Å) | 1.0000 | Ed ge=0.9794 | Peak=0.9796 | Remote=0.9719 |
| Resolution (Å) | 1.6 (1.66–1.6) | 1.95 (2.02–1.95) | ||
| No. of reflections | 39566 | 22201 | 22222 | 22468 |
| Rmergea(%) | 9.7 (44.5) | 7.6 (28.9) | 6.7 (28.8) | 6.9 (30.4) |
| I/ | 13.8 (1.5) | 18.7 (4.6) | 23.2 (4.6) | 22.4 (4.6) |
| Data completeness (%) | 97.3 (84.2) | 99.4 (97.5) | 99.5 (97.4) | 99.4 (97.6) |
| Redundancy | 4.4 (2.4) | 7.1 (5.7) | 7.1 (5.6) | 7.1 (5.7) |
|
Phasing | ||||
| No. sites | 4 | |||
| FOM | 0.41 | |||
| FOM after DM | 0.66 | |||
|
Refinement | ||||
| Rfactor(%) | 20.7 | |||
| Rfree(%) | 25.3 | |||
| Rms bond angle (°) | 1.731 | |||
| Rms bond length (Å) | 0.015 | |||
The values in parentheses relate to highest resolution shells.
Rmerge = Σ hkl | Ihkl − <Ihkl>|/Σ hkl Ihkl
The area of the interface involving the signal on μ4 is 426 Å2 whereas that of an YXXØ signal from the EGF receptor bound to μ2 is 393 Å2, as calculated by the PISA server (Krissinel and Henrick, 2007). The μ4-APP interface has substantial polar character, with seven direct hydrogen bonds (distance ≤ 3.1 Å) between peptide and protein (Fig. S2). The central portion of the peptide, comprising Tyr-687 to Phe-690, is in a β conformation. Residues 688–690 from the peptide form a β-sheet hydrogen-bonding pattern with the exposed edge of strand β4 spanning μ4 residues 253–257 (Fig. S2A). Where the N-terminus of the peptide pulls away from strand β4, a tightly bound water molecule makes a water-mediated hydrogen bond between the two backbones (Fig. S2A). The phenolic hydroxyl of the peptide Tyr-687 forms the shortest side-chain to side-chain hydrogen bond in the complex with the carboxylate of Glu-265 of μ4 (Fig. S2B). APP Glu-691, the penultimate visible residue in the peptide, forms a direct and a water-mediated hydrogen bond with the side-chain of μ4 Ser-257 (Fig. S2C). The main chain carbonyl group of Glu-691 is stabilized by a hydrogen bond with the side-chain of H is-256 (Fig. S2C). The peptide contains an unblocked C-terminal carboxylate, which forms a bidentate salt bridge with the Arg-283 guanidinium moiety (Fig. S2D).
All three aromatic side-chains in the peptide make substantial contacts with μ4. In addition to hydrogen bonding with μ4 Glu-265, the peptide Tyr-687 forms hydrophobic contacts with Leu-261 (Figs. 3A, 3B, S2B). The peptide Phe-689 is half-buried against the side chains of μ4 Phe-255, Val-259 and Leu-261 (Figs. 3A, 3B). The peptide Phe-690 is deeply buried and surrounded by the hydrocarbon portions of μ4 H is-256, Thr-280, and Arg-283 (Fig. 3A, 3B). The μ4 Arg-283 guanidinium group forms a cation-π interaction with the peptide Phe-690 phenyl ring (Figs. 3A, 3B, S2D). The residues involved in this interaction are well conserved in μ4 orthologs from A. thaliana to H. sapiens and, surprisingly, also in the μ subunits of other AP complexes (Fig. 3C). This conservation of the μ4 binding site, highlighted by analysis using ConSurf (Landau et al., 2005) (Fig. S2E), suggests that other μ subunits might also have a generally similar binding site, albeit with key differences in detail that explain their failure to bind the APP tail.
Consistent with a functional role for the signal-binding site on μ4, single mutation of Phe-255 to Ala, or Arg-283 to Asp, drastically reduced (Fig. 3D), and double mutation of these residues completely abrogated (Fig. 3E), binding to the APP tail in Y2H assays. The single Arg-283 to Asp mutation was sufficient to render the interaction undetectable by ITC (Fig. 1F). Single mutation of H is-256, Glu-265, Arg-283 to Ala had little or no effect on the interaction, but triple mutation of these residues greatly decreased the interaction in Y2H assays (Fig. 3, D and E). Similarly, mutation of Ser-257 to Ala had no detectable effect (Fig. 3D); however, this mutation together with H is-256 and Phe-255 to Ala completely abolished binding to the APP tail (Fig. 3E). This mutational analysis thus validated the identity of the binding site in the μ4 crystal structure as that responsible for the Y2H and in vitro interactions.
Disruption of the YKFFE-μ4 Interaction Shifts the Distribution of APP from Endosomes to the TGN
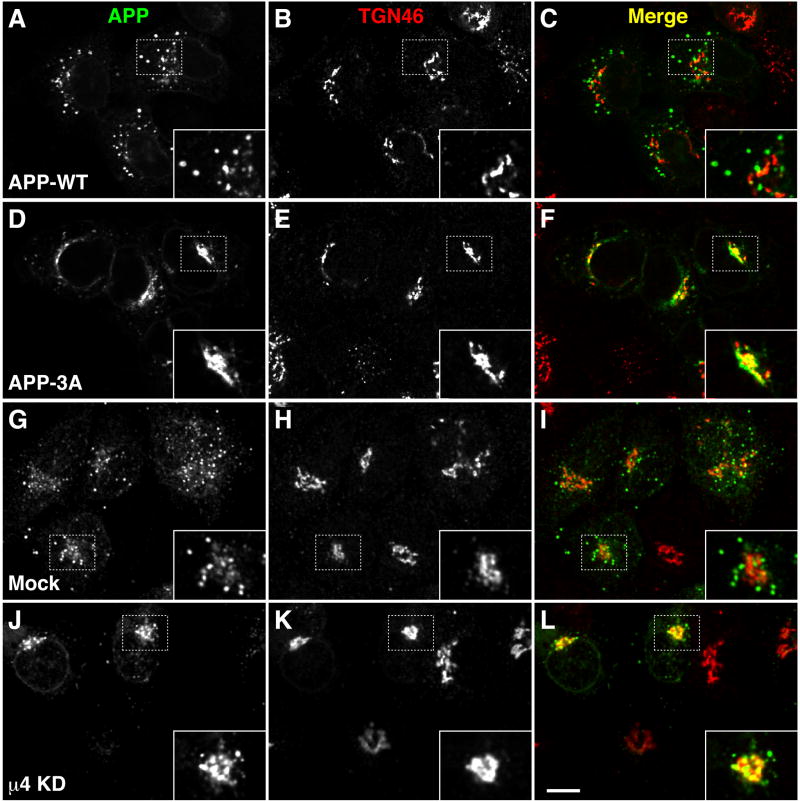
To determine whether the YKFFE-μ4 interaction was functional in vivo, we examined the effect of disrupting it on the intracellular distribution of APP analyzed by immunofluorescence microscopy of HeLa cells. Under normal conditions, APP localized mainly to endosomes (74 ± 8 %, n=11), whereas AP-4 was largely restricted to the TGN (Fig. S3A). We substituted three Ala residues for Phe-689, Phe-690 and Glu-691 in the APP tail in order to disrupt the μ4-interacting sequence, YKFFE (dashed box in Fig. 1B), while leaving intact the PTB-interacting sequence, NPTY (dashed underline in Fig. 1B). We observed that, in contrast to normal APP (APP-WT; Fig. 4, A–C), this mutant protein localized mostly to the TGN (68 ± 6 %, n=15) (APP-3A; Fig. 4, D–F). Similarly to mutation of the FFE sequence, depletion of μ4 by RNAi (Fig. S3B) caused redistribution of APP from endosomes (Fig. 4, G–I) to the TGN (70 ± 6 %, n=14) (Fig. 4, J–L; Fig. S3C), but did not alter the distribution of other transmembrane proteins such as the α chain of the interleukin-2 receptor (Tac) or β-secretase (BACE1) (data not shown). Inhibition of protein synthesis by treatment with cycloheximide (CHX) resulted in rapid disappearance of APP-WT staining in both mock- and μ4-depleted cells (Fig. S3C), indicating that APP localization to endosomes and the TGN reflected transient residence, rather than long-term accumulation, in those compartments. Taken together, these results demonstrated that disruption of the interaction of APP with μ4 caused a shift in the steady-state distribution of APP from endosomes to the TGN, pointing to a role of AP-4 in the transport of APP from the TGN to endosomes. In line with this interpretation, replacement of the μ4-specific YKFFE signal for the YXXØ-type YQRL signal of a Tac-TGN 38 chimera (Humphrey et al., 1993), redirected this protein from the TGN to endosomes (Fig. S3D).
Fig. 4.
Redistribution of APP from endosomes to the TGN upon disruption of its interaction with μ4. (A–F) HeLa cells were transfected with plasmids encoding either APP-CFP (WT) (A–C) or APP-CFP carrying the triple mutation, F689A, F690A and E691A (3A) (D–F). (G–L) HeLa cells were mock-transfected (G–I) or transfected twice with siRNA directed to μ4 (J–L), and then re-transfected with a plasmid encoding APP-CFP. Cells were stained for TGN46 and examined by confocal fluorescence microscopy. Merging red and green channels generated the third picture on each row; yellow indicates overlapping localization. Insets show 2X magnifications. Bars: 10 μm.
Disruption of the YKFFE-μ4 Interaction Decreases CTF Levels and Increases Aβ Secretion Due to Enhanced γ-secretase Cleavage
Next, we examined the effect of disrupting the YKFFE-μ4 interaction on the processing of APP. In the non-amyloidogenic pathway APP is initially cleaved near the middle of the Aβ region by α-secretases, leading to the production of a soluble, secreted N-terminal fragment (sAPPα) and a transmembrane C-terminal fragment (CTFα; also termed C83) (Fig. 1A). In contrast, in the amyloidogenic pathway APP is cleaved at the N-terminus of Aβ by β-secretase, yielding a slightly shorter sAPPβ fragment and a slightly longer CTFβ fragment (also termed C99) (Fig. 1A). Both CTFs are further cleaved within the transmembrane domain by γ-secretase into p3 and AICDγ (for CTFα), and Aβ and AICDγ (for CTFβ) fragments (Fig. 1A).
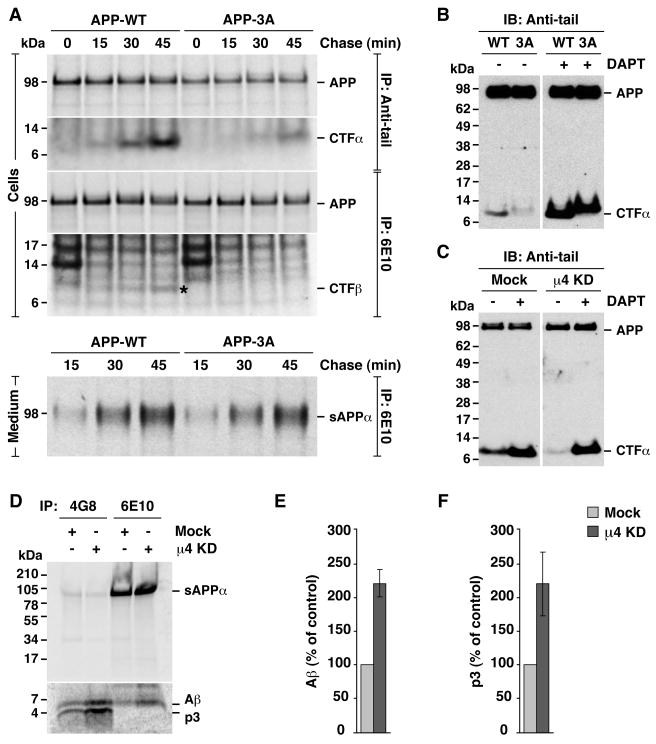
Metabolic labeling, pulse-chase analyses showed that mutation of the FFE sequence had no detectable effect on transport of APP from the ER to the Golgi complex, as evidenced by the normal appearance of a more slowly migrating species bearing complex carbohydrates (Fig. S4, A and B, m). Likewise, the mutation had no effect on APP expression at the cell surface, as assessed by biotinylation (Fig. S4C). Moreover, shedding of the sAPPα ectodomain fragment into the medium (Fig. 5A, medium) was not affected or slightly elevated by the mutation, indicating that cleavage mediated by α-secretases and secretion were not altered. Surprisingly, the levels of the other product of α-secretase cleavage, CTFα, were substantially decreased by the FFE mutation, as observed by both pulse-chase (Fig. 5A, cells) and immunoblot analysis (Fig. 5B) (CTFα derived from APP-3A has an intrinsically reduced electrophoretic mobility due to the three-amino acid substitution). Depletion of μ4 by RNAi also decreased the levels of CTFα, as detected by immunoblot analysis (Fig. 5C).
Fig. 5.
Disruption of the APP-μ4 interaction alters CTF levels and p3/ Aβ secretion. (A) HeLa cells transfected with HA-tagged wild-type APP (APP-WT) or APP with the triple mutation, F689A, F690A and E691A (APP-3A), were labeled for 15 min with [35S]methionine-cysteine and chased for 0–45 min at 37°C. APP species were immunoprecipitated (IP) from cell lysates or culture media with the indicated antibodies. Proteins were analyzed by SDS-PAGE and fluorography. (B–C) Anti-tail immunoblot (IB) analysis of APP and CTF species from cells expressing APP-WT or APP-3A (B), or from mock- or μ4-knockd own (KD) cells expressing APP-WT (C), in the absence (−) or presence (+) of 250 nM γ-secretase inhibitor, DAPT. (D) Mock- or μ4-KD cells expressing APP-WT were labeled for 4 h at 37°C with [35S]methionine-cysteine, and the culture medium was subjected to immunoprecipitation with antibodies 4G8 (recognizing both p3 and Aβ) and 6E10 (recognizing only Aβ). Proteins were analyzed by electrophoresis on Tricine 10–20% acrylamide gradient gels and fluorography. The positions of molecular mass markers are indicated on the left. (E, F) Mean ± SD from three experiments like that in D. Control: mock. See also Fig. S4.
Because the amyloidogenic pathway is minor in most cells, the products of β-secretase-mediated APP cleavage are more difficult to detect. Despite higher backgrounds, however, use of an antibody to an Aβ epitope (i.e., 6E10; Fig. 1A) revealed that mutation of the FFE sequence also decreased the levels of CTFβ (Fig. 5A, cells, asterisk). To enhance the production of CTFβ and thus increase the sensitivity of the assay, we introduced the pathogenic Swedish double mutation, K595N/M596L (Citron et al., 1992), in both APP-WT and APP-3A, and co-expressed the mutant proteins with excess β-secretase (BACE1) (Fig. S4D, left panel). Under these conditions, we also observed that mutation of the FFE sequence reduced the levels of CTFβ (Fig. S4D, right panel).
How could the levels of CTFα be reduced if α-secretase cleavage was not affected by the FFE mutation? We reasoned that turnover of CTFα by γ-secretase must be enhanced. Indeed, addition of the γ-secretase inhibitor, DAPT, abrogated the decrease in steady-state CTFα levels elicited by either mutation of the FFE sequence (Fig. 5B) or depletion of μ4 (Fig. 5C). Furthermore, depletion of μ4 increased the secretion of the p3 and Aβ peptides into the culture medium (Fig. 5, D–F), consistent with enhanced γ-cleavage caused by disruption of the YKFFE-μ4 interaction.
Increased AICDγ Production and Decreased Caspase Cleavage Caused by Disruption of the YKFFE-μ4 Interaction
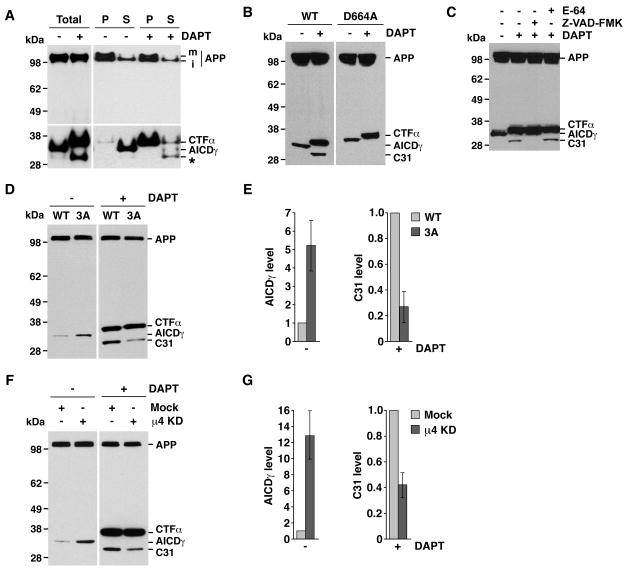
Enhanced γ-secretase cleavage should also lead to increased production of AICDγ (Fig. 1A). This fragment is normally difficult to detect because of its rapid turnover, but can be partially stabilized by addition of a C-terminal tag (Kaether et al., 2006). To test our prediction, we expressed in HeLa cells an APP construct tagged at the C-terminus with the cyan fluorescent protein (APP-CFP). Immunoblot analysis using an antibody to GFP revealed the presence of two bands in the total lysates (Fig. 6A, first lane). The upper band, enriched in the pellet (P) fraction, corresponded to membrane-bound, full-length APP-CFP (Fig. 6A). The lower band was more abundant in the soluble (S) fraction, as expected for cytosolic AICDγ-CFP (Fig. 6A). Inhibition of γ-secretase by DAPT precluded AICDγ-CFP formation and increased CTFα-CFP levels (Fig. 6A). Interestingly, DAPT treatment also increased the levels of a smaller, soluble fragment (Fig. 6A, asterisk). Formation of this fragment in DAPT-treated cells was prevented by mutation of Asp-664 in the APP tail (Fig. 6B) or by incubation with the inhibitor Z-VAD-FMK (Fig. 6C), properties that identify it as the previously described C31 caspase cleavage product (Lu et al., 2000).
Fig. 6.
Disruption of the APP-μ4 interaction alters AICDγ and C31 levels. All experiments were performed with HeLa cells expressing normal or mutant APP constructs tagged with CFP, incubated in the absence (−) or presence (+) of DAPT, and analyzed by SDS-PAGE and immunoblotting with antibody to GFP, with the variations indicated below. The positions of molecular mass markers and different APP species are indicated. (A) Cells expressing APP-CFP were homogenized to obtain pellet (P) and supernatant (S) fractions. m, mature; i, immature. (B) Cells expressing normal APP-CFP (WT) or APP-CFP carrying a D664A mutation. (C) Cells expressing normal APP-CFP were treated with 100 μM E-64, 200 μM Z-VAD-FMK and / or 250 nM DAPT. (D) Cells expressing normal APP-CFP (WT) or APP-CFP with the F689A, F690A and E691A mutation (3A). (E) Mean ± SD of AICDγ and C31 levels from three experiments such as that shown in D. (F) Mock- or μ4-depleted cells (μ4 KD) expressing APP-CFP. (G) Mean ± SD of AICDγ and C31 levels from three experiments such as that show n in F. See also Fig. S5.
Using this experimental setup, we found that mutation of the FFE sequence in APP (Fig. 6, D–E) or depletion of μ4 (Fig. 6, F–G) indeed increased AICDγ-CFP levels. This difference was abolished by incubation with DAPT, resulting in the accumulation of equal amounts of CTFs under all conditions (Fig. 6, D–G). Intriguingly, the levels of C31 fragment in DAPT-treated cells decreased upon disruption of this interaction for both normal APP (Fig. 6, D–G) and the APP Swedish mutant (Fig. S5A). Taken together, the results of the above biochemical analyses indicated that disruption of the YKFFE-μ4 interaction enhances γ-secretase cleavage of APP, resulting in decreased levels of both CTFα and CTFβ and elevated levels of the corresponding p3/Aβ and AICDγ products. In addition, disruption of this interaction decreases production of the C31 caspase cleavage product.
Similar results were obtained in experiments with H4 neuroglioma cells (Fig. S3C, Fig. S4, E–F, Fig. S5B), another well established model system for biochemical analysis of APP processing (Xie et al., 2005).
Non-amyloidogenic Disposal of CTF Fragments Promoted by AP-4
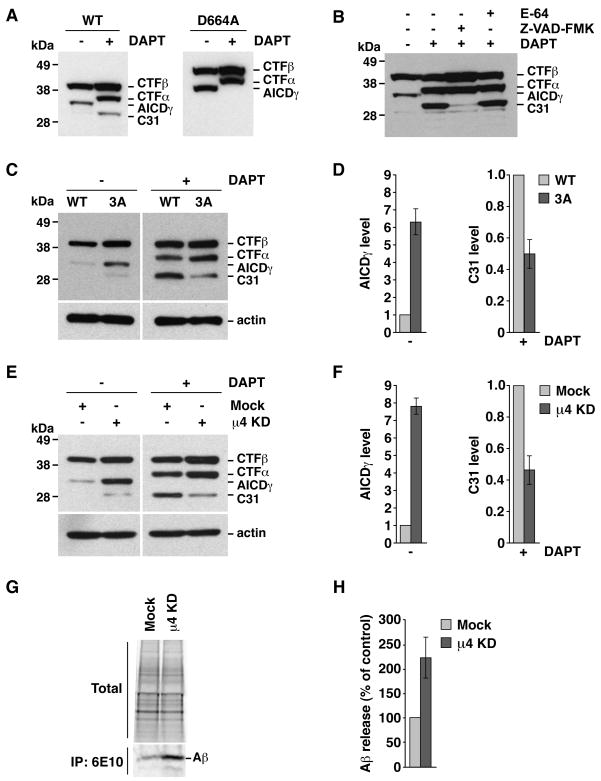
Because the substrate of γ-secretase is not full-length APP but the CTFs (Xia et al., 2000), we examined the effect of disrupting the YKFFE-μ4 interaction on the fate of a recombinant CTFβ construct that mimics β-secretase-cleaved APP (Kaether et al., 2006). Immunoblotting of HeLa cells expressing CTFβ tagged with GFP showed two species corresponding to intact CTFβ and AICDγ, indicating that this construct is indeed processed by γ-secretase (Fig. 7A). Treatment with DAPT prevented AICDγ formation and resulted in the appearance of CTFα and C31 (Fig. 7A). The identity of this latter fragment was confirmed by its absence upon mutation of the caspase cleavage site residue, Asp-664 (numbering corresponds to full-length APP) (Fig. 7A), or incubation with Z-VAD-FMK (Fig. 7B). Thus, CTFβ is a substrate for α-secretases and caspase. Mutation of the YKFFE sequence (Fig. 7, C–D) or depletion of μ4 (Fig. 7, E–F) increased the levels of AICDγ and decreased the levels of C31 generated from CTFβ. In addition, μ4 depletion increased Aβ secretion into the medium (Fig. 7, G–H). Thus, processing of CTFβ by γ-secretase and caspase is also dependent on the YKFFE-μ4 interaction, demonstrating a role for AP-4 in the sorting of CTFβ.
Fig. 7.
Altered processing of CTFβ upon disruption of the YKFFE-μ4 interaction. Experiments were performed as in Fig. 6, with the variations indicated below. The positions of molecular mass markers and different APP species are indicated. (A) Cells expressing normal CTFβ-GFP (WT) or CTFβ-GFP carrying a D664A mutation. (B) Cells expressing normal CTFβ-GFP were treated with 100 μM E-64, 200 μM Z-VAD-FMK and / or 250 nM DAPT. (C) Cells expressing normal CTFβ-GFP (WT) or CTFβ-GFP with the triple mutation, F689A, F690A and E691A (3A). (D) Mean ± SD of AICDγ and C31 levels from three experiments such as that shown in C. (E) Mock- or μ4-depleted cells (μ4 KD) expressing CTFβ-GFP. (F) Mean ± SD of AICDγ and C31 levels from three experiments such as that shown in E. (G) Mock- or μ4-depleted cells expressing normal CTFβ-GFP were labeled for 4 h at 37°C with [35S]methionine-cysteine, and the culture medium was subjected to immunoprecipitation with antibody 6E10 to Aβ. Proteins were analyzed by electrophoresis on Tricine 10–20% acrylamide gradient gels and fluorography. An aliquot of the culture media (total) is shown as loading control. (H) Mean ± SD from four experiments like that in G. Control: mock.
Discussion
The recognition of the YX[FYL][FL]E motif from the cytosolic tails of APP family members by the μ4 subunit of AP-4 represents a novel type of signal-adaptor interaction, characterized by the distinct properties of both the motif and the binding site.
The YX[FYL][FL]E motif encompasses an YXXØ motif, characteristic of signals that bind to the μ1, μ2 and μ3 subunits of the corresponding AP complexes (Ohno et al., 1996; Ohno et al., 1995). However, the YKFFE sequence from APP does not bind to these μ subunits, instead binding to the μ4 subunit of AP-4. This unusual recognition pattern stems from the distinctive features of the YX[FYL][FL]E motif. First, the N-terminal Tyr residue (herein designated position 0 to facilitate discussion of the data) is quantitatively important but not essential for interaction with μ4. Second, the presence of Phe, Tyr or Leu residues at position +2 is required for interaction with μ4, but strongly disfavors interactions with μ1, μ2 and μ3 (Ohno et al., 1998). This particular feature likely determines the lack of interaction of the APP tail with μ subunits other than μ4. Finally, another key determinant of specific recognition by μ4 is an additional Glu residue at position +4. Hence, the new motif defined here is a pentapeptide in which the [FYL], [FL] and E positions are more critical than the Tyr at position 0 for interaction with AP-4.
Other proteins reported to interact with AP-4 (Matsuda et al., 2008; Simmen et al., 2002; Yap et al., 2003) do not have sequences fitting the YX[FYL][FL]E motif. However, some of these proteins have scattered Tyr and Phe residues that are required for interaction with μ4 (Matsuda et al., 2008; Yap et al., 2003). In addition, a combinatorial screen for random sequences that bind to μ4 revealed a strong preference for peptides enriched in Tyr and Phe, most often as a pair (Aguilar et al., 2001). This indicates that Tyr and Phe residues in configurations other than YX[FYL][FL]E might also participate in AP-4 binding. It remains to be determined whether these represent variations on the motif or entirely different interaction determinants.
Given the unique features of the YX[FYL][FL]E motif, we expected that its binding site on μ4 would exhibit substantial differences relative to that of YXXØ signals on μ2 (Owen and Evans, 1998). We were surprised, however, that the μ4 binding site was distinct not just in terms of the residues and interactions involved, but also in its location on a face opposite to that of the μ2 binding site. This location of the μ4 binding site has important implications for the mechanism of signal recognition by AP-4. In the crystal structure of the AP-2 core, the signal-binding site on μ2 is partially occluded by contacts with the β2 subunit (Collins et al., 2002). Therefore, binding of YXXØ signals to μ2 requires a conformational change to displace the C-terminal domain of μ2 from the core and thus expose the signal-binding site (Collins et al., 2002). This conformational change could be triggered by phosphorylation of residues in the hinge that links the N- and C-terminal domains of μ2 (Olusanya et al., 2001; Ricotta et al., 2002). In contrast, modeling of the AP-4 core after the homologous AP-2 core predicts that the signal-binding site on μ4 is fully exposed on the surface of the complex and therefore does not require displacement of the C-terminal domain for binding. Hence, the signal-adaptor interactions involving μ2 and μ4 might also differ on how they are regulated.
A key criterion to establish the functional relevance of a signal-adaptor interaction in vivo is that its disruption should alter the localization and trafficking of signal-bearing cargo proteins. To test this for the interaction described here, we disrupted it by either mutating the signal or depleting cells of μ4. Similar results were obtained with both manipulations, ruling out indirect effects. A visible consequence of this disruption was a greatly reduced localization of APP to endosomes and increased labeling of the TGN. This labeling rapidly disappeared upon treatment of the cells with cycloheximide, indicating that it is not due to accumulation of APP within the TGN but to transient residence in this compartment prior to transport to the cell surface and secretase-mediated breakdown. Faint residual staining of endosomes could be seen in some cells upon disruption of the YKFFE-μ4 interaction; this could result from sorting mediated by interaction of the NPXY motif in APP with PTB-domain containing proteins at the plasma membrane or the TGN (Small and Gandy, 2006). Since AP-4 has been localized to the TGN by both immunofluorescence (Dell’Angelica et al., 1999; Hirst et al., 1999; Simmen et al., 2002) and immunoelectron microscopy (Hirst et al., 1999), the most parsimonious interpretation of our data is that AP-4 mediates biosynthetic sorting of a population of APP and/or CTFs from the TGN to endosomes.
Since the processing of APP by the non-amyloidogenic and amyloidogenic pathways is highly dependent on its intracellular itinerary (Small and Gandy, 2006), we anticipated that altering the trafficking of APP by perturbation of its interaction with AP-4 could change its processing pattern. We found that disruption of the YKFFE-μ4 interaction had no effect on APP export from the ER to the Golgi complex, transport to the plasma membrane, and α-secretase-catalyzed production of sAPPα, which occurs on the way from the TGN to the plasma membrane or at the plasma membrane itself (Schobel et al., 2008; Skovronsky et al., 2000). This disruption, however, caused enhanced cleavage of CTF products by γ-secretase, an enzyme that acts at various intracellular compartments, including the TGN, endosomes and the plasma membrane (Baulac et al., 2003; Kaether et al., 2006; Xia et al., 2000). As a consequence, there w as increased production of p3 and AICDγ fragments, and, most significantly, of the increased production of p3 and AICDγ fragments, and, most significantly, of the pathogenic Aβ peptide.
To date, the precise intracellular location where Aβ is generated remains controversial. Some studies have led to the conclusion that Aβ is produced in the Golgi complex and/or the late secretory pathway (Thinakaran et al., 1996; Xia et al., 2000; Xu et al., 1997), a notion that is consistent with the localization of at least a fraction of the β- and γ-secretases to this part of the pathway (Baulac et al., 2003; Skovronsky et al., 2000; Vassar et al., 1999; Xia et al., 2000) and with the detection of complexes between CTFs and γ-secretase in Golgi/TGN fractions (Xia et al., 2000). Others, however, have presented evidence in favor of Aβ production in endosomes (Haass et al., 1992; Koo and Squazzo, 1994; Perez et al., 1999; Perez et al., 1996). Our observations support the former scenario, by showing that decreased endosomal localization of APP results in increased γ-secretase-mediated cleavage and Aβ secretion.
AP-4-mediated diversion from the secretory pathway might serve the purpose of sequestering a fraction of APP or, more likely, removing CTFs away from γ-secretase, thus reducing the generation of Aβ. This diversion is likely a step towards the eventual transport of APP and/or CTFs to lysosomes for degradation by acid hydrolases (Caporaso et al., 1992; Haass et al., 1992). Our results also indicate that, conversely to Aβ, the C31 caspase-cleavage product of APP (Lu et al., 2000) arises upon AP-4-dependent transport to endosomes, consistent with the presence of caspase activity in association with endosomes (Cosulich et al., 1997).
In conclusion, our results suggest that AP-4 exerts a protective effect, guarding against excessive Aβ production. Altered expression of other trafficking factors has been associated with increased Aβ production and greater risk of AD (Small and Gandy, 2006). Defects in AP-4 should therefore be considered another potential risk factor for AD.
Experimental Procedures
Recombinant DNAs, Site-directed Mutagenesis and Y2H Assays
For all APP constructs generated in this study, a cDNA encoding an N-terminal, HA-tagged full-length human APP695 (GeneCopoeia, Germantown, MD) was used as template. To generate a GST-fusion construct with the C-terminal domain of human μ4, the sequence encoding residues 160–453 was obtained by PCR amplification and cloned in-frame into the EcoRI and SalI sites of pGST-Parallel-1 (Sheffield et al., 1999). The substitutions C235S and C431S were introduced to produce protein for ITC and protein crystallization. Single or multiple amino acid substitutions and stop codons were intriduced using the QuikChange mutagenesis kit (Stratagene, La Jolla, CA). Y2H assays were performed as previously described (Aguilar et al., 2001; Ohno et al., 1996; Ohno et al., 1995).
Transfection and RNAi
Transfections were carried out using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) for 1 h at 37°C in the absence of FBS. Cells were analyzed 8–16 h after transfection. For some experiments, cells were incubated for 16 h with 250 nM DAPT (Sigma-Aldrich, St. Louis, MO), 100 μM E-64 (EMD Chemicals, Gibbstown, NJ) or 200 μM Z-VAD-FMK (EMD Chemicals). RNAi of human μ4 was performed using the siRNA GUCUCGUUUCACAGCUCUG (Dharmacon, Lafayette, CO) (Janvier and Bonifacino, 2005).
Antibodies
Mouse monoclonal antibodies used were: 22C11 to N-terminal region of APP (Millipore, Billerica, MA); 4G8 and 6E10 to APP (Covance, Dedham, MA); clone 32 to the ε subunit of AP-4, and clone 14 to EEA1 (BD Biosciences, San Diego, CA); 7G7 to a Tac lumenal epitope (Humphrey et al., 1993); AC-40 to β-actin (Sigma-Aldrich); anti-GFP conjugated to horseradish peroxidase (Miltenyi Biotec, Auburn, CA). Polyclonal antibodies used were used: rabbit anti-GFP (R. Hegde, NICHD, NIH); rabbit anti-ε subunit of AP-4 (W. Smith, NICHD, NIH); rabbit anti-μ4 subunit of AP-4 (M. S. Robinson, University of Cambridge, UK); rabbit anti-APP tail (CT695, Invitrogen); rabbit anti-HA epitope (ABR-OPA1-10980; Affinity BioReagents, Golden, CO); sheep anti-TGN 46 and -TGN38 (Serotec, Raleigh, NC); human anti-EEA1 (A. González, Pontificia Universidad Católica, Chile).
Other Procedures
Immunofluorescence microscopy, SDS–PAGE, immunoblotting, immunoprecipitation and fluorography were performed as previously described (Mardones et al., 2007) (see also Supplemental Experimental Procedures).
Supplementary Material
Acknowledgments
We thank X. Zhu, and N. Tsai for excellent technical assistance, R. Doms, C. Dotti, A. González, C. Haass, R. Hegde, M. S. Robinson, and W. Smith for gifts of reagents, J. Sayer, J. Louis, and G. M. Clore for use of the iTC200 microcalorimeter, N. Noinaj for collecting the native data set, and the staffs of the SER-CAT and GM/CA-CAT beam lines at the APS, Argonne National Laboratory, for assistance with X-ray data collection. GM/CA-CAT has been funded in whole or in part with funds from the National Cancer Institute (Y1-CO-1020) and the National Institute of General Medical Science (Y1-GM-1104). Use of the APS was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract No. DE-AC02-06CH 11357. This work was funded by the Intramural Programs of NICHD and NIDDK, NIH.
Footnotes
Accession Number
Crystallographic coordinates have been deposited in the Protein Data Bank with accession code 3L81.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aguilar RC, Boehm M, Gorshkova I, Crouch RJ, Tomita K, Saito T, Ohno H, Bonifacino JS. Signal-binding specificity of the mu4 subunit of the adaptor protein complex AP-4. J Biol Chem. 2001;276:13145–13152. doi: 10.1074/jbc.M010591200. [DOI] [PubMed] [Google Scholar]
- Anliker B, Muller U. The functions of mammalian amyloid precursor protein and related amyloid precursor-like proteins. Neurodegener Dis. 2006;3:239–246. doi: 10.1159/000095262. [DOI] [PubMed] [Google Scholar]
- Baulac S, LaVoie MJ, Kimberly WT, Strahle J, Wolfe MS, Selkoe DJ, Xia W. Functional gamma-secretase complex assembly in Golgi/trans-Golgi network: interactions among presenilin, nicastrin, Aph1, Pen-2, and gamma-secretase substrates. Neurobiol Dis. 2003;14:194–204. doi: 10.1016/s0969-9961(03)00123-2. [DOI] [PubMed] [Google Scholar]
- Boehm M, Aguilar RC, Bonifacino JS. Functional and physical interactions of the adaptor protein complex AP-4 with ADP-ribosylation factors (ARFs) Embo J. 2001;20:6265–6276. doi: 10.1093/emboj/20.22.6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino JS, Traub LM. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu Rev Biochem. 2003;72:395–447. doi: 10.1146/annurev.biochem.72.121801.161800. [DOI] [PubMed] [Google Scholar]
- Caporaso GL, Gandy SE, Buxbaum JD, Greengard P. Chloroquine inhibits intracellular degradation but not secretion of Alzheimer beta/A4 amyloid precursor protein. Proc Natl Acad Sci U S A. 1992;89:2252–2256. doi: 10.1073/pnas.89.6.2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso GL, Takei K, Gandy SE, Matteoli M, Mundigl O, Greengard P, De Camilli P. Morphologic and biochemical analysis of the intracellular trafficking of the Alzheimer beta/A4 amyloid precursor protein. J Neurosci. 1994;14:3122–3138. doi: 10.1523/JNEUROSCI.14-05-03122.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri R, Lindwasser OW, Smith WJ, Hurley JH, Bonifacino JS. Downregulation of CD4 by human immunodeficiency virus type 1 Nef is dependent on clathrin and involves direct interaction of Nef with the AP2 clathrin adaptor. J Virol. 2007;81:3877–3890. doi: 10.1128/JVI.02725-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WJ, Goldstein JL, Brown MS. NPXY, a sequence often found in cytoplasmic tails, is required for coated pit-mediated internalization of the low density lipoprotein receptor. J Biol Chem. 1990;265:3116–3123. [PubMed] [Google Scholar]
- Citron M, Oltersdorf T, Haass C, McConlogue L, Hung AY, Seubert P, Vigo-Pelfrey C, Lieberburg I, Selkoe DJ. Mutation of the beta-amyloid precursor protein in familial Alzheimer’s disease increases beta-protein production. Nature. 1992;360:672–674. doi: 10.1038/360672a0. [DOI] [PubMed] [Google Scholar]
- Collins BM, McCoy AJ, Kent HM, Evans PR, Owen DJ. Molecular architecture and functional model of the endocytic AP2 complex. Cell. 2002;109:523–535. doi: 10.1016/s0092-8674(02)00735-3. [DOI] [PubMed] [Google Scholar]
- Cosulich SC, Horiuchi H, Zerial M, Clarke PR, Woodman PG. Cleavage of rabaptin-5 blocks endosome fusion during apoptosis. Embo J. 1997;16:6182–6191. doi: 10.1093/emboj/16.20.6182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell’Angelica EC. AP-3-dependent trafficking and disease: the first decade. Curr Opin Cell Biol. 2009 doi: 10.1016/j.ceb.2009.04.014. [DOI] [PubMed] [Google Scholar]
- Dell’Angelica EC, Mullins C, Bonifacino JS. AP-4, a novel protein complex related to clathrin adaptors. J Biol Chem. 1999;274:7278–7285. doi: 10.1074/jbc.274.11.7278. [DOI] [PubMed] [Google Scholar]
- Doray B, Lee I, Knisely J, Bu G, Kornfeld S. The gamma/sigma1 and alpha/sigma2 hemicomplexes of clathrin adaptors AP-1 and AP-2 harbor the dileucine recognition site. Mol Biol Cell. 2007;18:1887–1896. doi: 10.1091/mbc.E07-01-0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haass C, Koo EH, Mellon A, Hung AY, Selkoe DJ. Targeting of cell-surface beta-amyloid precursor protein to lysosomes: alternative processing into amyloid-bearing fragments. Nature. 1992;357:500–503. doi: 10.1038/357500a0. [DOI] [PubMed] [Google Scholar]
- Hirst J, Bright NA, Rous B, Robinson MS. Characterization of a fourth adaptor-related protein complex. Mol Biol Cell. 1999;10:2787–2802. doi: 10.1091/mbc.10.8.2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey JS, Peters PJ, Yuan LC, Bonifacino JS. Localization of TGN38 to the trans-Golgi network: involvement of a cytoplasmic tyrosine-containing sequence. J Cell Biol. 1993;120:1123–1135. doi: 10.1083/jcb.120.5.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janvier K, Bonifacino JS. Role of the endocytic machinery in the sorting of lysosome-associated membrane proteins. Mol Biol Cell. 2005;16:4231–4242. doi: 10.1091/mbc.E05-03-0213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janvier K, Kato Y, Boehm M, Rose JR, Martina JA, Kim BY, Venkatesan S, Bonifacino JS. Recognition of dileucine-based sorting signals from HIV-1 Nef and LIMP-II by the AP-1 gamma-sigma1 and AP-3 delta-sigma3 hemicomplexes. J Cell Biol. 2003;163:1281–1290. doi: 10.1083/jcb.200307157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaether C, Schmitt S, Willem M, Haass C. Amyloid precursor protein and Notch intracellular domains are generated after transport of their precursors to the cell surface. Traffic. 2006;7:408–415. doi: 10.1111/j.1600-0854.2006.00396.x. [DOI] [PubMed] [Google Scholar]
- Kelly BT, McCoy AJ, Spate K, Miller SE, Evans PR, Honing S, Owen DJ. A structural explanation for the binding of endocytic dileucine motifs by the AP2 complex. Nature. 2008;456:976–979. doi: 10.1038/nature07422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo EH, Squazzo SL. Evidence that production and release of amyloid beta-protein involves the endocytic pathway. J Biol Chem. 1994;269:17386–17389. [PubMed] [Google Scholar]
- Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Landau M, Mayrose I, Rosenberg Y, Glaser F, Martz E, Pupko T, Ben-Tal N. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 2005;33:W299–W302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu DC, Rabizadeh S, Chandra S, Shayya RF, Ellerby LM, Ye X, Salvesen GS, Koo EH, Bredesen DE. A second cytotoxic proteolytic peptide derived from amyloid beta-protein precursor. Nat Med. 2000;6:397–404. doi: 10.1038/74656. [DOI] [PubMed] [Google Scholar]
- Mardones GA, Burgos PV, Brooks DA, Parkinson-Lawrence E, Mattera R, Bonifacino JS. The trans-Golgi network accessory protein p56 promotes long-range movement of GGA/clathrin-containing transport carriers and lysosomal enzyme sorting. Mol Biol Cell. 2007;18:3486–3501. doi: 10.1091/mbc.E07-02-0190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda S, Miura E, Matsuda K, Kakegawa W, Kohda K, Watanabe M, Yuzaki M. Accumulation of AMPA receptors in autophagosomes in neuronal axons lacking adaptor protein AP-4. Neuron. 2008;57:730–745. doi: 10.1016/j.neuron.2008.02.012. [DOI] [PubMed] [Google Scholar]
- Ohno H, Aguilar RC, Yeh D, Taura D, Saito T, Bonifacino JS. The medium subunits of adaptor complexes recognize distinct but overlapping sets of tyrosine-based sorting signals. J Biol Chem. 1998;273:25915–25921. doi: 10.1074/jbc.273.40.25915. [DOI] [PubMed] [Google Scholar]
- Ohno H, Fournier MC, Poy G, Bonifacino JS. Structural determinants of interaction of tyrosine-based sorting signals with the adaptor medium chains. J Biol Chem. 1996;271:29009–29015. doi: 10.1074/jbc.271.46.29009. [DOI] [PubMed] [Google Scholar]
- Ohno H, Stewart J, Fournier MC, Bosshart H, Rhee I, Miyatake S, Saito T, Gallusser A, Kirchhausen T, Bonifacino JS. Interaction of tyrosine-based sorting signals with clathrin-associated proteins. Science. 1995;269:1872–1875. doi: 10.1126/science.7569928. [DOI] [PubMed] [Google Scholar]
- Olusanya O, Andrews PD, Swedlow JR, Smythe E. Phosphorylation of threonine 156 of the mu2 subunit of the AP2 complex is essential for endocytosis in vitro and in vivo. Curr Biol. 2001;11:896–900. doi: 10.1016/s0960-9822(01)00240-8. [DOI] [PubMed] [Google Scholar]
- Owen DJ, Evans PR. A structural explanation for the recognition of tyrosine-based endocytotic signals. Science. 1998;282:1327–1332. doi: 10.1126/science.282.5392.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez RG, Soriano S, Hayes JD, Ostaszewski B, Xia W, Selkoe DJ, Chen X, Stokin GB, Koo EH. Mutagenesis identifies new signals for beta-amyloid precursor protein endocytosis, turnover, and the generation of secreted fragments, including Abeta42. J Biol Chem. 1999;274:18851–18856. doi: 10.1074/jbc.274.27.18851. [DOI] [PubMed] [Google Scholar]
- Perez RG, Squazzo SL, Koo EH. Enhanced release of amyloid beta-protein from codon 670/ 671 “Swedish” mutant beta-amyloid precursor protein occurs in both secretory and endocytic pathways. J Biol Chem. 1996;271:9100–9107. doi: 10.1074/jbc.271.15.9100. [DOI] [PubMed] [Google Scholar]
- Ricotta D, Conner SD, Schmid SL, von Figura K, Honing S. Phosphorylation of the AP2 mu subunit by AAK1 mediates high affinity binding to membrane protein sorting signals. J Cell Biol. 2002;156:791–795. doi: 10.1083/jcb.200111068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schobel S, Neumann S, Hertweck M, Dislich B, Kuhn PH, Kremmer E, Seed B, Baumeister R, Haass C, Lichtenthaler SF. A novel sorting nexin modulates endocytic trafficking and alpha-secretase cleavage of the amyloid precursor protein. J Biol Chem. 2008;283:14257–14268. doi: 10.1074/jbc.M801531200. [DOI] [PubMed] [Google Scholar]
- Sheffield P, Garrard S, Derewenda Z. Overcoming expression and purification problem s of RhoGDI using a family of “parallel” expression vectors. Protein Expr Purif. 1999;15:34–39. doi: 10.1006/prep.1998.1003. [DOI] [PubMed] [Google Scholar]
- Simmen T, Honing S, Icking A, Tikkanen R, Hunziker W. AP-4 binds basolateral signals and participates in basolateral sorting in epithelial MDCK cells. Nat Cell Biol. 2002;4:154–159. doi: 10.1038/ncb745. [DOI] [PubMed] [Google Scholar]
- Skovronsky DM, Moore DB, Milla ME, Doms RW, Lee VM. Protein kinase C-dependent alpha-secretase competes with beta-secretase for cleavage of amyloid-beta precursor protein in the trans-golgi network. J Biol Chem. 2000;275:2568–2575. doi: 10.1074/jbc.275.4.2568. [DOI] [PubMed] [Google Scholar]
- Small SA, Gandy S. Sorting through the cell biology of Alzheimer’s disease: intracellular pathways to pathogenesis. Neuron. 2006;52:15–31. doi: 10.1016/j.neuron.2006.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens DJ, Banting G. Specificity of interaction between adaptor-complex medium chains and the tyrosine-based sorting motifs of TGN38 and lgp120. Biochem J. 1998;335:567–572. doi: 10.1042/bj3350567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thinakaran G, Teplow DB, Siman R, Greenberg B, Sisodia SS. Metabolism of the “Swedish” amyloid precursor protein variant in neuro2a (N2a) cells. Evidence that cleavage at the “beta-secretase” site occurs in the golgi apparatus. J Biol Chem. 1996;271:9390–9397. doi: 10.1074/jbc.271.16.9390. [DOI] [PubMed] [Google Scholar]
- Vassar R, Bennett BD, Babu-Khan S, Kahn S, Mendiaz EA, Denis P, Teplow DB, Ross S, Amarante P, Loeloff R, et al. Beta-secretase cleavage of Alzheimer’s amyloid precursor protein by the transmembrane aspartic protease BACE. Science. 1999;286:735–741. doi: 10.1126/science.286.5440.735. [DOI] [PubMed] [Google Scholar]
- Xia W, Ray WJ, Ostaszewski BL, Rahmati T, Kimberly WT, Wolfe MS, Zhang J, Goate AM, Selkoe DJ. Presenilin complexes with the C-terminal fragments of amyloid precursor protein at the sites of amyloid beta-protein generation. Proc Natl Acad Sci U S A. 2000;97:9299–9304. doi: 10.1073/pnas.97.16.9299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Romano DM, Tanzi RE. RNA interference-mediated silencing of X11alpha and X11beta attenuates amyloid beta-protein levels via differential effects on beta-amyloid precursor protein processing. J Biol Chem. 2005;280:15413–15421. doi: 10.1074/jbc.M414353200. [DOI] [PubMed] [Google Scholar]
- Xu H, Sweeney D, Wang R, Thinakaran G, Lo AC, Sisodia SS, Greengard P, Gandy S. Generation of Alzheimer beta-amyloid protein in the trans-Golgi network in the apparent absence of vesicle formation. Proc Natl Acad Sci U S A. 1997;94:3748–3752. doi: 10.1073/pnas.94.8.3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap CC, Murate M, Kishigami S, Muto Y, Kishida H, Hashikawa T, Yano R. Adaptor protein complex-4 (AP-4) is expressed in the central nervous system neurons and interacts with glutamate receptor delta2. Mol Cell Neurosci. 2003;24:283–295. doi: 10.1016/s1044-7431(03)00164-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.