Summary
While the Notch signaling pathway is one of the most intensely studied intracellular signaling pathways, the mechanisms by which Notch signaling regulates transcription remain incompletely understood. Here we report that B-cell leukemia/lymphoma 6 (BCL6), a transcriptional repressor, is a Notch-associated factor. BCL6 is necessary to maintain the expression of Pitx2 in the left lateral plate mesoderm during the patterning of left-right asymmetry in Xenopus embryos. For this process, BCL6 forms a complex with BCL6 co-repressor (BCoR) on the promoters of selected Notch target genes such as enhancer of split related 1. BCL6 also inhibits the transcription of these genes by competing for the Notch1 intracellular domain, preventing the co-activator Mastermind-like1 (MAM1) from binding. These results define a mechanism restricting Notch-activated transcription to cell-type-appropriate subsets of target genes, and elucidate its relevance in vivo, during left-right asymmetric development.
Introduction
Vertebrates show conserved anatomical left-right (LR) asymmetry of the internal organs such as the orientation of the cardiovascular system, visceral organs and the number of lung lobes, while their external bodies are bilaterally symmetrical (Levin, 2005; Palmer, 2004). Although many of the mechanisms involved in breaking LR symmetry during early development may not be conserved, the universal hallmark of vertebrate LR asymmetric development is left-side specific expression of genes such as Nodal, Lefty, and Pitx2 in the lateral plate mesoderm (LPM) (Boorman and Shimeld, 2002; Raya and Belmonte, 2006; Speder et al., 2007). Indeed, these genes play crucial roles during the patterning of LR asymmetry (Capdevila et al., 2000; Hamada et al., 2002).
The Notch signaling pathway is a well conserved signaling pathway in animals (Borggrefe and Oswald, 2009). Following an interaction between the Delta/Serrate/Lag-2 (DSL) ligand and the Notch receptor, the Notch receptor intracellular domain (NICD) is released from the membrane by two sequential proteolytic cleavages. NICD subsequently translocates into the nucleus and forms a complex with nuclear proteins including the C-promoter binding factor 1/Suppressor of Hairless/Lag-1 (CSL) transcriptional factor and the transcriptional co-activator, Mastermind-like (MAM), to activate the transcription of target genes. Notch signaling has been demonstrated to affect LR asymmetry in mice (Krebs et al., 2003; Raya et al., 2003), chick (Raya et al., 2004), and zebrafish (Kawakami et al., 2005; Raya et al., 2003). Previous studies in mice demonstrated that Notch signaling directly regulates early symmetric expression of Nodal through a node-specific enhancer (Adachi et al., 1999; Brennan et al., 2002; Norris and Robertson, 1999), which contains two functional binding sites for CSL (Krebs et al., 2003; Raya et al., 2003). Interestingly, although the expression of Pitx2 in the left LPM is initiated by Nodal (Shiratori et al., 2001), it can also be induced by down-regulation of Notch signaling even in the absence of Nodal function (Krebs et al., 2003; Raya et al., 2003), suggesting that the expression of Pitx2 is regulated by both Nodal-dependent and -independent mechanisms. Thus far, the regulatory mechanism governing Pitx2 expression remains incompletely understood.
B-cell leukemia/lymphoma 6 (BCL6) is a sequence-specific transcriptional repressor, which recruits a wide variety of co-repressors including BCoR (Huynh et al., 2000). BCL6 was originally identified via chromosomal translocations affecting band 3q27, which are common in B-cell non-Hodgkin lymphoma (Baron et al., 1993; Kerckaert et al., 1993; Ye et al., 1993). In fact, deregulated BCL6 expression is commonly observed in diffuse large B cell lymphomas and follicular lymphomas (Ohno, 2004; Pasqualucci et al., 2003). During normal B cell development, BCL6 is required for the formation of germinal centers (GC) (Dent et al., 1997; Ye et al., 1997) and maintains the expression of GC-specific genes by suppressing genes involved in B cell activation in response to DNA damage, cell cycle regulation, and plasma cell differentiation (Li et al., 2005; Niu et al., 2003; Phan and Dalla-Favera, 2004; Ranuncolo et al., 2007; Shaffer et al., 2001; Tunyaplin et al., 2004; Vasanwala et al., 2002). While the function of BCL6 in the formation of lymphoma and normal B cell development has been well studied, its roles during embryogenesis are poorly understood.
Here we report that BCL6 is a transcriptional repressor associated with Notch signaling during Xenopus LR patterning. By binding NICD, preventing MAM1 recruitment, and associating instead with BCoR, BCL6 inhibits certain Notch-induced target genes such as enhancer of split related 1 (ESR1). Target gene specificity is achieved by direct binding of BCL6 to relevant enhancer elements. This function helps maintain the expression of Pitx2 and thus LR asymmetry. Our studies elucidate crosstalk between Notch signaling and the BCL6/BCoR complex, and further show that BCL6 functions as a repressor of Notch signaling during LR patterning.
Results
Isolation of Notch-associated proteins
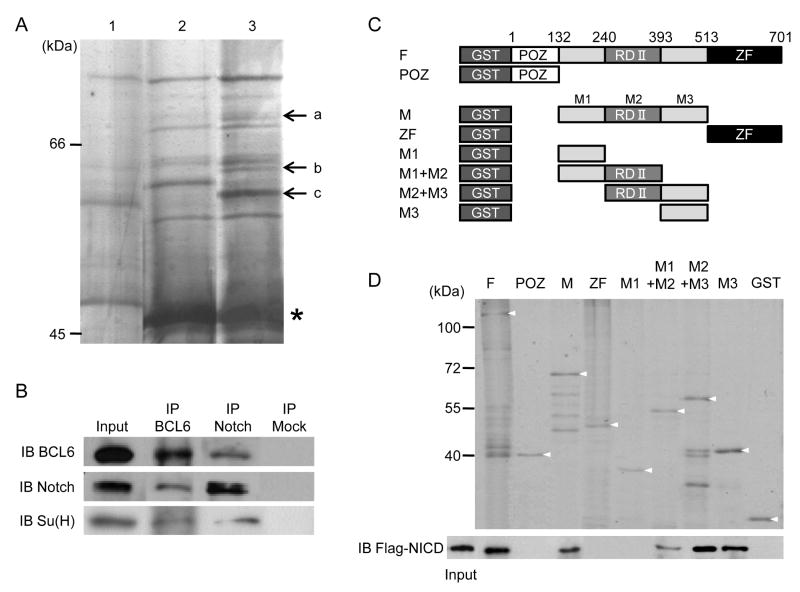
In studies to understand how Notch signaling regulates transcription during embryogenesis, we sought novel transcriptional regulators that can interact with NICD. A GST-fusion protein containing the ankyrin-like repeats domain of NICD protein (GST-ANK) was used to isolate interacting proteins by immunoprecipitation. The ANK domain was utilized because it is an important domain required for the transcriptional activation of Notch signaling and for interaction with the CSL transcriptional factor (Kato et al., 1997), MAM (Kurooka et al., 1998), the histone acetyltransferase complex (Tani et al., 2001) and Deltex (Diederich et al., 1994; Matsuno et al., 1995). Precipitation was performed with GST-ANK and protein extracts from 100 embryos at stages 15, 20 and 25. The co-precipitated proteins were separated by one dimensional (1D) gel electrophoresis, followed by silver staining (Figure 1 A). Three bands in lane 3 (GST-ANK + protein extract) were specific when compared with lane 1 (GST + protein extract) that shows GST-associated bacterial and embryonic proteins and lane 2 (GST-ANK alone) that shows GST-ANK-associated bacterial proteins. Via mass spectrometry analysis, we identified one of these bands indicated by “a” in Figure 1 A as BCL6. Deltex1, which is a regulator of Notch signaling (Diederich et al., 1994; Matsuno et al., 1995; Matsuno et al., 1998), was also identified from the same protein band, although the MASCOT score was not high (data not shown). To determine if BCL6 endogenously interacts with Notch1, we performed co-immunoprecipitation studies with α-Notch, which recognizes the intracellular domain of Notch1, or α-BCL6 antibody. Using protein extracts from Xenopus embryos, a specific endogenous association between Notch1 and BCL6 was observed (Figure 1 B). In addition, Suppressor of Hairless [Su(H)], Xenopus CSL, was also co-precipitated by α-BCL6 antibody (Figure 1 B).
Figure 1.
BCL6 interacts with the ANK domain of Notch1.
A: Lane 1: GST/embryonic protein extract, Lane 2: GST-ANK, and Lane 3: GST-ANK/embryonic protein extract. a: BCL6 and c: β-actin. GST-ANK is indicated by an asterisk. B: Protein extracts from stage 25 embryos were incubated with α-Notch1 or α-BCL6 antibody. Mouse IgG was used for a mock immunoprecipitation. C: GST-BCL6 constructs. The numbers on the top indicate the positions of amino acids. D: The top panel shows the expression of GST constructs and the bottom panel shows the interactions between Flag-tagged NICD (Flag-NICD) and GST-BCL6 constructs. Each arrowhead indicates an intact GST fusion protein.
To delineate the domain of BCL6 responsible for the interaction with Notch1, binding assays were performed. As the known functional domains of BCL6 are the POZ/BTB domain (POZ) at the N-terminus, the repression domain II (RDII) in the middle and the C2H2-type zinc finger domain (ZF) at the C-terminus (Albagli-Curiel, 2003; Chang et al., 1996), eight GST-BCL6 fusion constructs harbouring the individual domains were generated (Figure 1 C). Immunoprecipitation studies between in vitro translated Flag-tagged NICD protein and purified GST-fused BCL6 fragments were performed. The Notch-binding domain of BCL6 was localized to the region of BCL6 that harbored the RDII region (M2 in Figure 1 C) and M3 region (Figure 1 D). These regions also interacted with the ANK domain alone (Figure S1 A). In addition, interaction studies with in vitro translated proteins demonstrate that BCL6 appears to directly interact with NICD but not Su(H) (Figure S1 B).
BCL6 is required for the patterning of LR axis in Xenopus
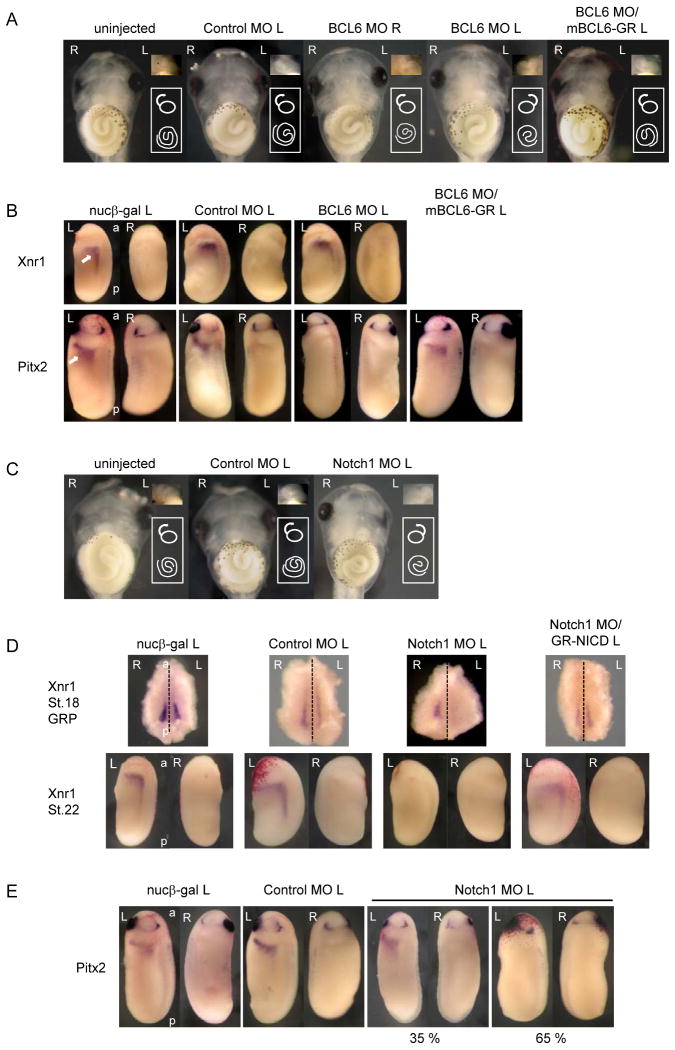
To determine functional roles for BCL6 in Notch signaling, the role of BCL6 during embryogenesis was first examined. BCL6 was expressed in ectodermal and mesodermal tissues through early embryogenesis (Figure S2 A and B). The injection of the highest dose (2 ng) of BCL6 RNA into one blastomere of 2-cell stage embryos or a single dorsal or ventral blastomere of 4-cell stage embryos did not elicit any morphological changes in the injected embryos (data not shown). We employed a Morpholino Antisense Oligo (MO) against BCL6 (BCL6 MO), which binds sequences encompassing the ATG site of directed transcripts and inhibits protein translation thus depleting the endogenous protein (Heasman et al., 2000). The injection of BCL6 MO significantly reduced endogenous BCL6 protein (Figure S2 C). BCL6 MO or a control MO (Control MO), which targets human beta-globin pre-mRNA and does not recognize BCL6 mRNA, was injected into a dorsal blastomere of 4-cell stage embryos. Green fluorescent protein (GFP) RNA was co-injected as a tracer (data not shown). When 40 ng of BCL6 MO was injected into a left dorsal blastomere of 4-cell stage embryos, striking abnormalities in the orientation of gut origin, gut coiling and the heart were observed (Figure 2 A and Table). The gut origin was left (27.8 %, n=115) and gut coiling was clockwise (34 %, n=115), while normal gut origin is right and normal gut coiling is counterclockwise. The orientation of the heart was also inverted in a number of these BCL6 MO-injected embryos (24.4 %, n=115). Phenotypes were scored according to Branford et al. (Branford et al., 2000). In contrast, there is no significant effect in the Control MO-injected or right-side BCL6 MO-injected embryos. In addition, the defects of gut extension were observed in about 30 % of the BCL6 MO-injected embryos and these embryos were not included when phenotypes were scored (data not shown). To show the specificity of the BCL6 MO effect, we co-injected BCL6 MO and a hormone inducible mutant BCL6 (mBCL6-GR) RNA whose translation initiation site was replaced by the Myc-tag and which is no longer recognized by BCL6MO, and examined gut and heart phenotypes. Except where noted otherwise, we consistently added dexamethasone (DEX) to the medium at stage 20, to activate a GR-fused protein. Thus activated, mBCL6-GR rescued gut origin (27.8 % to 4.3 %), gut coiling (34 % to 7.5 %) and the heart (24.4 % to 3.3 %) phenotypes to normal (Figure 2 A and Table). As the failure of LR asymmetric patterning causes the disorientation of gut origin, gut coiling and the heart (Branford et al., 2000), these results suggest that the expression of BCL6 in the left side of embryos is necessary for LR patterning.
Figure 2.
Related functions of BCL6 and Notch signaling during LR patterning.
A: 40 ng BCL6 MO, 40 ng Control MO or/and 2 ng mBCL6-GR was injected for each experiment. An arrow or a spiral indicates the orientation of the heart or the gut coiling, respectively. Ventral views are shown. B: The normal left-specific expression of Xnr1 or Pitx2 is indicated by an arrow in the nucβ-gal-injected embryo. C: 80 ng Notch1 MO or 80 ng Control MO was injected for each experiment. An arrow or a spiral indicates the orientation of the heart or the gut coiling, respectively. Ventral views are shown. D, E: 150 ng Notch1 MO, 150 ng Control MO or/and 1 ng GR-NICD RNA was injected for each experiment. The dotted line indicates the embryonic midline. The injected side is indicated by L (left) or R (right) beside the names of injected samples. L: left, R: right, a: anterior, p: posterior.
Table. Laterality scoringa in BCL6 MO, NBD-S or Notch1 MO injection.
| Injection | Injection Side | Cardiac Orientation, Forward | Cardiac Orientation, Reverse | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gut Origin Right | Gut Origin Left | Gut Origin Right | Gut Origin Left | |||||||
| CCWb | CWc | CCW | CW | CCW | CW | CCW | CW | n | ||
| uninjected | 95.4 | 2.3 | 0.8 | 1.5 | 0.0 | 0.0 | 0.0 | 0.0 | 130 | |
| <BCL6 MO> | ||||||||||
| 40 ng Control MO | Left | 96.1 | 1.3 | 2.6 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 76 |
| 20 ng Control MO | Left | 97.8 | 1.1 | 1.1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 89 |
| 40 ng BCL6 MO | Right | 90.5 | 1.9 | 1.9 | 1.0 | 1.0 | 0.0 | 1.9 | 1.9 | 105 |
| 20 ng BCL6 MO | Right | 90.7 | 1.9 | 3.7 | 0.9 | 0.9 | 0.0 | 0.9 | 0.9 | 108 |
| 40 ng BCL6 MO | Left | 47.0 | 11.3 | 7.8 | 9.6 | 7.0 | 7.0 | 4.3 | 6.1 | 115 |
| 20 ng BCL6 MO | Left | 59.5 | 20.2 | 6.0 | 9.5 | 1.2 | 2.4 | 0.0 | 1.2 | 84 |
| 40 ng BCL6 MO/2 ng mBCL6-GR | Left | 89.4 | 4.3 | 1.1 | 2.1 | 1.1 | 1.1 | 1.1 | 0.0 | 94 |
| 20 ng BCL6 MO/2 ng mBCL6-GR | Left | 85.7 | 3.1 | 2.0 | 1.0 | 1.0 | 2.0 | 2.0 | 3.1 | 98 |
| <NBD-S> | ||||||||||
| 1 ng NBD-S-GR | Left | 70.0 | 7.0 | 6.0 | 3.0 | 5.0 | 2.0 | 4.0 | 3.0 | 100 |
| 2 ng NBD-S-GR | Left | 55.0 | 11.0 | 9.0 | 4.0 | 4.0 | 3.0 | 8.0 | 4.0 | 98 |
| <Notch1 MO> | ||||||||||
| 150 ng Control MO | Left | 83.7 | 2.4 | 8.1 | 2.4 | 0.8 | 0.8 | 0.8 | 0.8 | 123 |
| 80 ng Control MO | Left | 91.3 | 3.6 | 2.2 | 2.9 | 0.0 | 0.0 | 0.0 | 0.0 | 138 |
| 150 ng Notch1 MO | Left | 38.7 | 33.1 | 8.9 | 6.5 | 2.4 | 4.0 | 3.2 | 3.2 | 124 |
| 80 ng Notch1 MO | Left | 57.1 | 22.7 | 4.2 | 1.7 | 3.4 | 3.4 | 4.2 | 3.4 | 119 |
Lateral scoring according to Branford et al. (2000).
Numbers indicate percentage of embryos displaying phenotype (total embryos as n)
CCW: counterclockwise
CW: clockwise
To further study the role of BCL6 in the patterning of LR asymmetry, we first characterized the role of BCL6 in the conserved Nodal-Pitx2 cascade that governs LR patterning. The expression of left-side specific genes, Xnr1 (a Nodal paralog) and Pitx2 (Lohr et al., 1997; Ohi and Wright, 2007; Schweickert et al., 2000; Vonica and Brivanlou, 2007), were tested in the BCL6-depleted embryos. BCL6 MO or Control MO was injected into a left dorsal blastomere of 4-cell stage embryos, respectively, and the expression of Xnr1 at stage 22 and Pitx2 at stage 25 in the left LPM was examined. Interestingly, the injection of BCL6 MO suppressed the expression of Pitx2 (100 %, n=28) but not Xnr1 (0 %, n=28) (Figure 2 B; see RT-PCR in Figure S2 D and E). As with the general embryonic LR defects, these gene expression patterns were also rescued by co-injection of mBCL6-GR (0 % to 97 %) (Figure 2 B). Although we were unable to detect BCL6 in the left LPM at stage 25 by whole mount in situ hybridization (Figure S2 A), RT-PCR clearly revealed left-right symmetric BCL6 expression in the LPM at this stage (Figure S2 F). Thus, BCL6 is required for the expression of Pitx2 but not Xnr1 in the left LPM.
Dual roles of Notch signaling during LR patterning are conserved in Xenopus
Previous studies in mice showed that Notch signaling initiates the symmetric expression of Nodal peri-nodally, while the down-regulation of Notch signaling acts independently of Nodal at later stages, to allow the expression of Pitx2 in the LPM (Krebs et al., 2003; Raya et al., 2003). This suggests that Notch signaling is involved in the regulation of both Nodal and Pitx2 expression at different developmental stages. In particular, Notch activity at the later stage, which could suppress the expression of Pitx2, may be a possible target of BCL6 during LR patterning. We therefore sought to confirm that Notch signaling has a conserved function in these aspects of LR patterning in Xenopus.
At stage 18, Xenopus Notch1 and Notch ligands, Delta1 and Serrate1, were expressed on the gastrocoel roof plate (GRP), which is analogous to the amniote node (Schweickert et al., 2007) (Figure S2 G); this where the expression of Xnr1 is initiated (Jones et al., 1995; Lustig et al., 1996) early during the acquisition of LR asymmetry. Notch1 and Serrate1were also expressed in the LPM at stage 25 similar to BCL6 (Figure S2 F) but Delta1 was hardly detected by RT-PCR (data not shown). The injection of Notch1 MO significantly reduced endogenous Notch1 protein (Figure S2 C) and the expression of Notch target genes (Figure S2 H). When 80 ng of Notch1 MO or Control MO was injected into a left dorsal blastomere of 4-cell stage embryos, the orientation of gut origin (13.5 %, n=119), gut coiling (31.2 %, n=119) and heart looping (14.4 %, n=119) was often inverted (Figure 2 C and Table). And again, as seen in the BCL6 MO experiments, defects in gut extension were observed in about 25 % of the Notch1 MO-injected embryos and these embryos were not included when phenotypes were scored (data not shown). Unlike BCL6 MO, however, the Notch1 MO suppressed the expression of Xnr1 on both sides of the GRP at stage 18 (left: 93 %, n=30; right: 89 %, n=28) (Figure 2 D and Figure S2 I) and in the left LPM at stage 22 (100 %, n=30) (Figure 2 D). Although Xnr1 expression in the GRP was not decreased in all cases, its expression completely disappeared from the LPM at stage 22. The effects of the Notch1 MO were rescued by a hormone inducible NICD (GR-NICD) RNA. When GR-NICD was activated by DEX at stage 12, Xnr1 expression in the GRP at stage 18 (left: 7 % to 92 %; right: 11 % to 86 %) and the left LPM at stage 22 (0 % to 60 %) were restored to normal levels (Figure 2 D and Figure S2 I). These data indicate that Xenopus Notch signaling promotes the expression of Xnr1 in the GRP during LR patterning.
The expression of Pitx2 in the Notch1 MO-injected embryos was next examined. As Xnr1 expression in the stage 22 LPM was not observed in the Notch1 MO-injected embryos and the expression of Pitx2 is initiated by Xnr1 (Ohi and Wright, 2007), we predicted that Pitx2 expression would be completely abolished in the Notch1 MO-injected embryos. However, Pitx2 expression was affected in only some of these embryos (65 %, n=31) (Figure 2 E). Interestingly, when Notch1 MO and GR-NICD RNA were co-injected for the rescue study, the expression of Pitx2 was not rescued and the number of embryos with suppressed Pitx2 increased (65 % to 100 %, data not shown). Accordingly, LR asymmetry defects induced by Notch1 MO were not rescued by co-injection of GR-NICD (data not shown). These findings suggest that, as in mice (Krebs et al., 2003; Raya et al., 2003), the expression of Xenopus Pitx2 could occur when Notch signaling was down-regulated in the absence of Xnr1 function and Notch signaling could suppress the expression of Pitx2. We therefore decided to test this hypothesis in more detail.
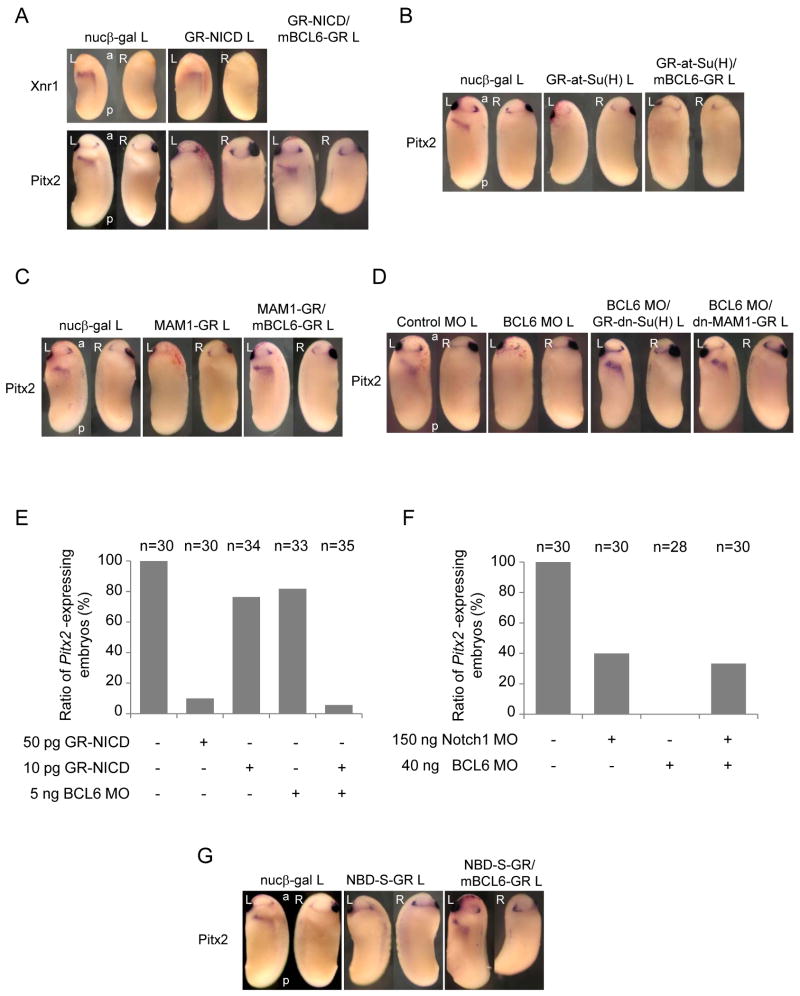
Indeed, when GR-NICD RNA was injected into a left dorsal blastomere of 4-cell stage embryos and GR-NICD was activated by DEX treatment at stage 20, Pitx2 expression was suppressed (90 %, n=30) (Figure 3 A). On the other hand, even when GR-NICD was activated at stage 12, the expression of Xnr1 remained unchanged (Figure 3 A). Although an increase in Xnr1 expression might have been expected, this result is consistent with the fact that over-expression of GR-NICD on the right side rarely induced the expression of Xnr1 (6 %, n=32) or Pitx2 (0 %, n=31) on the injected side (data not shown). It is unclear why NICD is insufficient to induce Xnr1 or Pitx2 in Xenopus, but is sufficient to do so in zebrafish (Raya et al., 2003); however, it is easy to imagine that other factors required for Xnr1 expression are not adequate expressed on the right side in Xenopus, and conversely that NICD over-expression in zebrafish may only exert early effects (on Nodal paralog expression) but may not last long enough to inhibit Pitx2. In any case, taken together, these findings suggest the possibility that BCL6 selectively antagonizes Notch-mediated inhibition of Pitx2 expression in Xenopus, and that this antagonism forms the basis for BCL6 requirements during LR asymmetric development.
Figure 3.
BCL6 maintains Pitx2 expression by inhibiting Notch signaling.
A-D: 50 pg GR-NICD, 100 pg GR-at-Su(H), 100 pg MAM1-GR, 2 ng GR-dn-Su(H), 2 ng dn-MAM1-GR or/and 2 ng mBCL6-GR was injected for each experiment. E, F: The expression of Pitx2 at stage 25 was tested by whole mount in situ hybridization and the ratios of Pitx2-expressing embryo number versus total tested embryo number are shown. Total numbers of each injection are shown as “n” on the top of each bar. G: 2 ng NBD-S-GR or/and mBCL6-GR was injected for each experiment. The injected side is indicated by L (left) beside the names of injected samples. L: left, R: right, a: anterior, p: posterior.
BCL6 inhibits Notch and maintains Pitx2 expression by interfering with MAM1
To test the possibility that BCL6 is necessary to suppress Notch activity and maintain Pitx2 expression, GR-NICD and mBCL6-GR RNA were co-injected into a left dorsal blastomere of 4-cell stage embryos and the expression of Pitx2 tested. BCL6 restored the expression of Pitx2 to normal levels (10 % to 58 %) (Figure 3 A), indicating that Notch signaling is indeed a likely target of BCL6 during LR patterning.
We next examined whether the suppression of Pitx2 by Notch signaling is mediated by Su(H) or MAM1. A hormone inducible active type of Su(H) fused to the VP16 activator domain [GR-at-Su(H)] (Rones et al., 2000), or a hormone inducible MAM1 (MAM1-GR) RNA was injected into a left dorsal blastomere of 4-cell stage embryos and the expression of Pitx2 examined. Both at-Su(H) (90 %, n=31) and MAM1 (91 %, n=32) suppressed the expression of Pitx2 (Figure 3 B and C). To test whether the suppression of Pitx2 by Su(H) or MAM1 is inhibited by BCL6, GR-at-Su(H) or MAM1-GR RNA was co-injected with mBCL6-GR. BCL6 rescued MAM1 effects on Pitx2 (9 % to 61 %) (Figure 3 C) but not at-Su(H) effects on Pitx2 (10 % to 13 %) (Figure 3 B), suggesting that BCL6 may interfere with specific aspects of transcriptional activation by the NICD/Su(H) complex. As at-Su(H) can activate transcription of Notch target genes without NICD, this result further suggests that BCL6 does not compete with Su(H) to bind to the CSL-binding sites in the promoters of target genes. To confirm the idea that BCL6's principal function in this context is to block Notch-dependent transcription, we used a hormone inducible dominant negative form of Su(H) [GR-dn-Su(H)], which has a mutation in the DNA-binding domain and can still interact with NICD (Rones et al., 2000; Wettstein et al., 1997), and a hormone inducible dominant negative form of MAM1 (dn-MAM1-GR), which has only the Notch-binding domain (Kiyota and Kinoshita, 2002). Co-injection of GR-dn-Su(H) or dn-MAM1-GR RNA with BCL6 MO restored Pitx2 expression to normal levels (8 % to 75 % with GR-dn-Su(H), n=24; 8 % to 84 % with dn-MAM1-GR, n=25) (Figure 3 D), indicating that blocking transcriptional outputs of Notch signaling can rescue BCL6 MO phenotypes.
To further confirm endogenous crosstalk between Notch signaling and BCL6, the following studies were performed. The maximum amounts of GR-NICD RNA (10 pg) and BCL6 MO (5 ng), which alone cannot sufficiently suppress the expression of Pitx2, showed a synthetic interaction, suppressing Pitx2 (Figure 3 E) and suggesting that endogenous BCL6 inhibits Notch activity. Moreover, the Notch1 MO was epistatic to the BCL6 MO (Figure 3 F), indicating that Notch signaling is an in vivo target of BCL6 for the expression of Pitx2 (Figure S3 A).
In order to design a reagent that would selectively block BCL6/Notch interactions, without affecting other BCL6 or Notch functions, a hormone inducible mutant BCL6 construct (NBD-S-GR), which contains only the M3 domain, was generated (Figure 1 C). To our knowledge, the M3 domain has not been reported to be required for the interaction between BCL6 and any other factors thus far, yet we find it is sufficient to interfere with the interaction between Notch1 and BCL6, while leaving the NICD transcriptional complex (Figure S3 B) and Notch activity (Figure S3 C and D) intact. NBD-S-GR RNA was injected into the left dorsal blastomere of 4-cell stage embryos and the expression of Pitx2 examined. The expression of Pitx2 was inhibited by NBD-S (87 %, n=31) and this inhibition was rescued by the co-injection of mBCL6-GR RNA (13 % to 76 %) (Figure 3 G). Note, the defects of LR asymmetry were also observed in the NBD-S-injected embryos (Table). To verify whether NBD-S enhances the ability of Notch to suppress Pitx2, the maximum amounts of GR-NICD (10 pg) and NBD-S-GR (100 pg), which alone cannot sufficiently suppress the expression of Pitx2, were co-injected, and again a synthetic interaction was observed (Figure S3 E). These findings together support the proposal that BCL6 maintains LR asymmetry by rendering Pitx2 expression resistant to the effects of Notch signaling.
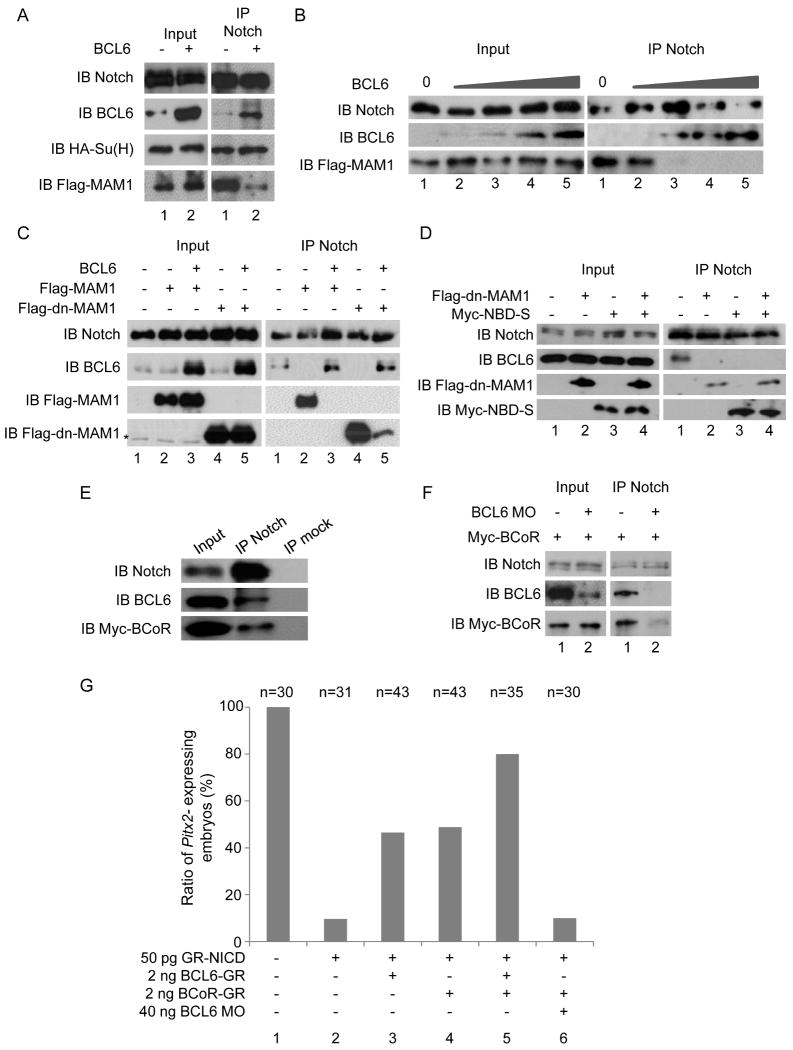
In order to determine the molecular mechanism of this BCL6 effect, we sought BCL6-dependent changes in the composition of Notch transcriptional complexes. BCL6 was over-expressed in embryos and co-immunoprecipitation with α-Notch1 antibody was performed. MAM1 but not Su(H) was replaced by the over-expressed BCL6 protein (Figure 4 A). Interaction studies with in vitro translated proteins demonstrate that this interference by BCL6 was dose-dependent (Figure 4 B). Conversely, the over-expression of MAM1 and dn-MAM1 displaced BCL6 from the transcriptional complex of Notch signaling (lanes 2 and 4 in Figure 4 C). Note, NBD-S and MAM1 (Figure S3 B) or dn-MAM1 (Figure 4 D) did not exclude each other from the transcriptional complex. It is possible that the NBD-S-binding site in the ANK domain of NICD may not overlap with the MAM1-binding site or/and NBD-S may interact with NICD more strongly than full-length of BCL6 because the truncation of other domains may lead to conformation change. These data together demonstrate that BCL6 competes with MAM1 for the ANK domain of NICD to inhibit the transcriptional activity of Notch.
Figure 4.
The mechanisms by which the BCL6/BCoR complex blocks Notch-dependent transcription.
A: HA-tagged Su(H), Flag-tagged MAM1 or/and BCL6 was expressed in embryos and protein extracts were isolated from 50 embryos at stage 10. Co-immunoprecipitation with α-Notch antibody was performed. B: Co-immunoprecipitation using in vitro synthesized proteins. C, D: Flag-tagged MAM1, Flag-tagged dn-MAM1, BCL6 or/and Myc-tagged NBD-S was expressed in embryos and protein extracts were isolated from 50 embryos at stage 10 for each experiment. E, F: Myc-tagged BCoR was expressed in embryos without (E) or with (F) the BCL6 MO, and protein extracts were isolated from 50 embryos at stage 10. Co-immunoprecipitation with α-Notch antibody was performed. α-vimentin antibody was used for a mock immunoprecipitation. G: The expression of Pitx2 at stage 25 was tested by whole mount in situ hybridization and the ratios of Pitx2-expressing embryo number versus total tested embryo number are shown. Total numbers of each injection are shown as “n” on the top of each bar.
BCL6 forms a complex with BCoR
As a previous study in Xenopus showed that BCoR is required for the expression of Pitx2 and LR patterning (Hilton et al., 2007) and BCoR was expressed in the LPM at stage 25 (Figure 2 C), we examined whether BCoR is present in the Notch/BCL6 complex. When immunoprecipitation with α-Notch1 antibody was performed, BCoR was precipitated with the Notch/BCL6 complex (Figure 4 E). When BCL6 was knocked down by BCL6 MO, the amount of BCoR precipitated by α-Notch1 antibody was reduced (Figure 4 F), suggesting that BCL6 recruits BCoR into the transcriptional complex of Notch signaling. To examine whether BCoR is functionally involved in the suppression of Notch signaling, the enhancement of BCL6 effect by BCoR in Pitx2 expression was tested. The number of Pitx2-expressing embryos in the co-injection of GR-NICD and mBCL6-GR was increased by the co-injection of BCoR (lanes 3 and 5 in Figure 4 G). Indeed, the over-expression of BCoR itself was sufficient to attenuate NICD's effects on Pitx2 (lanes 4 in Figure 4 G), and this BCoR activity was dependent on endogenous BCL6 (lanes 6 in Figure 4 G). Collectively, our data indicate that BCL6 inhibits Notch-dependent transcription by blocking NICD/Su(H) interactions with co-activator MAM1 and recruiting instead co-repressor BCoR.
ESR1 is a Notch target gene suppressed by BCL6 during LR patterning
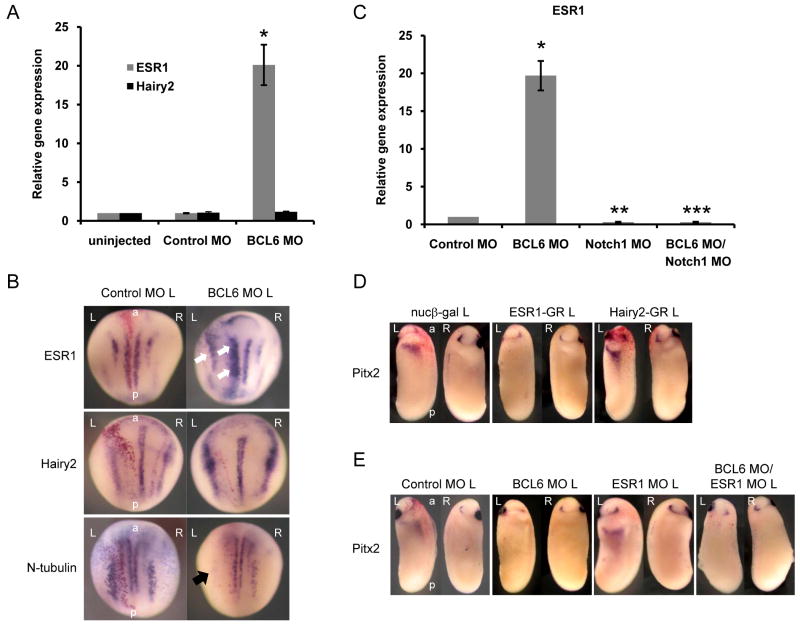
In an effort to refine our model for BCL6 action, we looked for direct target genes shared by BCL6 and Notch, where we might test the mechanistic model discussed above. The expression of Notch-activated genes in the LPM was therefore examined by RT-PCR. Interestingly, the expression of ESR1 (Lamar and Kintner, 2005; Wettstein et al., 1997) was barely detected, while Hairy2 (Davis et al., 2001) was expressed (Figure S4 A). To test whether BCL6 differentially regulates the transcription of selected Notch target genes, the expression of ESR1 and Hairy2 in the BCL6-depleted left LPM was tested by quantitative RT-PCR. The expression of ESR1 but not Hairy2 was increased in the BCL6-depleted LPM (Figure 5 A) and in the nervous system at stage 14 (Figure 5 B). This increase of ESR1 expression by BCL6 MO was decreased by the co-injection of Notch1 MO, suggesting that BCL6 directly regulates the transcriptional output of Notch signaling on ESR1 (Figure 5 C). In addition, an increase of ESR1 expression by NICD in the left LPM was reduced by the co-injection of BCL6 (Figure S4 B).
Figure 5.
ESR1 is a target of BCL6 during LR patterning and neural development.
A: BCL6 MO was injected into the left side of embryos and left LPM tissues were dissected from 20 embryos at stage 25 for quantitative RT-PCR. The injection side was traced by the co-injection of GFP (data not shown). * P<0.05, n=3. B: BCL6 MO was injected into a dorsal blastomere of four-cell stage embryos and embryos were fixed at stage 14. Increased ESR1 expression and decreased N-tubulin expressions are indicated by white and black arrows, respectively. C: BCL6 MO or/and Notch1 MO was injected into the left side of embryos and left LPM tissues were dissected from 20 embryos at stage 25 for quantitative RT-PCR. * P<0.01, n=3, **P<0.01, n=3, ***P<0.01, n=3. D: 1 ng ESR1-GR or Hairy2-GR was injected for each experiment. E: 40 ng ESR1 MO or/and BCL6 MO was injected for each experiment. The injected side is indicated by L (left) beside the names of injected samples. L: left, R: right, a: anterior, p: posterior.
Next, the possibility that ESR1 mediates Notch signaling to suppress the expression of Pitx2 was examined. A hormone inducible ESR1 (ESR1-GR) or Hairy2 (Hairy2-GR) RNA was injected into a left dorsal blastomere of 4-cell stage embryos and the expression of Pitx2 tested. ESR1 (74 %, n=34) but not Hairy2 (4 %, n=28) suppressed Pitx2 expression (Figure 5 D and Figure S4 C). To examine whether ESR1 is the primary mediator of Notch effects on Pitx2, ESR1 was knocked down using a MO (ESR1 MO) (Figure S4 D). However, the ESR1 MO was not able to rescue Pitx2 expression in BCL6 MO co-injected embryos, indicating that other Notch target genes that converge on Pitx2 are also suppressed by BCL6 in the left LPM (Figure 5 E and Figure S4 E).
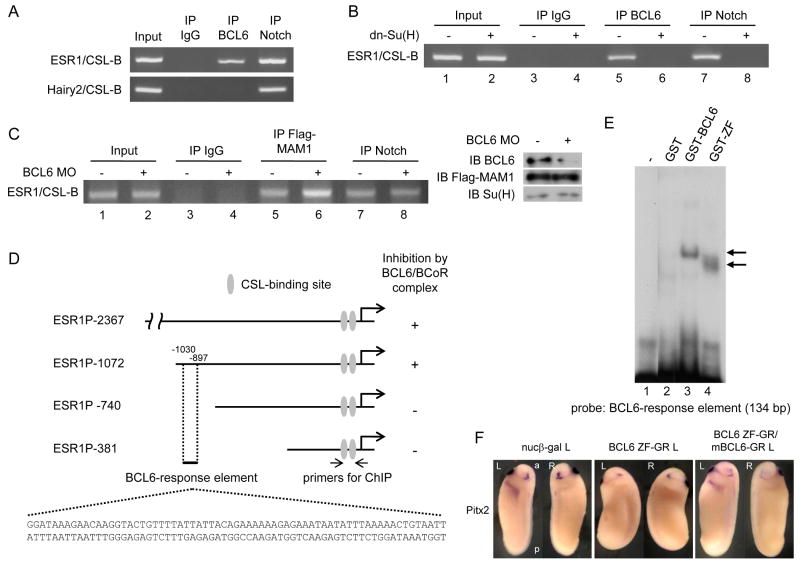
Chromatin immunoprecipitation (ChIP) assays with nuclear extracts isolated from stage 25 embryos confirmed that Notch1 is associated with the known CSL-binding sites at the ESR1 and Hairy2 genomic loci; however, BCL6 bound only the ESR1 CSL-binding element (Figure 6 A). We next examined whether BCL6 recruitment is dependent on the NICD. dn-Su(H) was used for this study, because over-expressed dn-Su(H) dominantly interacts with NICD but cannot bind the CSL-binding site (Wettstein et al., 1997). dn-Su(H) over-expression prevented both Notch1 (lanes 7 and 8 in Figure 6 B) and BCL6 (lanes 5 and 6 in Figure 6 B) from binding the ESR1 CSL-binding site. On the other hand, BCL6 MO increases MAM1 occupancy of ESR1 CSL-binding site (lanes 5 and 6 in Figure 6 C) without affecting NICD (lanes 7 and 8 in Figure 6 C). These data strongly suggest that BCL6 binds to the transcriptional complex present at the CSL-binding site of ESR1 through NICD and competes with MAM1. However, it still remains possible that BCL6 interacts directly with the ESR1 gene, at elements other than the CSL-binding site tested above.
Figure 6.
The mechanisms by which BCL6 shuts down the expression of selected Notch target genes.
A: ChIP assays were performed with nuclear extracts from stage 25 embryos using α-BCL6 antibody, α-Notch1 antibody or mouse IgG. Mouse IgG was used for a mock ChIP assay. B: 2 ng dn-Su(H) was injected into two-cell stage embryos and nuclear extracts were isolated at stage 10 for ChIP assays. C: BCL6 MO or/and 1 ng Flag-MAM1 was injected into two-cell stage embryos and nuclear extracts were isolated at stage 10 for ChIP assays. The levels of input proteins were confirmed by immunoblotting. D: Deleted fragments of X. tropicalis ESR1 gene were linked to the luciferase reporter. Numbers indicate the position of nucleotides from the initiation site. E: Incubation of GST-BCL6 or GST-ZF with a probe corresponding to a 134-bp (-1030/-897) element yielded one distinct retarded band indicated by an arrow. F: 2 ng BCL6 ZF-GR or/and mBCL6-GR was injected for each experiment. The injected side is indicated by L (left) beside the names of injected samples. L: left, R: right, a: anterior, p: posterior.
To search for a BCL6-response element, 2376 bp of the Xenopus tropicalis ESR1 genomic locus was amplified by PCR with primers designed using X. tropicalis genome sequences (UCSC Genome Bioinformatics; http://genome.ucsc.edu/) and linked to the luciferase reporter (pGL3-ESR1P-2376). After this construct was co-injected with NICD or/and BCL6/BCoR RNA into Xenopus embryos, the luciferase activity was measured. Increased luciferase activity by NICD was decreased by the co-injection of BCL6 and BCoR, suggesting this fragment of ESR1 gene includes the BCL6-response element (Figure 6 D and Figure S5 A). A deletion analysis of this genomic fragment revealed that the BCL6-response element was present between -1072 and -740 (Figure 6 D and Figure S5 A); however, consensus BCL6-binding sequences as published previously (Chang et al., 1996) were not found between -1072 and -740. Electrophoretic mobility shift assays (EMSA) were therefore performed with full-length or the C2H2-type zinc finger domain of BCL6 recombinant protein (GST-BCL6 or GST-ZF) to identify the BCL6-response element in this region of ESR1. Several probes for EMSA were designed in the candidate region of ESR1 (-1072/-740) and were radiolabeled by PCR. The -1030/-897 probe resulted in a BCL6-retarded band (lane 3: GST-BCL6 and lane 4: GST-ZF in Figure 6 E). Since BCL6 directly interacts with this probe, we will refer to the corresponding region of ESR1 as the BCL6-response element (see Figure 6 D). Indeed, the over-expression of the zinc finger domain of BCL6 (BCL6-ZF) could inhibit the expression of Pitx2, indicating that BCL6-ZF competes with endogenous BCL6 for the BCL6-binding site and inhibits the function of BCL6 by displacing endogenous BCL6 from the ESR1 locus (Figure 6 F). Since two non-overlapping fragments (5′ response element and 3′ response element) of the BCL6-response element (Figure S5 B), could each interact with GST-ZF, there are likely more than one BCL6-binding site in the BCL6-response element (Figure S5 C). These results indicate that direct binding of BCL6 to both the target locus and the NICD is required for its ability to shut down Notch target gene expression. These findings in turn suggest a mechanism by which selective inhibition of specific Notch-activated target genes is achieved.
Discussion
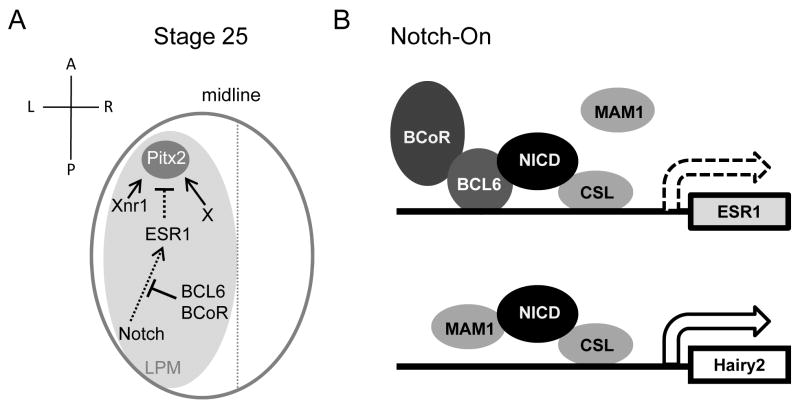
We have uncovered that BCL6 recruits BCoR and blocks the transcription of selected Notch target genes to maintain Pitx2 expression and LR asymmetry in Xenopus (Figure 7 A). It should be noted that mutations of human BCoR result in the Oculofaciocardiodental (OFCD) syndrome, which exhibits defective lateralization including dextrocardia, asplenia and intestinal malrotation (Hilton et al., 2007; Ng et al., 2004). These findings indicate that the dysfunction of BCL6 in mammals can likely lead to defects of LR asymmetry. However, the defects of LR asymmetry in BCL6-deficient mice have not been reported. BCL6-deficient mice displayed defective germinal center (GC) development and a selective defect in T-cell-dependent antibody responses (Ye et al., 1997), and also developed myocarditis and pulmonary vasculitis (Dent et al., 1997; Ye et al., 1997; Yoshida et al., 1999). Based on our findings and the observation in human syndrome, it remains possible that defects of LR asymmetry in BCL6-deficient mice may have been overlooked because defects of LR asymmetry are not lethal (Peeters and Devriendt, 2006). The re-examination of BCL6-deficient mice will be required to address this important question.
Figure 7.
A model for the regulation of Notch signaling by the BCL6/BCoR complex during LR patterning.
A: At stage 25, the BCL6/BCoR complex inhibits Notch's ability to suppress Pitx2 expression initiated by Xnr1-dependent and -independent signals and maintains LR asymmetry. B: Sequence-specific targeting of BCL6 to a subset of Notch-activated genes occurs by an unknown mechanism. Once recruited, however, BCL6 both competes MAM1 away from the locus and recruits BCoR, effectively blocking Notch-dependent transcription.
In mice, distinct asymmetric expression of Delta-like 1 (Dll1), Notch1 and Notch2 around the node (Bettenhausen et al., 1995; Krebs et al., 2003; Raya et al., 2003; Williams et al., 1995) and asymmetric Notch activation have not been reported. In Dll1 knockout or Notch1 and Notch2 double-knockout mice, the symmetric expression of Nodal in the peri-nodal region is completely abolished and these mice show defects of LR asymmetry (Krebs et al., 2003; Raya et al., 2003). Similar to the studies in mice, any LR asymmetry in the expression of Delta1, Serrate1 and Notch1 around the Xenopus GRP was not observed (Figure S2 G) and the symmetric expression of Xnr1 on the GRP was inhibited by the depletion of Xenopus Notch1 (Figure 2 D and Figure S2 I). These findings indicate that Xenopus Notch1 initiates symmetric Xnr1 expression around the GRP required for LR patterning and these mechanisms are conserved between mice and Xenopus. However, it still remains unclear whether asymmetric Notch activity exists around the GRP. It is possible that other signals, including the generation of a leftward fluid flow in or close to the GRP by the rotation of cilia (Schweickert et al., 2007), together could break the bilateral symmetry and induce the left-specific Xnr1 expression in the LPM. In contrast, study in chick have shown that the expression of Dll1 around the left side of Hensen's node is stronger than the right and asymmetric activity of Notch signaling on the left side of the node regulates the left-side expression of Nodal (Raya et al., 2004). It remains very likely that the precise role of Notch signaling in the patterning of LR asymmetry may be slightly different among species.
We have shown that the expression of Pitx2 on the left LPM is dually regulated by Nodal (Xnr1)-dependent and -independent manners. The expression of Pitx2 in the absence of Nodal function has been reported in Notch1 and Notch2 knockout mice (Krebs et al., 2003; Raya et al., 2003), mutations of mouse PDK2 (Pennekamp et al., 2002) and FURIN-deficient mice (Constam and Robertson, 2000). These findings suggest that Pitx2 expression is regulated in part by Nodal-independent mechanisms. As we have found that Pitx2 expression is significantly suppressed by loss of BCL6 or by the over-expression of NICD or ESR1 in Xenopus embryos, this indicates that a Notch-ESR1 signal could simultaneously inhibit Nodal-dependent and -independent signals to suppress the expression of Pitx2 (Figure S3 A). However, how Notch-ESR1 signal inhibits these signals still remains unsolved. Interestingly, the Nodal-dependent expression of mouse Pitx2 is controlled by a two-step mechanism during the patterning of LR asymmetry (Shiratori et al., 2001). Nodal signals acting in cooperation with the transcription factor FAST-1 are required to initiate left-side-specific expression of mouse Pitx2; however, the relevant left-side-specific enhancer is also dependent on Nkx2.5 to maintain activation. All these enhancer sequences are conserved at the Xenopus Pitx2 locus (Shiratori et al., 2001), suggesting that the left-specific expression of Xenopus Pitx2 is regulated by the same two-step mechanism. ESR1 may directly bind this left-side-specific enhancer of Pitx2 and shut down Pitx2 expression, although an indirect inhibition cannot be excluded. It will therefore be important to investigate in future studies which regulatory step of Pitx2 induction is inhibited by the Notch-ESR1 cascade and how ESR1 inhibits Pitx2 expression.
How does Notch signaling activate only the correct target genes in the LPM? One may posit that distinct repressors expressed in the LPM may play a crucial role to inhibit the transcription of unnecessary target genes during LPM development and BCL6 must be such a factor. Indeed, our studies show that the expression of ESR1 but not Hairy2 was selectively inhibited by BCL6 and that ESR1 but not Hairy2 inhibited the expression of Pitx2 (Figure 5). BCL6 directly interacts with the ESR1 cis-regulatory element (Figure 6), and competes with MAM1 for the ANK domain of Notch1 to shut down the transcription of ESR1 when Notch signaling is activated (Figure 7 B). Although 134 bp of the BCL6-response element at the ESR1 locus was identified, consensus BCL6-binding sequences (Chang et al., 1996) were not found in this element (Figure 6 D). It is possible that BCL6 may interact with the ESR1 element through slightly different binding sequences. Our study suggests that there are multiple BCL6-binding sites in the BCL6-response element of ESR1 (Figure S5 C). Therefore, a further analysis of this element remains necessary to identify the BCL6-binding site(s) and address how BCL6 is recruited to the ESR1 locus.
Our data define an important mechanism by which BCL6 constrains Notch signaling to provide cell-type-appropriate outputs. Since the expression of Notch1 overlaps with that of BCL6 in diverse ectodermal and mesodermal tissues including the eye, the nervous system, and the somites (Figure S2 A) – and indeed, both Notch signaling and BCL6 abnormalities have been implicated in leukemias – this regulatory mechanism may be important for other developmental, homeostatic, or pathophysiological processes.
Experimental Procedures
Embryo manipulations
Eggs were artificially fertilized by using testis homogenate and cultivated in 0.1× Marc's Modified Ringer's solution (MMR) (Peng, 1991). Embryos were staged according to Nieuwkoop and Faber (Nieuwkoop and Faber, 1967).
GST pull-down and Protein identification by mass spectrometry
GST fusion proteins were produced in E. coli strain BL21. The bacterial cells were disrupted by sonication in Phosphate Buffered Saline (PBS) with Protease inhibitor cocktail (EDTA-free Complete Mini, Roche Applied Science). To purify the GST fusion proteins, glutathione conjugated agarose beads (Sigma) were added to those samples and incubated at 4 °C for 1 hour. The beads were washed three times with 1 % Triton-X in PBS buffer. For our screen, GST fusion protein was additionally washed with the lysate buffer (20 mM Tris-HCl [pH 8.0], 5 mM MgCl2, 1 mM EDTA, 50 mM KCl, 0.1 % Triton X-100, 10% glycerol and 1mM dithiothreitol). Stage 15-25 embryos were homogenized in the lysate buffer containing protease inhibitors to isolate embryonic protein extracts. The beads with GST or GST-ANK protein were incubated with embryonic protein extracts at 4 °C for 4 hours. The samples were washed five times with the lysate buffer without glycerol. Following the washes, the proteins associated with GST or GST-ANK were eluted with the elution buffer (200 mM Tris-HCl [pH 8.0], 4 mM MgCl2, 0.8 mM EDTA, 40 mM KCl, 0.08 % Triton X-100, 0.8 mM dithiothreitol, 10 mM glutathione and protease inhibitors) at 4 °C for 1 hour. The eluted samples were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). The gel was stained with Silver Stain Plus kit (Bio-Rad). The candidate protein bands were excised from the gel, digested with trypsin and analyzed with matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS). The identification of candidate proteins was determined by the MASCOT search algorithm (http://www.matrixscience.com). The monoisotopic peptide masses were used to search the SwissProt database within a mass tolerance of ± 0.2 Da for Xenopus laevis protein, and one missed cleavage allowed.
Immunoprecipitation and Immunoblotting
Embryos were homogenized in the lysate buffer and embryonic protein extracts were used for immunoprecipitation. The embryonic protein extracts were incubated with an antibody at 4 °C overnight. α-Notch [Developmental Studies Hybridoma Bank (DSHB)], α-BCL6 (R&D Systems, Inc.), α-RBP-Jκ [for Su(H): Santa Cruz Biotechnology, Inc.], α-Flag (Sigma), α-HA (Santa Cruz Biotechnology, Inc.), α-Myc (Santa Cruz Biotechnology, Inc.) and α-vimentin (DSHB) antibodies were used for immunoprecipitation or immunoblotting. All in vitro translated proteins were synthesized by the TNT Coupled Reticulocyte Lysate System (Promega) for immunoprecipitation studies.
Microinjection of synthetic RNA and Morpholino Antisense Oligo
Capped synthetic mRNAs were generated by in vitro transcription with SP6 polymerase, using the mMessage mMachine kit (Ambion, Inc.). Morpholino Antisense Oligos (MO) were designed and produced by Gene Tools, LLC. For microinjections, embryos were transferred to 3% Ficoll 400 in 0.1× MMR and injected embryos were cultured in 0.1× MMR until the desired stage. In all injection studies, 100 pg of GFP RNA for observing phenotypes or 250 pg of nucβ-gal RNA (red color) for whole mount in situ hybridization was injected for a tracer of injection. For the activation of GR-fused protein, dexamethasone (DEX: final concentration 10 μM) was added to the medium. Details of plasmid construction and sequences of MOs are presented in the Supplemental Experimental Procedures.
ϐ-galactosidase staining and Whole mount in situ hybridization
Embryos were fixed with the gal fix solution (2 % formaldehyde, 0.2 % glutaraldehyde, 0.02 % Triton-X, 0.01 % sodium deoxycholate in PBS) on ice for 30 minutes. Galactosidase activity was visualized with the RedGal substrate (Research Organics) in the staining buffer (5 mM K3[Fe(CN)6], 5 mM K4[Fe(CN)6], 2 mM MgCl2 in PBS). After staining, embryos were re-fixed with the MEMFA (0.1 M MOPS, 2 mM EGTA [pH 8.0], 1 mM MgSO4, and 3.7% formaldehyde) for 30 minutes. Whole mount in situ hybridization was performed essentially as described previously (Harland, 1991; Takada et al., 2005) using Digoxigenin (Roche Applied Science)-labeled antisense RNA probes and BM purple (Roche Applied Science) for chromogenic reaction. The information of probes is presented in the Supplemental Experimental Procedures.
RT-PCR analysis
Total RNA was isolated by the method with 200 μg/ml Proteinase K described previously (Hilz et al., 1975). Reverse transcriptase reaction (RT) for the synthesis of cDNA was performed with Superscript II (Invitrogen) according to manufacturer's instructions. Specimens were analyzed for gene expression levels using regular PCR with Taq DNA polymerase (New England Biolabs) or iQ5 Real-Time PCR Detection System (Bio-Rad) with the QuantiTect™ SYBR Green PCR kit (Qiagen). The information of primers is presented in the Supplemental Experimental Procedures. All error bars shown are standard deviation (SD) from mean of triplicates.
Chromatin immunoprecipitation (ChIP)
ChIP assays were performed with a kit from Millipore (Kato et al., 2002; Sachs and Shi, 2000). Nuclei were isolated as described previously (Almouzni et al., 1994). The isolated nuclei were resuspended in 360 μl of the nucleus isolation buffer (0.25 M sucrose, 10 mM Tris-HCl [pH 7.5], 3 mM CaCl2, and Protease inhibitor cocktail). Proteins were crosslinked to DNA by adding formaldehyde (final concentration: 1 %) and incubated on ice for 10 min and at room temperature for 20 min. After centrifuging samples, the nuclei were resuspended in 200 μl of the lysis buffer (1 % SDS, 50 mM Tris-HCl [pH 8.1], 10 mM EDTA, Protease inhibitor cocktail) on ice for 10 min. The lysate was sonicated 10 times with 10-sec pulses by using a sonicator (Branson Sonifier 450, VWR) set to 50 % of maximum power to reduce DNA length to between 200 ∼ 1,000 bp. After debris was removed, DNA was quantified and adjusted to equal concentration for PCR. The information of primers is presented in the Supplemental Experimental Procedures. One milliliter of chromatin solution was used for each ChIP assay with α-BCL6, α-Notch1 or α-Flag antibody. One percentage of chromatin solution was stored for the input DNA.
Luciferase reporter assay
Luciferase reporter constructs of ESR1 gene were generated by sub-cloning different length of genomic fragments between -1 and -2367 into pGL3 basic vector (Promega) and pRL-CMV (Promega) was used for the internal control. Luciferase activity was measured using Dual Luciferase Reporter Assay System (Promega). All error bars shown are SD from mean of triplicates. X. tropicalis genomic DNA was gifted from Dr. K. Tamai.
Electrophoretic mobility shift assay (EMSA)
EMSA was performed as described (Huang et al., 1995). Recombinant proteins, which were eluted from glutathione conjugated agarose beads by pre-reaction buffer (50 mM Tris-HCl [PH 8.0], 50 mM KCl, 1 mM EDTA, 1 mM EGTA, 5 mM dithiothreitol, 20 % glycerol, 10 mM glutathione and protease inhibitors), were incubated for 10 min on ice in a 15 μl reaction volume containing 50 mM Tris-HCl (PH 8.0), 50 mM KCl, 7.5 mM Mg2Cl, 1 mM EDTA, 1 mM EGTA, 5 mM dithiothreitol, 20 % glycerol, 50 μg/ml poly (dI-dC) and protease inhibitors. 32P-labeled probe was added and incubation continued for 15 min on ice. Protein-DNA complex were separated by electrophoresis through 6 % native polyacrylamide gel.
Supplementary Material
Acknowledgments
We thank Drs. C. Kintner, M. Mercola, C. V. E. Wright, D. L. Turner, M. Levin, N. Ueno, T. Kinoshita, H. Sive, K. Tamai, R. Habas, and the NIBB for plasmids, and the DSHB for antibodies. We also thank Drs. J. Horabin and R. Habas for critical suggestions to the manuscript. This work was supported by the Bankhead-Coley Cancer Research Program, FSU CRC planning grant and NICHD (HD052526).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adachi H, Saijoh Y, Mochida K, Ohishi S, Hashiguchi H, Hirao A, Hamada H. Determination of left/right asymmetric expression of nodal by a left side-specific enhancer with sequence similarity to a lefty-2 enhancer. Genes & development. 1999;13:1589–1600. doi: 10.1101/gad.13.12.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albagli-Curiel O. Ambivalent role of BCL6 in cell survival and transformation. Oncogene. 2003;22:507–516. doi: 10.1038/sj.onc.1206152. [DOI] [PubMed] [Google Scholar]
- Almouzni G, Khochbin S, Dimitrov S, Wolffe AP. Histone acetylation influences both gene expression and development of Xenopus laevis. Developmental biology. 1994;165:654–669. doi: 10.1006/dbio.1994.1283. [DOI] [PubMed] [Google Scholar]
- Baron BW, Nucifora G, McCabe N, Espinosa R, 3rd, Le Beau MM, McKeithan TW. Identification of the gene associated with the recurring chromosomal translocations t(3;14)(q27;q32) and t(3;22)(q27;q11) in B-cell lymphomas. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:5262–5266. doi: 10.1073/pnas.90.11.5262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettenhausen B, Hrabe de Angelis M, Simon D, Guenet JL, Gossler A. Transient and restricted expression during mouse embryogenesis of Dll1, a murine gene closely related to Drosophila Delta. Development (Cambridge, England) 1995;121:2407–2418. doi: 10.1242/dev.121.8.2407. [DOI] [PubMed] [Google Scholar]
- Boorman CJ, Shimeld SM. The evolution of left-right asymmetry in chordates. Bioessays. 2002;24:1004–1011. doi: 10.1002/bies.10171. [DOI] [PubMed] [Google Scholar]
- Borggrefe T, Oswald F. The Notch signaling pathway: transcriptional regulation at Notch target genes. Cell Mol Life Sci. 2009;66:1631–1646. doi: 10.1007/s00018-009-8668-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branford WW, Essner JJ, Yost HJ. Regulation of gut and heart left-right asymmetry by context-dependent interactions between xenopus lefty and BMP4 signaling. Developmental biology. 2000;223:291–306. doi: 10.1006/dbio.2000.9739. [DOI] [PubMed] [Google Scholar]
- Brennan J, Norris DP, Robertson EJ. Nodal activity in the node governs left-right asymmetry. Genes & development. 2002;16:2339–2344. doi: 10.1101/gad.1016202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capdevila J, Vogan KJ, Tabin CJ, Izpisua Belmonte JC. Mechanisms of left-right determination in vertebrates. Cell. 2000;101:9–21. doi: 10.1016/S0092-8674(00)80619-4. [DOI] [PubMed] [Google Scholar]
- Chang CC, Ye BH, Chaganti RS, Dalla-Favera R. BCL-6, a POZ/zinc-finger protein, is a sequence-specific transcriptional repressor. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:6947–6952. doi: 10.1073/pnas.93.14.6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constam DB, Robertson EJ. Tissue-specific requirements for the proprotein convertase furin/SPC1 during embryonic turning and heart looping. Development (Cambridge, England) 2000;127:245–254. doi: 10.1242/dev.127.2.245. [DOI] [PubMed] [Google Scholar]
- Davis RL, Turner DL, Evans LM, Kirschner MW. Molecular targets of vertebrate segmentation: two mechanisms control segmental expression of Xenopus hairy2 during somite formation. Developmental cell. 2001;1:553–565. doi: 10.1016/s1534-5807(01)00054-5. [DOI] [PubMed] [Google Scholar]
- Dent AL, Shaffer AL, Yu X, Allman D, Staudt LM. Control of inflammation, cytokine expression, and germinal center formation by BCL-6. Science. 1997;276:589–592. doi: 10.1126/science.276.5312.589. [DOI] [PubMed] [Google Scholar]
- Diederich RJ, Matsuno K, Hing H, Artavanis-Tsakonas S. Cytosolic interaction between deltex and Notch ankyrin repeats implicates deltex in the Notch signaling pathway. Development (Cambridge, England) 1994;120:473–481. doi: 10.1242/dev.120.3.473. [DOI] [PubMed] [Google Scholar]
- Hamada H, Meno C, Watanabe D, Saijoh Y. Establishment of vertebrate left-right asymmetry. Nature reviews. 2002;3:103–113. doi: 10.1038/nrg732. [DOI] [PubMed] [Google Scholar]
- Harland RM. In situ hybridization: an improved whole-mount method for Xenopus embryos. Methods in cell biology. 1991;36:685–695. doi: 10.1016/s0091-679x(08)60307-6. [DOI] [PubMed] [Google Scholar]
- Heasman J, Kofron M, Wylie C. Beta-catenin signaling activity dissected in the early Xenopus embryo: a novel antisense approach. Developmental biology. 2000;222:124–134. doi: 10.1006/dbio.2000.9720. [DOI] [PubMed] [Google Scholar]
- Hilton EN, Manson FD, Urquhart JE, Johnston JJ, Slavotinek AM, Hedera P, Stattin EL, Nordgren A, Biesecker LG, Black GC. Left-sided embryonic expression of the BCL-6 corepressor, BCOR, is required for vertebrate laterality determination. Human molecular genetics. 2007;16:1773–1782. doi: 10.1093/hmg/ddm125. [DOI] [PubMed] [Google Scholar]
- Hilz H, Wiegers U, Adamietz P. Stimulation of proteinase K action by denaturing agents: application to the isolation of nucleic acids and the degradation of ‘masked’ proteins. European journal of biochemistry / FEBS. 1975;56:103–108. doi: 10.1111/j.1432-1033.1975.tb02211.x. [DOI] [PubMed] [Google Scholar]
- Huang HC, Murtaugh LC, Vize PD, Whitman M. Identification of a potential regulator of early transcriptional responses to mesoderm inducers in the frog embryo. The EMBO journal. 1995;14:5965–5973. doi: 10.1002/j.1460-2075.1995.tb00285.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh KD, Fischle W, Verdin E, Bardwell VJ. BCoR, a novel corepressor involved in BCL-6 repression. Genes & development. 2000;14:1810–1823. [PMC free article] [PubMed] [Google Scholar]
- Jones CM, Kuehn MR, Hogan BL, Smith JC, Wright CV. Nodal-related signals induce axial mesoderm and dorsalize mesoderm during gastrulation. Development (Cambridge, England) 1995;121:3651–3662. doi: 10.1242/dev.121.11.3651. [DOI] [PubMed] [Google Scholar]
- Kato H, Taniguchi Y, Kurooka H, Minoguchi S, Sakai T, Nomura-Okazaki S, Tamura K, Honjo T. Involvement of RBP-J in biological functions of mouse Notch1 and its derivatives. Development (Cambridge, England) 1997;124:4133–4141. doi: 10.1242/dev.124.20.4133. [DOI] [PubMed] [Google Scholar]
- Kato Y, Habas R, Katsuyama Y, Naar AM, He X. A component of the ARC/Mediator complex required for TGF beta/Nodal signalling. Nature. 2002;418:641–646. doi: 10.1038/nature00969. [DOI] [PubMed] [Google Scholar]
- Kawakami Y, Raya A, Raya RM, Rodriguez-Esteban C, Belmonte JC. Retinoic acid signalling links left-right asymmetric patterning and bilaterally symmetric somitogenesis in the zebrafish embryo. Nature. 2005;435:165–171. doi: 10.1038/nature03512. [DOI] [PubMed] [Google Scholar]
- Kerckaert JP, Deweindt C, Tilly H, Quief S, Lecocq G, Bastard C. LAZ3, a novel zinc-finger encoding gene, is disrupted by recurring chromosome 3q27 translocations in human lymphomas. Nature genetics. 1993;5:66–70. doi: 10.1038/ng0993-66. [DOI] [PubMed] [Google Scholar]
- Kiyota T, Kinoshita T. Cysteine-rich region of X-Serrate-1 is required for activation of Notch signaling in Xenopus primary neurogenesis. The International journal of developmental biology. 2002;46:1057–1060. [PubMed] [Google Scholar]
- Krebs LT, Iwai N, Nonaka S, Welsh IC, Lan Y, Jiang R, Saijoh Y, O'Brien TP, Hamada H, Gridley T. Notch signaling regulates left-right asymmetry determination by inducing Nodal expression. Genes & development. 2003;17:1207–1212. doi: 10.1101/gad.1084703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurooka H, Kuroda K, Honjo T. Roles of the ankyrin repeats and C-terminal region of the mouse notch1 intracellular region. Nucleic acids research. 1998;26:5448–5455. doi: 10.1093/nar/26.23.5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamar E, Kintner C. The Notch targets Esr1 and Esr10 are differentially regulated in Xenopus neural precursors. Development (Cambridge, England) 2005;132:3619–3630. doi: 10.1242/dev.01937. [DOI] [PubMed] [Google Scholar]
- Levin M. Left-right asymmetry in embryonic development: a comprehensive review. Mechanisms of development. 2005;122:3–25. doi: 10.1016/j.mod.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Li Z, Wang X, Yu RY, Ding BB, Yu JJ, Dai XM, Naganuma A, Stanley ER, Ye BH. BCL-6 negatively regulates expression of the NF-kappaB1 p105/p50 subunit. J Immunol. 2005;174:205–214. doi: 10.4049/jimmunol.174.1.205. [DOI] [PubMed] [Google Scholar]
- Lohr JL, Danos MC, Yost HJ. Left-right asymmetry of a nodal-related gene is regulated by dorsoanterior midline structures during Xenopus development. Development (Cambridge, England) 1997;124:1465–1472. doi: 10.1242/dev.124.8.1465. [DOI] [PubMed] [Google Scholar]
- Lustig KD, Kroll K, Sun E, Ramos R, Elmendorf H, Kirschner MW. A Xenopus nodal-related gene that acts in synergy with noggin to induce complete secondary axis and notochord formation. Development (Cambridge, England) 1996;122:3275–3282. doi: 10.1242/dev.122.10.3275. [DOI] [PubMed] [Google Scholar]
- Matsuno K, Diederich RJ, Go MJ, Blaumueller CM, Artavanis-Tsakonas S. Deltex acts as a positive regulator of Notch signaling through interactions with the Notch ankyrin repeats. Development (Cambridge, England) 1995;121:2633–2644. doi: 10.1242/dev.121.8.2633. [DOI] [PubMed] [Google Scholar]
- Matsuno K, Eastman D, Mitsiades T, Quinn AM, Carcanciu ML, Ordentlich P, Kadesch T, Artavanis-Tsakonas S. Human deltex is a conserved regulator of Notch signalling. Nature genetics. 1998;19:74–78. doi: 10.1038/ng0598-74. [DOI] [PubMed] [Google Scholar]
- Ng D, Thakker N, Corcoran CM, Donnai D, Perveen R, Schneider A, Hadley DW, Tifft C, Zhang L, Wilkie AO, et al. Oculofaciocardiodental and Lenz microphthalmia syndromes result from distinct classes of mutations in BCOR. Nature genetics. 2004;36:411–416. doi: 10.1038/ng1321. [DOI] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J. Normal table of Xenopus laevis (Daudin) North-Holland Publishing Co; Amsterdam, The Netherlands: 1967. [Google Scholar]
- Niu H, Cattoretti G, Dalla-Favera R. BCL6 controls the expression of the B7-1/CD80 costimulatory receptor in germinal center B cells. The Journal of experimental medicine. 2003;198:211–221. doi: 10.1084/jem.20021395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris DP, Robertson EJ. Asymmetric and node-specific nodal expression patterns are controlled by two distinct cis-acting regulatory elements. Genes & development. 1999;13:1575–1588. doi: 10.1101/gad.13.12.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohi Y, Wright CV. Anteriorward shifting of asymmetric Xnr1 expression and contralateral communication in left-right specification in Xenopus. Developmental biology. 2007;301:447–463. doi: 10.1016/j.ydbio.2006.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno H. Pathogenetic role of BCL6 translocation in B-cell non-Hodgkin's lymphoma. Histology and histopathology. 2004;19:637–650. doi: 10.14670/HH-19.637. [DOI] [PubMed] [Google Scholar]
- Palmer AR. Symmetry breaking and the evolution of development. Science. 2004;306:828–833. doi: 10.1126/science.1103707. [DOI] [PubMed] [Google Scholar]
- Pasqualucci L, Bereschenko O, Niu H, Klein U, Basso K, Guglielmino R, Cattoretti G, Dalla-Favera R. Molecular pathogenesis of non-Hodgkin's lymphoma: the role of Bcl-6. Leukemia & lymphoma. 2003;44 3:S5–12. doi: 10.1080/10428190310001621588. [DOI] [PubMed] [Google Scholar]
- Peeters H, Devriendt K. Human laterality disorders. European journal of medical genetics. 2006;49:349–362. doi: 10.1016/j.ejmg.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Peng HB. Xenopus laevis: Practical uses in cell and molecular biology. Solutions and protocols. Methods in cell biology. 1991;36:657–662. [PubMed] [Google Scholar]
- Pennekamp P, Karcher C, Fischer A, Schweickert A, Skryabin B, Horst J, Blum M, Dworniczak B. The ion channel polycystin-2 is required for left-right axis determination in mice. Curr Biol. 2002;12:938–943. doi: 10.1016/s0960-9822(02)00869-2. [DOI] [PubMed] [Google Scholar]
- Phan RT, Dalla-Favera R. The BCL6 proto-oncogene suppresses p53 expression in germinal-centre B cells. Nature. 2004;432:635–639. doi: 10.1038/nature03147. [DOI] [PubMed] [Google Scholar]
- Ranuncolo SM, Polo JM, Dierov J, Singer M, Kuo T, Greally J, Green R, Carroll M, Melnick A. Bcl-6 mediates the germinal center B cell phenotype and lymphomagenesis through transcriptional repression of the DNA-damage sensor ATR. Nature immunology. 2007;8:705–714. doi: 10.1038/ni1478. [DOI] [PubMed] [Google Scholar]
- Raya A, Belmonte JC. Left-right asymmetry in the vertebrate embryo: from early information to higher-level integration. Nature reviews. 2006;7:283–293. doi: 10.1038/nrg1830. [DOI] [PubMed] [Google Scholar]
- Raya A, Kawakami Y, Rodriguez-Esteban C, Buscher D, Koth CM, Itoh T, Morita M, Raya RM, Dubova I, Bessa JG, et al. Notch activity induces Nodal expression and mediates the establishment of left-right asymmetry in vertebrate embryos. Genes & development. 2003;17:1213–1218. doi: 10.1101/gad.1084403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raya A, Kawakami Y, Rodriguez-Esteban C, Ibanes M, Rasskin-Gutman D, Rodriguez-Leon J, Buscher D, Feijo JA, Izpisua Belmonte JC. Notch activity acts as a sensor for extracellular calcium during vertebrate left-right determination. Nature. 2004;427:121–128. doi: 10.1038/nature02190. [DOI] [PubMed] [Google Scholar]
- Rones MS, McLaughlin KA, Raffin M, Mercola M. Serrate and Notch specify cell fates in the heart field by suppressing cardiomyogenesis. Development (Cambridge, England) 2000;127:3865–3876. doi: 10.1242/dev.127.17.3865. [DOI] [PubMed] [Google Scholar]
- Sachs LM, Shi YB. Targeted chromatin binding and histone acetylation in vivo by thyroid hormone receptor during amphibian development. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:13138–13143. doi: 10.1073/pnas.260141297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweickert A, Campione M, Steinbeisser H, Blum M. Pitx2 isoforms: involvement of Pitx2c but not Pitx2a or Pitx2b in vertebrate left-right asymmetry. Mechanisms of development. 2000;90:41–51. doi: 10.1016/s0925-4773(99)00227-0. [DOI] [PubMed] [Google Scholar]
- Schweickert A, Weber T, Beyer T, Vick P, Bogusch S, Feistel K, Blum M. Cilia-driven leftward flow determines laterality in Xenopus. Curr Biol. 2007;17:60–66. doi: 10.1016/j.cub.2006.10.067. [DOI] [PubMed] [Google Scholar]
- Shaffer AL, Rosenwald A, Hurt EM, Giltnane JM, Lam LT, Pickeral OK, Staudt LM. Signatures of the immune response. Immunity. 2001;15:375–385. doi: 10.1016/s1074-7613(01)00194-7. [DOI] [PubMed] [Google Scholar]
- Shiratori H, Sakuma R, Watanabe M, Hashiguchi H, Mochida K, Sakai Y, Nishino J, Saijoh Y, Whitman M, Hamada H. Two-step regulation of left-right asymmetric expression of Pitx2: initiation by nodal signaling and maintenance by Nkx2. Molecular cell. 2001;7:137–149. doi: 10.1016/s1097-2765(01)00162-9. [DOI] [PubMed] [Google Scholar]
- Speder P, Petzoldt A, Suzanne M, Noselli S. Strategies to establish left/right asymmetry in vertebrates and invertebrates. Current opinion in genetics & development. 2007;17:351–358. doi: 10.1016/j.gde.2007.05.008. [DOI] [PubMed] [Google Scholar]
- Takada H, Hattori D, Kitayama A, Ueno N, Taira M. Identification of target genes for the Xenopus Hes-related protein XHR1, a prepattern factor specifying the midbrain-hindbrain boundary. Developmental biology. 2005;283:253–267. doi: 10.1016/j.ydbio.2005.04.020. [DOI] [PubMed] [Google Scholar]
- Tani S, Kurooka H, Aoki T, Hashimoto N, Honjo T. The N- and C-terminal regions of RBP-J interact with the ankyrin repeats of Notch1 RAMIC to activate transcription. Nucleic acids research. 2001;29:1373–1380. doi: 10.1093/nar/29.6.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tunyaplin C, Shaffer AL, Angelin-Duclos CD, Yu X, Staudt LM, Calame KL. Direct repression of prdm1 by Bcl-6 inhibits plasmacytic differentiation. J Immunol. 2004;173:1158–1165. doi: 10.4049/jimmunol.173.2.1158. [DOI] [PubMed] [Google Scholar]
- Vasanwala FH, Kusam S, Toney LM, Dent AL. Repression of AP-1 function: a mechanism for the regulation of Blimp-1 expression and B lymphocyte differentiation by the B cell lymphoma-6 protooncogene. J Immunol. 2002;169:1922–1929. doi: 10.4049/jimmunol.169.4.1922. [DOI] [PubMed] [Google Scholar]
- Vonica A, Brivanlou AH. The left-right axis is regulated by the interplay of Coco, Xnr1 and derriere in Xenopus embryos. Developmental biology. 2007;303:281–294. doi: 10.1016/j.ydbio.2006.09.039. [DOI] [PubMed] [Google Scholar]
- Wettstein DA, Turner DL, Kintner C. The Xenopus homolog of Drosophila Suppressor of Hairless mediates Notch signaling during primary neurogenesis. Development (Cambridge, England) 1997;124:693–702. doi: 10.1242/dev.124.3.693. [DOI] [PubMed] [Google Scholar]
- Williams R, Lendahl U, Lardelli M. Complementary and combinatorial patterns of Notch gene family expression during early mouse development. Mechanisms of development. 1995;53:357–368. doi: 10.1016/0925-4773(95)00451-3. [DOI] [PubMed] [Google Scholar]
- Ye BH, Cattoretti G, Shen Q, Zhang J, Hawe N, de Waard R, Leung C, Nouri-Shirazi M, Orazi A, Chaganti RS, et al. The BCL-6 proto-oncogene controls germinal-centre formation and Th2-type inflammation. Nature genetics. 1997;16:161–170. doi: 10.1038/ng0697-161. [DOI] [PubMed] [Google Scholar]
- Ye BH, Lista F, Lo Coco F, Knowles DM, Offit K, Chaganti RS, Dalla-Favera R. Alterations of a zinc finger-encoding gene, BCL-6, in diffuse large-cell lymphoma. Science. 1993;262:747–750. doi: 10.1126/science.8235596. [DOI] [PubMed] [Google Scholar]
- Yoshida T, Fukuda T, Hatano M, Koseki H, Okabe S, Ishibashi K, Kojima S, Arima M, Komuro I, Ishii G, et al. The role of Bcl6 in mature cardiac myocytes. Cardiovascular research. 1999;42:670–679. doi: 10.1016/s0008-6363(99)00007-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.