Abstract
Background
Results from large-scale epitope mapping using peptide microarray have been shown to correlate with clinical features of milk allergy.
Objectives
We sought to assess IgE and IgG4 epitope diversity and IgE affinity in different clinical phenotypes of milk allergy and identify informative epitopes that may be predictive of clinical outcomes of milk allergy.
Methods
Forty-one subjects were recruited from a larger study on the effects of ingesting heat-denatured milk proteins in milk-allergic individuals. Using food challenges, subjects were characterized as clinically reactive to all forms of milk (n = 17), tolerant to heated milk (HM) products (n = 16), or outgrown their milk allergy (n = 8). Eleven non-milk allergic, healthy volunteers served as controls. Peptide microarray was performed using the previously published protocol.
Results
Milk allergic subjects had increased epitope diversity as compared to those who outgrew their allergy. HM tolerant subjects had IgE binding patterns similar to those who had outgrown their allergy, but IgG4 binding patterns that were more similar to the allergic group. Binding to higher numbers of IgE peptides was associated with more severe allergic reactions during challenge. There was no association between IgG4 peptides and clinical features of milk allergy. Using a competitive peptide microarray assay, allergic patients demonstrated a combination of high and low affinity IgE binding whereas HM tolerant subjects and those who had outgrown their milk allergy had primarily low affinity binding.
Conclusions
Greater IgE epitope diversity and higher affinity as determined by peptide microarray were associated with clinical phenotypes and severity of milk allergy.
Keywords: Milk allergy, Peptide microarray, IgE pitope, IgE affinity, IgG4 epitope
INTRODUCTION
Cow’s milk hypersensitivity is a common disease affecting 2.5% of infants in the first year of life,[1] with approximately 60% of these milk disorders due to IgE-mediated mechanisms. Although the majority of children with IgE-mediated milk allergy develop tolerance by their teenage years,[2] 15—20% have lifelong allergy. The mechanisms responsible for tolerance are still not clearly understood; it has been shown that the presence of IgE antibodies to distinct allergenic epitopes of cow’s milk proteins can be used as a marker of persistent allergy.[3] Furthermore, a recent study demonstrated that the majority of milk allergic children can tolerate extensively heated forms of milk, and this subset of milk allergic patients are more likely to become tolerant to milk over time.[4]
The current diagnostic modalities for food allergy include skin prick testing and measurement of serum specific IgE levels. These results give an indication of the likelihood of clinical reactivity, however, individual results do not provide prognostic information or distinguish between the different phenotypes of food allergy. The importance of sequential epitope recognition in the persistence of cow’s milk allergy has been highlighted in several studies utilizing SPOTS membrane technology.[5–7] However, this method is time and labor intensive. Recently, peptide microarrays have been developed for large-scale epitope mapping using small quantities of serum.[8] Shreffler et al. [9] used the peptide microarray immunoassay to examine serum samples from peanut allergic patients and confirmed that antigenic areas identified by this method correlated with areas defined by SPOTS membrane mapping. Furthermore, epitope recognition correlated with peanut allergy severity.[9,10] Similarly, peptide microarray results have been shown to correlate with clinical features of milk allergy. Milk allergic and tolerant patients demonstrated different epitope recognition patterns, with allergic patients having higher ratios of IgE to IgG4 binding than those tolerant to milk.[11] Decreases in IgE binding and increases in IgG4 binding to milk peptides were correlated with clinical improvement in children undergoing oral immunotherapy with milk.[12] Differences in epitope diversity appear to be associated with clinical features of food allergy. Studies of milk allergenic epitopes have further demonstrated that certain milk IgE epitopes may be used as candidate biomarkers to predict the development of tolerance to milk.
In addition to epitope diversity and the concentration of epitope specific antibodies, IgE antibody affinity (the binding strength between antibodies and allergen or allergenic epitopes) may also be involved in the pathogenesis of allergic diseases. In this study, we sought to determine whether IgE and IgG4 epitope diversity and affinity of IgE antibodies were correlated with the different clinical phenotypes for milk allergy, including milk-allergic, milk-tolerant, and the intermediate group of milk allergic patients who can tolerate extensively heated forms of milk, i.e. baked-milk tolerant; and to identify informative epitopes that may be useful in predicting the clinical outcome of milk allergy.
METHODS
Patient sera
Sera previously used for mapping milk protein epitopes by SPOTS membrane technology [3] were used to confirm antigenic areas identified by peptide microarray.
To assess the association between epitope diversity and clinical phenotype of milk allergy, 41 subjects were recruited from a larger clinical study on the effects of ingesting heat-denatured milk proteins in milk-allergic individuals. Subjects were characterized based on the results of food challenges as “allergic” (reactive to all forms of milk products, n = 17), heated cow's milk (HM) tolerant (n = 16), or “outgrown” (n = 8) their milk allergy. The HM-tolerant group all tolerated heated milk in the form of muffins, waffles and pizza, but reacted to regular cow’s milk. Blood samples from subjects were obtained at the time of the initial baseline challenge. Eleven non-milk allergic, healthy volunteers served as controls. All research protocols were approved by the Mount Sinai Institutional Review Board, and informed consent was obtained for all subjects. Allergen-specific IgE levels and skin test data were obtained as previously described.[4]
Microarray
Peptides, Slides, Printing
A library of peptides, consisting of 20 amino acids overlapping by 17 (3-offset), corresponding to the primary sequences of αs1-, αs2-, β-, and κ-caseins, and β-lactoglobulin, was commercially synthesized. Peptides were resuspended in DMSO at 2 mg/mL, diluted 1:2 in Protein Printing Buffer (PPB, TeleChem International, Inc., Sunnyvale, CA) with 0.02% Sarkosyl to a final concentration of 1 mg/mL and printed on epoxy-derivatized glass slides (SuperEpoxy Substrate, TeleChem International, Inc.) using the NanoPrint™ Microarrayer 60 (TeleChem International, Inc.). PPB alone was used as negative controls and for background normalization, and fluorochrome-labeled bovine serum albumin (BSA) used for the purpose of grid alignment during analysis were included on each slide. All array elements were printed in duplicate (two sets of triplicates) to improve precision and to determine intra-assay variation.
Immunolabeling
Immunolabeling was performed as previously described with some modifications.[12,13] In brief, the slides were blocked with 400 µl of 1% human serum albumin (HSA) in phosphate-buffered saline containing 0.05% Tween 20 (PBS-T) for 60 minutes at room temperature, followed by incubation with 250 µl of patient serum diluted 1:5 in PBS-T/HSA for 24 hours at 4°C. Slides were then washed with PBS-T and incubated for 24 hours at 4°C with a cocktail of several monoclonal antibodies including three biotinylated monoclonal anti-human IgE antibodies: one from Invitrogen (Carlsbad, CA, USA) diluted 1:250, one from BD Biosciences Pharmingen (San Jose, CA, USA) diluted 1:250, and one as a gift from Phadia (Uppsala, Sweden), biotinylated in our laboratory and diluted 1:1000, and one monoclonal anti-human IgG4-FITC (SouthernBiotech, Birmingham, AL, USA) diluted 1:1000 in PBS-T/HSA. Slides were then incubated for 3 hours at 31°C with a cocktail of Anti-Biotin-Dendrimer_Oyster 550 (350) (Genisphere) and Anti-FITC_Dendrimer_Oyster 650 (350) (Genisphere, Hatfield, PA) in Dendrimer Buffer (Genisphere), both at 0.6 µg/ml with the addition of 0.02 µg/ml of salmon sperm DNA (Invitrogen), followed by wash with PBS-T, 15 mM Tris, 0.1X PBS, and 0.05X PBS. Slides were centrifuge dried and scanned using a ScanArray®Gx (PerkinElmer, Waltham, MA). Images were saved as TIF format.
Competition assay
The sera from seven milk allergic subjects and seven HM tolerant/outgrown subjects with binding to multiple milk peptides with the majority of binding having z-scores > 5 were selected for a competition assay. The competition assay was performed by immunolabeling the slide as described above with an additional incubation using a mixture of 1 mg/ml of casein (Sigma-Aldrich, St. Louis, MO) and 1.6 mg/ml of β-lactoglobulin (Sigma-Aldrich) at 16°C for 1 hour after serum incubation and before application of the 2nd antibody.
Data and Statistical Analysis
Fluorescence signal of each spot was digitized with the program ScanArray Express (Perkin Elmer), exported as comma-delimited text files and transformed to z-score as previously described.[11,12] An index z value of each peptide element was generated from the median of z-scores of the six replicate spots. An individual peptide sample was considered positive if its z-score exceeded 3, meaning that the signal was significantly above the background (p<0.003). In order to eliminate non-specific background noise, single positive peptides with negative binding by 4 adjacent peptides were filtered out by replacement with the median of itself and the 4 adjacent peptides.
In order to identify candidates of informative epitopes that would distinguish between the clinical phenotypes, we used the following procedure. The Wilcoxon test was used to determine statistical differences in reaction levels (signal intensity) to each peptide between groups. If two or more adjacent peptides demonstrated significantly different binding (<0.05) between groups, the amino acids spanning these peptides were considered a possible informative epitope.
The binding frequency of each group, which is the percentage of patients who had a z-score >3 at each peptide, was also calculated. For identification of informative epitopes, only areas showing a >30% difference in binding frequency between groups were considered.
Informative epitopes were identified as peptides that showed significantly different binding intensity between groups using the Wilcoxon test and >30% difference in binding frequency. Additional informative epitopes were identified based on IgE binding affinity. Amino acids of the informative epitopes were mapped based on the overlapping sequences of the identified peptides. Data presentation was performed using Microsoft Excel and TIGR Multiexperiment Viewer (TMeV).
RESULTS
Antigenic areas identified with the microarray correspond well with previously identified regions
Using the serum samples previously used for SPOTS membrane mapping, we confirmed that allergenic/antigenic areas identified with the peptide microarray correspond well with previously identified regions. Furthermore, microarray was able to detect binding in some areas where it was undetectable on SPOTS, indicating that microarray may be more sensitive than SPOTS membrane technology (data not shown).
IgE diversity corresponds to different phenotypes of milk allergy
Characteristics of the patient samples from the clinical study on the natural history of milk allergy are listed in Table 1. IgE from patients who have outgrown their milk allergy tended to bind fewer milk peptides as compared to IgE from those with persistent milk allergy (allergic patients) (median number of peptides bound by IgE = 3.5 vs 17, p=0.062), but there was no difference in IgG4 binding of peptides. IgE from HM tolerant individuals bound significantly fewer IgE peptides than the allergic group (median IgE peptides bound = 3 vs 17, p=0.019), and bind fewer peptides with IgG4 than either those who had outgrown their milk allergy or those who were still allergic (not statistically significant) (Table 2, Figure E1 and Figure E2 in Online Repository).
Table 1.
Patient characteristics
| Outgrown (n=8) |
Heated milk (HM) tolerant (n=16) |
Allergic (n=17) |
|
|---|---|---|---|
| Age (years) | |||
| Median | 5.8 | 5.7 | 7.3 |
| Mean | 7.9 | 6.6 | 7.7 |
| Range | 2.61 – 14.69 | 4.84 – 11.36 | 3.43 – 14.91 |
| Interquartile range (25–75%) | 3.38 – 12.94 | 5.49 – 6.95 | 4.66 – 9.77 |
| Gender (F/M) | 4/4 | 4/12 | 9/8 |
| Milk IgE (kU/L) | |||
| Median | 0.9* | 1.8* | 11.6* |
| Mean | 1.4 | 3.23 | 27.7 |
| Range | 0.36 – 3.23 | 0.35 – 14.7 | 0.77 – >100 |
| Interquartile range (25–75%) | 0.41 – 2.45 | 0.75 – 3.7 | 5.97 – 37.4 |
Statistically significant difference in milk IgE between the allergic and HM tolerant group (p=0.004) and between the allergic and outgrown group (p=0.02)
Table 2. Number of peptides bound by IgE and IgG4.
There was a significant difference between allergic subjects and HM tolerant subjects. There were no differences in IgG4 binding between groups.
| Outgrown | HM tolerant | Allergic | |
|---|---|---|---|
| Number of peptides bound by IgE | |||
| Mean | 12.3 | 14.6 | 46.6 |
| Median | 3.5 | 3* | 17* |
| Range | 0–66 | 0–103 | 0–186 |
| Number of peptides bound by IgG4 | |||
| Mean | 19 | 9.1 | 14.7 |
| Median | 9 | 6.5 | 13 |
| Range | 0–93 | 0–40 | 0–56 |
Peptide is positive if z-score > 3
Statistically different number of peptides bound by IgE between the HM tolerant and allergic group (p=0.019).
IgE and IgG4 binding frequency to each peptide by each group are shown in Figure 1. Individuals who were HM tolerant had IgE binding patterns similar to those who had outgrown their milk allergy, but IgG4 binding patterns that were more similar to the allergic group.
Figure 1.

Comparison of IgE and IgG4 binding frequency to peptides of five milk allergens between the different clinical phenotypes. The x-axis shows the overlapping peptides and the y-axis shows the percentage of patients within each group showing positive binding to each peptide. Binding for IgE is indicated above the x-axis line; binding for IgG4 is below the line. Candidates of informative epitopes are indicated with numbers.
IgE binding frequency was correlated with severity of symptoms during oral food challenge. Children in the allergic group who had a higher reaction grade during the challenge (anaphylaxis grade 4–5) bound a median of 89.5 IgE-binding peptides whereas children who reacted with anaphylaxis grades 1–2 bound a median of 4.5 IgE-binding peptides (p = 0.02). The HM tolerant group generally had milder reactions during their oral food challenges to milk (grades 1–2 only) and bound a median of 3 IgE-binding peptides. There was no correlation between number of IgG4 peptides and severity of allergic reactions during challenge or between number of IgE or IgG4 peptides bound and the eliciting dose during challenge.
In order to identify candidates of IgE or IgG4 informative epitopes, HM tolerant and outgrown individuals were combined as one group (HM tolerant/outgrown group, n=24) and compared with the allergic individuals (n=17). Only areas showing a >30% difference in binding frequency between groups were considered. Candidates of informative epitopes were identified as described in the Methods.
Eight areas which fulfilled the criteria listed in the Methods were identified as informative epitopes for IgE (Figure 1 and Figure 2). The majority were located in αs1-casein, correlating with SPOTS identified epitopes.[5–7] The corresponding IgE epitopes were identified on the protein sequence in Figure 3. There were no areas with >30% difference between groups for IgG4.
Figure 2.

Heat map of IgE binding to candidates of informative epitopes from the allergic patients compared to the HM-tolerant/outgrown patients.
Figure 3.

Amino acid sequence of milk allergens. Candidates of informative epitopes identified based on the binding frequency are in bold and underlined. Informative epitopes recognized by >50% of the allergic patients with high affinity IgE antibodies are highlighted gray. The numbers and letters indicated on top of each epitope correspond to the ones indicated in the other Figures.
Sera from non-milk allergic subjects were used as controls. Eight of the 11 sera had no detectable IgE binding, and 3 control subjects had IgE that bound a median of 9 milk peptides. The majority had IgG4 binding to milk peptides (median of 2.5 peptides).
Of note, within each clinical group, there was heterogeneity in terms of IgE and IgG4 binding to milk peptides, with each subject having a unique pattern of binding.
IgE affinity correlates to different phenotypes of milk allergy
In order to determine the relationship between IgE antibody affinity and clinical allergy phenotype, a competition assay was performed with a subset of patients who had binding to multiple milk epitopes with the majority of binding having z-scores > 5 (7 allergic, 5 HM tolerant, and 2 outgrown). The competition conditions, including the concentration of competitor proteins, temperature and length of incubation, were first determined using a serum pool of five highly milk allergic individuals (Figures E3-E5, Online repository). No competition effect could be observed from the replicate arrays incubated with PBS-T/HSA (data not shown).
Compared with the standard protocol, the competition assay using the caseins and beta-lactoglobulin virtually eliminated all IgE binding from HM tolerant/outgrown individuals (Figure 4). No competition effect was observed in arrays incubated with PBS-T/HSA. For patients in the allergic group, IgE binding was affected to a much lesser degree (Figure 5). For allergic patients, IgE antibodies to certain epitopes remained bound (Z score > 3) following competition and its Z score decreased <50% as compared to the standard protocol, suggesting that they are high affinity antibodies. In general, all allergic patients had a mixture of high and low affinity antibodies to different epitopes. There were three epitopes (A-C, Figure 6) recognized by >50% of the allergic patients whose specific IgE antibodies to these epitopes were of relatively high affinity. One epitope overlapped with the informative epitopes identified with the standard protocol (C in Figure 3). Two additional informative epitopes (A and B) with high antibody affinity are also indicated in Figure 3.
Figure 4.
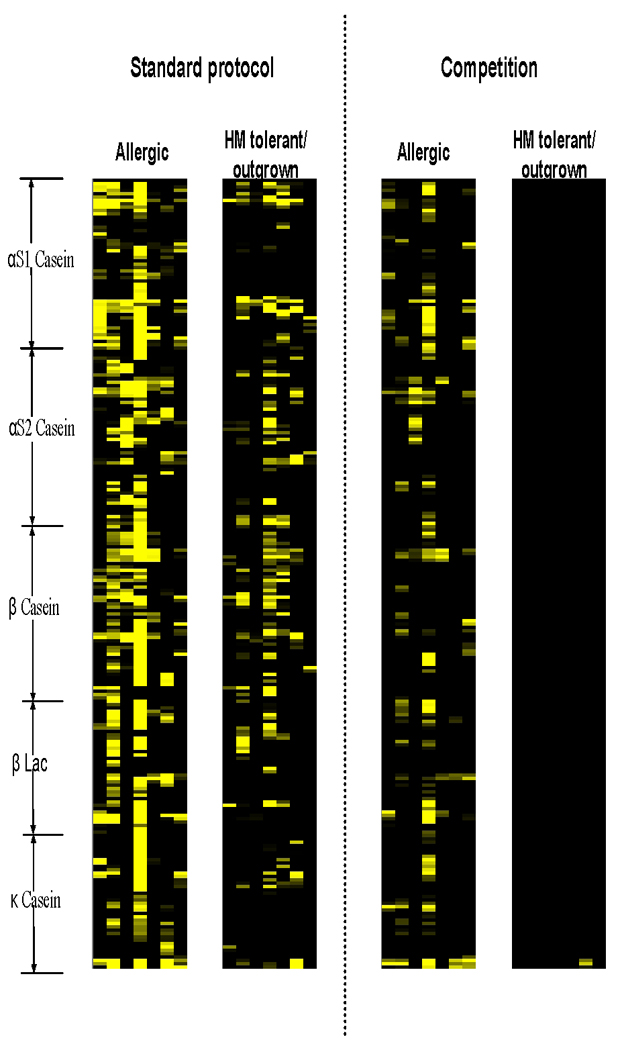
Heat map showing the effects of competition on 7 allergic and 7 HM-tolerant/outgrown patients.
Figure 5.

Scatter plot comparing IgE binding from 7 allergic (red) and 7 HM-tolerant/outgrown (blue) patients between the standard protocol (x-axis) and the competition assay (y-axis). Each spot represents IgE binding (represented in Z score) to one milk peptide.
Figure 6.

Comparison of IgE peptide binding frequency between groups using the standard protocol and competition assay. Informative epitopes recognized by >50% of the allergic patients with high affinity IgE antibodies are indicated with letters.
DISCUSSION
The aim of this study was to determine whether IgE and IgG4 milk allergen epitope recognition and IgE antibody affinity correlated with different phenotypes of milk allergy. First, we confirmed that antigenic areas identified with the peptide microarray correspond well with regions previously identified using SPOTS membrane technology, using stored serum from subjects participating in the original study.[3]. Next, we demonstrated that IgE epitope diversity on peptide microarray correlated with clinical phenotypes of milk allergy. Patients with persistent allergy had increased epitope diversity as compared to those with transient allergy (outgrown). Immunologic studies from the heated milk study suggest that those who are HM tolerant have a favorable prognosis and are less clinically reactive than those who are unable to tolerate heated milk.[4,14–15] Our microarray data are consistent with these results since IgE from HM tolerant subjects recognized fewer milk allergen peptides than those who were unable to tolerate HM, and were more similar to those who had outgrown their allergy. Furthermore, binding to increased numbers of IgE peptides correlated with severity of allergic reactions during oral food challenge.
IgG4 binding to milk peptides was detected in the majority of patients in all groups. The HM tolerant group tended to have the least amount of IgG4 binding to milk peptides. This was similar to the reported lower quantitative IgG4 levels to casein and β-lactoglobulin in the HM tolerant group [4]. Since strict avoidance is currently the mainstay of treatment, it is possible that IgG4 binding may increase as the HM tolerant group incorporates certain heated milk products into their diets. Alternatively, that as subjects develop tolerance to milk, they may lose IgG4 to some epitopes and develop IgG4 to other areas of the milk proteins.
Although several IgE epitopes bound by a significantly higher percentage of allergic subjects were identified in our study, no single informative epitope that reliably distinguished between the different phenotypes of milk allergy could be identified due to the heterogeneity of epitopes recognized by subjects within the different phenotypes, which is similar to observations in studies of the peanut microarray.[9,10] Previous studies have indicated that the number of epitopes recognized, rather than recognition of specific epitopes, may be more predictive of clinical features of food allergy.[19,20] However, an overlap in the number of epitopes recognized was also observed.
A possible explanation for the heterogeneity of IgE binding within the allergic group is the relatively young age of the study population (median age 7.3 years). A recent report indicated that some patients do not outgrow their milk allergy until their early teenage years.[2] Therefore, it is possible that some of the subjects currently classified as allergic will eventually outgrow their milk allergy, and thus should be re-classified in a different group.
Although epitope diversity to peanut allergens appears stable, [10] epitope diversity for milk allergens can change over time. In a study of patients undergoing oral immunotherapy with milk, children who were successfully desensitized demonstrated decreased IgE binding and increased IgG4 binding following treatment.[12] The opposite trend was noted in those who were unable to be desensitized. An important difference between these two studies is the therapeutic intervention in the milk study as opposed to the peanut study where subjects continued peanut avoidance. Therefore, it is possible that continued exposure to milk in the form of HM products in our patients will lead to similar immunologic effects on epitope binding patterns as seen in children undergoing oral immunotherapy. Alternatively, peanut allergy is generally more persistent, so characteristics of epitope recognition may be different for different food proteins.
A novel finding in our study is that IgE affinity is correlated with clinical features of milk allergy. This is the first time that antibody affinity has been shown to closely reflect clinical characteristics of milk allergy. Milk “allergic” patients have IgE antibodies that bind to certain peptide segments of milk proteins with high affinity and that are not competed off by native milk protein. In contrast, those who have transient allergy (outgrown) or are HM tolerant have low affinity binding that is completely competed away by native milk protein. These results suggest that IgE affinity may play a role in the development of tolerance as well.
Affinity maturation is an important step in the progressive differentiation of antigen-specific B cells, leading to antibody clones with higher affinity. Thus affinity may be one factor in determining the biological effects of IgE antibodies. However, the role of antibody affinity in allergy has not been well-described. One early report investigated the effect of IgE antibody affinity to Der p 2 on Der p 2-induced histamine release from human basophils and found that the sensitivity of histamine release was closely related to the affinity.[16] Recently, Christensen et al. [17] demonstrated that higher affinity IgE binding to Der p 2 was associated with higher levels of basophil activation. There is one published report investigating antibody avidity in peanut allergy. Using a thiocyanate ELISA elution assay, the authors found only a weak correlation between IgE and IgG avidity for Ara h 2 and symptom score during oral food challenge.[18] Thiocyanate elution works by non-specifically dissociating antigen–antibody complexes by interfering with non-covalent binding. In contrast, native milk protein was used in our study as a competitive inhibitor for IgE binding sites, which may be a more relevant indicator of antibody affinity in food allergy. Furthermore, when only antibodies with higher affinity were considered, the overlap observed in the epitope recognition pattern could be eliminated, resulting in a clearer distinction between groups.
Additional studies are necessary to confirm these findings in larger numbers of patients and to determine whether changes in epitope diversity and affinity occur over time as a result of therapeutic interventions and/or a part of the natural history of food allergy. This information may provide additional insight into the mechanisms of tolerance. On the other hand, if epitope diversity and affinity is found to be stable over time, peptide microarrays may be a useful diagnostic tool that can provide prognostic information regarding development of tolerance and severity of milk allergy based on the affinity profile and epitope diversity, respectively. The current diagnostic tools are unable to provide this type of prognostic information, and therefore the peptide microarray may represent a much needed advancement in diagnostics for the field of food allergy.
Acknowledgments
Funding source
Funded in part by a grant from the NIAID, AI-44236. Julie Wang, MD is funded in part by a grant from the National Institutes of Health/National Institute of Allergy and Infectious Diseases; AI083883. A. Nowak-Węgrzyn is supported in part by NIH NIAID AI 059318
ABBREVIATIONS
- IgE
immunoglobulin E
- IgG4
Immunoglobulin G4
- HM
heated milk
- HAS
human serum albumin
- PBS-T
phosphate-buffered saline containing 0.05% Tween 20
- PPB
protein printing buffer
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CLINICAL IMPLICATIONS
IgE epitope diversity and affinity correlate with clinical phenotypes and severity of milk allergy. Peptide microarray may be a useful diagnostic tool that can provide prognostic information regarding milk allergy.
CAPSULE SUMMARY
Using a peptide microarray assay, IgE epitope diversity and affinity were found to correlate with different clinical phenotypes and severity of milk allergy. This may be useful to provide prognostic information regarding milk allergy.
References
- 1.Sampson HA. Food allergy. Part 1: Immunopathogenesis and clinical disorders. J Allergy Clin Immunol. 1999;103:717–728. doi: 10.1016/s0091-6749(99)70411-2. [DOI] [PubMed] [Google Scholar]
- 2.Skripak JM, Matsui EC, Mudd K, Wood RA. The natural history of IgE-mediated cow's milk allergy. J Allergy Clin Immunol. 2007;120:1172–1177. doi: 10.1016/j.jaci.2007.08.023. [DOI] [PubMed] [Google Scholar]
- 3.Jarvinen KM, Beyer K, Vila L, Chatchatee P, Busse PJ, Sampson HA. B-cell epitopes as a screening instrument for persistent cow's milk allergy. J Allergy Clin Immunol. 2002;110:293–297. doi: 10.1067/mai.2002.126080. [DOI] [PubMed] [Google Scholar]
- 4.Nowak-Wegrzyn A, Bloom KA, Sicherer SH, Shreffler WG, Noone S, Wanich N, et al. Tolerance to extensively heated milk in children with cow's milk allergy. J Allergy Clin Immunol. 2008;122:342–347. doi: 10.1016/j.jaci.2008.05.043. [DOI] [PubMed] [Google Scholar]
- 5.Chatchatee P, Jarvinen KM, Bardina L, Beyer K, Sampson HA. Identification of IgE- and IgG-binding epitopes on alpha(s1)-casein: differences in patients with persistent and transient cow's milk allergy. J Allergy Clin Immunol. 2001;107:379–383. doi: 10.1067/mai.2001.112372. [DOI] [PubMed] [Google Scholar]
- 6.Järvinen KM, Chatchatee P, Bardina L, Beyer K, Sampson HA. IgE and IgG binding epitopes on alpha-lactalbumin and beta-lactoglobulin in cow's milk allergy. Int Arch Allergy Immunol. 2001;126:111–118. doi: 10.1159/000049501. [DOI] [PubMed] [Google Scholar]
- 7.Chatchatee P, Järvinen KM, Bardina L, Vila L, Beyer K, Sampson HA. Identification of IgE and IgG binding epitopes on beta- and kappa-casein in cow's milk allergic patients. Clin Exp Allergy. 2001;31:1256–1262. doi: 10.1046/j.1365-2222.2001.01167.x. [DOI] [PubMed] [Google Scholar]
- 8.Lin J, Bardina L, Shreffler WG, Andreae DA, Ge Y, Wang J, et al. Development of a novel peptide microarray for large scale epitope mapping of food allergens. J Allergy Clin Immunol. 2009;124:315–322. doi: 10.1016/j.jaci.2009.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shreffler WG, Beyer K, Chu TH, Burks AW, Sampson HA. Microarray immunoassay: association of clinical history, in vitro IgE function, and heterogeneity of allergenic peanut epitopes. J Allergy Clin Immunol. 2004;113:776–782. doi: 10.1016/j.jaci.2003.12.588. [DOI] [PubMed] [Google Scholar]
- 10.Flinterman AE, Knol EF, Lencer DA, Bardina L, den Hartog Jager CF, Lin J, et al. Peanut epitopes for IgE and IgG4 in peanut-sensitized children in relation to severity of peanut allergy. J Allergy Clin Immunol. 2008;121:737–743. doi: 10.1016/j.jaci.2007.11.039. [DOI] [PubMed] [Google Scholar]
- 11.Cerecedo I, Zamora J, Shreffler WG, Lin J, Bardina L, Dieguez MC, et al. Mapping of the IgE and IgG4 sequential epitopes of milk allergens with a peptide microarray-based immunoassay. J Allergy Clin Immunol. 2008;122:589–594. doi: 10.1016/j.jaci.2008.06.040. [DOI] [PubMed] [Google Scholar]
- 12.Pecora V, Nucera E, Schiavino D, Lombardo C, Bardina L, Lin J, et al. Evaluation of specific sequential IgE- and IgG4-binding epitopes, recognized in cow’s milk allergic patients during specific oral desensitization, using peptide microarray immunoassay. J Allergy Clin Immunol. 2009;123:S178. [Google Scholar]
- 13.Lin J, Bardina L, Shreffler WG. Reineke U, editor. Microarrayed allergen molecules for diagnostics of allergy. Methods in Molecular Biology-Epitope Mapping Protocols. 2009;524:259–272. doi: 10.1007/978-1-59745-450-6_19. [DOI] [PubMed] [Google Scholar]
- 14.Shreffler WG, Wanich N, Moloney M, Nowak-Wegrzyn A, Sampson HA. Association of allergen-specific regulatory T cells with the onset of clinical tolerance to milk protein. J Allergy Clin Immunol. 2009;123:43–52. doi: 10.1016/j.jaci.2008.09.051. [DOI] [PubMed] [Google Scholar]
- 15.Wanich N, Nowak-Wegrzyn A, Sampson HA, Shreffler WG. Allergen-specific basophil suppression associated with clinical tolerance in patients with milk allergy. J Allergy Clin Immunol. 2009;123:789–794. doi: 10.1016/j.jaci.2008.12.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mita H, Yasueda H, Akiyama K. Affinity of IgE antibody to antigen influences allergen-induced histamine release. Clin Exp Allergy. 2000;30:1583–1589. doi: 10.1046/j.1365-2222.2000.00921.x. [DOI] [PubMed] [Google Scholar]
- 17.Christensen LH, Holm J, Lund G, Riise E, Lund K. Several distinct properties of the IgE repertoire determine effector cell degranulation in response to allergen challenge. J Allergy Clin Immunol. 2008;122:298–304. doi: 10.1016/j.jaci.2008.05.026. [DOI] [PubMed] [Google Scholar]
- 18.El-Khouly F, Lewis SA, Pons L, Burks AW, Hourihane JO. IgG and IgE avidity characteristics of peanut allergic individuals. Pediatr Allergy Immunol. 2007;18:607–613. doi: 10.1111/j.1399-3038.2007.00542.x. [DOI] [PubMed] [Google Scholar]
- 19.Lewis SA, Grimshaw KE, Warner JO, Hourihane JO. The promiscuity of immunoglobulin E binding to peanut allergens, as determined by Western blotting, correlates with the severity of clinical symptoms. Clin Exp Allergy. 2005;35:767–773. doi: 10.1111/j.1365-2222.2005.02252.x. [DOI] [PubMed] [Google Scholar]
- 20.Peeters KA, Koppelman SJ, van Hoffen E, van der Tas CW, den Hartog Jager CF, Penninks AH, et al. Does skin prick test reactivity to purified allergens correlate with clinical severity of peanut allergy? Clin Exp Allergy. 2007;37:108–115. doi: 10.1111/j.1365-2222.2006.02628.x. [DOI] [PubMed] [Google Scholar]


