SUMMARY
Centrosomes and their component centrioles represent the principal microtubule organizing centers of animal cells. Here we show that the gene underlying Orofaciodigital Syndrome 1, Ofd1, is a component of the distal centriole that controls centriole length. In the absence of Ofd1, distal regions of centrioles, but not procentrioles, elongate abnormally. These long centrioles are structurally similar to normal centrioles, but contain destabilized microtubules with abnormal post-translational modifications. Ofd1 is also important for centriole distal appendage formation and centriolar recruitment of the intraflagellar transport protein Ift88. To model OFD1 Syndrome in embryonic stem cells, we replaced the Ofd1 gene with missense alleles from human OFD1 patients. Distinct disease-associated mutations cause different degrees of excessive or decreased centriole elongation, all of which are associated with diminished ciliogenesis. Our results indicate that Ofd1 acts at the distal centriole to build distal appendages, recruit Ift88, and stabilize centriolar microtubules at a defined length.
INTRODUCTION
Centrosomes organize the microtubule cytoskeleton of animal cells and play essential roles in mitosis in vertebrate cells (Badano et al., 2005; Mikule et al., 2007; Nigg, 2004). Centrioles, the functional hub of the centrosome, have a complex structure based upon a central core of microtubules arranged in a nine-fold triplet pattern. In a G1 or G0 cell, the centrosome consists of two centrioles, the mother and daughter, and pericentriolar material. In contrast to most cellular microtubules, which display dynamic instability and range widely in length, centriolar microtubules undergo regulated growth to a characteristic length, are extremely stable, and display numerous posttranslational modifications (PTMs) including acetylation and polyglutamylation (Bettencourt-Dias and Glover, 2009; Kochanski and Borisy, 1990). The centriole also exhibits polarity, with the microtubule minus ends defining proximal and the plus ends defining distal (Bornens, 2002).
The proximal and distal ends of centrioles are structurally and functionally distinct. By transmission electron microscopy (TEM), the distal lumen of both mother and daughter centrioles contains electron dense material (Vorobjev and Chentsov, 1980). Additionally, the mother centriole is longer than the daughter and possesses two sets of projections at the distal end called subdistal and distal appendages (Chretien et al., 1997; Paintrand et al., 1992).
The proximal end of the mother and daughter centrioles is the site at which a single new centriole, termed a procentriole, begins to grow during the process of centrosome duplication in S phase. The microtubules of procentrioles grow to a defined length before entry into mitosis, but these new centrioles must pass through the G2 phase of the next cell cycle before they achieve their full length. In the process of centriole maturation, the centriole grows and acquires the properties of a mother centriole, including appendages (Azimzadeh and Bornens, 2007).
The distal and subdistal appendages are required for ciliogenesis, another important centrosomal function (Graser et al., 2007; Ishikawa et al., 2005). Primary cilia project from the surface of many vertebrate cells and transduce signals essential for normal development and adult tissue homeostasis (Sharma et al., 2008). Recently, it has become clear that defects in cilia underlie a group of genetic syndromes known as ciliopathies. Ciliopathies include diseases such as polycystic kidney disease, Bardet-Biedl syndrome, and Orofaciodigital syndrome 1 (OFD1) (Badano et al., 2006; Christensen et al., 2007). OFD1 is X-linked, and the syndrome is lethal in males. Females present with a variable phenotype including malformations of the face, oral cavity and digits, and often polycystic kidney disease (Ferrante et al., 2001). It has not been clear how the Ofd1 gene product, which localizes to the centrosome, promotes primary cilia formation (Ferrante et al., 2006; Romio et al., 2004).
Here we show that Ofd1 associates with the distal ends of centriolar microtubules and constrains mother and daughter centriole elongation. We demonstrate that Ofd1 is necessary for distal appendage formation and Ift88 recruitment, two processes essential for cilium formation. We also model effects of Ofd1 human mutations in mouse embryonic stem (ES) cells, revealing that each disease-associated mutation differentially affects centriole length and ciliogenesis.
RESULTS
Ofd1 is required for centriole length control
To understand how Ofd1 contributes to normal centrosome structure and function, we characterized male murine ES cells with a gene trap mutation in the Ofd1 locus, Ofd1Gt cells. As Ofd1 is located on the X chromosome, these Ofd1Gt cells are hemizygous for Ofd1 and do not produce Ofd1 protein (Figure 1A).
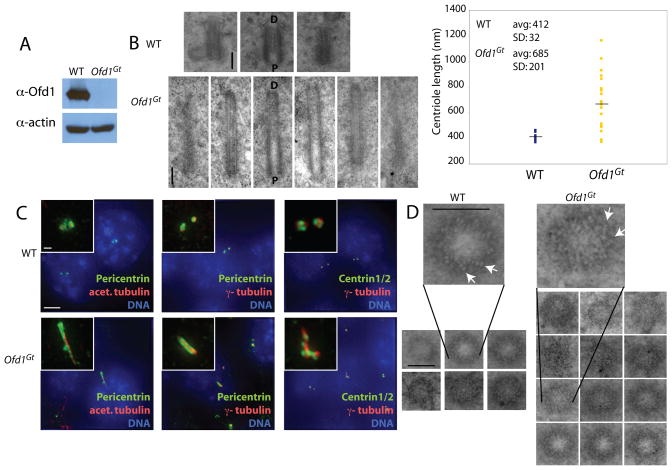
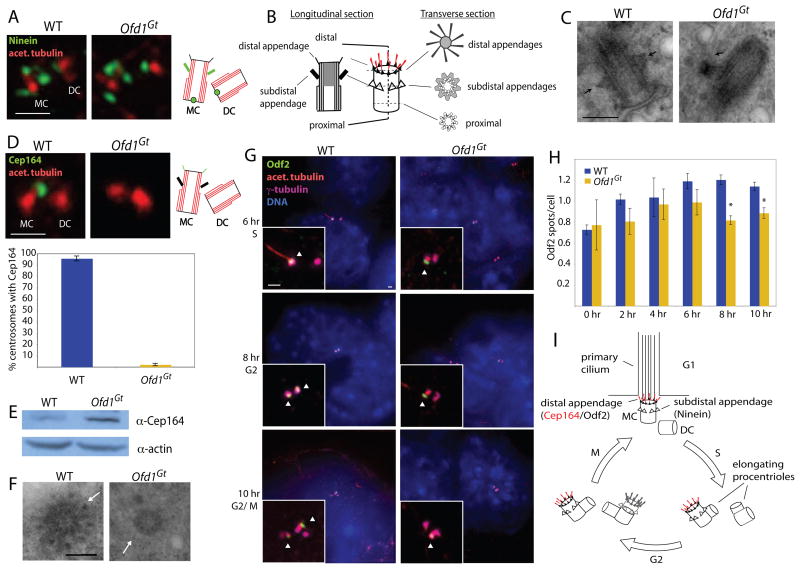
Figure 1. Ofd1 is essential for centriole length control.
(A) Immunoblot of cell lysate supernatants from wild type (WT) and Ofd1Gt cells. 15 μg protein loaded per lane. (B) Longitudinal TEM sections of WT and Ofd1Gt cell centrioles. Long centrioles (defined as > 600 nm) are seen in 35% of Ofd1Gt cells. P, proximal end and D, distal end of centriole. Graph shows centriole length data, collected from 9 WT and 23 Ofd1Gt centrioles. Each measured centriole was from a distinct cell. (C) Representative fluorescence micrographs of WT and Ofd1Gt cells showing centrosomes (Pericentrin and γ-tubulin), centrioles (Centrin and acetylated tubulin), and DNA (DAPI). (D) Transverse TEM sections of WT and Ofd1Gt cell centrioles. White arrows indicate triplet microtubules. Normal length centrioles are contained within a maximum of 8–10 transverse sections, whereas long centrioles span more than 10 sections. Scale bars indicate 200 nm (TEM), 5 μm, and 1 μm (inset). See also Figure S1.
TEM of asynchronously growing cells revealed abnormally long centriole-like structures in 35% of cells lacking Ofd1, but never in wild type cells (Figure 1B). The mean length of wild type centrioles was 412 nm, whereas the Ofd1 mutant centrioles averaged 685 nm, and were sometimes more than a micron long. In contrast to the wild type centrioles which showed little variation in length (SD = 32 nm), the mutant centrioles varied widely in length (SD = 201 nm). The long Ofd1 mutant centrioles had the microtubule composition and morphology of normal size centrioles, with distinct proximal and distal ends (Figure 1B), and recruited centrosomal and centriolar proteins including Pericentrin, acetylated tubulin, γ-tubulin, and Centrin (Figure 1C). The long Ofd1 mutant centrioles also possessed known centriole-specific proteins, including Ninein, CP110, and Cep97 (Kleylein-Sohn et al., 2007; Mogensen et al., 2000; Piel et al., 2000; Spektor et al., 2007) (Figure S1A-C). Thus, Ofd1 is essential for the regulation of centriole length.
Recent work has shown that CP110 is also required for centriole length control (Kohlmaier et al., 2009; Schmidt et al., 2009; Tang et al., 2009). CP110 and Cep97 showed normal levels and localization on both normal size and long centrioles of Ofd1 mutant cells, indicating that centriole elongation defects were not due to misregulation of the centriolar localization of these proteins. Transverse TEM sections showed that, like wild type centrioles, both the long and normal sized centrioles of Ofd1 mutant cells contained triplet microtubules (Figure 1D, Figure S1D).
Centrioles have critical roles in the mitotic and microtubule organizing functions of centrosomes, as well as in promoting procentriole formation. The doubling time of Ofd1Gt cells was not different from wild type cells (Figure S1E). Additionally, wild type and Ofd1Gt cells had similar cell cycle phase distributions (Figure S1F).
Furthermore, loss of Ofd1 did not affect the interphase microtubule array or mitotic spindle structures (Figure S1G). Ofd1Gt cells showed no changes in normal centrosome or procentriole number, indicating that Ofd1 is not required for centriole duplication (Figure S1G-I). Both TEM and localization of procentriole components showed that long Ofd1 mutant centrioles were associated with the normal number of procentrioles (Figure S1H-I), indicating that long mutant centrioles were capable of promoting normal centriole duplication. Microtubule nucleation and anchoring abilities of wild type and Ofd1Gt centrosomes were examined by using nocodazole treatment to depolymerize microtubules, and then observing microtubule regrowth and anchoring 30 seconds to 15 minutes following nocodazole removal (Figure S1J, and data not shown). Loss of Ofd1 did not affect microtubule nucleation or anchoring. Taken together, these data indicate that centriole duplication, microtubule organization and the mitotic functions of centrosomes do not depend on Ofd1.
Ofd1 localizes to the distal ends of all centrioles
To determine where within the centrosome Ofd1 localizes, we generated an antibody to Ofd1. The antibody recognized the centrosome of wild type cells, but not of Ofd1Gt cells (Figure 2A–B). Similarly, preimmune serum did not recognize the centrosome, confirming the specificity of the Ofd1 antibody (Figure 2A). In asynchronous cells, Ofd1 was present as two or four dots within the centrosome(s) (Figure 2B).
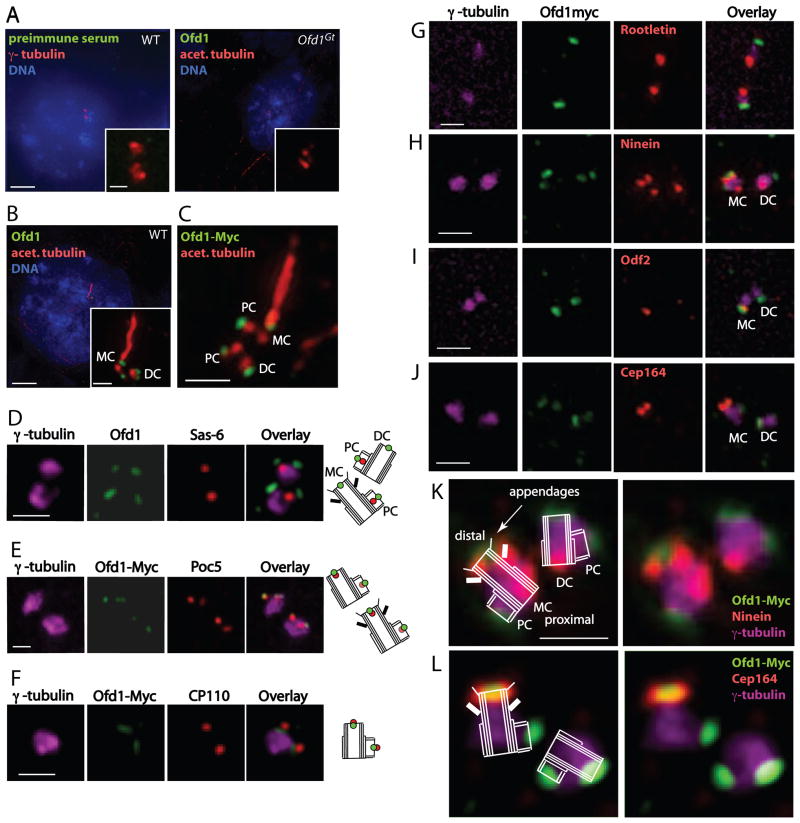
Figure 2. Ofd1 localizes to the distal ends of mother, daughter, and procentrioles.
(A–C) Representative micrographs of WT and Ofd1Gt cells showing centrosomes (γ-tubulin), centrioles and cilia (acetylated tubulin), DNA (DAPI), and other indicated antibodies. (D) WT cells showing centrosomes (γ-tubulin), Ofd1, and the proximal procentriole (Sas-6). (E) Ofd1Ofd1myc cells showing centrosomes (γ-tubulin), Myc (Ofd1-Myc), and the distal procentriole (Poc5). Poc5 localizes more strongly to mother or daughter centrioles than to procentrioles. (F) Ofd1Ofd1myc cells showing centrosomes (γ-tubulin), Myc (Ofd1-Myc), and the distal centriole and procentriole (CP110). (G) Ofd1Ofd1myc cells showing centrosomes (γ-tubulin), Myc (Ofd1-Myc), and the proximal centriole (Rootletin). (H) Ofd1Ofd1myc cells showing centrosomes (γ-tubulin), Myc (Ofd1-Myc), and mother centriole subdistal appendages (Ninein). The mother centriole is marked by 3 Ninein foci (2 on the subdistal appendages and one on the proximal end) whereas the daughter centriole is marked by one Ninein focus (on the proximal end). (I) Ofd1Ofd1myc cells showing centrosomes (γ-tubulin), Myc (Ofd1-Myc), and mother centriole appendages (Odf2). (J) Ofd1Ofd1myc cells showing Myc (Ofd1-Myc), mother centriole distal appendages (Cep164), and centrosomes (γ-tubulin). (K) – (L) Schematics showing Ofd1Ofd1myc cells stained for Myc (Ofd1-Myc), mother centriole subdistal (Ninein) or distal (Cep164) appendages, and centrosomes (γ-tubulin). MC, mother centriole. DC, daughter centriole. PC, procentriole. Scale bars (A)–(B) indicate 5 μm and 1 μm (inset), and 1 μm for (C)–(L). See also Figure S2.
Closer inspection of wild type ES cells revealed that Ofd1 was associated with mother, daughter and procentrioles (Figure S2A). In cells in which the mother centriole extended a cilium, Ofd1 localized to the base of the cilium (Figure 2B–C). Similar localization to centrioles and the cilium base was seen in fibroblast, kidney, bone and retinal cell lines of mouse or human origin (Figure S2B). To further assess Ofd1 centriolar association, we induced centriole overduplication by arresting U2OS cells in S phase (Habedanck et al., 2005). Consistent with findings in other cells, Ofd1 associated with all centrioles, including supernumerary centrioles, in arrested U2OS cells (Figure S2C).
We have developed a technology, called Floxin, to engineer ES cell gene trap loci (Singla et al., 2009). Floxin enables efficient targeted insertion of DNA constructs into gene trap loci. Using the Floxin approach, we introduced a carboxy-terminal Myc tagged version of wild type Ofd1 (Ofd1-Myc knock-in) at the endogenous locus (Ofd1Ofd1myc cells). Inserting an Ofd1-Myc gene into the native genomic context and under control of endogenous regulatory elements restored Ofd1 protein expression and prevented long centriole formation (Figure S2D–E). However, Ofd1-Myc Floxin cells expressed reduced levels of protein as compared to wild type (Singla et al., 2009). Localization of the Myc tag of Ofd1 confirmed that Ofd1 localized to centrioles and the cilium base (Figure 2C).
To ascertain where Ofd1 localizes on procentrioles, we examined three markers of procentrioles: Sas-6, Poc5, and CP110, which associate with the proximal, distal and very distal ends of procentrioles, respectively (Azimzadeh et al., 2009; Kleylein-Sohn et al., 2007; Strnad et al., 2007). Ofd1 localized to the procentriole distal end, in a domain between Poc5 and CP110 (Figure 2D–F).
We next examined the localization of Ofd1 on mother and daughter centrioles. Costaining with Rootletin or Poc1, which mark the proximal ends of mother and daughter centrioles, showed that Ofd1 localized to the opposite, distal ends (Bahe et al., 2005; Keller et al., 2008) (Figure 2G, Figure S2F). To ascertain the localization of Ofd1 more precisely, we examined Ofd1 localization relative to Ninein, Odf2, and Cep164, which are parts of the subdistal and distal centriole appendages (Graser et al., 2007; Ishikawa et al., 2005; Mogensen et al., 2000; Nakagawa et al., 2001; Piel et al., 2000) (Figure 2H–L). Ofd1 was located at the very distal ends of centrioles, at a more central position than the subdistal and distal appendages.
Taken together, these data reveal that Ofd1 localizes to the distal ends of all centrioles (mother, daughter, and procentrioles), closely associated with the centriole microtubule barrel. This is consistent with immuno-electron microscopy studies that showed Ofd1 to be associated with centrioles (Romio et al., 2004).
Ofd1 mutant centrioles contain destabilized microtubules
As Ofd1 is in close proximity to centriolar microtubules, we examined whether Ofd1 associates with microtubule components. We found that Ofd1 was best solubulized in a modified RIPA buffer containing sodium deoxycholate and NP-40 detergents (Figure 3A). Immunoprecipitation revealed that Ofd1 complexes contained γ-tubulin and acetylated tubulin, two forms of tubulin found in centriolar microtubules(Moudjou et al., 1996) (Figure 3B). Ofd1 complexes did not contain other proximal or distal centriolar proteins (Figure 3B, Figure S3A). Together with the above data that revealed Ofd1 localization at the centriole distal end, these data suggest that Ofd1 caps the distal ends of all centrioles in a complex intimately associated with centriolar microtubules.
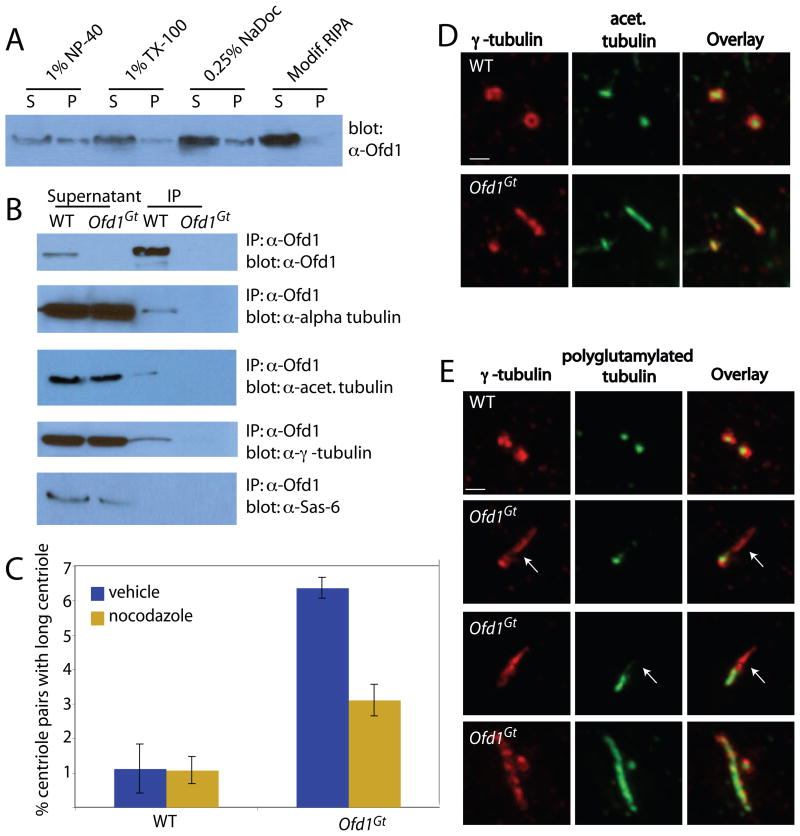
Figure 3. Ofd1 complexes contain centriolar microtubule components and control centriole microtubule stability.
(A) Immunoblot showing Ofd1 (detected with an Ofd1 antibody) in the supernatant (S) and pellet (P) of WT cells lysed with various detergents. (B) Immunoblots of Ofd1 complexes immunoprecipitated from WT or Ofd1Gt cell supernatant with an Ofd1 antibody. (C) Graph indicating percent of long centrioles in WT and Ofd1Gt cells. Cells were treated with nocodazole, fixed and stained for α- and γ-tubulin. γ-tubulin foci more than twice as long as they were wide were counted as long centrioles. Because immunofluorescent (IF) microscopy has lower resolution than TEM, a smaller percent of Ofd1Gt centrioles appeared long when assessed by IF (6–10% by IF versus 35% by TEM). (D) WT and Ofd1Gt cells stained for centrosomes (γ-tubulin) and acetylated tubulin. (E) WT and Ofd1Gt cells stained for centrosomes (γ-tubulin) and polyglutamylated tubulin (GT335). Arrows indicate areas of reduced or absent polyglutamylation. Scale bars indicate 1μm.
Centriolar microtubules are extremely stable, as reflected by their resistance to microtubule depolymerizing drugs (Kochanski and Borisy, 1990). To assess whether abnormal centriole length reflects altered centriolar microtubule dynamics, we treated cells with nocodazole. Whereas nocodazole did not affect the length of wild type centrioles, it shrank Ofd1 mutant centrioles (Figure 3B).
Stabilized microtubules are associated with PTMs such as acetylation and polyglutamylation (Bobinnec et al., 1998; Loktev et al., 2008). We therefore investigated whether microtubules of long Ofd1 mutant centrioles have aberrant PTMs. Although the microtubules of long centrioles were normally acetylated, polyglutamyl groups were reduced or absent from approximately 50% of long centrioles (Figure 3C–D). Thus, Ofd1 may constrain centriole length in part by affecting the dynamics of centriolar microtubules.
Ofd1 controls elongation of the distal centriole in G2
Procentrioles first elongate during S phase, while daughter centrioles elongate and mature during G2. To investigate if cell cycle phase influenced whether loss of Ofd1 was permissive for aberrant elongation, we blocked cells in G1, S, or G2 (Figure S3C). G1 arrested cells showed a lower proportion of long centrioles, while G2 arrested cells showed a higher proportion of long centrioles, indicating that elongation defects occurred predominantly during G2, the phase during which centriole maturation and daughter centriole elongation normally occurs (Figure 4A, Figure S3D).
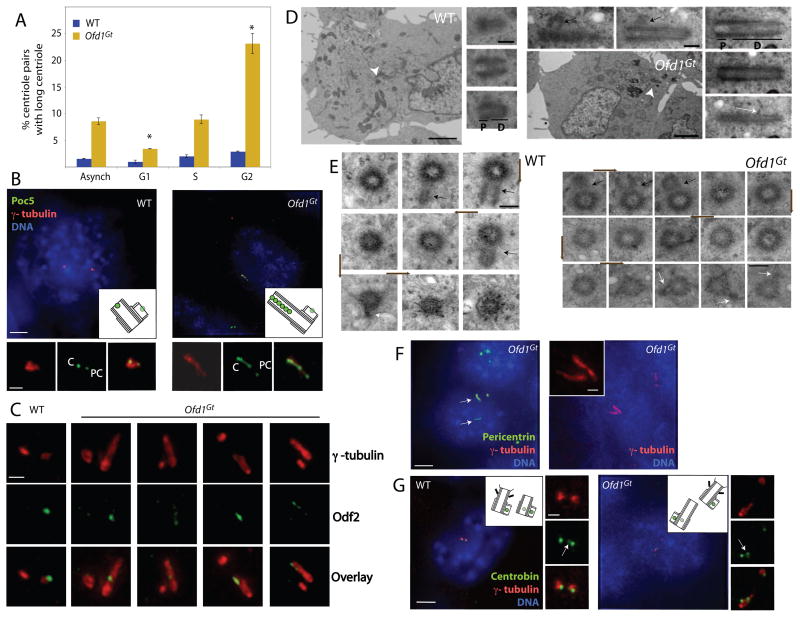
Figure 4. Ofd1 restrains growth of the distal domain of both mother and daughter centrioles in G2.
(A) Graph indicating the percent of long centrioles in WT and Ofd1Gt cells in different cell cycle phases. Asterisks indicate statistically significant differences compared to the asynchronous population (p < 0.01). (B) The distal centriole (Poc5), centrosomes (γ-tubulin), and DNA (DAPI) of WT and Ofd1Gt cells. Poc5 localizes more strongly to mother or daughter centrioles than to procentrioles. PC, procentriole. C, centriole. (C) Centriole appendages (Odf2) and centrosomes (γ-tubulin) of WT and Ofd1Gt cells. (D) Longitudinal and (E) transverse TEM sections of a WT and long Ofd1Gt centriole. Centriole proximal domain (P), distal domain (D), subdistal appendages (white arrows), procentrioles (black arrows). Arrowheads indicate centrioles in low magnification TEM images. Brown arrows show direction of section sequence. Normal length centrioles are contained within 8–10 sequential transverse sections, whereas long centrioles span more than 10 sections. (F) Ofd1Gt cells showing centrosomes (Pericentrin and γ-tubulin) and DNA (DAPI). (G) Daughter centrioles and procentrioles (Centrobin), centrosomes (γ-tubulin), and DNA (DAPI) of WT and Ofd1Gt cells. In S-G2 phase, Centrobin localizes more strongly to the procentrioles than to the daughter centriole. Arrows indicate daughter centrioles. Scale bars indicate 2 μm (TEM, low magnification), 200 nm (TEM, high magnification), 5 μm and 1 μm (inset). See also Figure S3.
To understand what part of the centriole was elongating abnormally, we examined the long centrioles for the presence of distal centriole components. Poc5 normally localizes to a small region in the centriole distal lumen (Azimzadeh et al., 2009). The Ofd1 mutant long centrioles displayed an expanded Poc5 region that comprised most of their length (Figure 4B). The appendage proteins Ninein and Odf2 usually localize to a small domain at the centriole subdistal and distal end, respectively (Mogensen et al., 2000; Nakagawa et al., 2001; Piel et al., 2000). Though sometimes found in the middle of long centrioles, expanded domains of Ninein and Odf2 were seen on many Ofd1 mutant long centrioles as well (Figure 4C, Figure S4C). The centriole distal end contains electron dense material within the lumen (Vorobjev and Chentsov, 1980). In TEM images, the electron dense distal end comprised most of the long Ofd1 mutant centrioles, whereas the electron sparse proximal end was of comparatively normal size (Figure 4D, Figure S3B). Centrin, which is normally present in the centriole distal lumen (Paoletti et al., 1996), was often present in discrete foci within the abnormal long centriole, suggesting that the elongated distal domain was structurally abnormal (Figure 1C, Figure S3E). Together, these data suggest that Ofd1 acts during G2 to restrain elongation specifically of the distal centriole.
Ofd1 controls elongation of mother and daughter centrioles
Because Ofd1 localized to all centrioles, we were interested to know if the distal ends of mother, daughter, and procentrioles showed equivalent length abnormalities in the absence of Ofd1. To determine if procentrioles showed length abnormalities, we re- examined the data regarding localization of Odf2. Odf2 is a component of appendages specifically found only on mother centrioles in G1-S and on both mother and maturing daughter centrioles in G2 (Nakagawa et al., 2001). Odf2 is never found on procentrioles. In asynchronous cells, 95% of long centrioles were positive for Odf2, suggesting that the long centrioles were not procentrioles (Figure 4C). TEM of long centrioles supported this conclusion, as Ofd1 mutant long centrioles showed appendages, procentriole nucleation, and microtubules anchored at the distal ends (Figure 4D–E, S1I, S3B, S4B), characteristics of mother and daughter centrioles not possessed by procentrioles (Piel et al., 2000).
As described above, during G1-S the mother centriole is the only Odf2-positive centriole in the cell. Therefore, the presence of a single long Odf2-positive centriole in Ofd1 mutant cells indicated that the long centriole was the mother centriole (Figure 4C, last 3 columns). Thus, the mother centriole depends upon Ofd1 for length control.
Although most Ofd1Gt cells had only one long centriole, they occasionally had two (Figure 4F), suggesting that daughter centrioles could also elongate aberrantly in the absence of Ofd1. In order to determine if daughter centrioles show Ofd1-dependent length perturbations, we assayed for the presence of the daughter centriole component Centrobin (Zou et al., 2005). Some long centrioles also contained Centrobin, indicating that long centrioles present in Ofd1Gt cells can display characteristics of daughter centrioles (Figure 4G). These experiments suggest that Ofd1 controls both mother and daughter centriole length.
Ofd1 is required for formation of distal, but not subdistal, appendages
As demonstrated above, Ofd1 localizes to a domain central to the distal appendages. We examined subdistal appendages in wild type and Ofd1Gt cells by immunofluorescent localization of Ninein, a subdistal appendage component, as well as by TEM. Ninein showed the normal localization to the proximal mother and daughter, as well as the subdistal mother centriole in Ofd1Gt cells (Figure 5A). The appendages are an important site of microtubule anchoring with characteristic TEM appearances depending on plane of section (Delgehyr et al., 2005; Paintrand et al., 1992) (Figure 5B). Based on both longitudinal and transverse sections, loss of Ofd1 did not affect subdistal appendage structure or microtubule anchoring (Figure 5C, Figure S4A).
Figure 5. Ofd1 is essential for distal appendage formation.
(A) Centrioles (acetylated tubulin) and mother centriole subdistal appendages (Ninein) of WT and Ofd1Gt cells. (B) Diagram depicting the TEM appearance of a mother centriole in transverse and longitudinal views. (C) TEM longitudinal views of WT and Ofd1Gt centrioles. Arrows indicate subdistal appendages. (D) Centrioles (acetylated tubulin) and mother centriole distal appendages (Cep164) of WT and Ofd1Gt cells. Graph shows percent of centrosome pairs showing Cep164 localization in WT and Ofd1Gt cells. (E) Immunblot showing Cep164 in the supernatants of WT and Ofd1Gt cell lysates. 20 μg protein loaded per lane. (F) TEM transverse views of WT and Ofd1Gt centrioles. Arrows indicate distal appendages. (Full serial reconstructions are included in Figure S4). (G) Centrioles and cilia (acetylated tubulin), centriole appendages (Odf2), centrosomes (γ-tubulin), and DNA (DAPI) of WT and Ofd1Gt cells at the indicated time after release from cell synchronization block. Arrowheads indicate centrioles positive for Odf2. (H) Quantification of Odf2 foci per cell in WT and Ofd1Gt cells at the indicated time after release from cell synchronization block. Asterisks indicate p< 0.05. (I) Diagram showing centriole duplication and maturation in coordination with the cell cycle. MC, mother centriole. DC, daughter centriole. Scale bar indicates 200 nm (TEM) and 1 μm. See also Figure S4.
Subdistal appendages were also present on long Ofd1 mutant centrioles, either in the middle of the long centriole or spread out along the elongated distal domain, as assayed by immunofluorescence localization or by TEM (Figure S1I, 4D–E, S4B–C). Though their proximal-distal position was sometimes abnormal, the structure of the subdistal appendages appeared normal on the long centrioles as well.
In contrast to the subdistal appendages, Ofd1Gt cells showed severe defects in distal appendage formation (Figure 5D). Loss of Ofd1 caused complete loss of the distal appendage component Cep164 from all centrioles. As Ofd1Gt cells expressed Cep164 (Figure 5E), but the distal appendages of Ofd1Gt centrioles appeared less electron dense by TEM (Figure 5F, Figure S4B), the delocalization of Cep164 in Ofd1Gt cells suggested a failure to form normal distal appendages. Whereas wild type distal appendages showed the characteristic elongated triangular shape with increased density on one end, loss of Ofd1 caused the appendages to appear thin and uniform along the length, on both normal (Figure 5F) and abnormal length centrioles (Figure S4B). Cep164 was not associated with immunoprecipitated Ofd1 complexes, suggesting that Ofd1 does not directly recruit Cep164 (Figure S3A), but rather is more generally required to promote normal distal appendage formation.
To establish the extent of the distal appendage defects, we examined centriolar localization of Odf2 in asynchronous Ofd1Gt cells. Odf2 is required for both subdistal and distal appendage formation and localizes at the base of the appendages (Ishikawa et al., 2005; Nakagawa et al., 2001). Unlike Cep164, Odf2 localized to the distal centriole in asynchronous Ofd1 mutant cells (data not shown). To ascertain if the distal appendage defects might be temporally related to the centriole elongation defects, cells were synchronized and Odf2 localization followed as the cell cycle progressed. At 0 hr, cells were in G1, with one mother and one daughter centriole. As the cells progressed through G2/M at 8–10 hr, the daughter matured by gaining appendages and Odf2 localization (Figure 5I, Figure S4D). This process was observed by monitoring the presence of cells with two Odf2 spots, indicating two mature centrioles. During G2, the same phase in which centriole elongation defects occur, Ofd1Gt cells had significantly fewer cells with two Odf2-positive centrioles (Figure 5G–H). No differences in acquisition of the subdistal appendage marker Ninein was observed, suggesting that the maturation defect is confined to the distal appendages (data not shown). Restoring Ofd1 protein expression in Ofd1Ofd1myc cells restored normal localization of Cep164 and Odf2 to the mother centriole (Figure 2I, J).
Together, these data indicated that Ofd1 is required for recruitment of distal appendage proteins. Odf2 is associated with both distal and subdistal appendage structures and participates in their formation (Ishikawa et al., 2005; Nakagawa et al., 2001). Cep164, on the other hand, is only associated with the distal appendages (Graser et al., 2007). It seems that some Odf2 protein, perhaps the pool associated with the subdistal appendages, is recruited normally in Ofd1 mutant cells, whereas the distal appendage-specific protein Cep164 is not.
Ofd1 is required for the recruitment of Ift88 to the centrosome
As distal appendages may also be important for docking of intraflagellar transport (IFT) proteins during the process of ciliogenesis (Deane et al., 2001), we examined Ift recruitment. Two components of Ift complex B, Ift20 and Ift80, localized normally to centrosomes in Ofd1Gt cells (Follit et al., 2006; Lucker et al., 2005) (Figure 6A–B). Similarly, Kif3a, part of the anterograde Ift motor, localized to centrosomes in both wild type and Ofd1Gt cells (Figure 6C). Ift88, another Ift complex B component, is present at the centrosome and along the cilium (Haycraft et al., 2007). Immunofluorescence staining and quantification revealed that, in contrast to Ift20, Ift80 and Kif3a, Ift88 failed to associate with centrosomes in Ofd1Gt cells (Figure 6D–E). This defect is not due to differences in protein expression, as wild type and Ofd1Gt cells expressed Ift88 at similar levels (Figure 6F). Restoring Ofd1 protein expression in Ofd1Ofd1myc cells restored normal localization of Ift88 to the centrosome (Figure 6G–H). Together, these data suggest that loss of Ofd1 caused a specific loss of Ift88 from the centrosome.
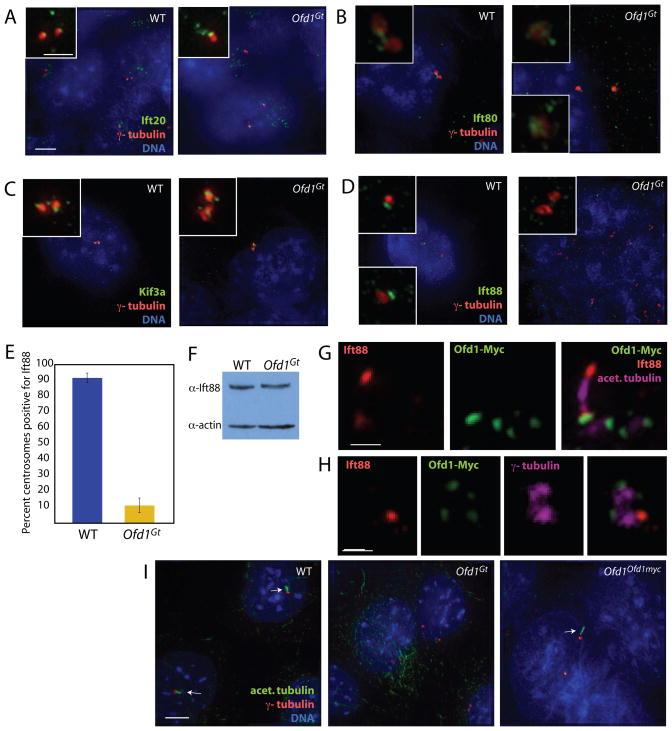
Figure 6. Ofd1 is required for centrosomal recruitment of Ift88, but not Ift20, Ift80, or Kif3a.
(A) The intraflagellar transport protein Ift20, centrosomes (γ– tubulin), and DNA (DAPI) of WT and Ofd1Gt cells. IFT20 localizes to the Golgi and near the centrosome (Follit et al., 2006). (B) Ift80, centrosomes (γ–tubulin), and DNA (DAPI) of WT and Ofd1Gt cells. (C) Anterograde kinesin motor component Kif3A, centrosomes (γ–tubulin), and DNA (DAPI) of WT and Ofd1Gt cells. (D) Ift88, centrosomes (γ–tubulin), and DNA (DAPI) of WT and Ofd1Gt cells. (E) Graph showing percent of centrosome pairs with Ift88 localization in WT and Ofd1Gt cells. (F) Immunblot showing Ift88 in the supernatants of WT and Ofd1Gt cell lysates. 20 μg protein loaded per lane. (G) Ift88, Ofd1-Myc (Myc), and centrioles and cilia (acetylated tubulin) of Ofd1Ofd1myc cells. (H) Ift88, Ofd1-Myc (Myc), and centrosomes (γ-tubulin) of Ofd1Ofd1myc cells. (I) Centrioles and cilia (acetylated tubulin), centrosomes (γ-tubulin), and DNA (DAPI) of WT, Ofd1Gt and Ofd1Ofd1myc cells. Arrows indicate cilia. Scale bar indicates 5 μm or 1 μm (inset, (G)- (H)). See also Figure S5.
To determine if loss of centrosomal Ift88 was due to a general disruption of trafficking to the centrosome, we investigated localization of three proteins known to be important for this function: Dynactin, Pericentrin, and Cep290 (Jurczyk et al., 2004; Kim et al., 2008; Quintyne and Schroer, 2002; Tsang et al., 2008). All localized normally in cells lacking Ofd1 (Figure S5), indicating that the requirement for Ofd1 in the recruitment of Ift88 does not reflect a general disruption of centrosomal trafficking.
As both Ofd1 and Ift88 localize to centrosomes, we were interested to determine if Ofd1 co-localized with Ift88 to the same regions of this organelle. In ciliated cells, Ift88 colocalized with Ofd1 at the base of the cilium (Figure 6G). When cells were not ciliated, Ift88 and Ofd1 colocalized at the distal end of the mother centriole (Jurczyk et al., 2004) (Figure 6H).
Both Ift88 and centriole appendages are required for ciliogenesis(Ishikawa et al., 2005; Pazour et al., 2000). Consistent with this, and previous data showing that Ofd1 is required for ciliogenesis (Ferrante et al., 2006), Ofd1Gt cells did not make primary cilia (Figure 6I). Collectively these data revealed that Ofd1 is required for length control of the distal mother and daughter centriole, recruitment of distal appendages and Ift88, and ciliogenesis.
OFD1 syndrome-associated mutations cause centriole length defects and disrupt normal ciliogenesis
Ofd1 protein has an amino-terminal Lis1 homology (LisH) domain and five coiled-coil domains that are important for centrosomal localization (Romio et al., 2004). To identify how human mutations affected Ofd1 function, we generated ES cell lines expressing five disease-associated missense mutations (Figure S6A): S75F and A80T affect the LisH domain, S437R affects the second coiled-coil domain, and G139S and KDD359-361FSY affect intervening highly conserved residues (Ferrante et al., 2001; Rakkolainen et al., 2002; Romio et al., 2003; Thauvin-Robinet et al., 2006). Using the Floxin system, we inserted Myc-tagged murine Ofd1 alleles containing the homologous mutations into the Ofd1 locus. Because the cells are hemizygous for Ofd1, they express only the inserted mutant allele under control of the endogenous promoter.
Cells were examined for Ofd1 protein expression and centrosomal localization. All disease-associated alleles were expressed, though three (S75F, A80T and KDD359-361FSY) reduced protein levels (Figure 7A, Table 1). These finding are consistent with previous studies in HEK293 cells that found that the A80T mutation reduced protein half-life (Gerlitz et al., 2005). Because KDD359-361FSY mutates the region of the protein that the Ofd1 antibody is expected to recognize, protein levels were also compared by immunoprecipitating and blotting against the carboxy-terminal Myc tag. The same decrease was seen by this method (Figure S6B). Low levels of the S75F and A80T mutant proteins prohibited accurate assessment of centriolar localization in ES cells. The S75F mutant protein has been shown previously to localize normally to centrioles in HEK293 cells, and deletion of the LisH domain does not affect Ofd1 localization (Romio et al., 2004). None of the other disease associated mutations altered centriolar localization (Figure 7B).
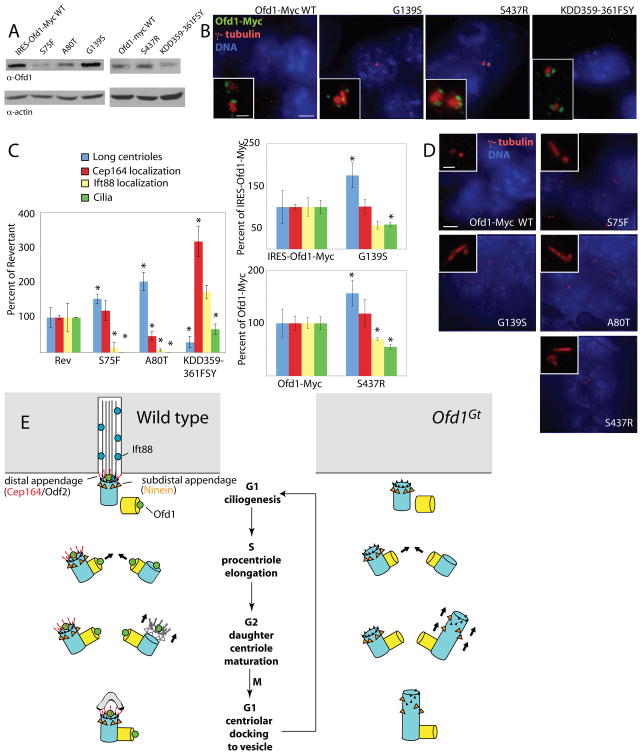
Figure 7. Missense Ofd1 mutations found in human patients affect centriole length control and ciliogenesis.
(A) Immunoblots showing Ofd1 (detected with an Ofd1 antibody) in the supernatants of lysates from cells of the indicated genotypes. 20 μg protein loaded per lane. (B) Cells of the indicated genotypes stained for Ofd1-Myc (Myc), centrosomes (γ-tubulin), and DNA (DAPI). (C) Graphs comparing the frequencies of long centriole formation, centriolar localization of Cep164 and Ift88, and ciliogenesis of cells with Ofd1 alleles to Ofd1Rev, Ofd1IRESOfd1myc, or Ofd1Ofd1myc cells. Asterisks indicate statistically significant differences (p < 0.05). (D) Cells of the indicated genotypes showing centrosomes (γ-tubulin) and DNA (DAPI). (E) A model of Ofd1-dependent control of centriole structure. In wild type cells, the mother centriole distal appendages contain Odf2 and Cep164, and Ift88 is recruited at the distal centriole for entry into the primary cilium. Ofd1 binds to the distal ends of centriolar microtubules, stabilizes centrioles at the proper length during maturation and recruits Ift88 and distal appendage proteins. In the absence of Ofd1, both mother and daughter centrioles show microtubule destabilization and unrestrained elongation of the distal domain during G2, the phase during which centriole maturation occurs. Without Ofd1, subdistal and distal appendages may be present in the middle of the long centriole, or distributed along the elongated portion. The inability to make primary cilia may be due to centriole elongation defects, distal appendage defects, Ift88 recruitment defects, inability to dock to a vesicular membrane (Sorokin, 1962), or a combination of these. See also Figure S7.
Table 1. Summary of phenotypes caused by OFD1 syndrome-associated mutations.
Ofd1 protein expression quantified by immunoblot. Percentages reflect comparison to wild type (for the Gene trap line), Ofd1IRESOfd1myc cells (for the S75F, A80T, G139S mutants) or Ofd1Ofd1myc cells (for the S437R, KDD359-361FSYmutants), as appropriate. Ofd1 centrosomal localization, as assessed by immunofluorescence. NA, not assessed. Percent of cells showing long centrioles, centrosomal Cep164 and Ift88 localization, and cilia, as assessed by immunofluorescent staining and normalized to wild type cells. Asterisks indicate a statistically significant difference (p < 0.05) when compared to Ofd1IRESOfd1myc or Ofd1Ofd1myc cells (for G139S and S437R mutants, respectively), or Ofd1Rev cells (for S75F, A80T, and KDD359-361FSY mutants). Errors are standard deviations. See also Figure S6.
| Cell Line | Percent of Ofd1 protein expression, compared to control line | Does Ofd1 localize to the centrosome? | Percent of long centrioles, compared to wild type cells | Percent of centrosomes with Cep164 localization, compared to wild type cells | Percent of centrosomes with Ift88 localization, compared to wild type cells | Percent of cells with cilia, compared to wild type cells |
|---|---|---|---|---|---|---|
| Gene trap | 0 | No | 475 ± 34 | 2 ± 1 | 12 ± 5 | 0 |
| S75F | 22 | NA | 232 ± 20 * | 28 ± 7 | 3 ± 4 * | 0 * |
| A80T | 51 | NA | 307 ± 39 * | 11 ± 3 * | 2 ± 1 * | 0 * |
| G139S | 112 | Yes | 175 ± 30 * | 93 ± 15 | 22 ± 3 | 20 ± 1 * |
| S437R | 103 | Yes | 146 ± 23 * | 100 ± 22 | 32 ± 1 * | 35 ± 3 * |
| KDD359-361FSY | 34 | Yes | 45 ± 25 * | 75 ± 10 | 47 ± 5 | 35 ± 8 * |
To assess how disease-associated mutations affected Ofd1 function, centrioles were examined for length defects, Cep164 and Ift88 recruitment. A quantitative summary of this data is presented in Table 1 and Figure S6C with all lines compared to wild type ES cells. However, the significance of these differences cannot be determined directly from this type of comparison, as Ofd1-Myc Floxin cells express reduced levels of Ofd1 (Singla et al., 2009). This reduction does not affect centriole length control or Cep164 recruitment, but does reduce Ift88 recruitment and ciliogenesis (Figure S6C). Ofd1Rev cells (Singla et al., 2009) expressed wild type Ofd1 at still lower levels, lower than any of the disease allele lines (data not shown). This reduced level did not affect centriole size, but did affect recruitment of Cep164 and Ift88, as well as ciliogenesis. (Figure S6C).
To understand how disease mutations affect Ofd1 function independent of protein stability, disease-allele carrying cells were compared to a line that expressed similar levels of wild type Ofd1 (Figure 7C). The G139S and S437R Ofd1 mutant lines expressed protein levels comparable to the Floxin Ofd1-Myc line. The S75F, A80T, and KDD359-361FSY mutant lines expressed protein levels comparable to Ofd1Rev. Comparing these lines suggested that the deficits described below are not entirely attributable to decreased protein levels.
Four Ofd1 mutations decreased the ability of Ofd1 to restrain centriole elongation, resulting in abnormally long centrioles (Figure 7D). Quantification of long centrioles indicated that the hypomorphic mutations affected Ofd1 function to different degrees, indicating that the mutations represent an allelic series (Figure S6C). Mutations in the LisH domain caused the most profound centriole elongation. In contrast, the KDD359-361FSY mutation decreased the number of long centrioles below that observed in cells expressing similar levels of wild type Ofd1, suggesting that this mutation may shorten centrioles (Figure 7C).
Cep164 recruitment was affected by A80T, one mutation affecting the LisH domain, but none of the other mutations (Figure S6D, Table 1).
The LisH mutations blocked Ift88 recruitment. Of the other mutations only S437R affected Ift88 recruitment, suggesting that the second coiled coil of Ofd1 is particularly important for recruiting Ift88 (Figure S6E, Table 1).
The LisH mutations also blocked ciliogenesis, whereas the carboxy-terminal mutations caused decreased ciliogenesis (Figure 7C, S6F).
Thus, phenotyping a variety of human disease mutations reveals that distinct Ofd1 domains contribute to genetically separable Ofd1 functions. Although the disease associated missense mutations represented alleles with varying degrees of stability and function, all compromised the ability of Ofd1 to regulate centriole length and ciliogenesis.
DISCUSSION
Taken together, our results reveal that Ofd1 is a critical component of the distal centriole required for centriole length control, distal appendage formation and Ift88 recruitment. Ofd1 localizes to all centriole distal ends, and Ofd1 complexes with α- and γ-tubulin. Ofd1 mutant cells show instability of centriolar microtubules, suggesting that Ofd1 functions as a cap to stabilize centriolar microtubules at a defined length.
Daughter centrioles normally elongate and gain subdistal and distal appendages during centriole maturation in G2 phase. Ofd1 regulation of centriolar size is most critical in G2, and only mother and daughter centriole distal ends show excessive elongation in the absence of Ofd1. Thus, Ofd1 capping may be specifically required for stabilization and length control of centrioles during centriole maturation (Figure 8A).
Loss of Ofd1 does not affect subdistal appendage structure or function. Distal appendage formation, on the other hand, is severely perturbed, suggesting that centriole stability may be a prerequisite for assembly of distal, but not subdistal, appendages.
Ofd1 control of centriole length and distal structure are separable functions
OFD1 patients do not show a tight genotype-phenotype correlation, making it difficult to assign a relationship between OFD1 mutations and disease severity (Feather et al., 1997; Prattichizzo et al., 2008). Use of the Floxin system allowed us to create a panel of ES cell models that express alleles orthologous to human disease alleles. Expression of the disease alleles from the endogenous Ofd1 locus allows for direct comparison of the effects of the mutation on gene function. The phenotyping of these cellular models indicated that the human disease-associated mutations form an allelic series which, from weakest to strongest, are KDD359-361FSY, S347R, G139S, S75F and A80T.
We found that the LisH domain is essential for all Ofd1 functions. In contrast, mutations carboxy-terminal to the LisH domain and in the coiled-coil domain do not abolish Cep164 localization, Ift88 recruitment, or ciliogenesis, but do affect centriole length. Whereas it is likely that many manifestations of OFD1 are due to defective ciliogenesis, the additional Ofd1 roles discovered by modeling the disease in ES cells raise the possibility that some OFD1 phenotypes may be due to centriolar, not ciliary, dysfunction. Centrosomes have many important developmental functions, including roles in cell migration and fate determination (Higginbotham and Gleeson, 2007). It will be interesting to investigate if and how centriolar length dysfunction contributes to OFD1 pathogenesis.
As loss of Ofd1 also causes defects in distal appendage formation and centriolar recruitment of Ift88, it is possible that these phenotypes are secondary to abnormal centriole elongation. Alternatively, the requirement for Ofd1 in distal appendage formation and Ift88 recruitment may reflect separate function(s) from its role in centriole length control. In support of this possibility, neither Cep164 nor Ift88, proteins respectively critical for distal appendage formation and ciliogenesis, have been implicated in centriole length control (Graser et al., 2007; Pazour et al., 2000). Moreover, our finding that the G139S and KDD359-361FSY substitutions disrupted centriole length control without changing Ift88 or Cep164 localization indicates that length abnormalities do not necessarily result in the other Ofd1 null phenotypes. These missense mutations also reveal that distinct domains of Ofd1 are involved in centriole length control and recruitment of distal appendages and Ift88. Interestingly, cells expressing G139S and KDD359-361FSY mutant forms of Ofd1 also show decreased ciliogenesis, suggesting that control of centriole length itself may be essential for ciliogenesis.
CPAP, CP110, and Ofd1 have different roles in centriole length control
The proteins CPAP, Poc1, and CP110 also have functions in centriole length control (Keller et al., 2008; Kohlmaier et al., 2009; Schmidt et al., 2009; Tang et al., 2009), summarized in Figure S7A. CPAP is part of the proximal centriole and is required for procentriole formation (Kleylein-Sohn et al., 2007). In contrast, Ofd1 is part of the distal centriole and is not required for procentriole formation. Abnormal centrioles caused by CPAP overexpression can display procentriole characteristics and show incomplete centriolar walls. Loss of Ofd1 affects mother and daughter centrioles, but not procentrioles, and does not affect the integrity of centriole walls.
Overexpression of CPAP induces long centrioles that do not display a normal proximal to distal polarity, as CPAP and other proximal centriole proteins are present along the length of the centriole (Kohlmaier et al., 2009; Schmidt et al., 2009; Tang et al., 2009). Also, the abnormal elongated portion of CPAP-associated centrioles can initiate procentriole formation, a function of the proximal centriole. Long CPAP-associated centrioles do not possess an elongated appendage domain, as appendages are located in the middle of the long centriole. These findings suggest that CPAP overexpression induces the formation of an elongated domain at the distal end of the centriole that possesses proximal characteristics.
In contrast to long CPAP-associated centrioles, long Ofd1 mutant centrioles do not nucleate extra procentrioles and display expanded localization of distal centriole proteins. These findings suggest that loss of Ofd1 results in elongation of a distal centriole-like domain. The extensive differences between the CPAP overexpression phenotype and the Ofd1 loss-of-function phenotype argue that CPAP and Ofd1 may regulate the elongation of different domains within the centriole. CPAP may regulate the elongation of proximal domains, whereas Ofd1 is required to regulate the elongation of the distal domain.
The functions of CP110 and Ofd1 are similarly distinct. Although not studied as extensively as CPAP overexpression, the elongated centrioles of CP110 depleted cells show morphological similarities to the abnormal CPAP centrioles. Depletion of CP110, like CPAP overexpression and unlike loss of Ofd1, affects procentriole length, suggesting distinct roles for CP110 and Ofd1.
Ofd1 controls elongation of a distinct centriole distal domain
The microtubule pattern of centrioles shows a change from a triplet arrangement at the proximal end to a doublet arrangement at the distal end. This shift in microtubule pattern occurs approximately where the subdistal appendages attach to the centriole (Paintrand et al., 1992). Perhaps Ofd1 controls elongation specifically of distal centriole doublet microtubules, while other proteins like CP110 regulate triplet microtubule length.
The LisH domain may be involved in the regulation of microtubule dynamics (Emes and Ponting, 2001). We favor a model for Ofd1 function in which the coiled-coil domains mediate Ofd1 centrosomal localization, and the LisH domain then stabilizes centriole doublet microtubules during elongation, allowing posttranslational modification of centriolar microtubules and construction of distal appendages. After centriole maturation, Ofd1 remains at the centriole distal end, where the second coiled-coil domain is important for recruitment of Ift88.
Conclusion
Ofd1 orthologs are present in the genomes of many animals, including single celled organisms such as Tetrahymena (Figure S7B). This conservation suggests that Ofd1 is part of an ancient mechanism for regulating centriole structure and length and reveals the importance of centriole length control in centrosome function.
EXPERIMENTAL PROCEDURES
Cell Lines and Cell Culture
Ofd1Gt (RRF427) E14 ES cell line was obtained from BayGenomics. Ofd1rev, Ofd1Ofd1myc, and cell lines with human mutations were created as described previously (Singla et al., 2009). Cells were cultured on 0.1% gelatin in GMEM supplemented with 10% FBS, glutamine, pyruvate, NEAA, βME, and LIF.
3T3 (ATCC) and POC1-GFP U2OS (gift of Dr. Wallace Marshall) cells were cultured in DMEM supplemented with 10% FBS and antibiotics. IMCD3 (ATCC) and hTERT-RPE1 (gift of Dr. Wallace Marshall) were cultured in DMEM:F12 supplemented with 10% FBS and antibiotics.
cDNA constructs and cloning
Ofd1 cDNA was cloned as described previously (Singla et al., 2009). Missense mutations were created using Quik Change II XL site directed mutagenesis kit (Stratagene). Final products were confirmed by sequencing.
Creation of Floxin cell lines
Missense mutations S75F and A80T occur in exon 1 of Ofd1, while G139S occurs in exon 3. The gene trap insertion in Ofd1Gt cells is in intron 3 of the Ofd1 genomic locus. Full length cDNA for Ofd1-Myc-S75F, Ofd1-Myc-A80T, and Ofd1-Myc-G139S, alleles in which the mutation occurs in exons upstream of the gene trap insertion site, was cloned into the vector pFloxin-IRES (Genbank EU916835). Missense mutations S437R and KDD359-361FSY occur in exons downstream of the gene trap insertion site. cDNA for exons 4-23 of Ofd1-Myc-S437R and Ofd1-Myc-KDD359-361FSY was cloned into the vector pFloxin (Genbank EU916834). pFloxin and pFloxin-IRES constructs were electroporated into Ofd1Rev cells as previously described. Cells were selected with 300 μg/mL G418 (Invitrogen) and colonies were transferred to 48 well plates after 6 days. Correct integration was verified by genomic PCR.
Antibodies
Antibodies to Ofd1 were generated by Covance, Inc. Rabbits were immunized with the peptide [H]-CDTYDQKLKTELLKYQLELKDDYI–[NH2] corresponding to amino acids 340-362 of murine Ofd1. Antibody was used at 1:5000 for Western blotting and for immunofluorescence, 1:2000 in murine cells, 1:1000 in human cells. See supplemental experimental procedures for other antibody information.
Immunofluorescence and Microscopy
For ES cell ciliation studies: ES cells were plated on coverslips coated with 1% matrigel (BD) and treated with 0.5 mM mimosine (Sigma) overnight to arrest cells. Cells were fixed 5′ in 4% PFA, washed in PBS, and fixed 2–3′ in −20° 100% methanol. The cells were washed in PBS with 0.1% Triton-X100 (PBST), blocked in 2% BSA in PBST, and incubated with primary antibodies in block for 1 hr at RT. The cells were washed in PBST, incubated with secondary antibodies in block for 30′ at RT, and mounted with Vectashield hardset with DAPI (Vector labs).
POC1-GFP U2OS S-phase arrest: Cells were plated on coverslips and treated with 3.2 μg/mL aphidocolin (Sigma) for 72 hr, then fixed in 100% methanol and washed and processed as above.
For cell synchronization studies: Cells were synchronized using thymidine-mimosine block(Fujii-Yamamoto et al., 2005). Briefly, cells were plated on coated coverslips in 2.5 mM thymidine, incubated for 12 hr, released into regular media for 6 hr, then blocked in 0.5 mM mimosine for 6 hr. Timepoints were taken after release from mimosine block. Cells for FACS were collected as described below. For IF, cells were fixed in 100% methanol, then washed and processed as above.
For all other experiments, cells were plated on coverslips and fixed in 100% methanol, then washed and processed as above.
Slides were viewed on a Deltavision microscope (Applied Precision) and image processing was completed with Deltavision and Metamorph (Molecular Devices) software. Images are maximum projections of Z-stacks.
Immunoblots and Quantification
Cells were grown in flasks, trypsinized, collected, and washed once in PBS. Cell pellets were lysed in buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1:200 dilution protease inhibitor cocktail (Calbiochem)) containing 1% NP-40, 1%Triton-X-100, 0.25% sodium deoxycholate, or 1%NP-40 and 0.25% sodium deoxycholate (modified RIPA) for 30 minutes at 4 degrees. Lysates were centrifuged for 15 minutes, 16,000 rcf, at 4 degrees. Cleared supernatants were transferred to a new tube and 6X reducing sample buffer was added to the pellet.
Ofd1 protein expression was quantified by densitometry and normalized to actin.
Immunoprecipitations
Cells were grown in flasks, trypsinized, collected, and washed once in PBS. Cell pellets were lysed in modified RIPA buffer for 30 minutes at 4 degrees. Lysates were centrifuged for 15 minutes, 16,000 rcf, at 4 degrees. Protein concentration of the cleared supernatant was determined by Bradford assay. Supernatants were standardized to 1.6 mg/mL concentration, 3 mg total protein, and pre-cleared with protein G agarose beads (Invitrogen) for 2 hours. Beads were removed and supernatants were incubated overnight with 1.6 μg Ofd1 antibody. Next day, complexes were captured with protein G beads for 1 hr. Beads were washed 4 times with modified RIPA and proteins eluted with 6X reducing sample buffer.
Population Doubling Studies, FACS, and Microtubule Regrowth Assays
Population doubling: Cell lines were grown in T25 flasks, counted and replated every 3 days.
FACS
Cells from a confluent T75 flask were collected and stained with propidium iodide. Samples were analyzed on a BD FACsort (Beckton Dickinson), 40,000 events collected per sample. FlowJo software (TreeStar) was used to perform cell cycle analysis. Microtubule regrowth assays: Cells were plated on coated coverslips and treated with 1 μM nocodazole for 1 hr in culture to depolymerize microtubules. Cells were fixed with 100% methanol at 0″, 30″, 1′, 2′, 10′, and 15′ after nocodazole washout, and processed for IF as described above.
Electron Microscopy
Cells were plated on 8 well Permanox slides (Nunc), fixed in 3% glutaraldehyde in 0.1M phosphate buffer (PB) for 30′ at room temperature, then washed 3 times in 0.1M PB. Cells were postfixed in 2% osmium for 2 hr, dehydrated and embedded in Araldite (Durcupan, Fluka). Serial ultrathin sections (70nm) were cut with a diamond knife, stained with lead citrate and examined under a FEI Tecnai Spirit electron microscope. Percent of Ofd1Gt cells with long centrioles was determined using information from centrioles in both longitudinal and transverse sections. Quantitative centriole length measurements were performed on longitudinal sections only using ImageTool software.
Centriole Dynamics and Cell Cycle Studies
Cells were plated on coated coverslips, then treated with 10 μg/mL nocodazole for 1 hr in culture, or 0.5 mM mimosine, 3.2 μg/mL aphidocolin, or 2 μM camptothecin (Sigma) overnight. Cells were fixed in 100% methanol and processed for IF as described above.
Statistics
All error bars represent one standard deviation. For immunofluorescene quantifications, at least 200 cells were counted on each of duplicate coverslips in at least two separate experiments. Student’s unpaired t-test was used to determine statistical significance with a p value of less than 0.05.
Supplementary Material
Acknowledgments
The authors thank Wallace Marshall and Elizabeth Blackburn for use of Deltavision microscopes, Lani Keller, Juliette Azimzadeh, Hiroaki Ishikawa and the members of the Reiter lab for critical discussions and reading of this manuscript. We also thank the centrosome and cilia communities for generously sharing antibodies. This work was funded by grants from the National Science Foundation (V.S.), NIH (RO1AR054396), CIRM (RN2-00919), the Burroughs Wellcome Fund, the Packard Foundation, the Leona M. and Harry B. Helmsley Charitable Trust, and the Sandler Family Supporting Foundation (J.F.R.).
Footnotes
AUTHOR CONTRIBUTIONS
V.S. and J.R. conceived and designed the experiments. V.S., M.R.R., and J.M.G.V. performed the experiments. V.S. and J.R. wrote the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Azimzadeh J, Bornens M. Structure and duplication of the centrosome. J Cell Sci. 2007;120:2139–2142. doi: 10.1242/jcs.005231. [DOI] [PubMed] [Google Scholar]
- Azimzadeh J, Hergert P, Delouvee A, Euteneuer U, Formstecher E, Khodjakov A, Bornens M. hPOC5 is a centrin-binding protein required for assembly of full-length centrioles. J Cell Biol. 2009;185:101–114. doi: 10.1083/jcb.200808082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badano JL, Mitsuma N, Beales PL, Katsanis N. The ciliopathies: an emerging class of human genetic disorders. Annu Rev Genomics Hum Genet. 2006;7:125–148. doi: 10.1146/annurev.genom.7.080505.115610. [DOI] [PubMed] [Google Scholar]
- Badano JL, Teslovich TM, Katsanis N. The centrosome in human genetic disease. Nat Rev Genet. 2005;6:194–205. doi: 10.1038/nrg1557. [DOI] [PubMed] [Google Scholar]
- Bahe S, Stierhof YD, Wilkinson CJ, Leiss F, Nigg EA. Rootletin forms centriole-associated filaments and functions in centrosome cohesion. J Cell Biol. 2005;171:27–33. doi: 10.1083/jcb.200504107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettencourt-Dias M, Glover DM. SnapShot: centriole biogenesis. Cell . 2009;136:188–188. e181. doi: 10.1016/j.cell.2008.12.035. [DOI] [PubMed] [Google Scholar]
- Bobinnec Y, Khodjakov A, Mir LM, Rieder CL, Edde B, Bornens M. Centriole disassembly in vivo and its effect on centrosome structure and function in vertebrate cells. J Cell Biol. 1998;143:1575–1589. doi: 10.1083/jcb.143.6.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornens M. Centrosome composition and microtubule anchoring mechanisms. Curr Opin Cell Biol. 2002;14:25–34. doi: 10.1016/s0955-0674(01)00290-3. [DOI] [PubMed] [Google Scholar]
- Chretien D, Buendia B, Fuller SD, Karsenti E. Reconstruction of the centrosome cycle from cryoelectron micrographs. J Struct Biol. 1997;120:117–133. doi: 10.1006/jsbi.1997.3928. [DOI] [PubMed] [Google Scholar]
- Christensen ST, Pedersen LB, Schneider L, Satir P. Sensory cilia and integration of signal transduction in human health and disease. Traffic. 2007;8:97–109. doi: 10.1111/j.1600-0854.2006.00516.x. [DOI] [PubMed] [Google Scholar]
- Deane JA, Cole DG, Seeley ES, Diener DR, Rosenbaum JL. Localization of intraflagellar transport protein IFT52 identifies basal body transitional fibers as the docking site for IFT particles. Curr Biol. 2001;11:1586–1590. doi: 10.1016/s0960-9822(01)00484-5. [DOI] [PubMed] [Google Scholar]
- Delgehyr N, Sillibourne J, Bornens M. Microtubule nucleation and anchoring at the centrosome are independent processes linked by ninein function. J Cell Sci. 2005;118:1565–1575. doi: 10.1242/jcs.02302. [DOI] [PubMed] [Google Scholar]
- Emes RD, Ponting CP. A new sequence motif linking lissencephaly, Treacher Collins and oral-facial-digital type 1 syndromes, microtubule dynamics and cell migration. Hum Mol Genet. 2001;10:2813–2820. doi: 10.1093/hmg/10.24.2813. [DOI] [PubMed] [Google Scholar]
- Feather SA, Winyard PJ, Dodd S, Woolf AS. Oral-facial-digital syndrome type 1 is another dominant polycystic kidney disease: clinical, radiological and histopathological features of a new kindred. Nephrol Dial Transplant. 1997;12:1354–1361. doi: 10.1093/ndt/12.7.1354. [DOI] [PubMed] [Google Scholar]
- Ferrante MI, Giorgio G, Feather SA, Bulfone A, Wright V, Ghiani M, Selicorni A, Gammaro L, Scolari F, Woolf AS, et al. Identification of the gene for oral-facial-digital type I syndrome. Am J Hum Genet. 2001;68:569–576. doi: 10.1086/318802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrante MI, Zullo A, Barra A, Bimonte S, Messaddeq N, Studer M, Dolle P, Franco B. Oral-facial-digital type I protein is required for primary cilia formation and left-right axis specification. Nat Genet. 2006;38:112–117. doi: 10.1038/ng1684. [DOI] [PubMed] [Google Scholar]
- Follit JA, Tuft RA, Fogarty KE, Pazour GJ. The intraflagellar transport protein IFT20 is associated with the Golgi complex and is required for cilia assembly. Mol Biol Cell. 2006;17:3781–3792. doi: 10.1091/mbc.E06-02-0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii-Yamamoto H, Kim JM, Arai K, Masai H. Cell cycle and developmental regulations of replication factors in mouse embryonic stem cells. J Biol Chem. 2005;280:12976–12987. doi: 10.1074/jbc.M412224200. [DOI] [PubMed] [Google Scholar]
- Gerlitz G, Darhin E, Giorgio G, Franco B, Reiner O. Novel functional features of the Lis-H domain: role in protein dimerization, half-life and cellular localization. Cell Cycle. 2005;4:1632–1640. doi: 10.4161/cc.4.11.2151. [DOI] [PubMed] [Google Scholar]
- Graser S, Stierhof YD, Lavoie SB, Gassner OS, Lamla S, Le Clech M, Nigg EA. Cep164, a novel centriole appendage protein required for primary cilium formation. J Cell Biol. 2007;179:321–330. doi: 10.1083/jcb.200707181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habedanck R, Stierhof YD, Wilkinson CJ, Nigg EA. The Polo kinase Plk4 functions in centriole duplication. Nat Cell Biol. 2005;7:1140–1146. doi: 10.1038/ncb1320. [DOI] [PubMed] [Google Scholar]
- Haycraft CJ, Zhang Q, Song B, Jackson WS, Detloff PJ, Serra R, Yoder BK. Intraflagellar transport is essential for endochondral bone formation. Development. 2007;134:307–316. doi: 10.1242/dev.02732. [DOI] [PubMed] [Google Scholar]
- Higginbotham HR, Gleeson JG. The centrosome in neuronal development. Trends Neurosci. 2007;30:276–283. doi: 10.1016/j.tins.2007.04.001. [DOI] [PubMed] [Google Scholar]
- Ishikawa H, Kubo A, Tsukita S, Tsukita S. Odf2-deficient mother centrioles lack distal/subdistal appendages and the ability to generate primary cilia. Nat Cell Biol. 2005;7:517–524. doi: 10.1038/ncb1251. [DOI] [PubMed] [Google Scholar]
- Jurczyk A, Gromley A, Redick S, San Agustin J, Witman G, Pazour GJ, Peters DJ, Doxsey S. Pericentrin forms a complex with intraflagellar transport proteins and polycystin-2 and is required for primary cilia assembly. J Cell Biol. 2004;166:637–643. doi: 10.1083/jcb.200405023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller LC, Geimer S, Romijn E, Yates J, 3rd, Zamora I, Marshall WF. Molecular Architecture of the Centriole Proteome: The Conserved WD40 Domain Protein POC1 Is Required for Centriole Duplication and Length Control. Mol Biol Cell. 2008 doi: 10.1091/mbc.E08-06-0619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Krishnaswami SR, Gleeson JG. CEP290 interacts with the centriolar satellite component PCM-1 and is required for Rab8 localization to the primary cilium. Hum Mol Genet. 2008;17:3796–3805. doi: 10.1093/hmg/ddn277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleylein-Sohn J, Westendorf J, Le Clech M, Habedanck R, Stierhof YD, Nigg EA. Plk4-induced centriole biogenesis in human cells. Dev Cell. 2007;13:190–202. doi: 10.1016/j.devcel.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Kochanski RS, Borisy GG. Mode of centriole duplication and distribution. J Cell Biol. 1990;110:1599–1605. doi: 10.1083/jcb.110.5.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlmaier G, Loncarek J, Meng X, McEwen BF, Mogensen MM, Spektor A, Dynlacht BD, Khodjakov A, Gonczy P. Overly long centrioles and defective cell division upon excess of the SAS-4-related protein CPAP. Curr Biol. 2009;19:1012–1018. doi: 10.1016/j.cub.2009.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loktev AV, Zhang Q, Beck JS, Searby CC, Scheetz TE, Bazan JF, Slusarski DC, Sheffield VC, Jackson PK, Nachury MV. A BBSome subunit links ciliogenesis, microtubule stability, and acetylation. Dev Cell. 2008;15:854–865. doi: 10.1016/j.devcel.2008.11.001. [DOI] [PubMed] [Google Scholar]
- Lucker BF, Behal RH, Qin H, Siron LC, Taggart WD, Rosenbaum JL, Cole DG. Characterization of the intraflagellar transport complex B core: direct interaction of the IFT81 and IFT74/72 subunits. J Biol Chem. 2005;280:27688–27696. doi: 10.1074/jbc.M505062200. [DOI] [PubMed] [Google Scholar]
- Mikule K, Delaval B, Kaldis P, Jurcyzk A, Hergert P, Doxsey S. Loss of centrosome integrity induces p38-p53-p21-dependent G1-S arrest. Nat Cell Biol. 2007;9:160–170. doi: 10.1038/ncb1529. [DOI] [PubMed] [Google Scholar]
- Mogensen MM, Malik A, Piel M, Bouckson-Castaing V, Bornens M. Microtubule minus-end anchorage at centrosomal and non-centrosomal sites: the role of ninein. J Cell Sci. 2000;113(Pt 17):3013–3023. doi: 10.1242/jcs.113.17.3013. [DOI] [PubMed] [Google Scholar]
- Moudjou M, Bordes N, Paintrand M, Bornens M. gamma-Tubulin in mammalian cells: the centrosomal and the cytosolic forms. J Cell Sci. 1996;109(Pt 4):875–887. doi: 10.1242/jcs.109.4.875. [DOI] [PubMed] [Google Scholar]
- Nakagawa Y, Yamane Y, Okanoue T, Tsukita S, Tsukita S. Outer dense fiber 2 is a widespread centrosome scaffold component preferentially associated with mother centrioles: its identification from isolated centrosomes. Mol Biol Cell. 2001;12:1687–1697. doi: 10.1091/mbc.12.6.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg EAe. Centrosomes in Development and Disease. Wiley-VCH; 2004. [Google Scholar]
- Paintrand M, Moudjou M, Delacroix H, Bornens M. Centrosome organization and centriole architecture: their sensitivity to divalent cations. J Struct Biol. 1992;108:107–128. doi: 10.1016/1047-8477(92)90011-x. [DOI] [PubMed] [Google Scholar]
- Paoletti A, Moudjou M, Paintrand M, Salisbury JL, Bornens M. Most of centrin in animal cells is not centrosome-associated and centrosomal centrin is confined to the distal lumen of centrioles. J Cell Sci. 1996;109(Pt 13):3089–3102. doi: 10.1242/jcs.109.13.3089. [DOI] [PubMed] [Google Scholar]
- Pazour GJ, Dickert BL, Vucica Y, Seeley ES, Rosenbaum JL, Witman GB, Cole DG. Chlamydomonas IFT88 and its mouse homologue, polycystic kidney disease gene tg737, are required for assembly of cilia and flagella. J Cell Biol. 2000;151:709–718. doi: 10.1083/jcb.151.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piel M, Meyer P, Khodjakov A, Rieder CL, Bornens M. The respective contributions of the mother and daughter centrioles to centrosome activity and behavior in vertebrate cells. J Cell Biol. 2000;149:317–330. doi: 10.1083/jcb.149.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prattichizzo C, Macca M, Novelli V, Giorgio G, Barra A, Franco B. Mutational spectrum of the oral-facial-digital type I syndrome: a study on a large collection of patients. Hum Mutat. 2008;29:1237–1246. doi: 10.1002/humu.20792. [DOI] [PubMed] [Google Scholar]
- Quintyne NJ, Schroer TA. Distinct cell cycle-dependent roles for dynactin and dynein at centrosomes. J Cell Biol. 2002;159:245–254. doi: 10.1083/jcb.200203089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakkolainen A, Ala-Mello S, Kristo P, Orpana A, Jarvela I. Four novel mutations in the OFD1 (Cxorf5) gene in Finnish patients with oral-facial-digital syndrome 1. J Med Genet. 2002;39:292–296. doi: 10.1136/jmg.39.4.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romio L, Fry AM, Winyard PJ, Malcolm S, Woolf AS, Feather SA. OFD1 is a centrosomal/basal body protein expressed during mesenchymal-epithelial transition in human nephrogenesis. J Am Soc Nephrol. 2004;15:2556–2568. doi: 10.1097/01.ASN.0000140220.46477.5C. [DOI] [PubMed] [Google Scholar]
- Romio L, Wright V, Price K, Winyard PJ, Donnai D, Porteous ME, Franco B, Giorgio G, Malcolm S, Woolf AS, Feather SA. OFD1, the gene mutated in oral-facial-digital syndrome type 1, is expressed in the metanephros and in human embryonic renal mesenchymal cells. J Am Soc Nephrol. 2003;14:680–689. doi: 10.1097/01.asn.0000054497.48394.d2. [DOI] [PubMed] [Google Scholar]
- Schmidt TI, Kleylein-Sohn J, Westendorf J, Le Clech M, Lavoie SB, Stierhof YD, Nigg EA. Control of centriole length by CPAP and CP110. Curr Biol. 2009;19:1005–1011. doi: 10.1016/j.cub.2009.05.016. [DOI] [PubMed] [Google Scholar]
- Sharma N, Berbari NF, Yoder BK. Ciliary dysfunction in developmental abnormalities and diseases. Curr Top Dev Biol. 2008;85:371–427. doi: 10.1016/S0070-2153(08)00813-2. [DOI] [PubMed] [Google Scholar]
- Singla V, Hunkapiller J, Santos N, Seol AD, Norman AR, Wakenight P, Skarnes WC, Reiter JF. Floxin, a resource for genetically engineering mouse ESCs. Nature Methods. 2009 doi: 10.1038/nmeth.1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorokin S. Centrioles and the formation of rudimentary cilia by fibroblasts and smooth muscle cells. J Cell Biol. 1962;15:363–377. doi: 10.1083/jcb.15.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spektor A, Tsang WY, Khoo D, Dynlacht BD. Cep97 and CP110 suppress a cilia assembly program. Cell. 2007;130:678–690. doi: 10.1016/j.cell.2007.06.027. [DOI] [PubMed] [Google Scholar]
- Strnad P, Leidel S, Vinogradova T, Euteneuer U, Khodjakov A, Gonczy P. Regulated HsSAS-6 levels ensure formation of a single procentriole per centriole during the centrosome duplication cycle. Dev Cell. 2007;13:203–213. doi: 10.1016/j.devcel.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang CJ, Fu RH, Wu KS, Hsu WB, Tang TK. CPAP is a cell-cycle regulated protein that controls centriole length. Nat Cell Biol. 2009;11:825–831. doi: 10.1038/ncb1889. [DOI] [PubMed] [Google Scholar]
- Thauvin-Robinet C, Cossee M, Cormier-Daire V, Van Maldergem L, Toutain A, Alembik Y, Bieth E, Layet V, Parent P, David A, et al. Clinical, molecular, and genotype-phenotype correlation studies from 25 cases of oral-facial-digital syndrome type 1: a French and Belgian collaborative study. J Med Genet. 2006;43:54–61. doi: 10.1136/jmg.2004.027672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang WY, Bossard C, Khanna H, Peranen J, Swaroop A, Malhotra V, Dynlacht BD. CP110 suppresses primary cilia formation through its interaction with CEP290, a protein deficient in human ciliary disease. Dev Cell. 2008;15:187–197. doi: 10.1016/j.devcel.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorobjev IA, Chentsov YS. The ultrastructure of centriole in mammalian tissue culture cells. Cell Biol Int Rep. 1980;4:1037–1044. doi: 10.1016/0309-1651(80)90177-0. [DOI] [PubMed] [Google Scholar]
- Zou C, Li J, Bai Y, Gunning WT, Wazer DE, Band V, Gao Q. Centrobin: a novel daughter centriole-associated protein that is required for centriole duplication. J Cell Biol. 2005;171:437–445. doi: 10.1083/jcb.200506185. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.