SUMMARY
The endothelial specific receptor tyrosine kinase Tie2, and the orphan receptor tyrosine kinase, Tie1, are essential for endothelial cell proliferation, migration, and survival during angiogenesis. Despite their considerable similarity, experiments with Tie1 or Tie2 deficient mice highlight distinct functions for these two receptors in vivo. However, Tie1 cooperates with Tie2 during Angiopoietin signaling, demonstrating a degree of functional overlap between the two receptor-systems. Tie2 is further unique among receptor tyrosine kinases with respect to its structurally homologous ligands. Angiopoietin-2 and -3 can function as agonists or antagonists depending upon the local environment, while Angiopoietin-1 and -4, are constitutive agonists. To address the role of Tie1 in Angiopoietin-mediated Tie2 signaling, and determine the basis for the unique behavior of the individual Angiopoietins, we used an in vivo FRET-based proximity assay to monitor Tie1 and Tie2 localization and association in the presence and absence of ligands. Here, we provide direct evidence for Tie1-Tie2 complex formation on the endothelial cell surface and identify molecular surface areas essential for receptor-receptor recognition. We further demonstrate that the Tie1-Tie2 interactions are dynamic, inhibitory, and differentially modulated by Angiopoietin-1 and -2, thereby providing a discrete molecular mechanism for the observed variations in Angiopoietin function. Based upon the available data, we propose a unified model for Angiopoietin-induced Tie2 signaling which highlights the essential role of Tie1 in vascular homeostasis.
INTRODUCTION
Tie1 and Tie2 are type 1 transmembrane protein receptor tyrosine kinases (RTKs)(Ward and Dumont, 2002; Yancopoulos et al., 2000). Along with the vascular endothelial growth factor (VEGF) receptor, these are the only known endothelial cell-specific RTKs. Studies with dominant negative and null mice reveal that loss of Tie2 function results in embryonic death because of a failure of the vasculature system to expand(Dumont et al., 1994; Sato et al., 1995). Thus, it was proposed that Tie2 is not required for the differentiation of endothelial cells, but rather for their maintenance and proliferation(Dumont et al., 1994). Mice lacking Tie1 also die in utero, most likely a result of pulmonary edema(Puri et al., 1995). Although the vasculature remains intact, the integrity of vessel endothelial cells is compromised. Accordingly, Tie1 null mice display defects in vessel integrity demonstrated by localized hemorrhaging and the presence of an underdeveloped heart. However, the exact role of Tie1 in angiogenesis remains unknown. Considerable evidence also identifies the angiopoietins and the Tie2 receptor as important regulators of tumor-induced angiogenesis and, therefore, cancer growth and metastasis(Lin et al., 1998; Lin et al., 1997; Oliner et al., 2004).
The two receptors are remarkably similar with two amino-terminal immunoglobulin (Ig) domains followed by three epidermal growth factor (EGF) repeats, a third Ig domain and finally, three fibronectin type-III repeats in the extracellular region. Both Ties also contain a catalytic carboxyl-terminal tyrosine kinase domain responsible for intracellular signaling. Although binding studies originally identified both the Ig and EGF domains of Tie2 as necessary and sufficient for angiopoietin binding(Barton et al., 2005; Fiedler et al., 2003), more recent mutagenesis and crystallographic data revealed the ligand-binding site as the second Ig domain(Barton et al., 2006).
Initially described as an orphan receptor(Dumont et al., 1992), Tie2 was subsequently shown to interact with all four of the angiopoietins (Ang1-Ang4)(Davis et al., 1996; Maisonpierre et al., 1997; Valenzuela et al., 1999). The different angiopoietins, although having high sequence homology, elicit different responses from the receptor tyrosine kinase Tie2. Indeed, Ang1 is a constitutive receptor agonist while Ang2 is a context-dependent one(Davis et al., 1996; Gale et al., 2002; Maisonpierre et al., 1997). The hypothesis that differential presentation and binding of the various angiopoietins to Tie2 is responsible for their distinct biological effects appears unlikely in light of previous evidence that Ang1 and Ang2 bind to the same region of Tie2(Barton et al., 2005; Barton et al., 2006; Maisonpierre et al., 1997).
Now, it appears more likely that other cell-specific surface receptors exist that could help transduce the angiopoietin signals and modulate their functional potential. This hypothesis was recently supported by several reports defining a potential role for Tie1 in Tie2 signaling(Kim et al., 2006; Saharinen et al., 2005; Yuan et al., 2007). Tie1, although a close sequence homologue of Tie2, does not interact directly with the angiopoietins and its in vivo ligands are yet to be identified(Maisonpierre et al., 1997). Nevertheless, the angiopoietins may affect Tie1 function(Kim et al., 2006; Saharinen et al., 2005; Yuan et al., 2007). As observed by co-immunoprecipitation analysis, Tie1 and Tie2 appear to associate on the cell surface with receptor phosphorylation correlating with activation. A recent study using catalytically inactive Tie2 demonstrates that Tie1 phosphorylation can occur in trans and is dependent on a functional Tie2(Kim et al., 2006; Yuan et al., 2007).
Although great strides have been made in the elucidation of the structural and molecular mechanisms of the Ang-Tie signaling, many essential interactions and functional roles have yet to be determined. Of fundamental importance is the precise role of the Tie1 receptor in Tie2 signaling. It remains unclear when and how Tie1 and Tie2 associate on the cell surface. Furthermore, the ultimate biological significance of these interactions has yet to be determined, particularly with regard to the specific signaling characteristics of the individual angiopoietins.
To better understand the role and mechanism of action of Tie1 in angiopoietin mediated signaling, we examined Tie1 and Tie2 receptor localization and association in the presence and absence of Angiopoietin ligand on the cell surface via a FRET-based proximity assay. Based upon our studies, we propose a new model for the role of the Tie1 in angiopoietin-induced Tie2 signaling.
RESULTS
Pre-existent Tie1-Tie2 Complexes Observed By FRET Imaging On The Cell Surface
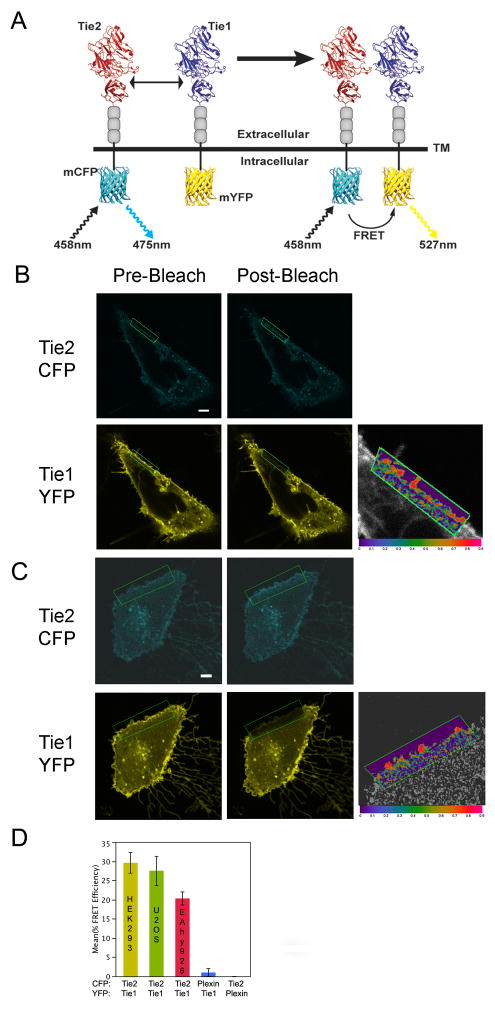
To examine the potential role of Tie1 as a co-receptor and evaluate Tie1-Tie2 interactions on the cell surface, we monitored receptor-receptor interactions in vivo by tagging the individual proteins with CFP and YFP, and following their localization and association during both agonist and antagonist angiopoietin signaling by confocal microscopy coupled with fluorescence (Förster) resonance energy transfer (FRET). Specifically, for assessing Tie1 and Tie2 receptor spatial proximity during angiopoietin signaling, we exploit the FRET methodology recently utilized by the Tsien and Springer groups to analyze the role of lipid modifications in membrane partitioning and Integrin signaling, respectively(Kim et al., 2003; Zacharias et al., 2002). We fused the monomeric enhanced green fluorescent protein (eGFP) variants mCFP and mYFP to the carboxyl termini of Tie2 and Tie1 respectively in place of the catalytic tyrosine kinase domain. As energy transfer between donor (CFP) and acceptor (YFP) only occurs over short distances (≤ 10nm), emission of YFP following excitation of CFP is observed only when Tie1 and Tie2 are in close proximity, presumably as a receptor/co-receptor complex (illustrated conceptually in Figure 1A). FRET efficiency is calculated by subtracting the background FRET (determined by the acceptor-photobleaching method) from the experimental FRET efficiency(Wouters et al., 2001).
Figure 1. Association of Tie1 and Tie2 on the cell surface as analyzed by the FRET-based spatial proximity assay.
A) Schematic representation of the spatial proximity assay used to monitor Tie1-Tie2 interactions in vivo. Receptor association allows measurable FRET between CFP and YFP via confocal microscopy. Ligand dimerization is shown for simplicity. B–C) Tie1-YFP and Tie2-CFP co-expression and association in U2OS (B), and EA.hy 926 (C) cells (See also Figure S1 for co-expression and localization in HEK293 cells). The majority of the receptor-fluorophore fusion localizes predominantly to the plasma membrane (although a fraction is also observed in the secretory system). D) Graphical representation of average FRET values for wild-type Tie1 and Tie2, as well as control receptors (Control experiments were conducted in U2OS cells and displayed in Figure S2). Data are represented as mean +/− SEM. *For all images CFP will be displayed as cyan and YFP as yellow. Photobleaching experiments were restricted to, and FRET values calculated from, the region within the green box. Both pre- and post-photobleaching images are displayed as left and right panels, respectively. FRET efficiency is displayed as an absolute range from high (red-1.0) to low (purple-0.0) on a magnified overlay of a CFP/YFP merged image for orientation purposes only. Light gray bar in each image indicates 10 μm.
Using our FRET-based proximity assay, we clearly observe Tie1-Tie2 association on the cell surface indicating the two receptors are within less than 100Å of one another. Interestingly, our observations are in the absence of angiopoietin ligand, further demonstrating that Tie1 and Tie2 are in a pre-existent complex prior to ligand recognition. Figure 1B–C illustrates a representative image of the fluorescence intensity and membrane localization of Tie2-CFP/Tie1-YFP, which we will denote as wild-type for our discussions. Both receptors localize with uniform diffuse staining predominantly on the plasma membrane. Using the acceptor-photobleaching technique for measuring FRET efficiency, we observed Tie1-Tie2 association with an average overall efficiency of 30%, 27%, and 19% under optimal conditions in HEK293, U2OS, and EA.hy 926 cells, respectively (Supp. Figure 1, Figures 1B–C). In accordance with the variables identified by Springer and colleagues(Kim et al., 2003), the length of linker between receptor transmembrane domain and amino-terminus of fluorescent protein was varied to identify the optimal combination of receptor length, and fluorophore-receptor pair (data not shown). Furthermore, for our experiments, HEK293 and U2OS cells were chosen in addition to EA.hy 926 endothelial cells, based upon their lack of endogenous Tie receptors as well as ease of transfection and cellular imaging(Yuan et al., 2007).
We found, as expected, that increased linker lengths significantly decreased overall FRET efficiency(Kim et al., 2003). Among several constructs tested, one pair with eleven and ten residues for Tie1 and -2 respectively, within the linker, consistently yielded reliable results (data not shown) in all three independent cell lines (EA.hy 926, HEK293, and U2OS). Importantly, both CFP and YFP variants carry the A206K, L221K, and F223R mutations thought to significantly decrease the chance of fluorescent protein multimerization. As a control, we have also co-expressed each receptor with both fluorophores to account for any non-specific interactions that may occur between them(Zacharias et al., 2002). No FRET signal is observed under these circumstances (data not shown). Similarly, to evaluate possible effects of over-expression, Tie receptors were co-expressed with the functionally unrelated receptor plexin-A1 (both CFP and YFP variants) (Supplemental Figures 2A and 2B). Under these circumstances we observed between 0 and 2% FRET efficiency. The five experimental conditions are graphically compared in Figure 1D and summarized in Table S1. Together, these findings validate the overall specificity of the assay and reveal the conservation of Tie receptor interactions within both epithelial and endothelial cell lines while simultaneously demonstrating that protein overexpression does not contribute significantly to FRET efficiency.
Homology Modeling Of Tie1
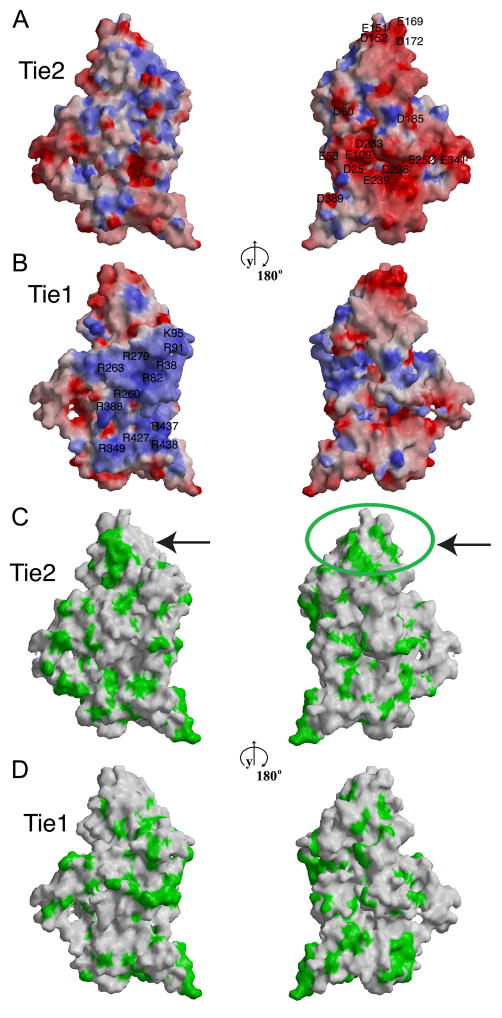
To understand the potential structural differences between Tie1 and Tie2 that mediate their distinct biological properties and identify structural elements that may contribute to Tie1-Tie2 interactions, we modeled the structure of Tie1 using the experimentally-determined 2.5Å Tie2 structure(Barton et al., 2006) and the program MODELLER(Marti-Renom et al., 2000). Tie1 and Tie2 are highly homologous – sharing 39% amino acid identity – and, not surprisingly Tie1 can be easily modeled on the structure of Tie2. A schematic representation of the molecular surfaces of Tie2 and of the Tie1 model, color coded according to electrostatic potential and hydrophobicity of the exposed amino acid side chains, is presented in Figure 2. The hydrophobic surface features of Tie2 and Tie1 are very similar overall. Interestingly, two patches of exposed hydrophobic residues, indicated with arrows, are present at the tip of Tie2, but are absent in the equivalent Tie1 region. Indeed, these overlap with the binding site of the Tie2-specific ligand Angiopoietin-2 (Ang2) indicated with a green circle(Barton et al., 2006), providing the structural explanation for the distinct ligand-binding properties of the two Tie receptors.
Figure 2. Comparison of the molecular surface properties of Tie2 and Tie1.
Molecular surface representation of Tie2 (A–C) and of the modeled Tie1 (B–D) structure color-coded according to electrostatic surface potential (A, B) or hydrophobicity (C, D). Red and blue represent electrostatic potentials in the range of −11 to +11 kBT, where kB is the Boltzman constant and T is the temperature (293 K). Green represents surface residues with hydrophobic side chains. The left and right panels are related by 180° rotation about the y-axis. Arrows point to the previously determined ligand-binding region within the tip of the second immunoglobulin domain in Tie2.
Comparison of the surface electrostatic potentials of Tie1 and Tie2 (Figures 2A and 2B) reveals that Tie2 has a slight negative overall charge with a theoretical pI value, if isolated in solution, of 6.9. Tie1, on the other hand has a positive overall charge with a corresponding pI value of 9.3. Tie2 has one expansive negatively-charged surface area, which results from the approximation of several exposed aspartic and glutamic acids in Ig1 and EGF1, including D25, E53, D60, E109, D236, D239, D252 and D283 (Figure 2A, right). The corresponding Tie1 surface area is overall neutral in charge. Tie1, on the other hand, contains one large positively charged area on its surface, located on the opposite side of the molecule (Figure 2B, left). Its electrostatic surface potential results from the approximation of several arginines and lysines in Ig1, EGF2, and Ig3, including R38, R82, K95, R91, R260, R279, R263, R349, R388, R427, R437, and R438.
Identification Of Residues Involved In Tie1-Tie2 Interactions
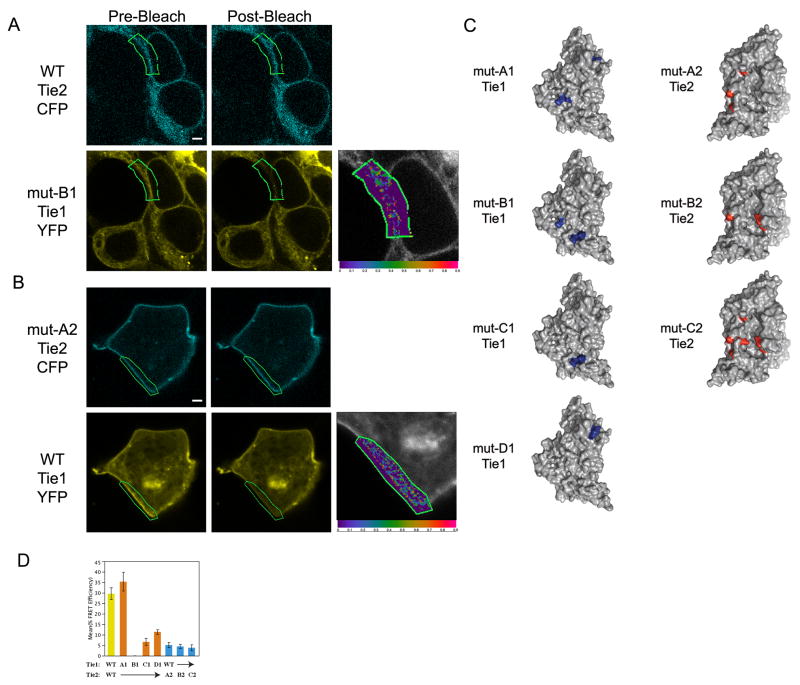
The presence of large patches of oppositely charged molecular surfaces in Tie1 and Tie2 suggest that these areas might be involved in Tie1-Tie2 recognition. To evaluate their roles in mediating receptor-receptor interactions, we utilized site-directed mutagenesis in combination with the proximity assay utilized in Figure 1. Various ‘regions’ within the charged patches were targeted by mutagenesis through the construction of Tie variants containing multiple amino acid substitutions and analyzed for their ability to associate via FRET. Our experience has shown that single site mutations often have little or no effect on interactions involving relatively large protein-protein interfaces(Barton et al., 2003; Barton et al., 2005; Barton et al., 2006) and, therefore we made multiple mutations simultaneously (typically two or three), following a charge reversal strategy (replaced basic residues with acidic amino acids, and vice versa). As all of these mutations are in surface exposed residues, they likely do not affect the overall folding of the proteins. In agreement, all mutant receptors are well expressed, processed, and localize correctly to the cell surface (Figure 3A, 3B, and data not shown). A list of all seven mutant receptor combinations, including averaged FRET efficiencies, is included in Table S1 and schematically displayed in Figure 3C.
Figure 3. Complementary electrostatic surface patches mediate Tie1-Tie2 association.
HEK293 cells were transfected with A) wild type Tie2-CFP and mutant Tie1-YFP B1, or B) Tie1-YFP wild type and mutant Tie2-CFP A2, and monitored by confocal microscopy. Individual mutations are abbreviated for clarity and are; A1: Tie1(K95E, R260E, R263E), B1: Tie1(R260E, R437E, R438E), C1: Tie1(R437E, R438E), D1: Tie1(R91E, K95E, R427E), A2: Tie2(E53K, D236K, E239K), B2: Tie2(E53K, D60K, D389K), and C2: Tie2(E53K, D60K, E109K, D236K, E239K, D389K). C) Molecular surface representation of Tie2 and modeled Tie1 structure color-coded according to their corresponding mutations. D) The average FRET efficiencies from the pairing of wild-type and mutant receptors. Data are represented as mean +/− SEM. * Images are as described in Figure 1.
Upon examination of our individual receptor mutants, it is clear that we have identified several charged surface residues involved in their association. Indeed, three of the four Tie1 mutants (B1, C1, D1) and all three Tie2 variants (A2, B2, C2) exhibit significantly lower FRET efficiencies than the wild-type receptors. A representative image of the receptor pair Tie1-YFP-B1/wild-type-Tie2-CFP, is displayed in Figure 3A. Even under ideal conditions, we failed to observe significant FRET when these two receptors are co-expressed in either HEK293 or U2OS cell lines (0% FRET efficiency). Similarly, Tie2-CFP-A2 exhibits proper localization to the membrane, yet fails to associate with wild-type Tie1-YFP to any significant extent (5% FRET efficiency - Figure 3B). Together, the data suggest a complex interaction between Tie1 and Tie2 involving a broad surface area within their basic and acidic regions, respectively. Indeed, several different mutant receptor combinations with a wild-type co-receptor result in the loss of receptor-receptor association as illustrated by low or absent FRET efficiencies, demonstrating that the charged surface patches play a role in the Tie1-Tie2 association (See Figures 3D and Table S1). Moreover, analysis of the Tie1 mutants suggests that residues R260, R437, and R438 in the bottom half of the ectodomain (See Figure 2B and Figure 3C) play a central role in receptor association (compare mutants A1, B1, and C1 in Figure 3D). Finally, the identification of mutant-wild-type receptor pairs that fail to demonstrate appreciable FRET further demonstrates the exquisite specificity of this proximity assay.
Tie1 And Tie2 Interactions Are Direct
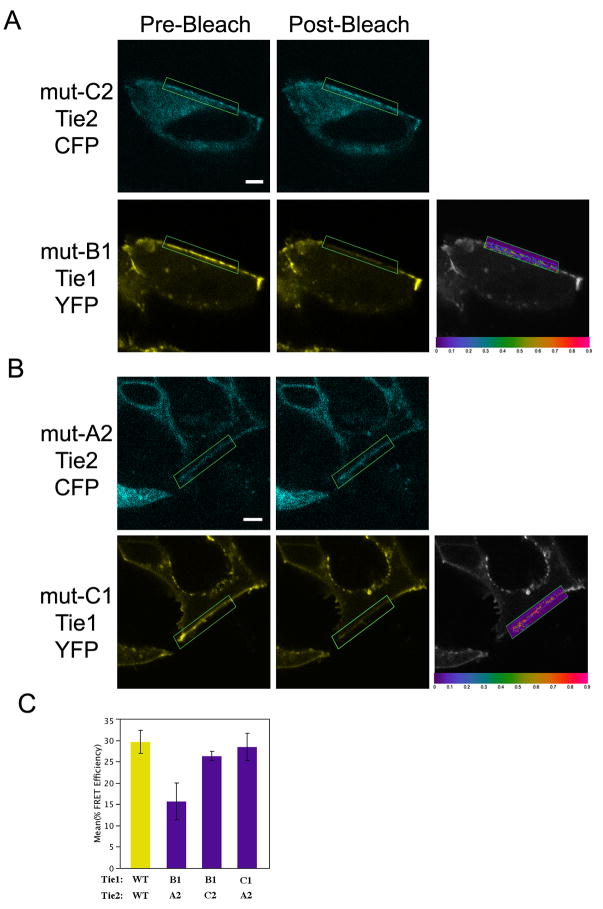
To validate the notion that Tie1-Tie2 interactions are direct, and not mediated by an unidentified binding partner, we assayed for receptor complementation - as illustrated by the restoration of wild-type FRET efficiencies - between two individual mutant receptors. HEK293 cells were cotransfected with all nine possible combinations of mutant Tie1 and Tie2 receptors. Given the size and complexity of the potential interface involved, only a subset of receptor combinations would be predicted to have molecular surfaces of sufficient complementarity to rescue the FRET efficiency to near wild-type levels. Indeed, from all tested receptor combinations we identified only two mutant receptor pairs; Tie1-YFP-B1/Tie2-CFP-C2, and Tie1-YFP-C1/Tie2-CFP-A2, which display significant receptor association (representative FRET efficiencies for each receptor pair are shown in Figure 4C and Table S1). Individually, each of these mutated receptors is incapable of interacting with their counterpart wild-type receptor or other mutant receptors, yet when co-expressed, display FRET efficiencies nearly identical to that of wild-type receptors (Figure 4A and 4B). Averaged FRET efficiencies from several independent experiments for each receptor pair are graphed and compared to wild-type in Figure 4C. FRET efficiencies between wild-type receptors and these two mutant pairs are not significantly different. This demonstration of “allelic suppression” excludes the possibility of an unknown binding partner, and provides strong support for a direct interaction between Tie1 and -2 involving the complementary charged surface areas within the receptor ectodomains.
Figure 4. Tie1 and Tie2 form a direct interaction as demonstrated by allelic suppression.
HEK293 cells were transfected with the following mutant receptor pairs A) Tie1-YFP B1 and Tie2-CFP C2 or B) Tie1-YFP C1 and Tie2-CFP A2, and monitored by confocal microscopy. C) The average FRET efficiencies from experiments in A–B. Data are represented as mean +/− SEM. * Images are as described in Figure 1.
Different Angiopoietins Differentially Modulate The Tie1-Tie2 Interaction
Despite the high level of sequence identity between Tie2 and Tie1, only Tie2 can form high-affinity complexes with all four known angiopoietins, while Tie1 does not bind any of them(Davis et al., 2003; Ramsauer and D’Amore, 2002). Nevertheless, the identification of pre-existing Tie1-Tie2 complexes suggests that Tie1 plays a role in Tie2 signaling. Interestingly, our crystal structure of Tie2 bound to Ang2(Barton et al., 2006) and to Ang1 (unpublished data) show that all angiopoietins bind Tie2 in a similar conformation, excluding the possibility of differential Tie2 activation resulting from alternate Tie2/angiopoietin structural arrangements. However, our structural analysis does suggest that the different angiopoietin ligands could present distinct molecular surfaces outside of the receptor-binding interface that could influence the Tie1-Tie2 receptor complexes(Barton et al., 2005). Based upon our findings we hypothesized that the observed direct Tie2/Tie1 interactions are inhibitory and the ability, or inability, of individual angiopoietins to effectively destabilize the Tie1/Tie2 complexes at the cell surface would define their respective agonistic or antagonistic roles.
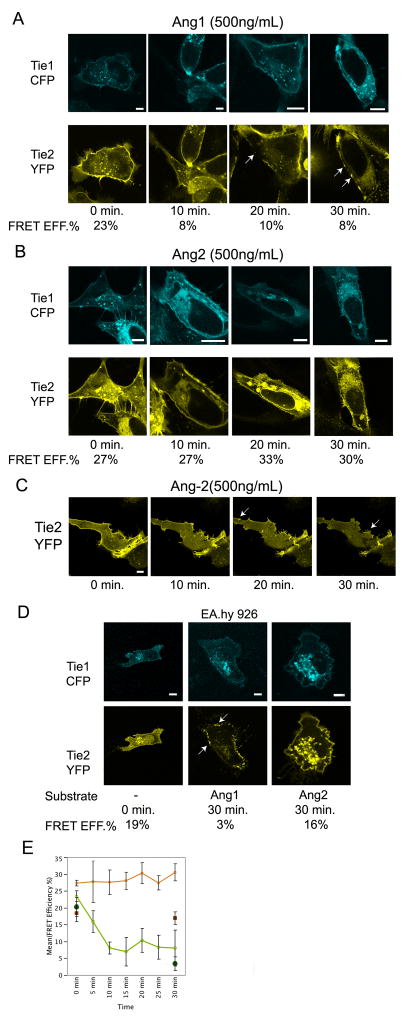
To assess the role of Tie1 in Tie2 clustering and activation following angiopoietin exposure, receptor association, localization, and activation was monitored in the presence of Angiopoietin-1 or -2. Specifically, Tie1-CFP and Tie2-YFP were expressed in U2OS and EA.hy 926 cells and stimulated with varying concentrations of Angiopoietin-1 or –2, and followed by confocal microscopy over the course of 30 or 60 minutes. As shown in Figure 5A (and via time-lapse imaging in the Supplemental Data), addition of Ang1 to the culture media led to a drastic reduction of Tie1-Tie2 association in U2OS cells within 10 minutes as measured by FRET. This timeframe corresponds to previously determined rates of Tie2 activation(Bogdanovic et al., 2006; Yuan et al., 2007). Furthermore, the loss in FRET signal was followed by a dramatic change in Tie2 localization (Compare t=0 and t=20 in lower panel of Figure 5A). Specifically, the majority of the membrane-associated Tie2 migrates and forms discrete foci, reminiscent of the Tie2 punctate staining observed by others in human lung microvascular endothelial cells(Fukuhara et al., 2008; Saharinen et al., 2008). Thirty minutes post Ang1 addition cells that contain discrete Tie2 foci are still observed (lower panel of Figure 5A). As expected from our FRET analysis, Tie1 localization remains undisturbed following Ang-1 stimulation (compare bottom and top panels of Figure 5A). EA.hy 926 endothelial cells under similar conditions behave correspondingly. In these cells Tie2 readily localizes to the membrane, yet transforms to punctate staining with similar kinetics to HEK293 cells upon addition of Ang-1 (Figure 5D middle panels). FRET efficiency rapidly decreases (from 19% to 3%) further demonstrating disruption of Tie1-Tie2 interactions.
Figure 5. Ang-1, but not Ang-2, promotes Tie2 clustering and dissociation of Tie1-Tie2 complexes.
A–B) U2OS cells were transfected with both Tie2-YFP and Tie1-CFP and monitored by confocal microscopy over a period of 30 minutes. At time=0, either 500ng/ml of Ang-1 (panel A), or 500 ng/ml of Ang-2 (panel B) was added to the growth media. Listed below each representative image are averaged FRET values correlating to the listed time point. Arrowheads indicate regions of clustered Tie2. C) U2OS cells were singly transfected with Tie2-YFP and monitored by confocal microscopy over a period of 30 minutes following addition of 500 ng/ml of Ang-2 to the growth media. D) EA.hy 926 cells were transfected with both Tie2-YFP and Tie1-CFP and monitored by confocal microscopy over a period of 30 minutes. At time=0, either 500ng/ml of Ang-1 (middle panels), or 500 ng/ml of Ang-2 (right panels) was added to the growth media. Listed below each representative image are averaged FRET values correlating to the listed time point. E) The average FRET efficiencies from experiments in A, B, and C. Green and Orange lines represent Ang1, or Ang2 addition for U2OS cells, respectively, while closed Green circles and closed Orange squares represent Ang1, or Ang2 addition for EA.hy 926 cells, respectively. Data are represented as mean +/− SEM.
Alternatively, when Tie1 and Tie2 expressing U2OS or EA.hy 926 cells are treated with similar concentrations of Ang-2, no changes in the Tie1-Tie2 association and/or Tie2 localization are observed (Compare Figures 5A and 5B and right bottom panels in Figure 5D). FRET efficiencies and receptor localization do not significantly change over a thirty-minute period as graphically illustrated in Figure 5E. This is in sharp contrast with the ability of Ang-2 to induce changes in Tie2 localization in the absence of Tie1. Under these conditions, Tie2 displays punctate staining within 10–20 minutes identical to that observed for Ang-1 (Figure 5C and Supplementary video). Thus, our data demonstrates a unique difference between individual angiopoietins to affect Tie1 –Tie2 association and subsequent Tie2 clustering.
Tie1 is an Inhibitory Co-Receptor
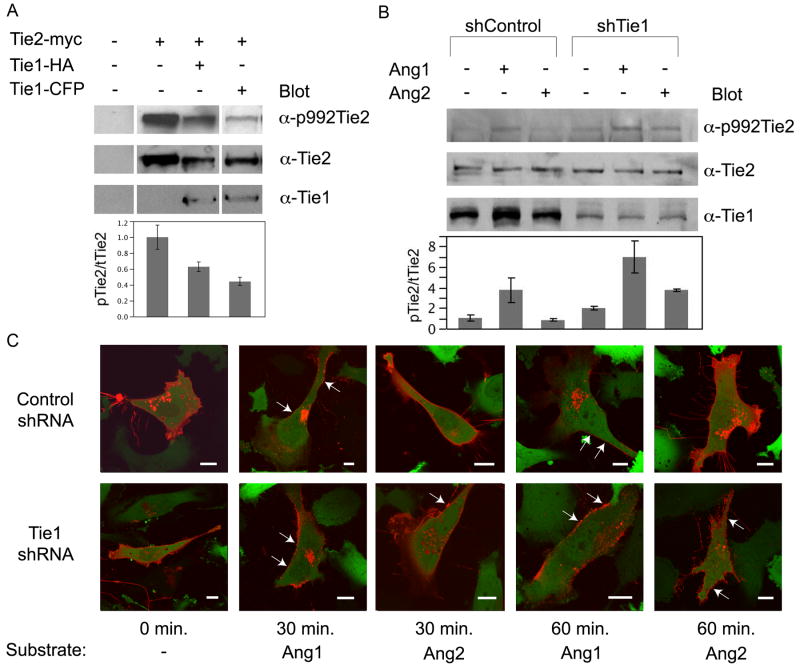
To further assess the physiological role of Tie1, particularly with its capability to manipulate Tie2 activation, we followed Tie2 phosphorylation in the presence or absence of Tie1 and Angiopoietin in both HEK293, and HUVEC’s. Initially, HEK293 cells, which lack endogenous Tie receptors, were transiently transfected with full-length myc-tagged Tie2, full-length HA-tagged Tie1, Tie1-CFP lacking a tyrosine kinase domain, or a combination of the three. As seen in Figure 6A, western blotting of crude lysates with anti-Tie1 and anti-Tie2 antibodies, respectively, clearly demonstrate robust Tie1 and Tie2 expression. Interestingly, ectopically expressed Tie2 displays a low level of basal activation in 293 cells as observed by the anti-phosphotyrosine 992 Tie2 antibody (top panel Figure 6A). However, as predicted, addition of Tie1 dramatically decreased Tie2 basal activation (~50% relative to Tie2 alone). Interestingly, addition of chimeric Tie1-CFP lacking an intracellular tyrosine kinase domain also decreases Tie2 activation to levels comparable to those observed with full-length Tie1 (Figure 6A). This finding further demonstrates that Tie ectodomain interactions are necessary and sufficient for Tie2 inhibition.
Figure 6. Tie1 is as an inhibitory co-receptor for Tie2.
A) HEK293 cells were transfected with full-length Tie2-myc alone or in combination with full-length Tie1-HA, or Tie1-CFP, harvested and lysates probed with anti-Tie1, anti-Tie2, and anti-phosphotyrosine 992 Tie2. The relative ratio of phosphorylated Tie2 (top panel) to total Tie2 (middle panel) was determined and the calculated value was arbitrarily set equal to 1.0 for Tie2 expressed alone. Values are graphically displayed below each respective lane. *Samples are from non-adjacent lanes of the same blot. B) Human umbilical vein endothelial cells were infected with an shRNA control or Tie1 knockdown lentivirus and selected with puromycin for stable integration (See Figure S3 for extent of Tie1 silencing). Asynchronous cultures were subjected to four hours of serum withdrawal, followed by stimulation with vehicle (buffer), 500ng/ml Angiopoietin-1, or 500ng/ml Angiopoietin-2. Fifteen minutes following addition of ligand, cells were harvested and subjected to western blotting with anti-Tie2 (middle panel), anti-Tie1 (bottom panel), and the phosphotyrosine specific anti-Tie2 (992) (top panel), as indicated. The relative ratio of phosphorylated Tie2 (top panel) over total Tie2 (middle panel) was determined and the calculated value was arbitrarily set equal to 1.0 for unstimulated Tie2 from shControl cells. Values are graphically displayed below each respective lane. C) Control (top panels) or Tie1 knockdown (bottom panels) stable EA.hy 926 cells were transiently transfected with full-length Tie2 fused to mCherry and imaged via confocal microscopy forty-eight hours post transfection. Cells were stimulated with Angiopoietin-1 or -2 and mCherry expression was followed over the course of sixty minutes. GFP expression (green) indicates stable viral integration and was overlaid onto mCherry (red) for orientation purposes. Arrows indicate areas of punctate Tie2 staining. Light gray bar in each image indicates 10 μm.
Using an alternative approach to examine the role of endogenous Tie1, HUVEC’s were infected with recombinant lentivirus encoding a non-specific (control) or Tie1-specific miR-shRNA. Stable cell lines were selected with puromycin and Tie receptor expression and receptor activation was analyzed following ligand addition by means of western blot analysis. In an shRNA control cell line, which displays wild-type levels of Tie1, Tie2 activation can be readily observed within 15 minutes following the addition of Ang-1 (Figure 6B). In contrast, and in agreement with others, addition of Ang-2 fails to significantly alter receptor phosphorylation and activation (Davis et al., 2003; Maisonpierre et al., 1997).
Alternatively, HUVEC Tie1-shRNA stable cell lines, which express significantly (approximately 70%) less Tie1 as seen in control (non-specific shRNA) cells, behave considerably different. In the absence Tie1, and in agreement with our findings in epithelial cells, Tie2 displays approximately 2 fold greater basal activation than within control cells (Figure 6B). Furthermore, despite the observation that Tie2 appears partially active in Tie1 silenced cells, addition of Ang-1 also leads to ~3.5 fold greater receptor activation. However, in the absence of Tie1, Ang-2 is now also capable of stimulating Tie2 activation. Indeed, despite the observation that Tie2 appears partially active in Tie1 silenced cells, addition of Ang-1 or Ang-2 leads to greater receptor activation (~3.5 and 2 fold, respectively). Cumulatively, these observations are consistent with an inhibitory role for endogenous Tie1 and indicate its presence is necessary to distinguish the agonist/antagonist role for Ang-2.
The Tie2 Kinase Domain is Unnecessary for Receptor Clustering and Tie1 Interaction
Despite a clear demonstration of Tie receptor ectodomain interactions by FRET analysis, it remained unclear if the intracellular kinase domain could influence receptor localization and clustering in response to ligand. Therefore, to evaluate the role of the Tie2 tyrosine kinase domain in Tie interactions, a full-length version of Tie2 with a carboxyl terminal mCherry domain was expressed in control and Tie1 silenced EA.hy 926 cells (extent of Tie1 knockdown can be observed in Figure S3). Under these conditions, endogenous Tie1 will interact with, and influence Tie2 localization, if indeed the tyrosine kinase domains are essential. Accordingly, Tie2-mCherry localization was monitored by confocal microscopy following both Angiopoietin-1 and -2 addition. Figure 6C includes images of cells prior to, 30, and 60 minutes post ligand addition. As observed with Tie2-CFP chimeras lacking the tyrosine kinase domain, full-length Tie2-mCherry exhibits punctate staining within 30 minutes in the presence of Ang-1 in both control and Tie1 silenced cells. Similarly, Tie2 clustering is observed within 60 minutes addition of Ang-2 in the absence of Tie1 (Figure 6C-bottom panels). A slight difference in time between the observed clustering in Ang-1 and -2 induced cells is likely due to the residual Tie1 (~25% of wild-type levels) that remains in Tie1 knockdown cells. Although these studies do not rule out other potential roles for the Tie tyrosine kinase domains, these findings demonstrate that kinase domain interactions do not significantly influence the clustering and localization of Tie2.
DISCUSSION
Until now, the molecular basis for the Tie1 and the differential angiopoietin functions has remained elusive. However, using a powerful in vivo proximity assay, we now identify a clear role for Tie1 in Tie2 signaling, and demonstrate a direct, inhibitory interaction between these receptors. We further identify, via structure-based site-directed mutagenesis, the precise molecular surface regions of the Tie1 and Tie2 ectodomains, which mediate this critical interaction. Finally, we reveal that this interaction is dynamic and differentially modulated by the different angiopoietin ligands thereby providing a molecular mechanism for the observed differences in angiopoietin function.
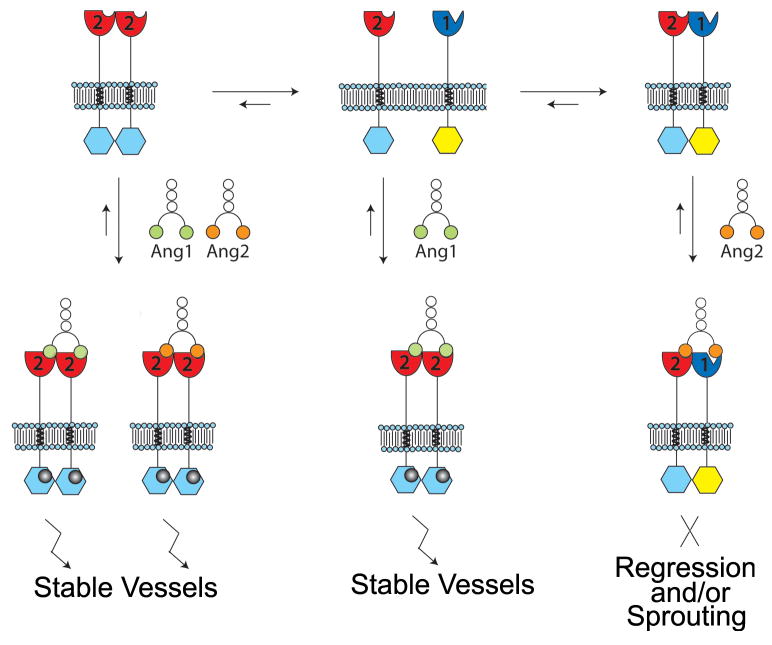
Based upon previous studies and the data presented here, we propose a model for Tie2 signaling as illustrated in Figure 7. Specifically, in cells expressing both Tie1 and Tie2, the receptors form heterodimers within endothelial cells via ectodomain electrostatic interactions that inhibit Tie2 activation and clustering. Binding of Angiopoietin-1 to Tie2 promotes heterodimer dissociation, Tie2 clustering, and signaling initiation. On the contrary, Angiopoietin-2, is unable to dissociate the inhibitory Tie2/Tie1 complexes upon binding Tie2 and, therefore, does not induce Tie2 clustering and signaling, yet behaves as a competitive antagonist by blocking further binding of Ang-1. Alternatively, for cells that do not express Tie1, all angiopoietins promote Tie2 clustering and activation. Our model explicitly proposes that the balance of Tie1 and Tie2 expression modulates the functional potential of Angiopoietin-2, and by analogy, vascular homeostasis.
Figure 7. Model for Angiopoietin-mediated Tie2 signaling.
Expression of Tie2 in the absence of Tie1 at sites of vessel quiescence and maturity. In the absence of Tie1, Tie2 can be activated with either Ang-1 or Ang-2. Both ligands stimulate receptor clustering, tyrosine kinase activity, and down-stream signaling events, effectively become unresponsive to vessel sprouting and branching cues. Within sites of active angiogenesis, Tie1 and Tie2 associate to form a complex prior to ligand stimulation. Upon addition of Ang-2, Tie1 and Tie2 association and localization remains unchanged. Under these conditions, Ang-2 fails to activate the Tie2 receptor and opposes the activation of downstream signaling generated by Ang-1. However, upon addition of Ang-1, the opposite is observed. Ang-1 stimulates Tie2 clustering, tyrosine kinase activity, and down-stream signaling events similar to that observed in the absence of Tie1. *Angiopoietins are depicted as dimers for illustration purposes although they are known to exist as higher-order clusters.
While it is known that Tie2 is expressed ubiquitously throughout the vascular endothelium(Dumont et al., 1994; Dumont et al., 1992), Tie1 expression is significantly restricted to vascular bifurcations and branching points following embryogenesis(Porat et al., 2004). Furthermore, Tie1 is transcriptionally up-regulated by hypoxia, VEGF stimulation, and in areas of wound healing and tumor growth and development (i.e. sites of neo-vascularization)(Korhonen et al., 1992; McCarthy et al., 1998). Therefore, the adult vasculature is composed of regions of alternating (high and low, or absent) Tie1 expression. Regions that are actively involved in angiogenesis, and require the antagonistic function of Ang-2, are, interestingly, the very same regions where Tie1 expression is observed. Under these conditions, the majority of Tie1 and Tie2 form signaling-incompetent heterotypic complexes on the cell surface (Figure 7). Their association is dynamic and mediated via an electrostatic interaction between charged residues within their ectodomains, as well as the presence or absence of activating ligand (i.e. Ang-1). Cells expressing both receptors are responsive to the survival, migration, and chemotactic cues caused by activation and inactivation of Tie2 via Ang-1 and Ang-2 and are, therefore, able to promote the required vessel branching and sprouting necessary for angiogenesis.
Alternatively, Tie1 is absent in vascular regions that are stable and quiescent, such as mature vessels. The absence of Tie1 negates the functional differences between Ang-1 and Ang-2 and allows endothelial cells to respond to either ligand in a similar fashion (both as agonist), and foster the same phenotypic response-quiescence and survival (Figure 7). In this regard, Tie1 serves as a selectivity factor, designating when and how Ang-2 functions. In general, this could provide greater cellular adhesion with the underlying vessel support cells, and provide yet another mechanism to stabilize the adult vasculature and prevent aberrant vessel sprouting and branching that could lead to pathogenesis.
Consistent with our proposed model, in the absence of Tie1 (Figure 6), Tie2 exhibits basal phosphorylation and remains partially activated despite the absence of ligand, yet it can become further clustered and activated by both the agonist Ang-1 as well as antagonist, Ang-2(Davis et al., 2003; Maisonpierre et al., 1997). Alternatively, in the presence of Tie1, the ligand-independence and basal Tie2 activity are attenuated (Figure 6A and B). In agreement, loss of Tie1 (via shRNA), eliminates the functional differences between Ang-1 and Ang-2 in HUVEC (Figure 6B). Under these conditions, endothelial cells respond to both ligands similarly. In agreement, Yuan et al. observed that siRNA toward Tie1 significantly increased the downstream activation of Tie2 signaling components, therefore concluding that Tie1, in it’s normal role, antagonizes Tie2 function(Yuan et al., 2007). An analogous conclusion was drawn by Patan(Patan, 1998) following histological assessment of Tie1 and Tie2 knockout mice phenotypes. In addition, Kim et al. demonstrate Ang2-induced Tie2 activation following down-regulation of Tie1 via siRNA in HUVECs(Kim et al., 2006) and Nguyen et al.(Nguyen et al., 2007) observed differential responses of lymphatic versus venous or arterial endothelial cells to Angiopoietin-1 and -2. In line with this observation, our model would predict that lymphatic endothelial cells, thought to be the primary target of the Ang-2 agonist (as shown by knockout and over-expression experiments), would display lower levels of Tie1 protein, although this has yet to be investigated.
Some have observed Tie2 to preferentially localize at sites of cell-cell contacts (Fukuhara et al., 2008; Saharinen et al., 2008). Indeed, our expression constructs with and without the intracellular tyrosine kinase domain demonstrate related localization patterns, and we, therefore, conclude that clustering to cell-cell junctions is likely mediated by ectodomain interactions and not through the Tie2 kinase domain. While Tie2 clustering to cell junctions presents an appealing means to localize specific signaling events, the exact mechanism by which this occurs remains unclear.
It is interesting to note that our model for Tie2 activation differs from others. Using Tie1 truncation mutants and a TrkA/Tie2 fusion protein, Marron and colleagues, for example, suggest an association between Tie1 and Tie2 that is mediated by their cytoplasmic kinase domains (Marron et al., 2000), while we observe a dynamic interaction between these receptors in the absence of the catalytic kinase domains and any subsequent down-stream signaling events. Moreover, our structure-based receptor mutagenesis, combined with localization and FRET analysis strongly supports receptor interactions through their extracellular domains. Interestingly, despite having documented receptor association, Marron and colleagues did not observe significant phosphorylation of Tie1(Marron et al., 2000). However, during the course of Tie2 activation, others have observed Tie1 phosphorylation (Saharinen et al., 2005; Yuan et al., 2007). It appears in some circumstances Tie1 phosphorylation correlates with Tie2 activation, although it remains unknown if this is as a result of stable or of transient interactions caused by high local concentrations of Tie2, i.e. within signaling clusters(Milner et al., 2008; Saharinen et al., 2005). Furthermore, the physiological significance of Tie1 phosphorylation remains unknown.
Finally, it should be noted that our studies also contrast with those by Saharinen et al. in which Tie1 and Tie2 association is proposed to occur in the presence of Ang1 as suggested by co-localization experiments(Saharinen et al., 2008). Previous reports by the same group indicate stimulation of Tie1-Tie2 co-immunoprecipitation following Ang-1 exposure (Saharinen et al., 2005). Although we observe dramatic changes in Tie2 localization, clustering, and association with Tie1 upon binding of Ang-1, we do not see a concomitant change in Tie1 localization. Furthermore, the extent of Tie2 clustering we observe, is significantly greater, and FRET analysis reveals nearly immediate dissociation of the Tie1-Tie2 complexes upon receptor stimulation. Since our analysis of receptor complex formation is at a significantly higher resolution (<10nm versus >1μm) and more direct, we feel the lack of change in Tie1 localization and loss of FRET documents Ang-1 induced Tie2-Tie1 complex dissociation rather than association. Furthermore, our data and proposed function of Tie1 correlate with the observed phenotypes of Tie and Ang knock-out mice, Tie receptor expression patterns, and recent data (including data presented here), confirming the inhibitory effects of Tie1 on Tie2 activation(Kim et al., 2006; Patan, 1998; Yuan et al., 2007). Collectively, our results indicate that the balance of expression and dynamic interaction between Tie1 and Tie2 provides an effective means of controlling receptor activation, and by analogy vascular homeostasis, using a single set of structurally similar ligands. The exquisite level of molecular control clearly highlights the importance of restricting Tie2 activation in order to maintain vascular homeostasis and to prevent pathogenesis; therefore, the Tie1-Tie2 interface may serve as an attractive therapeutic target and may be more relevant than the currently scrutinized Ang2-Tie2 interaction.
EXPERIMENTAL PROCEDURES
Homology model of Tie1
The Tie1 homology structure was modeled using the program MODELLER (Marti-Renom et al., 2000) and the experimentally determined 2.5Å Tie2 ligand-binding domain crystal structure (PDB 2GY5)(Barton et al., 2006). Although initial alignments between Tie1 and Tie2 were prepared using CLUSTALX(Larkin et al., 2007), use of the sequence/secondary structure alignment implementation in MODELLER yielded better results (as assessed below) and was therefore adopted for final calculations. Four distinct models were calculated which satisfied basic spatial and stereochemical restraints, however, the model used for interpretation and illustration purposes had the lowest discrete optimized protein energy (DOPE) assessment score and MODELLER objective function (Shen and Sali, 2006). Stereochemical analysis via PROCHECK (CCP4, 1994) revealed main chain parameters better than or within the typical range of values for experimentally determined protein structures at 2.5Å resolution.
Cloning and Gene Expression
The sequences encoding the human Tie1 (IMAGE 5767075) and Tie2 (IMAGE 5228999) genes were cloned as interchangeable receptor-fluorophore fusion cassettes in both pcDNA3.1(+) hygromycin and neomycin resistance vectors for expression in human embryonic kidney 293 cells and human osteosarcoma cell line U2OS. Briefly, NheI and EcoRI sites were appended to the amino and carboxy-terminus via PCR using the following oligonucleotides: Tie1-Nhe; gctagcATGgtctggcgggtgccc, Tie2-Nhe; gctagcATGgactctttagccagcttag, Tie1-EcoR1; gaattcggtgcgtctccgatgcaggcagc and Tie2-EcoRI; gaattctctcctttgcacatttgccctc (restriction sites are underlined and initiating methionine is in capital letters). After insertion of receptor DNA into the pcDNA vectors, GFP cassettes were cloned down-stream and in-frame with EcoRI and XhoI yielding an open reading frame consisting of Tie receptor fused to either C- or YFP. Monomeric CFP and YFP variants (containing dimeric suppression mutations-obtained from Dr Timothy Springer) were utilized as templates for fluorophore incorporation via PCR using the following oligonucleotides; C/YFP-EcoRI; gaattcatggtgagcaagggcgaggag, C/YFP-XhoI; ctcgagttatctagatccggtggatcc. Although several initial constructs were constructed and tested for optimal linker length between receptor and fluorophore, ultimately, one receptor pair was chosen for further study in which the Tie receptors were fused to either CFP or YFP after residues 796 and 780 of Tie1 and Tie2 respectively, leaving the transmembrane domain intact as well as ten or eleven additional cytoplasmic residues, but eliminating the carboxyl-terminal tyrosine kinase catalytic domain. Altering fluorophores on individual receptors did not appear to make any significant difference and were, therefore, used interchangeably.
Full-length Tie2-mCherry was constructed by fusing the fluorophore to the carboxy terminus of Tie2 via PCR stitching using the partially complementary oligonucleotides mCherry/TKD; gttctgctgaagaagcggccggtgcatctggttctatggccatcatcaaggag, TKD/mCherry; ctccttgatgatggccatagaaccagatgcaccggccgcttcttcagcagaac, and mCherry Xho; gctcactcgagctacttgtacagctcgtcc. The final construct was cloned as an NheI-XhoI fragment into pcDNA3.1(+) hygromycin.
Mutations within Tie1 and Tie2 coding regions were introduced by site directed mutagenesis (Quikchange Multi, Stratagene) following manufacturers recommendations. To confirm the presence of the desired mutations, both DNA strands were sequenced using standard di-deoxy sequencing chemistry (Cornell University Bioresource Center).
Cell Manipulations and Transfections
HEK293, U2OS, and EA.hy 926 (a gift from Dr Cora-Jean Edgell) cells were grown in DMEM (Invitrogen) supplemented with 10% fetal bovine serum, 100U/mL penicillin, and 100 μg/mL streptomycin. Human umbilical vein endothelial cells (HUVEC) were grown in EBM II media according to recommendations (Clonetics). Cells were consistently transfected at 80–90% confluence in 35mm glass bottom culture dishes (MatTek) using Lipofectamine 2000 (HEK293) or FuGENE HD (U2OS) or FuGENE 6 (EA.hy 926) according to manufacturer’s recommendations (Invitrogen and Roche). For co-expression experiments, equimolar concentrations of Tie1 and Tie2 vector DNA were used.
shRNA Knockdown of Tie1
Recombinant lentivirus encoding a Tie1 or control shRNA were constructed according to manufacturers recommendations (Open Biosystems). Stable EA.hy 926 or early passage HUVEC’s were selected in the presence of 0.8 ug/ml puromycin 48 hours post viral infection. After seven days, stable cells were monitored for Tie1 and Tie2 expression via western blot analysis (antibodies listed below).
Tie2 Activation Assays
For analysis of Tie2 activation in HEK293, cells were transfected with combinations of full-length Tie1-HA, and/or Tie2-myc tagged vectors. Forty-eight hours post-transfection, cells were lysed in HBST (20mM Hepes pH 7.4, 150mM NaCl, 0.1% Triton X-100) in the presence of 0.5mM sodium orthovanadate and subjected to western blotting. For Tie2 activation in HUVEC and EA.hy, post-confluent cells were serum starved for 2 hours prior to the addition of 500ng/ml Angiopoietin-1, -2, or vehicle (PBS). Fifteen minutes prior to ligand addition, sodium orthovanadate (Sigma) was added to 1mM in the culture medium. Thirty and sixty minutes following ligand addition, cells were harvested as stated above. Endogenous Tie1 and -2 were analyzed with anti-Tie2 (Santa Cruz Biotechnology), anti-phosphotyrosine-specific 992 Tie2 (R&D Systems), or anti-Tie1 (R&D Systems and Santa Cruz Biotechnology). Quantitative values for band intensities were obtained from western blots using the program ImageJ (Abramoff et al., 2004). Briefly, the integrated pixel intensity was determined for each band of interest using an identically sized rectangular masking box. The background was similarly determined from an identical region of the blot from lanes lacking the protein of interest - except in cases where no such lane control could be used. Under these conditions, background was calculated from a blank region above each band of interest. Finally, the background was subtracted uniformly from the experimental values to obtain the final raw values. All statistical calculations were determined using JMP software version 7.0 (SAS).
Cellular Imaging
The spatial proximity assay was based upon the work of Dr Roger Tsien (Zacharias et al., 2002) and Dr Timothy Springer and colleagues(Kim et al., 2003). Live cell imaging was performed 24–48 hour post-transfection on a Leica TCS-SP2 AOBS confocal laser scanning microscope equipped with blue diode (405nm), Argon (458, 476, 488, 514nm). green HeNe (543nm), orange HeNe (594nm), and red HeNe (633nm) lasers, an HCX PI Apo 63x/1.3 n.a. glycerin-immersion objective lens, a motorized XY stage (Märzhäuser), and an environmentally controlled (temperature, humidity, and CO2) stage incubator (PeCon). Fluorophore-receptor fusions were imaged using excitation and emission wavelengths of 458nm and 514 nm for CFP and YFP respectively and fluorescence emissions were detected with SP window settings of 465–505nm and 525–600nm (for CFP and YFP, respectively). FRET efficiencies were determined using the acceptor photobleaching methodology using Leica software (ver. 2.61). Briefly, regions of interest were chosen for analysis based on extent of fluorophore expression, localization, and uniformity. For acceptor photobleaching, YFP within the region of interest (ROI-green box within images) was consistently photobleached (with the AOTF ramped up to 100% transmission of the 514nm laser line) from 50% to 70% reduction in fluorescence intensity as monitored by Leica software. Care was taken to exclude cells for analysis that displayed significantly higher, or lower, fluorescent intensity than the “average” cell. To eliminate bias, cells were also chosen based on similar levels of CFP and YFP fusion protein. Similar to the work by Kim et al. (Kim et al., 2003), cells that displayed significant drift in the x-y focal plane were discarded for FRET analysis. Due to significant cellular drift, discrete regions, rather than whole cells were subjected to photobleaching to accelerate FRET analysis. Pre- and post-bleach images were recorded for both donor (CFP) and acceptor (YFP) and FRET efficiency was calculated as: FRETEff=(Dpost−Dpre)/Dpost for all Dpost > Dpre where Dpre and Dpost is the donor intensity before and after photobleaching respectively. JMP Software (SAS) was used for all statistical analysis. mCherry-receptor fusions and GFP were imaged as stated above with the exception that excitation wavelengths were 594nm and 488nm, respectively, and fluorescence emissions were detected with SP window settings of 605–700nm and 500–560nm (for mCherry and GFP, respectively).
Supplementary Material
Acknowledgments
This research was supported by grants from the National Institutes of Health 1RO1CA127501 to W.A.B. and 1RO1HL077249 to D.B.N., as well as pilot project funding from the Massey Cancer Center and School of Medicine (VCU) to W.A.B. Microscopy was performed at the VCU-Dept. of Neurobiology & Anatomy Microscopy Facility, supported, in part, with funding from NIH-NINDS Center core grant 5P30NS047463. We would also like to gratefully acknowledge the helpful suggestions and support of Drs. Rick Moran and Gordon Ginder.
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental data includes; three figures, three tables, and three time-lapse videos.
References
- Abramoff MD, Magelhaes PJ, Ram SJ. Image Processing with ImageJ. Biophotonics International. 2004;11:36–42. [Google Scholar]
- Barton WA, Liu BP, Tzvetkova D, Jeffrey PD, Fournier AE, Sah D, Cate R, Strittmatter SM, Nikolov DB. Structure and axon outgrowth inhibitor binding of the Nogo-66 receptor and related proteins. The EMBO journal. 2003;22:3291–3302. doi: 10.1093/emboj/cdg325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton WA, Tzvetkova D, Nikolov DB. Structure of the angiopoietin-2 receptor binding domain and identification of surfaces involved in Tie2 recognition. Structure. 2005;13:825–832. doi: 10.1016/j.str.2005.03.009. [DOI] [PubMed] [Google Scholar]
- Barton WA, Tzvetkova-Robev D, Miranda EP, Kolev MV, Rajashankar KR, Himanen JP, Nikolov DB. Crystal structures of the Tie2 receptor ectodomain and the angiopoietin-2-Tie2 complex. Nature structural & molecular biology. 2006;13:524–532. doi: 10.1038/nsmb1101. [DOI] [PubMed] [Google Scholar]
- Bogdanovic E, Nguyen VP, Dumont DJ. Activation of Tie2 by angiopoietin-1 and angiopoietin-2 results in their release and receptor internalization. Journal of cell science. 2006;119:3551–3560. doi: 10.1242/jcs.03077. [DOI] [PubMed] [Google Scholar]
- Davis S, Aldrich TH, Jones PF, Acheson A, Compton DL, Jain V, Ryan TE, Bruno J, Radziejewski C, Maisonpierre PC, Yancopoulos GD. Isolation of angiopoietin-1, a ligand for the TIE2 receptor, by secretion-trap expression cloning. Cell. 1996;87:1161–1169. doi: 10.1016/s0092-8674(00)81812-7. [DOI] [PubMed] [Google Scholar]
- Davis S, Papadopoulos N, Aldrich TH, Maisonpierre PC, Huang T, Kovac L, Xu A, Leidich R, Radziejewska E, Rafique A, et al. Angiopoietins have distinct modular domains essential for receptor binding, dimerization and superclustering. Nature structural biology. 2003;10:38–44. doi: 10.1038/nsb880. [DOI] [PubMed] [Google Scholar]
- Dumont DJ, Gradwohl G, Fong GH, Puri MC, Gertsenstein M, Auerbach A, Breitman ML. Dominant-negative and targeted null mutations in the endothelial receptor tyrosine kinase, tek, reveal a critical role in vasculogenesis of the embryo. Genes & development. 1994;8:1897–1909. doi: 10.1101/gad.8.16.1897. [DOI] [PubMed] [Google Scholar]
- Dumont DJ, Yamaguchi TP, Conlon RA, Rossant J, Breitman ML. tek, a novel tyrosine kinase gene located on mouse chromosome 4, is expressed in endothelial cells and their presumptive precursors. Oncogene. 1992;7:1471–1480. [PubMed] [Google Scholar]
- Fiedler U, Krissl T, Koidl S, Weiss C, Koblizek T, Deutsch U, Martiny-Baron G, Marme D, Augustin HG. Angiopoietin-1 and angiopoietin-2 share the same binding domains in the Tie-2 receptor involving the first Ig-like loop and the epidermal growth factor-like repeats. The Journal of biological chemistry. 2003;278:1721–1727. doi: 10.1074/jbc.M208550200. [DOI] [PubMed] [Google Scholar]
- Fukuhara S, Sako K, Minami T, Noda K, Kim HZ, Kodama T, Shibuya M, Takakura N, Koh GY, Mochizuki N. Differential function of Tie2 at cell-cell contacts and cell-substratum contacts regulated by angiopoietin-1. Nature cell biology. 2008;10:513–526. doi: 10.1038/ncb1714. [DOI] [PubMed] [Google Scholar]
- Gale NW, Thurston G, Hackett SF, Renard R, Wang Q, McClain J, Martin C, Witte C, Witte MH, Jackson D, et al. Angiopoietin-2 is required for postnatal angiogenesis and lymphatic patterning, and only the latter role is rescued by Angiopoietin-1. Developmental cell. 2002;3:411–423. doi: 10.1016/s1534-5807(02)00217-4. [DOI] [PubMed] [Google Scholar]
- Kim KL, Shin IS, Kim JM, Choi JH, Byun J, Jeon ES, Suh W, Kim DK. Interaction between Tie receptors modulates angiogenic activity of angiopoietin2 in endothelial progenitor cells. Cardiovasc Res. 2006;72:394–402. doi: 10.1016/j.cardiores.2006.08.002. [DOI] [PubMed] [Google Scholar]
- Kim M, Carman CV, Springer TA. Bidirectional transmembrane signaling by cytoplasmic domain separation in integrins. Science. 2003;301:1720–1725. doi: 10.1126/science.1084174. [DOI] [PubMed] [Google Scholar]
- Korhonen J, Partanen J, Armstrong E, Vaahtokari A, Elenius K, Jalkanen M, Alitalo K. Enhanced expression of the tie receptor tyrosine kinase in endothelial cells during neovascularization. Blood. 1992;80:2548–2555. [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics (Oxford, England) 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lin P, Buxton JA, Acheson A, Radziejewski C, Maisonpierre PC, Yancopoulos GD, Channon KM, Hale LP, Dewhirst MW, George SE, Peters KG. Antiangiogenic gene therapy targeting the endothelium-specific receptor tyrosine kinase Tie2. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:8829–8834. doi: 10.1073/pnas.95.15.8829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin P, Polverini P, Dewhirst M, Shan S, Rao PS, Peters K. Inhibition of tumor angiogenesis using a soluble receptor establishes a role for Tie2 in pathologic vascular growth. The Journal of clinical investigation. 1997;100:2072–2078. doi: 10.1172/JCI119740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maisonpierre PC, Suri C, Jones PF, Bartunkova S, Wiegand SJ, Radziejewski C, Compton D, McClain J, Aldrich TH, Papadopoulos N, et al. Angiopoietin-2, a natural antagonist for Tie2 that disrupts in vivo angiogenesis. Science. 1997;277:55–60. doi: 10.1126/science.277.5322.55. [DOI] [PubMed] [Google Scholar]
- Marron MB, Hughes DP, Edge MD, Forder CL, Brindle NP. Evidence for heterotypic interaction between the receptor tyrosine kinases TIE-1 and TIE-2. The Journal of biological chemistry. 2000;275:39741–39746. doi: 10.1074/jbc.M007189200. [DOI] [PubMed] [Google Scholar]
- Marti-Renom MA, Stuart AC, Fiser A, Sanchez R, Melo F, Sali A. Comparative protein structure modeling of genes and genomes. Annual review of biophysics and biomolecular structure. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- McCarthy MJ, Crowther M, Bell PR, Brindle NP. The endothelial receptor tyrosine kinase tie-1 is upregulated by hypoxia and vascular endothelial growth factor. FEBS letters. 1998;423:334–338. doi: 10.1016/s0014-5793(98)00122-7. [DOI] [PubMed] [Google Scholar]
- Milner CS, Hansen TM, Singh H, Brindle NP. Roles of the receptor tyrosine kinases Tie1 and Tie2 in mediating the effects of angiopoietin-1 on endothelial permeability and apoptosis. Microvasc Res. 2008 doi: 10.1016/j.mvr.2008.09.003. [DOI] [PubMed] [Google Scholar]
- Nguyen VP, Chen SH, Trinh J, Kim H, Coomber BL, Dumont DJ. Differential response of lymphatic, venous and arterial endothelial cells to angiopoietin-1 and angiopoietin–2. BMC cell biology. 2007;8:10. doi: 10.1186/1471-2121-8-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliner J, Min H, Leal J, Yu D, Rao S, You E, Tang X, Kim H, Meyer S, Han SJ, et al. Suppression of angiogenesis and tumor growth by selective inhibition of angiopoietin-2. Cancer Cell. 2004;6:507–516. doi: 10.1016/j.ccr.2004.09.030. [DOI] [PubMed] [Google Scholar]
- Patan S. TIE1 and TIE2 receptor tyrosine kinases inversely regulate embryonic angiogenesis by the mechanism of intussusceptive microvascular growth. Microvasc Res. 1998;56:1–21. doi: 10.1006/mvre.1998.2081. [DOI] [PubMed] [Google Scholar]
- Porat RM, Grunewald M, Globerman A, Itin A, Barshtein G, Alhonen L, Alitalo K, Keshet E. Specific induction of tie1 promoter by disturbed flow in atherosclerosis-prone vascular niches and flow-obstructing pathologies. Circulation research. 2004;94:394–401. doi: 10.1161/01.RES.0000111803.92923.D6. [DOI] [PubMed] [Google Scholar]
- Puri MC, Rossant J, Alitalo K, Bernstein A, Partanen J. The receptor tyrosine kinase TIE is required for integrity and survival of vascular endothelial cells. The EMBO journal. 1995;14:5884–5891. doi: 10.1002/j.1460-2075.1995.tb00276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsauer M, D’Amore PA. Getting Tie(2)d up in angiogenesis. The Journal of clinical investigation. 2002;110:1615–1617. doi: 10.1172/JCI17326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saharinen P, Eklund L, Miettinen J, Wirkkala R, Anisimov A, Winderlich M, Nottebaum A, Vestweber D, Deutsch U, Koh GY, et al. Angiopoietins assemble distinct Tie2 signalling complexes in endothelial cell-cell and cell-matrix contacts. Nature cell biology. 2008;10:527–537. doi: 10.1038/ncb1715. [DOI] [PubMed] [Google Scholar]
- Saharinen P, Kerkela K, Ekman N, Marron M, Brindle N, Lee GM, Augustin H, Koh GY, Alitalo K. Multiple angiopoietin recombinant proteins activate the Tie1 receptor tyrosine kinase and promote its interaction with Tie2. The Journal of cell biology. 2005;169:239–243. doi: 10.1083/jcb.200411105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato TN, Tozawa Y, Deutsch U, Wolburg-Buchholz K, Fujiwara Y, Gendron-Maguire M, Gridley T, Wolburg H, Risau W, Qin Y. Distinct roles of the receptor tyrosine kinases Tie-1 and Tie-2 in blood vessel formation. Nature. 1995;376:70–74. doi: 10.1038/376070a0. [DOI] [PubMed] [Google Scholar]
- Shen MY, Sali A. Statistical potential for assessment and prediction of protein structures. Protein Sci. 2006;15:2507–2524. doi: 10.1110/ps.062416606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela DM, Griffiths JA, Rojas J, Aldrich TH, Jones PF, Zhou H, McClain J, Copeland NG, Gilbert DJ, Jenkins NA, et al. Angiopoietins 3 and 4: diverging gene counterparts in mice and humans. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:1904–1909. doi: 10.1073/pnas.96.5.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward NL, Dumont DJ. The angiopoietins and Tie2/Tek: adding to the complexity of cardiovascular development. Seminars in cell & developmental biology. 2002;13:19–27. doi: 10.1006/scdb.2001.0288. [DOI] [PubMed] [Google Scholar]
- Wouters FS, Verveer PJ, Bastiaens PI. Imaging biochemistry inside cells. Trends in cell biology. 2001;11:203–211. doi: 10.1016/s0962-8924(01)01982-1. [DOI] [PubMed] [Google Scholar]
- Yancopoulos GD, Davis S, Gale NW, Rudge JS, Wiegand SJ, Holash J. Vascular-specific growth factors and blood vessel formation. Nature. 2000;407:242–248. doi: 10.1038/35025215. [DOI] [PubMed] [Google Scholar]
- Yuan HT, Venkatesha S, Chan B, Deutsch U, Mammoto T, Sukhatme VP, Woolf AS, Karumanchi SA. Activation of the orphan endothelial receptor Tie1 modifies Tie2-mediated intracellular signaling and cell survival. Faseb J. 2007;21:3171–3183. doi: 10.1096/fj.07-8487com. [DOI] [PubMed] [Google Scholar]
- Zacharias DA, Violin JD, Newton AC, Tsien RY. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science. 2002;296:913–916. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.