Abstract
Background
Deposits of abnormally hyperphosphorylated tau are a hallmark of several dementias, including Alzheimer disease (AD), and about 10% of familial frontotemporal dementia (FTD) cases are caused by mutations in the tau gene. As a known tau kinase, GSK3B is a promising candidate gene in the remaining cases of FTD and in AD, for which tau mutations have not been found.
Objective
To examine the promoter of GSK3B and all 12 exons, including the surrounding intronic sequence, in patients with FTD, patients with AD, and aged healthy subjects to identify single-nucleotide polymorphisms associated with disease.
Design, Setting, and Participants
Single-nucleotide polymorphism frequency was examined in a case-control cohort of 48 patients with probable AD, 102 patients with FTD, 38 patients with primary progressive aphasia, and 85 aged healthy subjects. Results were followed up in 2 independent AD family samples consisting of 437 multiplex families with AD (National Institute of Mental Health Genetics Initiative AD Study) or 150 sibships discordant for AD (Consortium on Alzheimer’s Genetics Study).
Results
Several rare sequence variants in GSK3B were identified in the case-control study. An intronic polymorphism (IVS2−68G>A) occurred at more than twice the frequency among patients with FTD (10.8%) and patients with AD (14.6%) than in aged healthy subjects (4.1%). The polymorphism showed association with disease in both follow-up samples independently, although only the Consortium on Alzheimer’s Genetics sample showed the same direction of association as the case-control sample.
Conclusions
To our knowledge, this is the first evidence that a gene known to be involved in tau phosphorylation, GSK3B, is associated with risk for primary neurodegenerative dementias. This supports previous work in animal models suggesting that such genes are therapeutic targets.
Alzheimer disease (AD) Accounts for approximately 45% of all adult-onset neurodegenerative dementias, whereas frontotemporal dementia (FTD) is responsible for about 10%.1,2 Despite the obvious clinical and pathologic distinctions between these conditions, all cases of AD and almost half of FTD cases manifest protein deposits consisting of insoluble, hyperphosphorylated tau that form filamentous inclusions.3 However, mutations in the tau gene are found in only about 15% of familial FTD cases, and tau mutations have not been identified in AD or sporadic FTD.2–6 Therefore, understanding the factors that lead to tau pathologic findings in these diseases in the context of non-mutant tau is of major importance.
An accumulating body of biological evidence in both model organisms and human AD and FTD suggests that factors related to tau phosphorylation are biologically plausible susceptibility genes.7–10 Two proline-directed kinases, glycogen synthase kinase (GSK3B [GenBankNC_000003.10]) and cyclin-dependent kinase-5 (CDK5/P25 [GenBankNC_000007.12]), are major tau kinases.9 In AD, pathologically aggregated forms of tau are hyperphosphorylated on residues that overlap with GSK3B and CDK5/P25 targets; in animal models, tau hyperphosphorylation by either of these kinases accelerates neurodegeneration in the context of both wild-type and mutant tau.8–10 Several different inhibitors of GSK3B activity block neurodegeneration in vitro, and GSK3B-mediated Wnt signaling also can mediate amyloid peptide toxicity in vitro.7,11,12 Lastly, in human postmortem brain, GSK3B and CDK5/P25 are physically associated with a pathologic hallmark of AD, neurofibrillary tangles.13
Here we investigate the role of GSK3B in dementia susceptibility using a staged approach in several different cohorts of patients with AD and FTD. To our knowledge, these data provide the first genetic evidence implicating GSK3B in neurodegenerative dementia.
METHODS
SUBJECTS
The protocols and consent forms used to recruit subjects for this study were approved by institutional review boards. Subject age, ethnicity, and sex are summarized in Table 1 and were fairly matched across the cohorts. Aging healthy subjects were collected as part of an institutional review board–approved protocol at the University of California, Los Angeles Alzheimer’s Disease Research Center. Subjects with probable and definite AD were diagnosed using standard National Institute of Neurological and Communicative Disorders and Stroke/Alzheimer’s Disease and Related Disorders Association criteria for the diagnosis of AD.14 Subjects with FTD were collected under the auspices of the University of California, Los Angeles Alzheimer’s Disease Research Center or the Emory Alzheimer’s Disease Research Center and diagnosed according to the modified Lund-Manchester criteria as described in previous studies.15 The autopsy confirmation rate in this sample was more than 90%.16 From the Emory Alzheimer’s Disease Research Center, the 39 patients with FTD and 1 patient with AD (age range, 44–83 years; mean age, 65.1 years; median age, 65 years; 84% white; 13% African American; and 3% Asian–Pacific Islander) were matched to their own set of 39 control subjects who were of the same sex and same or similar age. Primary progressive aphasia (PPA) (n = 38) was diagnosed according to the criteria of Mesulam17,18 and Sobrido et al,19 and pathologic confirmation was not available. Pathologic findings and clinical characteristics of PPA overlap considerably with FTD, although between one-fifth and one-third may have pathologically proven AD. Thus, it made sense to consider PPA separately and combined with the 2 other dementia groups.
Table 1.
Breakdown of Case-Control Samples With Descriptive Statistics
| Characteristic | UCLA Samplea | Emory Sampleb | Combined UCLA and Emory Samplec | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| AD (n = 47) |
FTD (n = 64) |
PPA (n = 38) |
Control (n = 46) |
AD (n = 1) |
FTD (n = 38) |
Control (n = 39) |
AD and FTD (n = 150) |
AD, FTD, and PPA (n = 188) |
Control (n = 85) |
|
| Age, mean, y | 76.3 | 63.0 | 68.6 | 72.8 | 57.0 | 65.3 | 65.0 | 67.7 | 67.9 | 69.4 |
| Race, No. (%) | ||||||||||
| White | 40 (85) | 63 (98) | 35 (92) | 45 (98) | 1 (100) | 32 (84) | 36 (92) | 136 (91) | 171 (91) | 81 (95) |
| Other | 7 (15) | 1 (2) | 3 (8) | 1 (2) | 0 | 6 (16) | 3 (8) | 14 (9) | 17 (8) | 4 (5) |
| Sex, % | ||||||||||
| Male | 16 (34) | 34 (53) | 18 (47) | 22 (48) | 1 (100) | 23 (61) | 22 (56) | 74 (49) | 92 (49) | 44 (52) |
| Female | 31 (66) | 30 (47) | 20 (53) | 24 (52) | 0 | 15 (39) | 17 (44) | 76 (51) | 96 (51) | 41 (48) |
| A allele frequency | 13.8 | 12.5 | 10.5 | 5.4 | 50.0 | 7.9 | 2.6 | 12.0 | 11.7 | 4.1 |
| G allele frequency | 86.2 | 87.5 | 89.5 | 94.6 | 50.0 | 92.1 | 97.4 | 88.0 | 88.3 | 95.9 |
Abbreviations: AD, Alzheimer disease; FTD, frontotemporal dementia; PPA, primary progressive aphasia; UCLA, University of California, Los Angeles.
There were 149 cases and 46 control subjects.
There were 39 cases and 39 control subjects.
There were 188 cases and 85 control subjects.
One of the 2 AD replication samples consisted of multiplex families with AD (ie, ≥2 affected siblings; 94% white) diagnosed using standard National Institute of Neurological Disorders and Stroke criteria.20 The sample comprised 1439 individuals from 437 multiplex families in which all affected individuals had onset at age 50 years or later (n = 994 who were affected, with a mean [SD] age at onset of 72.4 [7.7] years and an age range of 50–97 years; n = 411 who were unaffected; and n = 34 with phenotype unknown). Pedigrees were classified as late onset (320 families) when all of the sampled affected individuals had onset at age 65 years or later and as early or mixed (117 families) otherwise. The clinical diagnosis of AD was confirmed in 94% of the approximately 300 autopsied cases.21
The other AD family sample was independently ascertained and consisted of discordant sibling pairs collected under the auspices of the Consortium on Alzheimer’s Genetics (CAG), a collaborative effort of the Memory Disorders Unit at Massachusetts General Hospital, the Massachusetts Alzheimer’s Disease Research Center, Northwestern University Feinberg School of Medicine, University of California at Los Angeles, University of California at San Diego, and the University of Rochester Medical Center.22 The CAG sample included 334 individuals from 150 sibships in which all of the affected individuals had onset at age 50 years or later (n = 154 who were affected, with a mean [SD] age at onset of 69.9 [8.8] years and an age range of 50–88 years; and n = 180 who were unaffected; 99% white). Most sibships comprised just 1 discordant sib pair, but in 27 families there were more than 2 siblings available.
SEQUENCING AND SINGLE-NUCLEOTIDE POLYMORPHISM DETECTION
Using standard procedures, DNA was extracted either from blood or brain tissue. Polymerase chain reaction (PCR)–amplified fragments of relevant genomic regions were purified and bidirectional sequencing was performed using standard protocols (Applied Biosystems, Foster City, California; Li-COR Biotechnologies, Lincoln, Nebraska). To identify possible disease-associated variants, all of the 12 exons, surrounding intronic regions, and 2955 base pairs of the putative promoter of GSK3B were sequenced in 138 chromosomes (24 patients with FTD, 20 patients with probable AD, and 25 nondemented, mostly white subjects aged >65 years).23
GENOTYPING
For single-nucleotide polymorphism (SNP) genotyping in the case-control cohorts, PCR reactions were carried out in 20-μL volumes and used to differentiate alternative nucleotides of SNPs by allele-specific PCR. The primers used for sequencing are listed in eTable 1 (http://www.archneurol.com). Primers used for amplification prior to enzyme digestion and those used for allele-specific PCR are listed in eTable 2. The PCR components in the 50-μL reactions used for restriction enzyme digestion or sequencing were identical in proportion except that the concentration of primers was increased to 0.4μM. For the intron 3 SNP, DraI digests were performed in 1 × New England Biolabs buffer 4 at 37°C according to the manufacturer’s instructions. For the second exon 9 SNP, BfuAI was used in 1 × New England Biolabs buffer 3 at 50°C according to the manufacturer’s instructions. Products were electrophoresed on a 2% agarose gel and visualized with ethidium bromide for analysis.
Table 2.
Allele Frequency of Single-Nucleotide Polymorphisms in GSK3B Among a Cohort of Patients With Alzheimer Disease, Patients With Frontotemporal Dementia, and Aged Healthy Subjects
| Reference No. |
SNP | Systematic Location Namea |
Healthy Subjectsb |
Patients With ADb |
Patients With FTDb |
|---|---|---|---|---|---|
| 1 | −1726A>T (promoter)c | g.26310103A>T | 14.1 (13/92) | 11.7 (11/94) | 10.2 (13/128) |
| 2 | −251G>T (promoter)c | g.26308630G>T | <1.1 (0/92) | 1.1 (1/94) | <0.8 (0/128) |
| 3 | IVS2 + 69T>C | g.26215970T>C | <2.0 (0/50) | 6.5 (4/62) | <1.0 (0/108) |
| 4d | IVS2−68G>A | g.26161412G>A | 5.4 (5/92) | 13.8 (13/94) | 12.5 (16/128) |
| 5 | IVS3−16A>T | g.26137492A>T | <1.0 (0/100) | 1.9 (2/108) | <1.0 (0/104) |
| 6 | Exon 4 + 90G>Ae | g.26137387G>A | <1.1 (0/92) | <1.6 (0/62) | <0.9 (0/116) |
| 7 | IVS4−45G>A | g.26130212G>A | 2.3 (1/44) | <2.4 (0/40) | 2.4 (1/42) |
| 8 | IVS5 + 55T>C | g.26129982T>C | <2.2 (0/44) | 2.5 (1/40) | <2.3 (0/42) |
| 9 | IVS7 + 121G>Ae | g.26119627G>A | <2.4 (0/40) | <2.7 (0/36) | <2.8 (0/34) |
| 10 | IVS8 + 202C>T | g.26090204C>T | <2.0 (0/50) | <2.4 (0/40) | 2.5 (1/40) |
| 11 | IVS8 + 209T>C | g.26090197T>C | 2.5(1/50) | 5.0 (2/40) | <2.4 (0/40) |
| 12 | IVS8 + 227G>A | g.26090179G>A | <2.0 (0/50) | 5.0 (2/40) | <2.4 (0/40) |
| 13 | IVS8−84T>C | g.26080705T>C | <2.1 (0/48) | <2.4 (0/40) | 2.5 (1/40) |
| 14 | Exon 10 + 105A>G | g.26077494A>G | <1.1 (0/92) | <1.6 (0/62) | 0.9 (1/116) |
| 15 | Exon 10 + 162T>C | g.26077437T>C | 1.1 (1/92) | 2.2 (2/92) | <0.9 (0/116) |
Abbreviations: AD, Alzheimer disease; FTD, frontotemporal dementia; SNP, single-nucleotide polymorphism.
Systematic location names are given for each SNP based on the genomic sequence of GenBank NT_005612.14 as of March 4, 2004, build 34 (http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?db=nucleotide&val=37550867).
Values are expressed as percentage (number of chromosomes in which the SNP was found over the total number successfully tested). Sample sizes were expanded after the initial screening for SNPs in the promoter or in exons as well as for intron SNPs with a frequency greater than 5% in healthy subjects.
The SNPs in the promoter are numbered with respect to the putative transcription start site (+1).
P = .03 comparing healthy subjects vs patients with AD and FTD.
These SNPs were found only in a spinocerebellar ataxia comparison group.
In the sib-pair cohorts, APOE (GenBank NC_000019.8) was genotyped using phosphorus 33–labeled restriction fragment length polymorphism analysis as previously described.20 The IVS2−68G>A polymorphism in GSK3B was genotyped using fluorescence-polarization–detected single-base extension following the same protocol as previously described.24 On average, genotyping efficiency for the GSK3B intron 2 SNP was 97.3% with no discrepant genotypes based on approximately 10% of duplicate samples.
STATISTICAL ANALYSIS
Case-Control
Association was tested using a 2 × 2 table of alleles by disease categories. Fisher exact test was used to assess the significance of the allele frequency differences between cases and control subjects for the 2 identified polymorphisms that occurred in control subjects at a frequency of 5% or greater. Two-sided P values are reported. Linkage disequilibrium between the 2 common SNPs (rare allele >5%) was tested with a 3 × 3 contingency table association analysis. As several cells had small expected values, an exact test as programmed in the StatXact software (Statcon, Witzenhausen, Germany) was used.
Sib Pair
Hardy-Weinberg equilibrium was tested using the Haploview program (http://www.broad.mit.edu/personal/jcbarret/haploview/index.php). To test GSK3B IVS2−68G>A for genetic association, we used FBAT version 1.5.1 software (Departments of Bio-statistics and Environmental Health and Program for Population Genetics, Harvard School of Public Health, Boston, Massachusetts), a program for family-based association testing that can accommodate a variety of different scenarios while not being susceptible to bias due to population admixture.25 For all analyses, we used the empirical variance function of the program to account for multiple affected individuals per pedigree, and we used an equal-weight offset correction to incorporate genotypes from both affected and unaffected individuals (see the FBAT Web site, http://www.biostat.harvard.edu/∼fbat/default.html, for more details). Effect sizes were estimated only in strata found to be associated by conditional logistic regression stratified on family.26
RESULTS
SNP DISCOVERY AND CASE-CONTROL COHORTS
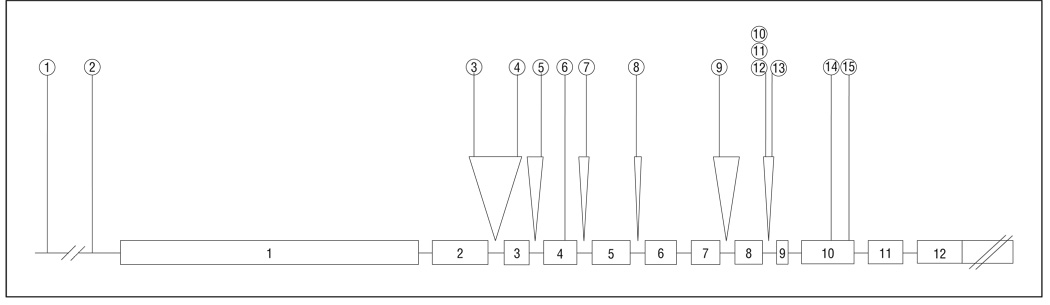
Thirteen SNPs with minor allele frequency estimates less than 5% in this initial screen were identified (rare variants) (Table 2 and Figure), none of which were previously described. Two SNPs with minor allele frequencies greater than 5% in control subjects (common variants) were identified. Of these 2 more common SNPs, the one in the promoter (−1726A>T) had been previously identified as not associated with AD.27 The other (IVS2−68G>A), not in linkage disequilibrium with the first, was located in intron 2, was 68 nucleotides upstream from exon 3, and had not been previously described. Although this sequence did not correspond to known splice enhancer sites, it did occur within a region of high homology with rodent in which 10 of 11 surrounding nucleotides were conserved.
Figure.
Schematic representation of GSK3B showing the location of all single-nucleotide polymorphisms found In this assay. Rectangles indicate exons; straight lines, introns.
To assess the association of these 2 SNPs with disease, subjects were analyzed using a case-control design. The groups consisted of 47 patients with probable AD, 64 patients with FTD, 38 patients with PPA, and 46 aged healthy control subjects. Each variant was genotyped either by restriction fragment length polymorphism analysis when a restriction site was altered or by allele-specific PCR as described in the “Methods” section. Both SNPs were in Hardy-Weinberg equilibrium. The promoter SNP (A-1726T) showed no significant association with disease status and was observed at a frequency of 14.1% in healthy subjects, 11.7% in patients with AD, 14.5% in patients with PPA, and 10.2% in patients with FTD, similar to a previous report.27
The IVS2−68G>A A variant frequency for the original group of 149 total affected subjects (ie, 64 subjects with FTD, 47 subjects with AD, and 38 subjects with PPA combined) was more than twice the frequency found in the aged healthy control group (12.4% vs 5.4%, respectively). Thirty-eight additional patients with FTD, 1 additional patient with AD, and 39 matched control subjects from another center (Emory) were added to the cohort to increase the power. The minor allele frequencies for this SNP in the disease group were 14.6% for AD (14 of 96 patients), 10.8% for FTD (22 of 204 patients), 10.5% for PPA (8 of 76 patients), and 4.1% for the control group (7 of 170 subjects) (Table 1). Because this polymorphism occurred at a similar frequency in the disease cohorts, they were combined to provide greater power to detect an effect when compared with the healthy control subjects. Using a 2-sided Fisher exact test and considering either the total group or only the white subjects to avoid differences due to race, a significant difference between the combined disease and control groups was observed (P = .004 for all, Bonferroni corrected P = .008; P = .009 for white subjects only, Bonferroni corrected P = .01) (Table 3). Because there was no pathologic confirmation for the PPA group, the analysis of only white subjects was also performed considering only the FTD and AD cases, which remained significant (P = .007; Bonferroni corrected P = .01) for allelic and genotypic (P = .05) association between this SNP and disease status. Effects size estimates revealed a significant increase in risk for AD (odds ratio [OR] = 5.2; 95% confidence interval [CI], 1.8–15.1; 2-sided Mantel-Haenszel P = .05) and for the FTD and AD disease groups combined (OR = 3.4; 95% CI, 1.3–8.6; 2-sided Mantel-Haenszel P = .05).
Table 3.
GSK3B IVS2−68G>A Allele Frequencies in Cases and Control Subjectsa
| Participants | G Allele | A Allele |
|---|---|---|
| Healthy | 163 (156) | 7 (6) |
| Affected, with AD and FTD | 264 (242) | 36 (30) |
| All affected, with AD, FTD, and PPA | 332 (306) | 44 (36) |
Abbreviations: AD, Alzheimer disease; FTD, frontotemporal dementia: PPA, primary progressive aphasia.
Alleles for AD, FTD, and PPA were combined as described In the text. Values are expressed as allele frequencies in total samples (allele frequencies in white samples only). Affected groups (both excluding and Including PPA) were significantly different from the healthy control subjects at the P < .005 level using a 2-tailed Fisher exact test.
FAMILY-BASED COHORTS
To minimize the possibility of a type I error or error due to population admixture, we tested this significant association in 2 independent family-based cohorts (see “Methods”). Unfortunately, no similar large, family-based cohorts of the rarer conditions, FTD or PPA, were available.
There were no significant deviations from Hardy-Weinberg equilibrium in either of the family samples (P > .99 for both samples). Minor allele (ie, A allele) frequencies were 11.1% in the National Institute of Mental Health (NIMH) (multiplex affected) families and 10.6% in the CAG (discordant siblings) families. Testing the same SNP for association in the CAG sample revealed marginal association in the total sample (P = .06), which was more pronounced and significant in sibships with at least 1 affected APOE ε4 carrier (P = .04) (Table 4). The association observed in the CAG families was based on an overtransmission of the A allele to affected individuals in concordance with the case-control study in stage 1. The frequency of the A allele was 0.10 in the cases vs 0.07 in the unaffected siblings, consistent with the increase of the A allele in cases relative to the control subjects observed in the case-control sample. Accordingly, effect size estimates showed a statistically significant increase in AD risk for carriers of the A allele vs G/G carriers (ORAPOE ε4-positive = 4.18; 95% CI, 1.14–15.33). Although the CAG sample is a family-based sample that controls for potential confounding by population stratification, it represents simplex AD (no first-degree relatives with dementia), which is in contrast with the NIMH sample that represents multiplex AD (≥2 affected siblings). Thus, this finding in the CAG sample supports the association of the GSK3B IVS2−68G>A polymorphism with disease status in what are predominantly sporadic, or simplex, cases of dementia.
Table 4.
Association Analyses of IVS2−68G>A and Alzheimer Disease in 2 Independent Family Samples
| AD Patient Group | NIMH Samplea | CAG Sampleb | ||||
|---|---|---|---|---|---|---|
| Informative, No.c | Z Scored | P Value | Informative, No.c | Z Scored | P Value | |
| Total | 41 | −2.4 | .02 | 23 | 1.9 | .06 |
| Late onset | 23 | −1.2 | .23 | 18 | 1.8 | .07 |
| Early or mixed onset | 18 | −2.3 | .02 | 5 | NA | NA |
| APOE ε4/4 positive | 18 | −1.4 | .15 | 4 | NA | NA |
| APOE ε4 positive | 38 | −2.4 | .02 | 14 | 2.0 | .04 |
| APOE ε4 negative | 3 | NA | NA | 7 | NA | NA |
Abbreviations: AD, Alzheimer disease; CAG, Consortium on Alzheimer’s Genetics; NA, not applicable; NIMH, National Institute of Mental Health.
There were 437 multiplex families with AD.
There were 150 sibships discordant for AD.
Number of families Informative for the Individual test statistics (as determined by FBAT version 1.5.1 software [Departments of Biostatistics and Environmental Health and Program for Population Genetics, Harvard School of Public Health, Boston, Massachusetts]).
z Score for minor allele (positive values Indicate overtransmlsslon to affected individuals).
To determine whether the GSK3B intron 2 association held true for individuals with a stronger family history, we also examined the multiplex AD family sample (NIMH). This revealed a significant undertransmission of the A allele to affected individuals in the unstratified (total) sample of NIMH families (P = .02) (Table 4), suggesting an effect in the opposite direction of the association observed in the CAG and case-control samples. Stratification by onset age and APOE ε4 allele revealed that this preferential transmission was most pronounced in families with early or mixed onset ages or at least 1 affected APOE ε4 carrier (Table 4), similar to the CAG sample. Effect size estimates using conditional logistic regression revealed a significant decrease in risk in carriers of the A/A vs the G/G genotype only in the former 2 samples (total: OR = 0.43; 95% CI, 0.24–0.76; APOE ε4 positive: OR = 0.40; 95% CI, 0.22–0.73). However, even in this sample with a strong family history of dementia, the A allele was observed at an increased frequency (8% in cases and 9% in unaffected siblings) relative to the frequency observed in the aged healthy control cohort (about 5%).
COMMENT
Here we screened the GSK3B gene for association with AD, FTD, and PPA based on previous functional evidence from animal models implicating tau kinases as potential susceptibility genes for neurodegenerative dementias involving tau in humans.28 This study suggests for the first time a significant genetic association of a variant within GSK3B and 2 dementia phenotypes in several different independent populations using both case-control and family-based methods. These genetic findings are consistent with the tau pathologic findings observed in these diseases and the known biological role of GSK3B as a tau kinase.
To assess genetic variability in different case and control cohorts, we performed a comprehensive mutation screening by sequencing all known exons, adjacent intronic regions, and about 3 kilobases of the known promoter region. One of 2 SNPs with minor allele frequencies greater than 5% showed a significant association with the neurodegenerative disease groups relative to healthy control subjects. Whereas both family samples showed evidence for significant genetic association with IVS2−68G>A, the direction of the effects was discrepant. The −68A variant was over-transmitted in both samples representing sporadic disease, the case-control and CAG cohorts.
This represents an independent observation, or replication, of the case-control finding involving an a priori biological hypothesis. This same polymorphism also showed an association in another family-based cohort, the large NIMH cohort of sib pairs with AD. The variant that was slightly overtransmitted in the NIMH cohort was the more common variant (−68G).
There are several possible explanations for the opposite associations observed with the rare alleles in the 2 sibling-based samples. First, all of the 4 observed associations (FTD, AD, CAG, and NIMH) may be spurious. This is unlikely because we have used several different samples and observed the increase of the A allele in affected subjects in 3 of the 4 samples. Second and most likely, the opposite transmission pattern of the minor allele with respect to disease status may also be due to linkage disequilibrium with an as-yet unidentified genetic variant either within the GSK3B gene or nearby. Future studies should focus on the identification of linkage disequilibrium blocks within the GSK3B gene in this and other independent samples so as to further narrow the region of functional significance associated with dementia.
It should also be noted that the case-control and family samples were collected using different ascertainment schemes. The case-control subjects primarily represent sporadic cases. The NIMH families were ascertained based on the presence of at least 2 AD cases in first-degree relatives of the same pedigree, whereas the CAG families were ascertained on the presence of at least 1 sib pair discordant for AD. This could lead to the sampling of genetically distinct populations, ie, samples that are governed by different genetic risk factors and risk alleles as we have recently observed in another complex neuropsychiatric disorder, autism.29 Similarly, significant associations with opposite alleles of the same gene across different samples have been reported with several other AD candidate genes in the past.30,31 It is noteworthy in this context, however, that in both of the family-based samples analyzed here, possession of the APOE ε4 allele and ε4/4 genotype leads to an increase in AD risk.22 In this regard, the fact that APOE has been shown to induce GSK3B in vitro is interesting and suggests that potential biochemical interactions between APOE and GSK3B are worth further investigation.32
This is the first gene related to tau phosphorylation that has been identified as a potential genetic risk factor for AD or FTD, consistent with the concept that pathways related to tau phosphorylation are important therapeutic targets to consider in these diseases. The role of GSK3B as a tau kinase and the association of other tau kinase pathways with neurodegeneration in cell and animal models suggest that these other genes involved in tau phosphorylation and dephosphorylation, such as CDK5 and PIN1 (GenBank NC_000019.8), may also provide appealing targets for drug discovery.7,28 It is plausible that the mechanisms by which GSK3B variants could increase the risk for neurodegeneration are not limited to tau phosphorylation.11 Whatever ultimate mechanisms are involved downstream of GSK3B, specific inhibitors of GSK function are increasingly attractive pharmacologic interventions in AD and related dementias, an approach supported by the suggested genetic role uncovered in this study.
Supplementary Material
Acknowledgments
Funding/Support: This work was supported by grants AG16570 (Ms Schaffer and Dr Geschwind), AG19724 (Dr Miller), AG13854 (Dr Mesulam), and AG025688 (Dr Levey) from the National Institute on Aging, grant MH60009 from the National Institute of Mental Health (Dr Tanzi), The John Douglas French Alzheimer’s Foundation (Drs Wiedau-Pazos and Geschwind), the Cure Alzheimer’s Fund (Dr Tanzi), and the Turken Family Scholarship Award from the Alzheimer’s Association of Los Angeles (Dr Geschwind). At the time of this work, Dr Bertram was a fellow of the Harvard Center for Neurodegeneration and Repair and was a fellow of the Deutsche Forschungsgemeinschaft.
Footnotes
Financial Disclosure: Ms Schaffer and Dr Geschwind have a patent pending for Genetic Risk Factor for Neurodegenerative Disease, UC reference No. 2004-191-2.
Additional Information: The eTables are available at http://www.archneurol.com.
Author Contributions: Study concept and design: Schaffer, Bertram, Miller, Wiedau-Pazos, Jackson, Cummings, Levey, Tanzi, and Geschwind. Acquisition of data: Schaffer, Bertram, Miller, Mullin, Weintraub, Johnson, Bigio, Mesulam, Levey, and Tanzi. Analysis and interpretation of data: Schaffer, Bertram, Cantor, Levey, Tanzi, and Geschwind. Drafting of the manuscript: Schaffer, Bertram, Tanzi, and Geschwind. Critical revision of the manuscript for important intellectual content: Schaffer, Bertram, Miller, Mullin, Weintraub, Johnson, Bigio, Mesulam, Wiedau-Pazos, Jackson, Cummings, Cantor, Levey, Tanzi, and Geschwind. Statistical analysis: Schaffer, Bertram, Cantor, and Tanzi. Obtained funding: Bertram, Cummings, and Geschwind. Administrative, technical, and material support: Schaffer, Bertram, Miller, Mullin, Weintraub, Johnson, Bigio, Mesulam, Cummings, Levey, Tanzi, and Geschwind. Study supervision: Bertram, Tanzi, and Geschwind.
Additional Contributions: We thank all of the subjects for participating in this study. Kirk Wilhelmsen, MD, PhD, provided helpful contributions in our early genetic studies of FTD.
REFERENCES
- 1.Nussbaum RL, Ellis CE. Alzheimer’s disease and Parkinson’s disease. N Engl J Med. 2003;348(14):1356–1364. doi: 10.1056/NEJM2003ra020003. [DOI] [PubMed] [Google Scholar]
- 2.Bird T, Knopman D, VanSwieten J, et al. Epidemiology and genetics of frontotemporal dementia/Pick’s disease. Ann Neurol. 2003;54(5):S29–S31. doi: 10.1002/ana.10572. [DOI] [PubMed] [Google Scholar]
- 3.Lee VM, Goedert M, Trojanowski JQ. Neurodegenerative tauopathies. Annu Rev Neurosci. 2001;24:1121–1159. doi: 10.1146/annurev.neuro.24.1.1121. [DOI] [PubMed] [Google Scholar]
- 4.Goedert M, Spillantini MG. Tau gene mutations and neurodegeneration. Biochem Soc Symp. 2001;(67):59–71. doi: 10.1042/bss0670059. [DOI] [PubMed] [Google Scholar]
- 5.Houlden H, Baker M, Adamson J, et al. Frequency of tau mutations In three series of non-Alzheimer’s degenerative dementia. Ann Neurol. 1999;46(2):243–248. doi: 10.1002/1531-8249(199908)46:2<243::aid-ana14>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 6.Sobrido MJ, Miller BL, Havlioglu N, et al. Novel tau polymorphisms, tau haplotypes, and splicing in familial and sporadic frontotemporal dementia. Arch Neurol. 2003;60(5):698–702. doi: 10.1001/archneur.60.5.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brion JP, Anderton BH, Authelet M, et al. Neurofibrillary tangles and tau phosphorylation. Biochem Soc Symp. 2001;(67):81–88. doi: 10.1042/bss0670081. [DOI] [PubMed] [Google Scholar]
- 8.Noble W, Olm V, Takata K, et al. Cdk5 is a key factor in tau aggregation and tangle formation in vivo. Neuron. 2003;38(4):555–565. doi: 10.1016/s0896-6273(03)00259-9. [DOI] [PubMed] [Google Scholar]
- 9.Cruz JC, Tseng HC, Goldman JA, Shih H, Tsai LH. Aberrant Cdk5 activation by p25 triggers pathological events leading to neurodegeneration and neurofibrillary tangles. Neuron. 2003;40(3):471–483. doi: 10.1016/s0896-6273(03)00627-5. [DOI] [PubMed] [Google Scholar]
- 10.Jackson GR, Wiedau-Pazos M, Sang TK, et al. Human wild-type tau interacts with wingless pathway components and produces neurofibrillary pathology in Drosophila. Neuron. 2002;34(4):509–519. doi: 10.1016/s0896-6273(02)00706-7. [DOI] [PubMed] [Google Scholar]
- 11.Kim HS, Kim EM, Lee JP, et al. C-terminal fragments of amyloid precursor protein exert neurotoxicity by inducing glycogen synthase kinase-3beta expression. FASEB J. 2003;17(13):1951–1953. doi: 10.1096/fj.03-0106fje. [DOI] [PubMed] [Google Scholar]
- 12.Pérez M, Hernández F, Lim F, Díaz-Nido J, Avila J. Chronic lithium treatment decreases mutant tau protein aggregation in a transgenic mouse model. J AIzheimers Dis. 2003;5(4):301–308. doi: 10.3233/jad-2003-5405. [DOI] [PubMed] [Google Scholar]
- 13.Ishizawa T, Sahara N, Ishiguro K, et al. Co-localization of glycogen synthase kinase-3 with neurofibrillary tangles and granulovacuolar degeneration in transgenic mice. Am J Pathol. 2003;163(3):1057–1067. doi: 10.1016/S0002-9440(10)63465-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM. Clinical diagnosis of Alzheimer’s disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer’s Disease. Neurology. 1984;34(7):939–944. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- 15.Geschwind DH, Robidoux J, Alarcon M, et al. Dementia and neurodevelopmental predisposition: cognitive dysfunction in presymptomatic subjects precedes dementia by decades in frontotemporal dementia. Ann Neurol. 2001;50(6):741–746. doi: 10.1002/ana.10024. [DOI] [PubMed] [Google Scholar]
- 16.Geschwind D, Karrim J, Nelson SF, Miller B. The apolipoprotein E epsilon4 allele is not a significant risk factor for frontotemporal dementia. Ann Neurol. 1998;44(1):134–138. doi: 10.1002/ana.410440122. [DOI] [PubMed] [Google Scholar]
- 17.Mesulam MM. Primary progressive aphasia: a language-based dementia. N Engl J Med. 2003;349(16):1535–1542. doi: 10.1056/NEJMra022435. [DOI] [PubMed] [Google Scholar]
- 18.Mesulam MM. Primary progressive aphasia. Ann Neurol. 2001;49(4):425–432. [PubMed] [Google Scholar]
- 19.Sobrido MJ, Abu-Khalil A, Weintraub S, et al. Possible association of the tau H1/H1 genotype with primary progressive aphasia. Neurology. 2003;60(5):862–864. doi: 10.1212/01.wnl.0000049473.36612.f2. [DOI] [PubMed] [Google Scholar]
- 20.Blacker D, Haines JL, Rodes L, et al. ApoE-4 and age at onset of Alzheimer’s disease: the NIMH genetics initiative. Neurology. 1997;48(1):139–147. doi: 10.1212/wnl.48.1.139. [DOI] [PubMed] [Google Scholar]
- 21.Blacker D, Bertram L, Saunders AJ, et al. Results of a high-resolution genome screen of 437 Alzheimer’s disease families. Hum Mol Genet. 2003;12(1):23–32. doi: 10.1093/hmg/ddg007. [DOI] [PubMed] [Google Scholar]
- 22.Bertram L, Hiltunen M, Parkinson M, et al. Family-based association between Alzheimer’s disease and variants in UBQLN1. N Engl J Med. 2005;352(9):884–894. doi: 10.1056/NEJMoa042765. [DOI] [PubMed] [Google Scholar]
- 23.Schaffer B, Wiedau-Pazos M, Geschwind DH. Gene structure and alternative splicing of glycogen synthase kinase 3 beta (GSK-3beta) in neural and non-neural tissues. Gene. 2003;302(1–2):73–81. doi: 10.1016/s0378-1119(02)01092-2. [DOI] [PubMed] [Google Scholar]
- 24.Saunders AJ, Bertram L, Mullin K, et al. Genetic association of Alzheimer’s disease with multiple polymorphisms in alpha-2-macroglobulin. Hum Mol Genet. 2003;12(21):2765–2776. doi: 10.1093/hmg/ddg310. [DOI] [PubMed] [Google Scholar]
- 25.Rabinowitz D, Laird N. A unified approach to adjusting association tests for population admixture with arbitrary pedigree structure and arbitrary missing marker information. Hum Hered. 2000;50(4):211–223. doi: 10.1159/000022918. [DOI] [PubMed] [Google Scholar]
- 26.Witte JS, Gauderman WJ, Thomas DC. Asymptotic bias and efficiency in case-control studies of candidate genes and gene-environment interactions: basic family designs. Am J Epidemiol. 1999;149(8):693–705. doi: 10.1093/oxfordjournals.aje.a009877. [DOI] [PubMed] [Google Scholar]
- 27.Russ C, Lovestone S, Powell JF. Identification of sequence variants and analysis of the role of the glycogen synthase kinase 3 beta gene and promoter in late onset Alzheimer’s disease. Mol Psychiatry. 2001;6(3):320–324. doi: 10.1038/sj.mp.4000852. [DOI] [PubMed] [Google Scholar]
- 28.Geschwind DH. Tau phosphorylation, tangles, and neurodegeneration: the chicken or the egg? Neuron. 2003;40(3):457–460. doi: 10.1016/s0896-6273(03)00681-0. [DOI] [PubMed] [Google Scholar]
- 29.Sebat J, Lakshmi B, Malhotra D, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316(5823):445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bertram L, McQueen MB, Mullin K, Blacker D, Tanzi RE. Systematic metaanalyses of Alzheimer disease genetic association studies: the AlzGene database. Nat Genet. 2007;39(1):17–23. doi: 10.1038/ng1934. [DOI] [PubMed] [Google Scholar]
- 31.Lin PI, Vance JM, Pericak-Vance MA, Martin ER. No gene is an island: the flip-flop phenomenon. Am J Hum Genet. 2007;80(3):531–538. doi: 10.1086/512133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cedazo-Mínguez A, Popescu BO, Blanco-Millán JM, et al. Apolipoprotein E and beta-amyloid (1–42) regulation of glycogen synthase kinase-3beta. J Neurochem. 2003;87(5):1152–1164. doi: 10.1046/j.1471-4159.2003.02088.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.