Abstract
Maintenance of genomic integrity is important for cellular viability and proliferation. During DNA replication, cells respond to replication stress by activating checkpoint pathways that stabilize replication forks and prevent cell cycle progression. The Saccharomyces cerevisiae F-box protein Dia2 is a ubiquitin ligase component required for genomic stability and may help replication complexes negotiate damaged DNA or natural fragile sites. We recently implicated Dia2 in the replication stress response. We demonstrated that Dia2 is targeted for ubiquitin-mediated proteolysis and that activation of the S-phase checkpoint inhibits Dia2 protein turnover. S-phase checkpoint mutants fail to stabilize the Dia2 protein and checkpoint mutants that lack Dia2 exhibit increased sensitivity to replication stress. We also showed that Dia2 protein turnover is not the result of an autocatalytic mechanism. Instead, an N-terminal 20 amino acid motif that is also required for nuclear localization is necessary for Dia2 proteolysis. Dia2 mutants lacking this motif but modified with an exogenous strong nuclear localization signal are both nuclear and stable and disrupt cell cycle dynamics. In summary, our studies suggest that inhibition of Dia2 proteolysis is a novel target of the S-phase checkpoint. We think that this work will help to identify the mechanisms that function downstream of checkpoint activation and that intersect with cell cycle control pathways.
Introduction
Accurate DNA replication is critical to faithful chromosome segregation and cell viability. The progression of DNA synthesis may be hindered by the presence of damaged DNA or genotoxic stresses, which can lead to replication fork stalling or fork collapse. Such events have the potential to induce genomic instability, one of the hallmarks of cancer cells. To maintain genomic stability, checkpoint responses are activated during replication stress or in response to defects during DNA replication (reviewed in [1,2]). Activation of checkpoint pathways promotes cell viability by stabilizing replication forks, suppressing the firing of late replication origins and inhibiting the progression of S phase [3-7]. By delaying further progress through S phase, checkpoint activation allows time for the replication defects to be resolved, thus preventing damage to chromosomes.
Checkpoint mechanisms are often defective in cancer cells, suggesting that the absence of these surveillance pathways contributes to a proliferative advantage in tumor formation (reviewed in [8-11]). Therefore, determining how checkpoint mechanisms intersect with DNA replication and cellular proliferation pathways has implications for understanding cancer biology. There has been remarkable progress in identifying key proteins required for activation of checkpoint signaling pathways [1,2], but little is known about the molecular targets of these pathways and how they inhibit progression through S phase. The goal of this commentary is to describe recent work from my laboratory [12] that suggests the proteolytic regulation of the budding yeast ubiquitin ligase component Dia2, a protein previously determined to be required for genomic stability [13-15], is a target of the replication checkpoint pathway.
Discussion
Dia2, DNA replication, and genomic stability
Dia2 functions as a ubiquitin ligase specificity adaptor and is required for genomic stability [13,14]. In the absence of Dia2, cells are hypersensitive to DNA damaging agents, accumulate DNA damage foci and exhibit increased chromosome loss and rearrangements, particularly at repetitive regions such as the rDNA locus [13-15]. The replication checkpoint is constitutively active in dia2 null cells, as evidenced by hyperphosphorylation of the checkpoint kinase Rad53 [14,15]. The dia2 deletion strain shows synthetic interactions with a number of mutants in DNA replication and checkpoint proteins [13-16]. Together, these phenotypes indicate that Dia2 is important to maintain a stable genome and that it is likely to have a role in DNA replication or the replication stress response. To better understand the role of Dia2, we undertook a study examining the proteolytic regulation of the Dia2 protein.
Proteolytic regulation of F-box proteins
SCF complexes are multi-component ubiquitin ligases that function in many cellular processes, including cell cycle control and signaling pathways (reviewed in [17,18]). F-box proteins function as specificity adaptors for SCF ubiquitin ligases by binding both the substrate protein to be ubiquitinated and interacting with the rest of the complex [19,20]. The conserved F-box domain is required to bind the Skp1 component in the complex [19,20]. There are many F-box proteins; thus there are many individual SCF complexes and the specificity of each is determined by which F-box protein is bound. A convenient way to regulate the activity of SCF ubiquitin ligases, therefore, is to control the availability of the F-box protein.
Most F-box proteins are found at lower abundance than the rest of the SCF complex components. This is often accomplished by proteolytic regulation of the F-box protein itself [21,22]. One major mechanism is that of autoubiquitination, in which the F-box protein is tagged with ubiquitin and targeted for destruction by interacting with the SCF core complex in the absence of a substrate protein. This mechanism has been proposed to be a means to keep SCF ubiquitin ligase activity in check when the substrate protein is not available [21-23]. In vitro, many F-box proteins can serve as substrates in autoubiquitination reactions [24], but the extent to which this process occurs in vivo is unclear. By contrast, a few F-box proteins are destabilized when SCF components are impaired [25-28]. Moreover, some F-box proteins are targeted for degradation by SCF-independent pathways. For example, the human F-box protein Skp2 is a cell cycle-dependent substrate of the APC ubiquitin ligase [29].
We observed that Dia2 is an unstable protein, targeted for degradation by the ubiquitin proteasome system, in a cell cycle dependent manner. We demonstrated that the abundance of the Dia2 protein is low in G1, but then increases as cells enter S phase [12], consistent with a role for Dia2 in DNA replication or the replication stress response. To determine whether Dia2 protein turnover was the result of an autoubiquitination mechanism, we examined the half-life of the Dia2 protein in scf mutants [12]. We found that Dia2 was not stabilized in scf mutants. Instead, Dia2 appeared even less stable in skp1 and cdc53 mutants, consistent with the possibility that an intact SCF complex promotes Dia2 stability. In addition, we showed that a Dia2 mutant lacking the F-box domain has the same half-life as wildtype Dia2 [12]. Together, these data indicate that Dia2 proteolysis is not controlled by an autoubiquitination mechanism.
A standard structure-function approach was used to determine which domain in Dia2 is required for its turnover. We demonstrated that a 20 amino acid motif in the N-terminal TPR repeat region of Dia2 is required for its proteolysis [12]. Remarkably, we found that this same motif is required for localization of Dia2 to the nucleus. The addition of a strong NLS to Dia2 mutants lacking the 20 amino acid motif led to stable, nuclear forms of Dia2. Overexpression of these stable, nuclear forms of Dia2 disrupted cell cycle progression, leading to an accumulation of cells in G1 phase with a concomitant decrease in G2/M phase cells [12]. These results suggest that failure to degrade the Dia2 protein interferes with cell cycle control.
Activation of the replication checkpoint inhibits Dia2 proteolysis
To explore the cell cycle dynamics of Dia2 protein degradation, the stability of Dia2 was determined in cells arrested in G1, S and G2/M phases of the cell cycle. We showed that the Dia2 protein is most unstable in G1, but is significantly stabilized in cells arrested in S phase [12]. Since the S-phase arrest relies on activation of the S-phase checkpoint, we examined whether checkpoint activation was required for stabilization of Dia2 by analyzing the turnover of the Dia2 protein in a number of checkpoint mutants. Strikingly, we found that the Dia2 protein was unstable when the S-phase checkpoint was defective [12]. This result suggests that inhibiting Dia2 protein turnover might be a functional consequence of activation of the S-phase checkpoint.
We reasoned that if Dia2 protein levels are regulated by the S-phase checkpoint, then removing Dia2 from checkpoint mutants should exacerbate the phenotypes of checkpoint mutants. We examined a series of dia2Δ checkpoint double mutants for their sensitivity to hydroxyurea, which induces replication stress. These double mutants exhibited significantly increased sensitivity to replication stress under these conditions [12], consistent with the hypothesis that Dia2 protein turnover is required for the cellular response to replication stress.
Conclusions
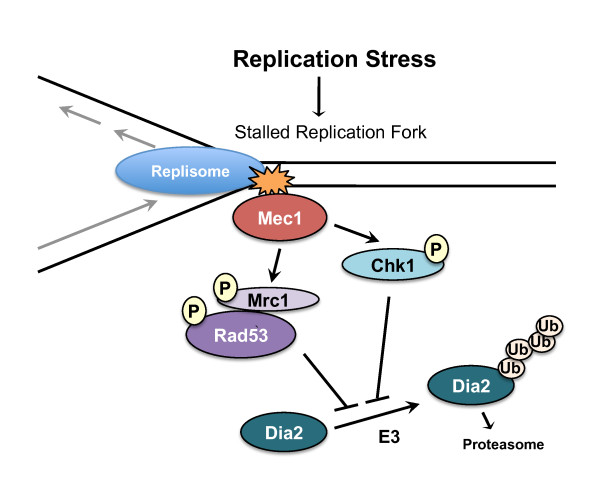
We demonstrated that activation of the S-phase checkpoint inhibits Dia2 protein degradation and we propose that SCFDia2 activity is required for the replication stress response (see model in Figure 1). Previous work had suggested that Dia2 is required to help replication complexes traverse areas of the genome that are difficult to replicate and prone to DNA damage. Our studies suggest that as replication complexes encounter difficulties and activate the checkpoint, Dia2 is stabilized, presumably to form an active SCFDia2 complex. This model predicts that Dia2 might interact with components of the checkpoint pathway. Consistent with this hypothesis, recent studies have determined that Dia2 associates with replisome complexes [30-32]. Strikingly, Dia2 binds to Mrc1 [31,32], a replication protein that also has a critical role in activation of the S-phase checkpoint [3,7]. Mrc1 is important for activation of the Rad53 checkpoint effector kinase [3,7]. Our results suggest that stabilization of Dia2 occurs downstream of Rad53 activation. Altogether, the data point to Dia2 protein stabilization as a target of the S-phase checkpoint.
Figure 1.
Model for S-phase checkpoint regulation of Dia2 ubiquitin-mediated destruction. In response to replication stress, replication forks stall and activate S-phase checkpoint pathways, leading to the inhibition of Dia2 protein turnover. Our data suggest that Dia2 stabilization is downstream of both Rad53 and Chk1 activation and work from other groups indicate that Dia2 binds to Mrc1 [31,32].
How Dia2 protein turnover is controlled by the S-phase checkpoint is an open question. Two straightforward possibilities are: 1) that activation of the checkpoint pathway prevents Dia2 from being targeted for ubiquitin-dependent proteolysis or 2) that the ubiquitination pathway that controls Dia2 protein turnover may be inactivated when the S-phase checkpoint is triggered. Our study did not address the identity of the pathway responsible for Dia2 protein degradation, so future work will be necessary to distinguish between these two possibilities.
Our work adds to the number of ubiquitin ligase components that have been shown to be regulated by ubiquitin-mediated proteolysis. For Dia2, we demonstrated that this regulation is not the result of autoubiquitination, but rather the activity of an independent pathway. It is intriguing that the 20-amino acid motif that is responsible for Dia2 protein instability is also critical for its nuclear localization. Nuclear localization signals are rich in positively-charged amino acids such as lysine residues, which also serve as the acceptor sites for ubiquitin modification. This may simply be an interesting coincidence, but it is tempting to speculate that the pathways responsible for ubiquitination and nuclear localization of the Dia2 protein may counteract each other.
By studying the proteolytic regulation of a ubiquitin ligase component, we have identified a novel target of the S-phase checkpoint. We think that this work serves as a starting point to elucidate the cellular pathways and key players that function downstream of checkpoint activation and intersect with cell cycle control mechanisms. We anticipate that determining the molecular mechanisms involved in checkpoint-mediated cell cycle inhibition is likely to expand our understanding of tumor biology, as checkpoints are frequently defective in cancer cells.
Competing interests
The author declares that they have no competing interests.
Authors' contributions
D.M.K. devised and wrote the manuscript.
Acknowledgements
The author thanks members of the Koepp laboratory for discussion and comments on the manuscript. This work was support by NIH R01 GM076663.
References
- Branzei D, Foiani M. The checkpoint response to replication stress. DNA Repair (Amst) 2009;8:1038–1046. doi: 10.1016/j.dnarep.2009.04.014. [DOI] [PubMed] [Google Scholar]
- Zegerman P, Diffley JF. DNA replication as a target of the DNA damage checkpoint. DNA Repair (Amst) 2009;8:1077–1088. doi: 10.1016/j.dnarep.2009.04.023. [DOI] [PubMed] [Google Scholar]
- Alcasabas AA, Osborn AJ, Bachant J, Hu F, Werler PJ, Bousset K, Furuya K, Diffley JF, Carr AM, Elledge SJ. Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat Cell Biol. 2001;3:958–965. doi: 10.1038/ncb1101-958. [DOI] [PubMed] [Google Scholar]
- Tercero JA, Diffley JF. Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature. 2001;412:553–557. doi: 10.1038/35087607. [DOI] [PubMed] [Google Scholar]
- Katou Y, Kanoh Y, Bando M, Noguchi H, Tanaka H, Ashikari T, Sugimoto K, Shirahige K. S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature. 2003;424:1078–1083. doi: 10.1038/nature01900. [DOI] [PubMed] [Google Scholar]
- Lopes M, Cotta-Ramusino C, Pellicioli A, Liberi G, Plevani P, Muzi-Falconi M, Newlon CS, Foiani M. The DNA replication checkpoint response stabilizes stalled replication forks. Nature. 2001;412:557–561. doi: 10.1038/35087613. [DOI] [PubMed] [Google Scholar]
- Osborn AJ, Elledge SJ. Mrc1 is a replication fork component whose phosphorylation in response to DNA replication stress activates Rad53. Genes Dev. 2003;17:1755–1767. doi: 10.1101/gad.1098303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann WK. Initiating the uninitiated: replication of damaged DNA and carcinogenesis. Cell Cycle. 2007;6:1460–1467. [PubMed] [Google Scholar]
- Hook SS, Lin JJ, Dutta A. Mechanisms to control rereplication and implications for cancer. Curr Opin Cell Biol. 2007;19:663–671. doi: 10.1016/j.ceb.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dash BC, El-Deiry WS. Cell cycle checkpoint control mechanisms that can be disrupted in cancer. Methods Mol Biol. 2004;280:99–161. doi: 10.1385/1-59259-788-2:099. [DOI] [PubMed] [Google Scholar]
- Nojima H. G1 and S-phase checkpoints, chromosome instability, and cancer. Methods Mol Biol. 2004;280:3–49. doi: 10.1385/1-59259-788-2:003. [DOI] [PubMed] [Google Scholar]
- Kile AC, Koepp DM. Activation of the S-phase checkpoint inhibits degradation of the F-box protein Dia2. Mol Cell Biol. 2010;30:160–171. doi: 10.1128/MCB.00612-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koepp DM, Kile AC, Swaminathan S, Rodriguez-Rivera V. The F-box protein Dia2 regulates DNA replication. Mol Biol Cell. 2006;17:1540–1548. doi: 10.1091/mbc.E05-09-0884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake D, Luke B, Kanellis P, Jorgensen P, Goh T, Penfold S, Breitkreutz BJ, Durocher D, Peter M, Tyers M. The F-box protein Dia2 overcomes replication impedance to promote genome stability in Saccharomyces cerevisiae. Genetics. 2006;174:1709–27. doi: 10.1534/genetics.106.057836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X, Ye P, Yuan DS, Wang X, Bader JS, Boeke JD. A DNA integrity network in the yeast Saccharomyces cerevisiae. Cell. 2006;124:1069–1081. doi: 10.1016/j.cell.2005.12.036. [DOI] [PubMed] [Google Scholar]
- Sarin S, Ross KE, Boucher L, Green Y, Tyers M, Cohen-Fix O. Uncovering novel cell cycle players through the inactivation of securin in budding yeast. Genetics. 2004;168:1763–1771. doi: 10.1534/genetics.104.029033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang XL, Wade Harper J. SCF-mediated protein degradation and cell cycle control. Oncogene. 2005;24:2860–2870. doi: 10.1038/sj.onc.1208614. [DOI] [PubMed] [Google Scholar]
- Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper JW, Elledge SJ. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86:263–274. doi: 10.1016/S0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- Feldman RM, Correll CC, Kaplan KB, Deshaies RJ. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell. 1997;91:221–230. doi: 10.1016/S0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- Galan JM, Peter M. Ubiquitin-dependent degradation of multiple F-box proteins by an autocatalytic mechanism. Proc Natl Acad Sci USA. 1999;96:9124–9129. doi: 10.1073/pnas.96.16.9124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, Howley PM. Ubiquitination and degradation of the substrate recognition subunits of SCF ubiquitin-protein ligases. Mol Cell. 1998;2:571–580. doi: 10.1016/S1097-2765(00)80156-2. [DOI] [PubMed] [Google Scholar]
- Cope GA, Deshaies RJ. Targeted silencing of Jab1/Csn5 in human cells downregulates SCF activity through reduction of F-box protein levels. BMC Biochem. 2006;7:1. doi: 10.1186/1471-2091-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kus BM, Caldon CE, Andorn-Broza R, Edwards AM. Functional interaction of 13 yeast SCF complexes with a set of yeast E2 enzymes in vitro. Proteins. 2004;54:455–467. doi: 10.1002/prot.10620. [DOI] [PubMed] [Google Scholar]
- Mathias N, Johnson S, Byers B, Goebl M. The abundance of cell cycle regulatory protein Cdc4p is controlled by interactions between its F box and Skp1p. Mol Cell Biol. 1999;19:1759–1767. doi: 10.1128/mcb.19.3.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton EE, Willems AR, Sa D, Kuras L, Thomas D, Craig KL, Tyers M. Cdc53 is a scaffold protein for multiple Cdc34/Skp1/F box protein complexes that regulate cell division and methionine biosynthesis in yeast (vol 12, pg 914, 1998) Genes Dev. 1998;12:3144–3144. doi: 10.1101/gad.12.5.692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smothers DB, Kozubowski L, Dixon C, Goebl MG, Mathias N. The abundance of met30p limits SCFMet30p complex activity and is regulated by methionine availability. Mol Cell Biol. 2000;20:7845–7852. doi: 10.1128/MCB.20.21.7845-7852.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fey JP, Lanker S. Delayed accumulation of the yeast G(l) cyclins Clnl and Cln2 and the F-box protein Grrl in response to glucose. Yeast. 2007;24:419–429. doi: 10.1002/yea.1472. [DOI] [PubMed] [Google Scholar]
- Bashir T, Dorrello NV, Amador V, Guardavaccaro D, Pagano M. Control of the SCF(Skp2-Cks1) ubiquitin ligase by the APC/C(Cdh1) ubiquitin ligase. Nature. 2004;428:190–193. doi: 10.1038/nature02330. [DOI] [PubMed] [Google Scholar]
- Swaminathan S, Kile AC, MacDonald EM, Koepp DM. Yra1 is required for S phase entry and affects Dia2 binding to replication origins. Mol Cell Biol. 2007;27:4674–4684. doi: 10.1128/MCB.02052-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morohashi H, Maculins T, Labib K. The amino-terminal TPR domain of Dia2 tethers SCF(Dia2) to the replisome progression complex. Curr Biol. 2009;19:1943–1949. doi: 10.1016/j.cub.2009.09.062. [DOI] [PubMed] [Google Scholar]
- Mimura S, Komata M, Kishi T, Shirahige K, Kamura T. SCF(Dia2) regulates DNA replication forks during S-phase in budding yeast. Embo J. 2009;28:3693–3705. doi: 10.1038/emboj.2009.320. [DOI] [PMC free article] [PubMed] [Google Scholar]