Abstract
Plasmodium falciparum causes the most severe form of malaria in humans. An important class of drugs in malaria treatment is the sulfone/sulfonamide group, of which sulfadoxine is the most commonly used. The target of sulfadoxine is the enzyme dihydropteroate synthase (DHPS), and sequencing of the DHPS gene has identified amino acid differences that may be involved in the mechanism of resistance to this drug. In this study we have sequenced the DHPS gene in 10 isolates from Thailand and identified a new allele of DHPS that has a previously unidentified amino acid difference. We have expressed eight alleles of P. falciparum PPPK-DHPS in Escherichia coli and purified the functional enzymes to homogeneity. Strikingly, the Ki for sulfadoxine varies by almost three orders of magnitude from 0.14 μM for the DHPS allele from sensitive isolates to 112 μM for an enzyme expressed in a highly resistant isolate. Comparison of the Ki of different sulfonamides and the sulfone dapsone has suggested that the amino acid differences in DHPS would confer cross-resistance to these compounds. These results show that the amino acid differences in the DHPS enzyme of sulfadoxine-resistant isolates of P. falciparum are central to the mechanism of resistance to sulfones and sulfonamides.
Keywords: malaria, sulfadoxine, drug resistance
The sulfonamide/sulfone group of compounds is used extensively in the treatment of bacterial diseases including infections caused by Streptococcus spp., Neisseria spp., and Mycobacterium spp as well as many parasitic infections involving Pneumocystis, Toxoplasma, Cryptosporidium, and Plasmodium. Members of this group of chemotherapeutic compounds, such as sulfadoxine and dapsone, inhibit the enzyme dihydropteroate synthase (DHPS) (1), a component of the folate biosynthetic pathway. The enzyme DHPS catalyzes the condensation of p-aminobenzoic acid (pABA) with 6-hydroxymethyldihydropterin pyrophosphate to yield 7,8-dihydropteroate (1). Plasmodia generate the vast majority of their folates de novo, and hence inhibition of DHPS by these drugs leads to depletion of dTTP and decreased DNA synthesis (2). However, there is also some evidence for folate salvage by the parasite (3). The human host lacks most of this biosynthetic pathway, and consequently many effective chemotherapeutic agents have been developed that target some of the enzymes involved in the synthesis of pyrimidines.
In the case of Plasmodium falciparum, the causative agent of the most lethal form of human malaria, sulfadoxine has been used extensively for prophylaxis. Usually, this has been in combination with the antifolate drug, pyrimethamine, which inhibits the enzyme dihydrofolate reductase (DHFR) (4), because the combination of these drugs shows a marked synergism in their antimalarial effect (5). These drugs have become less useful as the parasite develops resistance (6); however, despite this they are still used extensively in Africa where resistance is not yet widespread.
The DHPS gene has been cloned from P. falciparum and encodes a putative bifunctional enzyme with 7,8-dihydro-6-hydroxymethylpterin pyrophosphokinase (PPPK), the enzyme before DHPS in the folate pathway (7, 8). All the protozoan DHPS enzymes sequenced so far have been either bifunctional with both PPPK and DHPS domains, as in P. falciparum (7, 8), or trifunctional as in Pneumocystis carinii with another enzyme from the folate pathway dihydroneopterin aldolase (DHNA), PPPK, and DHPS activities (9). The Toxoplasma gondii DHPS enzyme is also likely to be bifunctional (10).
Identification of amino acid differences in DHPS between sulfadoxine-sensitive and -resistant isolates of P. falciparum has suggested that mutations of this enzyme may be involved in the mechanism of resistance to this antimalarial drug (7, 8); however, no direct evidence for this has yet been obtained. A point mutation in a highly conserved region of the DHPS gene of E. coli confers sulfonamide resistance (11), and a 2-aa insertion in DHPS of both Neisseria meningitidis and Streptococcus pneumoniae has also been shown to confer sulfonamide resistance (12, 13). However, in both N. meningitidis and Streptococcus pyogenes, sulfonamide resistance may be a result of multiple mutations in DHPS genes, which suggests they have arisen not by mutation of the sensitive gene but by intraspecies or interspecies transfer of genetic material (13, 14).
To test whether the amino acid differences found in DHPS between sulfadoxine-resistant and -sensitive isolates of P. falciparum are important in the drug resistance phenotype, we have expressed eight different PPPK-DHPS alleles in E. coli, purified the active enzymes, and determined their kinetic parameters. We show that mutations within the P. falciparum DHPS gene are responsible for the mechanism of sulfadoxine resistance in P. falciparum.
MATERIALS AND METHODS
Parasites and DNA.
The origin of P. falciparum clones and strains is shown in Table 1. The PR145, 336, 338, 342, 345, 347, 327, 343, 352, and 346 isolates were kindly provided by Sodsri Thaithong of the WHO Collaborating Centre on the Biological Characterization of Malaria Parasites, Chulalongkorn University, Bangkok, Thailand. Parasites were maintained in asynchronous cultures as described (15).
Table 1.
Genotypes and sulfadoxine sensitivity of P. falciparum lines and genotypes of DHPS constructs
| Isolate | Origin | Sulfadoxine | Amino acid
|
Construct | Reference | ||||
|---|---|---|---|---|---|---|---|---|---|
| 436 | 437 | 540 | 581 | 613 | |||||
| D10, HB3, ItG2, M24, NF7 | PNG, Africa, Brazil | S | S | A | K | A | A | D10-C | (7, 8) |
| 3D7 | Africa | R | S | G | K | A | A | 3D7-C | (7, 22) |
| Tak9/96 | SE Asia | R | A | G | K | A | A | Tak9/96-C | (22) |
| K1 | SE Asia | R | S | G | K | G | A | K1-C | (7) |
| W2mef, Dd2 | SE Asia | R | F | G | K | A | S | W2mef-C | (7) |
| PR145, 336, 338, 342, 345, 347 | Thailand | ND | A | G | E | A | A | PR145-C | This publication |
| PR327, 343, 352 | Thailand | ND | S | G | E | G | A | This publication | |
| PR346 | Thailand | ND | A | G | E | G | A | This publication | |
| S | A | K | G | A | D10-C/G581 | ||||
| S | G | E | A | A | 3D7-C/E540 | ||||
ND, not determined.
Expression Constructs.
The full-length PPPK-DHPS gene from P. falciparum was amplified from D10 cDNA in two halves using a unique AccI restriction site at position 963 in the cDNA sequence where the A of the start codon is position 1. The four oligonucleotides were: P1 (5′-ATATATATACCATGGAAACTATACAAGAA) with the introduced NcoI site underlined, P2 (5′-CTTTTAATACATATATCCTTTTAT), P3 (5′-TTAGAGATCCACAAGAAATAATAAAC), and P4 (5′-GTTGAATTCCATGTTTGCACTTTCC) with the introduced EcoRI site underlined. The two halves of the PPPK-DHPS gene were cloned into the NcoI and EcoRI sites of the pTrc expression vector (Invitrogen) to express the native PPPK-DHPS enzyme. A region of instability, encompassing a stretch of 12 nucleotides at position 1551–1562, was mutated to the sequence AAAGAAGAAGAA by PCR mutagenesis. This did not change the encoded amino acids but stabilized the sequence in E. coli. The PPPK-DHPS D10 construct (D10-C) was then used as the starting plasmid for all subsequent constructs (Table 1). The 3D7 (3D7-C) and W2 mef (W2 mef-C) alleles of PPPK-DHPS were constructed by amplifying DNA fragments, which contained the required mutations, from genomic DNA and subcloning into the D10 construct by using appropriate restriction enzymes. Similarly, the K1 (K1-C), Tak 9/96 (Tak 9/96-C), and PR145 (PR145-C) PPPK-DHPS alleles were made by substituting amplified fragments into the 3D7 construct. The constructs D10-C/G581 and 3D7-C/E540 (Table 1) were obtained as intermediate steps toward the final K1-C and PR145-C constructs.
Peptide Synthesis and Antibody Production.
A 15-residue peptide (Chiron) corresponding to amino acids 631–644 of the DHPS domain was conjugated to diphtheria toxoid to allow antibody production in rabbits.
Purification of PPPK-DHPS.
Cultures of each recombinant clone were grown at 37°C and induced with 0.5 mM isopropyl β-d-galactopyranoside (IPTG) at 30°C. Cells were pelleted, resuspended in sonication buffer (50 mM phosphate, pH 8.0/300 mM NaCl/1 mM EDTA/2 mM DTT/0.5 mM phenylmethylsulfonyl fluoride (PMSF)/1 μg/ml leupeptin/1 μg/ml pepstatin A/10 mg/ml DNaseI/0.1 mg/ml RNase A/0.1 mg/ml lysozyme), and stirred. Cells were sonicated and centrifuged at 6,000 × g for 10 min. The supernatant pH was adjusted to 6.2 with solid Mes, dialyzed against 10 mM Mes-NaOH, pH 6.2 (cation exchange buffer), centrifuged (10 min, 18,000 × g), and the supernatant was separated on a SP-Sepharose HPLC chromatography column (Pharmacia). Proteins were eluted with a gradient of 0–0.45 M NaCl in cation-exchange buffer. Fractions containing PPPK-DHPS were combined, and the buffer was exchanged with an Amicon stirred-cell nitrogen-pressure ultrafiltration unit (YM10 membrane) with gel filtration buffer (5 mM Mes-NaOH/0.5 M NaCl/1 mM DTT/0.1 mM Na2EDTA/0.2 mM leupeptin/0.2 mM pepstatin A/10 mg/ml PMSF, pH 6.2), and proteins were concentrated to <3 ml. Proteins were separated on a Sephacryl S-300 HR column (Pharmacia) equilibrated with gel filtration buffer. Fractions containing PPPK-DHPS were combined, concentrated by ultrafiltration, and diluted 1:7 with anion-exchange buffer (10 mM Tris⋅HCl/5 mM DTT/0.1 mg/ml PMSF/1 μM leupeptin/1 μM pepstatin, pH 8.6) before loading onto a Mono Q HR 5/5 column (Pharmacia). Proteins were eluted with a gradient of 0–0.35 M NaCl in anion-exchange buffer. At this pH the addition of DTT was essential. The purified protein was stored in gel filtration buffer by ultrafiltration/buffer exchange as described above and finally concentrated to 0.3–0.5 ml. PPPK-DHPS was detected in fractions by activity assay, probing Western blotted protein with affinity-purified anti-DHPS antibody, or on SDS/PAGE stained for protein.
For purification of the D10-C enzyme, the final recovery was 11.1%, which represented 0.04% of the initial protein. Routinely, around 0.2 mg of pure enzyme was obtained from 400 mg of starting protein.
Enzyme Assays.
The PPPK-DHPS enzymes were assayed using a modification of a published procedure (16). The assay was dependent on the presence of both PPPK and DHPS activities, but the DHPS activity component was isolated by measurement of the conversion of [3H]pABA to [3H]dihydropteroate. Enzyme activity was found to be dependent on protein concentration; hence, BSA was included in assay reaction mixtures. All assays were done at 37°C, and kinetic assays were done in triplicate. For the estimations of Ki and Km, reactions (80 μl containing 1 mg/ml enzyme, 100 mM Tris⋅HCl (pH 8.5), 10 mM MgCl2, 20 mM 2-mercaptoethanol, 80 mM 6-hydroxymethylpterin, 5 mM ATP, 1 mg/ml BSA, pH 8.5) were commenced by the addition of various amounts of 4 Ci/mmol [3H]pABA (American Radiolabeled Chemicals, St. Louis) and either dimethyl sulfoxide (DMSO) or sulfadoxine (Hoffmann–LaRoche) in DMSO. Reactions were sampled at appropriate time points, and [3H]dihydropteroate was separated from [3H]pABA on PEI-cellulose (Merck) with 0.2 M LiCl. The origins were excised and [3H]dihydropteroate was counted by liquid scintillation. Initial rates of reaction were estimated with the kinetics program enzfitter, by using linear regression and first-order nonlinear regression analysis, as appropriate. Ki and Km values were calculated from the vo values using the program ez-fit (DuPont). Ki values for dapsone, sulfamethoxazole, and sulfathiazole were calculated from the IC50 data using the equation v = Vmax[s]([s] + (1 + I.Ki−1)) − 1, where v = reaction velocity, Vmax = maximum reaction rate, [s] = substrate concentration, Km = Michaelis constant, I = inhibitor concentration, and Ki = inhibitory constant. One unit of enzyme activity is equal to the formation of 1 μmol [3H]dihydropteroate per minute.
Western Blotting.
Protein samples were analyzed by SDS/PAGE and electrophoretically transferred to nitrocellulose as described (17). Bound antibody was detected with horseradish peroxidase-coupled goat anti-rabbit IgG and developed by ECL, a luminescence-based method (Amersham).
RESULTS
Amino Acid Differences in DHPS.
Sequence analysis of the DHPS gene from sulfadoxine-resistant and -sensitive isolates of P. falciparum has shown that there are amino acid differences at four positions, and it has been suggested that they may be involved in the mechanism of resistance to sulfadoxine (7, 8). To determine whether other differences in the DHPS gene could occur, we sequenced isolates obtained from Thailand, a geographic area where there has been extensive use of sulfadoxine and pyrimethamine. The PPPK-DHPS gene from 10 Thai isolates (PR145, 336, 338, 342, 345, 346, 347, 327, 343, and 352) was obtained by PCR and sequenced. The DHPS gene from the 10 isolates encoded a Glu at amino acid 540 (Glu-540) rather than Lys-540, which has been described for all other isolates that have so far been sequenced (Table 1), and also a Gly-437 and Ala-613. The DHFR gene from the Thai isolates was also sequenced across the amino acids responsible for pyrimethamine and cycloguanil resistance (18–21). All had the same sequence for the DHFR gene: Ala-16, Ile-51, Arg-59, Asn-108, and Leu-164, indicating that all had the genotype that would confer resistance to high levels of pyrimethamine (18–21).
Expression of Functional P. falciparum PPPK-DHPS in E. coli.
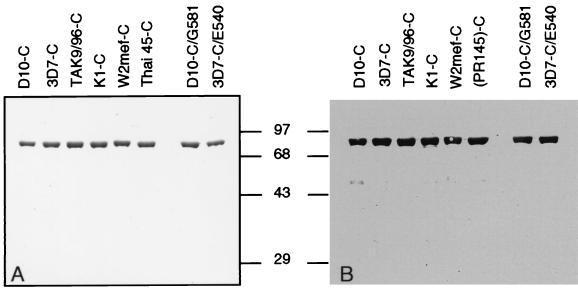
To measure the effects of amino acid changes on the DHPS enzyme we expressed the full-length PPPK-DHPS gene in E. coli. Six plasmid constructs were made that express the D10 (D10-C), 3D7 (3D7-C), Tak9/96 (Tak9/96-C), KI (KI-C), W2 mef (W2 mef-C), and PR145 (PR145-C) alleles of PPPK-DHPS in E. coli (Table 1). In addition, two constructs were made to investigate the role of Gly-581 (D10-C/G581) and Glu-540 (3D7-C/E540) in resistance to sulfadoxine (Table 1). The eight constructs were transformed into E. coli and expression was analyzed by SDS/PAGE and an anti-DHPS antibody. A band of 83-kDa was recognized in all constructs, suggesting that the PPPK-DHPS constructs were expressing full-length PPPK-DHPS protein in E. coli (Fig. 1).
Figure 1.
Expression of eight alleles of P. falciparum PPPK-DHPS in E. coli. Eight different alleles of PPPK-DHPS were cloned into the pTrc vector and transformed into E. coli. Extracted proteins were separated by SDS/PAGE and stained with Coomassie blue (A) or transferred to nitrocellulose and probed with affinity-purified anti-DHPS antibodies (B). The order of the tracks in each panel from left to right is pTrc, D10-C, 3D7-C, Tak9/96-C, K1-C, W2 mef-C, PR145, D10-C/G581, and 3D7-C/E540.
Purification of P. falciparum PPPK-DHPS from E. coli.
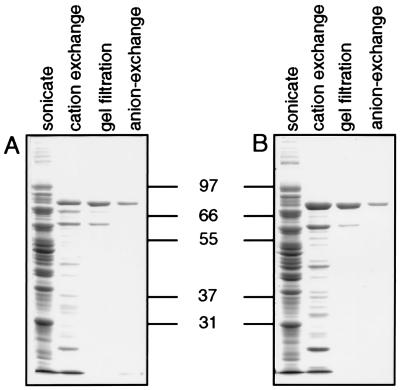
Functional P. falciparum PPPK-DHPS was isolated from E. coli by using sequential steps of SP-Sepharose cation exchange, Sephacryl S300HR gel filtration, and Mono Q high-resolution anion-exchange chromatography. The molecular mass estimated by gel filtration ranged from 207 to 246 kDa (mean 222 kDa), suggesting that the PPPK-DHPS enzyme is a multimer of two or three subunits. Purification was monitored throughout the procedure by the PPPK-DHPS enzyme assay, and immunoblots of fractions were monitored using the anti-DHPS antibody (Fig. 2). Enzyme activity copurified with a protein of 83 kDa when separated by SDS/PAGE. Visualization by Coomassie blue staining of chromatography fractions on SDS/PAGE throughout the purification of the D10-C and K1-C enzymes showed a single protein band after anion-exchange chromatography, suggesting that the enzymes had been purified essentially to homogeneity (Fig. 2). Active PPPK-DHPS enzyme was purified from the eight different alleles of PPPK-DHPS using the same procedure. Analysis by SDS/PAGE and Coomassie blue staining suggested that all were purified to homogeneity (Fig. 3A). The purified proteins have the predicted size from the sequence of the gene for the P. falciparum PPPK-DHPS enzyme (7, 8), contain both PPPK and DHPS enzyme activities, and are recognized by an anti-DHPS antibody (Fig. 3B). Therefore, we have purified from E. coli the active P. falciparum PPPK-DHPS bifunctional recombinant enzyme.
Figure 2.
Purification of functional recombinant PPPK-DHPS enzyme from P. falciparum in E. coli. Four liters of either D10-C or K1-C were grown and the PPPK-DHPS activity purified by sequential steps of cation-exchange, gel-filtration, and anion-exchange chromatography. Purification was monitored by enzyme activity and detection with anti-DHPS antibodies. Aliquots of D10-C (A) or K1-C (B) chromatography aliquots were separated by SDS/PAGE, and protein was detected by staining with Coomassie blue. Tracks are as indicated and are in both panels E. coli sonicate, pooled-positive cation-exchange, gel-filtration, and anion-exchange chromatography fractions.
Figure 3.
Purified recombinant PPPK-DHPS from eight different alleles. The eight alleles of PPPK-DHPS were purified using the protocol described in Material and Methods, and aliquots were separated by SDS/PAGE and either stained with Coomassie blue (A) or electroblotted to nitrocellulose and probed with the anti-DHPS antibodies (B). In both panels the tracks contain the sample as indicated and include, in order, D10-C, 3D7-C, Tak9/96-C, K1-C, W2 mef-C, PR145-C, D10-C/G581, and 3D7-C/E540.
Kinetic Constants of DHPS Activity and Inhibitory Constants for Sulfadoxine.
The kinetic parameters of DHPS activity of the eight PPPK-DHPS enzymes were obtained to determine the effect of the different amino acid changes on the catalytic properties. The Km values for pABA were found to vary more than 10-fold, with D10-C having the lowest and W2 mef-C, the highest (Table 2), whereas Vmax for all enzymes were similar. The different amino acid changes in PPPK-DHPS had little effect on catalytic turnover, kcat, varying from 0.024 s−1 for D10-C to 0.058 s−1 for 3D7-C/E540. The kcat/Km, or specificity constant of the different enzymes, also varied for the different enzymes, which is essentially a measure of catalytic efficiency for pABA, by up to 9-fold (Table 2).
Table 2.
Kinetic parameters of the PPPK-DHPS enzymes
| Enzyme | Km, nM | Ki, μM | Vmax, mU/mg protein | kcat, s−1 | kcat/Km, s−1M−1 × 106 |
|---|---|---|---|---|---|
| D10-C | 16.4 ± 1.3 | 0.14 ± 0.01 | 17.2 ± 0.3 | 0.024 | 1.46 |
| 3D7-C | 24.2 ± 3.3 | 1.39 ± 0.23 | 29.8 ± 1.0 | 0.042 | 1.71 |
| Tak9/96-C | 88.9 ± 9.1 | 19.9 ± 4.0 | 33.0 ± 0.9 | 0.046 | 0.52 |
| K1-C | 77.4 ± 7.2 | 16.2 ± 2.7 | 27.6 ± 0.7 | 0.038 | 0.50 |
| W2mef-C | 193.3 ± 13.9 | 112 ± 10 | 33.7 ± 0.8 | 0.047 | 0.24 |
| PR145-C | 112.8 ± 5.8 | 98.3 ± 7.5 | 34.4 ± 0.5 | 0.048 | 0.42 |
| D10-C/G581 | 20.0 ± 2.3 | 0.55 ± 0.06 | 32.4 ± 0.9 | 0.045 | 2.25 |
| 3D7-C/E540 | 105 ± 10 | 27.7 ± 6.1 | 41.4 ± 1.1 | 0.058 | 0.55 |
mU, milliunits.
To test whether the amino acid changes in the DHPS enzyme are important in the mechanism of resistance to sulfadoxine, we measured the inhibitory constant for this drug. Inhibition was by a competitive mechanism for all enzymes, and the Ki for sulfadoxine of the eight PPPK-DHPS enzymes was found to vary significantly by up to 811-fold (Table 2). The enzyme with the lowest Ki was D10-C, which is an enzyme present in sulfadoxine-sensitive isolates (8), and the highest was with W2 mef-C, which is present in highly sulfadoxine-resistant isolates (8). 3D7-C and Tak9/96-C are PPPK-DHPS enzymes from isolates that have been previously defined as sulfadoxine-sensitive (8). However, the Ki of these enzymes would suggest that these isolates may show some resistance to this drug. Indeed, recent analysis of the sulfadoxine sensitivity of 3D7 has shown that this isolate is resistant to medium levels of the drug (22). The PR145-C enzyme, which represents the genotype of DHPS found in 6 of the 10 Thai isolates examined, has a Ki of 98.3 μM, suggesting these isolates are highly resistant to sulfadoxine. Comparison of the Ki of the different enzymes shows that there is a relationship between the amino acid differences found in the DHPS enzyme of P. falciparum isolates and resistance to sulfadoxine. Therefore, mutations in the DHPS enzyme are important in the mechanism of resistance to this drug.
Comparison of the kinetic properties of the different enzymes not only shows that the amino acid differences are important in sulfadoxine resistance but also indicates the effect they have on the magnitude of the Km and Ki. The PPPK-DHPS enzyme 3D7-C has Gly-437 rather than Ala-437 in D10-C, and the Ki was 10-fold higher for 3D7-C compared with the D10-C enzyme. However, this amino acid change has a minor effect on the Km. Because this alteration (Gly-437) was present in almost all sulfadoxine-resistant parasites, it suggests that this may be one of the first mutations commonly selected under sulfadoxine pressure. The D10-C/G581 has Gly-581 rather than Ala-581 as in D10-C, and this amino acid change has only a 4-fold effect on the Ki of sulfadoxine, suggesting that this amino acid change by itself plays a small role in resistance. However, when Gly-581 is combined with Gly-437, as seen with K1-C, the Ki increased approximately 10-fold compared with the single amino acid change either at Gly-581 or Gly-437, showing that the two amino acid changes are synergistic in their effect on Ki. Similarly, comparison of the Ki for the enzyme D10-C with 3D7-C, Tak9/96-C, and PR145-C shows that it increases from 0.14 to 1.39, 19.9, and 98.3 μM, respectively. The differences between each consecutive enzyme is a single amino acid change (Table 1), and each alteration results in an increase in the Ki of the enzyme for sulfadoxine in approximately 10-fold increments. Concomitant with the change in Ki is a corresponding increase in the Km of the enzymes for pABA. The effect of the amino acid changes on Ki of the enzymes suggests that an accumulation of mutations in DHPS, as a result of selective pressure with sulfadoxine in the field, has occurred, resulting in an enzyme with not only much greater Ki, which would confer resistance to this drug, but also with increased Km.
Interaction of DHPS with Sulfa Analogues.
Various sulfonamides and the sulfone dapsone were tested for their ability to inhibit the eight different recombinant PPPK-DHPS enzymes. The Ki of the sulfonamides (sulfadoxine, sulfamethoxazole, and sulfathiazole) varied for each enzyme by approximately 5-fold, with sulfathiazole being the most potent among them (Fig. 4). The sulfone dapsone was found to be the most potent compound tested, which is in agreement with previous results obtained by assaying DHPS enzyme activity in extracts of P. falciparum isolates (23).
Figure 4.
Comparison of the Ki values of the purified PPPK-DHPS enzymes for sulfadoxine with sulfathiazole, sulfamethoxazole, and dapsone. The correlation coefficients are sulfadoxine:sulfathiazole, 0.99; sulfadoxine:sulfamethoxazole, 1.0; and sulfadoxine:dapsone, 0.96; and the slopes for each plot are 0.99, 0.87, and 0.89, respectively.
The different amino acid changes in the eight DHPS enzymes had similar effects on the magnitude of the Ki for the sulfone and sulfonamide compounds (Fig. 4). The slope of the plots for Ki of sulfadoxine vs. the Ki for the other compounds gave no significant deviation from linearity with a slope of approximately one, suggesting that each amino acid change had an equal effect on the affinity of binding of each of the inhibitory compounds and that each inhibitor is competing for the same binding site in the active site of the enzyme. Therefore, P. falciparum parasites that are resistant to sulfadoxine are likely to show cross-resistance with these different sulfone and sulfonamide compounds.
DISCUSSION
The enzyme dihydropteroate synthase is the target of sulfone and sulfonamide drugs (1), which are used extensively in the control of many infections including P. falciparum. Sulfadoxine is the major sulfonamide used for the prophylaxis of P. falciparum infections, and this drug inhibits the enzyme DHPS (23) by direct competition for the substrate binding site. Identification of amino acid differences in the DHPS enzyme of sulfadoxine-resistant compared with sulfadoxine-sensitive P. falciparum isolates suggested that the mechanism of resistance may involve mutations in DHPS that alter the affinity of binding of the drug (7, 8). We have shown in this work that the amino acid differences in DHPS decrease the efficacy of inhibition by sulfadoxine and that such mutations are important for resistance of P. falciparum toward this drug.
Sequencing of the DHPS gene from Thai isolates has identified a new allele that was likely to be linked to sulfadoxine resistance. These isolates originate from a geographic area in which there previously has been a heavy use of the combination of sulfadoxine and pyrimethamine, which would favor selection for drug resistance. The DHFR gene of the same Thai isolates have mutations that confer pyrimethamine resistance (18, 19, 24), and therefore, it is highly likely that the differences identified in the DHPS gene of the Thai isolates have evolved in parallel to the DHFR mutations and are important in the mechanism of resistance to sulfadoxine. This was strongly supported by our data showing that the recombinant PPPK-DHPS enzyme from these Thai isolates has an approximately 800-fold higher Ki for sulfadoxine compared with the same enzyme from a sulfadoxine-sensitive isolate.
Expression and purification of functional PPPK-DHPS enzyme from P. falciparum in E. coli allowed us to directly address the role of the different amino acids found in alleles of DHPS in the mechanism of resistance to sulfadoxine by analyzing the kinetic properties of the enzyme. The recombinant enzyme was found to have a molecular mass of approximately 222 kDa, suggesting that it consists of a dimer or trimer. This is in agreement with the previously reported size of 200–250 kDa for the P. berghei PPPK-DHPS enzyme (25) and also the P. chabaudi enzyme (190 kDa) (16). The D10-C recombinant PPPK-DHPS enzyme had an apparent Km of 16.4 nM for pABA compared with a previously measured value for the unpurified enzyme in extracts of P. falciparum of 8.1 μM (23). Measurement of Km for T. gondii (10) and P. berghei (25) PPPK-DHPS enzymes in cell extracts and in purified form showed that the apparent Km decreased by a factor of 10 upon purification. It was suggested that the conformation of the enzyme was altered during purification, which allowed a greater access to the active site and thereby enhanced the apparent affinity (decreased Km) of the substrate for the enzyme (10). However, it is more likely that an inhibitor or a modulator is present in the unpurified enzyme preparations, which would explain the lower Km values in more purified extracts. The low Km for pABA would suggest that the P. falciparum enzyme would be capable of efficient catalysis at very low concentrations of pABA.
The amino acid changes in the PPPK-DHPS enzymes had a large effect on both the apparent Km for pABA and the Ki for sulfadoxine of the PPPK-DHPS enzyme. Because sulfadoxine is a structural analogue of pABA, the increasing Ki and Km for each amino acid change suggests that most of the alterations are affecting the interaction of the homologous moiety of the inhibitor, rather than the side group, with the active site of the enzyme. The DHPS enzyme, when compared with other organisms, is not highly conserved (7, 8); however, the mutations linked to sulfadoxine resistance in the DHPS enzyme of P. falciparum are located in or very near to small regions of highly conserved amino acids, and it is likely that many of the mutations identified are at or very close to the pABA binding site in the catalytic site of the DHPS enzyme. The large effect of the amino acid mutations on Km of the DHPS enzyme for pABA would suggest that a parasite expressing some of these mutant enzymes would be unable to efficiently utilize this substrate at low cytosolic pABA concentrations. However, the parasites that express this enzyme do not show any obvious deleterious growth effects, suggesting that they are able to cope with a less efficient enzyme or that the concentration of pABA is sufficiently high even compared with the highest Km.
The single amino acid change, Ala-437 to Gly-437, is found in all of the DHPS sequences from sulfadoxine-resistant isolates, suggesting that this may be the first mutation in response to drug pressure. The 3D7 and Tak9/96 isolates are resistant to medium levels of sulfadoxine, and subsequent selection would result in additional mutations that have an additive effect on the Ki and, therefore, the level of resistance of the parasite to the drug. This is similar to the results found in resistance of pyrimethamine for the enzyme DHFR in P. falciparum (24).
The amino acid changes in DHPS have a profound effect on the Ki of sulfadoxine, proving that these alterations are important in the mechanism of resistance to this drug. The P. falciparum line 3D7, which has a DHPS sequence identical to D10 except for the single alteration Gly-437, has been previously classed as sensitive (8). However, more recent data have shown that the 3D7 parasite line was resistant to low levels of sulfadoxine (22). This is consistent with a higher Ki of the 3D7-C PPPK-DHPS enzyme compared with D10-C. A major problem with the classification of P. falciparum with respect to resistance to sulfadoxine is the difficulty of the in vitro parasite IC50 assay. This is exemplified by the large variation in IC50 that has been reported for this drug, probably as a result of differing concentrations of pABA and folate in the growth medium that can confound the assay (8, 26). The line Tak9/96 has also been classified as sensitive to sulfadoxine (8), but because of the high Ki of the Tak9/96-C enzyme it is likely that it is also resistant to sulfadoxine. The PPPK-DHPS enzymes PR145-C and W2 mef-C are from highly sulfadoxine-resistant parasites, and both have Ki values that are close to three orders of magnitude higher than that seen for the enzyme from sulfadoxine-sensitive parasites. This provides strong evidence that these mutations in the DHPS enzyme are important in the mechanism of resistance to sulfadoxine by implicating a reduced binding affinity of the drug.
The inhibition of recombinant DHPS with a range of sulfonamides and sulfones demonstrated that they exhibited a broad range of inhibitory potencies, with sulfathiazole and dapsone being the most potent of the compounds tested. Sulfadoxine, the DHPS inhibitor used in prophylaxis regimens for P. falciparum, was found to be the least potent, which is in agreement with previous studies (23). However, this does not account for differences such as membrane transport, drug stability, pharmacokinetics, and pharmacodynamic properties, which are important in vivo. Nevertheless, it is clear that the amino acid mutations in DHPS had an approximately equal effect on the Ki of all of the sulfones and sulfonamides tested, suggesting that they are all competing for the pABA binding site. Therefore, it is likely that the different P. falciparum isolates with DHPS genotypes of sulfadoxine-resistant parasites would be cross-resistant to the other sulfone and sulfonamide compounds tested here.
This study clearly has shown that the different amino acids seen in the DHPS enzyme of sulfadoxine-resistant P. falciparum are important in the mechanism of resistance to this compound and other sulfone and sulfonamide drugs. However, it does not rule out involvement or alterations in other genes contributing to the level of resistance. Analysis of the genetic cross between a sulfadoxine-resistant and a sulfadoxine-sensitive parent has shown that mutations in DHPS are important in the cloned line tested, but it has also implicated a second gene (22). It is difficult to extrapolate the profound differences in Ki of sulfadoxine of the purified PPPK-DHPS enzymes to the effect the amino acid changes would have in vivo on the magnitude of resistance to the drug, but it is highly likely that they are of major importance in the level of resistance to sulfadoxine that is seen in P. falciparum.
Acknowledgments
We thank S. Caruana and M. Swift for culture of P. falciparum parasites and Dr. S. Thaithong for providing the isolates from Thailand. We also thank Drs. H. Ginsburg, L. Tilley, and B. Scopes for helpful advice. This research was supported by the Australian National Health and Medical Research Council, the United Nations Development Program/World Bank/World Health Organization Special Program for Research, and Training in Tropical Diseases. A.F.C. is supported by an International Research Scholar’s award from the Howard Hughes Medical Institute.
ABBREVIATIONS
- DHFR
dihydrofolate reductase
- DHNA
dihydroneopterin aldolase
- DHPS
7,8-dihydropteroate synthetase
- pABA
p-aminobenzoic acid
- PPPK
7,8-dihydro-6-hydroxymethylpterin pyrophosphokinase
References
- 1.Brown G M. Adv Enzymol Relat Areas Mol Biol. 1971;35:35–77. doi: 10.1002/9780470122808.ch2. [DOI] [PubMed] [Google Scholar]
- 2.Schellenberg K A, Coatney G R. Biochem Pharmacol. 1961;6:143–152. doi: 10.1016/0006-2952(61)90158-7. [DOI] [PubMed] [Google Scholar]
- 3.Krungkrai J, Webster H K, Yuthavong Y. Mol Biochem Parasitol. 1989;32:25–38. doi: 10.1016/0166-6851(89)90126-6. [DOI] [PubMed] [Google Scholar]
- 4.Ferone R. J Biol Chem. 1970;245:850–854. [PubMed] [Google Scholar]
- 5.Chulay J D, Watkins W M, Sixsmith D G. Am J Trop Med Hyg. 1984;33:325–330. doi: 10.4269/ajtmh.1984.33.325. [DOI] [PubMed] [Google Scholar]
- 6.Landgraf B, Kollaritsch H, Wiedermann G, Wernsdorder W H. Trans R Soc Trop Med Hyg. 1994;88:440–442. doi: 10.1016/0035-9203(94)90424-3. [DOI] [PubMed] [Google Scholar]
- 7.Triglia T, Cowman A F. Proc Natl Acad Sci USA. 1994;91:7149–7153. doi: 10.1073/pnas.91.15.7149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brooks D, Wang P, Read M, Watkin W, Sims P, Hyde J. Eur J Biochem. 1994;224:397–405. doi: 10.1111/j.1432-1033.1994.00397.x. [DOI] [PubMed] [Google Scholar]
- 9.Volpe F, Dyer M, Scaife J G, Darby G, Stammers D K, Delves C J. Gene. 1992;112:213–218. doi: 10.1016/0378-1119(92)90378-3. [DOI] [PubMed] [Google Scholar]
- 10.Allegra C J, Boarman D, Kovacs J A, Morrison P, Beaver J, Chabner B A, Masur H. J Clin Invest. 1990;85:371–379. doi: 10.1172/JCI114448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dallas W S, Gowen J E, P H, R, Cox M J, Dev I K. J Bacteriol. 1992;174:5961–5970. doi: 10.1128/jb.174.18.5961-5970.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lopez P, Espinosa M, Greenberg B, Lacks S A. J Bacteriol. 1987;169:4320–4326. doi: 10.1128/jb.169.9.4320-4326.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fermer C, Kristiansen B-E, Skold O, Swedberg G. J Bacteriol. 1995;177:4669–4675. doi: 10.1128/jb.177.16.4669-4675.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Swedberg G, Fermer C, Skold O. Adv Exp Med Biol. 1993;33:555–559. doi: 10.1007/978-1-4615-2960-6_113. [DOI] [PubMed] [Google Scholar]
- 15.Trager W, Jensen J B. Nature (London) 1978;273:621–622. doi: 10.1038/273621a0. [DOI] [PubMed] [Google Scholar]
- 16.Walter R D, Konigk E. Methods Enzymol. 1980;66:564–570. doi: 10.1016/0076-6879(80)66508-2. [DOI] [PubMed] [Google Scholar]
- 17.Cowman A F, Karcz S, Galatis D, Culvenor J G. J Cell Biol. 1991;113:1033–1042. doi: 10.1083/jcb.113.5.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cowman A F, Morry M J, Biggs B A, Cross G A M, Foote S J. Proc Natl Acad Sci USA. 1988;85:9109–9113. doi: 10.1073/pnas.85.23.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peterson D S, Walliker D, Wellems T E. Proc Natl Acad Sci USA. 1988;85:9114–9118. doi: 10.1073/pnas.85.23.9114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Foote S J, Galatis D, Cowman A F. Proc Natl Acad Sci USA. 1990;87:3014–3017. doi: 10.1073/pnas.87.8.3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peterson D S, Milhous W K, Wellems T E. Proc Natl Acad Sci USA. 1990;87:3018–3022. doi: 10.1073/pnas.87.8.3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang P, Read M, Sims P F G, Hyde J E. Mol Microbiol. 1997;23:979–986. doi: 10.1046/j.1365-2958.1997.2821646.x. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Y, Meshnick S R. Antimicrob Agents Chemother. 1991;35:267–271. doi: 10.1128/aac.35.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sirawaraporn W, Sirawaraporn R, Cowman A F, Yuthavong Y, Santi D V. Biochemistry. 1990;29:10779–10785. doi: 10.1021/bi00500a009. [DOI] [PubMed] [Google Scholar]
- 25.Ferone R. J Protozool. 1973;20:459–463. doi: 10.1111/j.1550-7408.1973.tb00926.x. [DOI] [PubMed] [Google Scholar]
- 26.Watkins W M, Sixsmith D G, Chulay J D, Spencer H C. Mol Biochem Parasitol. 1985;14:55–61. doi: 10.1016/0166-6851(85)90105-7. [DOI] [PubMed] [Google Scholar]