Abstract
This review focuses on the role of ADAM-17 in disease. Since its debut as the tumor necrosis factor converting enzyme or TACE, ADAM-17 has been reported to be an indispensible regulator of almost every cellular event from proliferation to migration. The central role of ADAM-17 in cell regulation is rooted in its diverse array of substrates: cytokines, growth factors, and their receptors as well as adhesion molecules are activated or inactivated by their cleavage with ADAM-17. It is therefore not surprising that ADAM-17 is implicated in numerous human diseases including cancer, heart disease, diabetes, rheumatoid arthritis, kidney fibrosis, Alzheimer’s disease, and is a promising target for future treatments. The specific role of ADAM-17 in the pathophysiology of these diseases is very complex and depends on the cellular context. To exploit the therapeutic potential of ADAM-17, it is important to understand how its activity is regulated and how specific organs and cells can be targeted to inactivate or activate the enzyme.
Keywords: ectodomain shedding, growth factor, inflammation, signaling, proteolysis, TNFα
INTRODUCTION
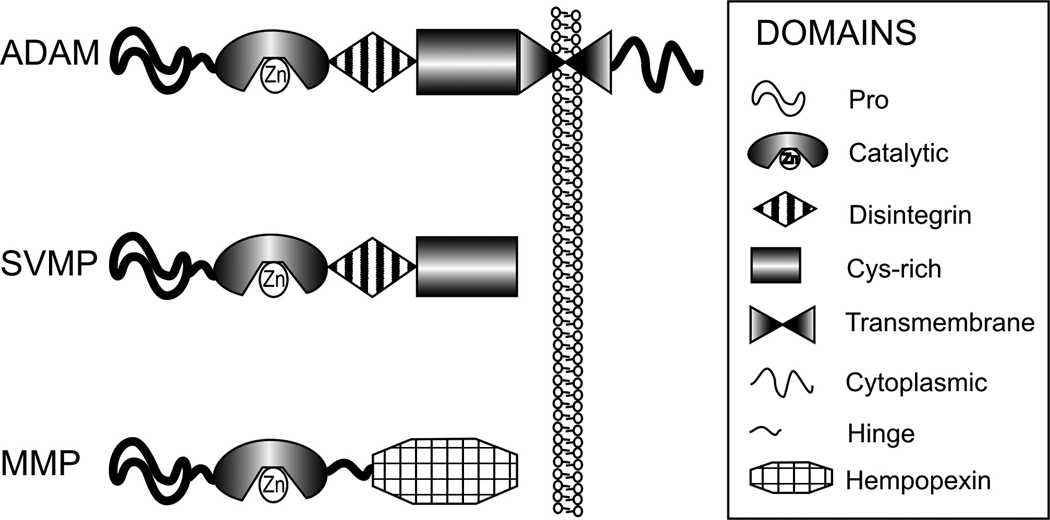
ADAM enzymes (a disintegrin and metalloproteinase) are Zn2+-dependent, modular cell surface proteins which belong to the adamalysin protein family. They are closely related to other metalloenzymes such as ADAM-TSs (ADAMs with thrombospondin domains), matrix metalloproteinases (MMPs) and snake venom metalloproteinases (SVMP). The structure of ADAMs closely resembles the Class III snake venom metalloenzymes or reprolysines (discussed in detail in (Calvete et al., 2007)) and it forms the basis of the many ADAM enzyme functions. The typical ADAM modules and their comparison to modules present in MMP, and SVMP are depicted in Figure 1.
Fig 1.
ADAMs can have both adhesive and proteolytic properties and can therefore participate in cellular adhesion and proteolytic cleavage of various cell surface molecules. Therefore, ADAMs are also important mediators of cell signaling events which determine cellular fate, proliferation, and growth (Blobel, 2005, Edwards et al., 2008). Because of these features, ADAMs are important contributors to physiological and pathophysiological processes and may be potential therapeutic targets in various diseases. In the mammalian genome, 40 ADAMs were identified, and in the human genome, 21 ADAMs have been described. However, of these 21 molecules, only 13 are proteolytically active (Edwards et al., 2008). Some ADAMs lose their metalloenzyme domain during intracellular maturation and some lack the necessary Zn-binding (HEXXHXXGXXH) sequences present in their catalytic domain to be proteolytically active (ADAM-1–7, 22, 23, 29, 31, 32). These proteolytically inactive ADAMs participate in intercellular communication through their adhesive properties rather than by activating cell surface molecules through proteolytic cleavage or “shedding”.
Some ADAM enzymes can be expressed in somatic cells and some are present primarily in the testis, which suggests tissue-specific function (reviewed in detail in (Edwards et al., 2008)). The importance of ADAMs is underscored by the fact that orthologs of some of the most vital ADAM genes (ADAM-10 and -17) can be found in yeast Schizosaccharomyces pombre (Nakamura et al., 2004), in Drosophila melanogaster, and in primitive chordates such as Ciona intestinalis and the zebrafish Danio rerio (Huxley-Jones et al., 2007).
Our review focuses on ADAM-17—one of the most well studied ADAM enzymes. However, there are several excellent recent reviews available which discuss the importance and roles of other ADAMs (Reiss and Saftig, 2009, Edwards et al., 2008, Charrier-Hisamuddin et al., 2008, Higashiyama and Nanba, 2005, Huovila et al., 2005, Blobel, 2005).
ADAM-17 makes history
ADAM-17 was discovered in 1997 and described simultaneously by two research groups as the enzyme that releases membrane bound tumor necrosis factor (TNF)-α precursor to a soluble form (Black et al., 1997, Moss et al., 1997b). This discovery was significant because TNFα is critical in inflammatory processes (Feldmann et al., 1995), and there had been a lengthy search to identify the enzyme responsible for solubilizing this cytokine. Investigators hypothesized that the sheddase enzyme that cleaves cell-surface-bound pro-TNFα. was a metalloenzyme because hydroxamate-type inhibitors attenuated surface-bound pro-TNF-α release from cells (McGeehan et al., 1994, Mohler et al., 1994, Mohler et al., 1993). However, the discovery that this cytokine-releasing enzyme belongs to the adamalysins provided the first evidence for physiological catalytic activity of ADAMs (Black et al., 1997, Moss et al., 1997b) as previously ADAMs were merely regarded as adhesion molecules.
ADAM-17 (synonyms: CD156b; cSVP; MGC71942; TACE) was described as a protein of 824 amino acids (accession number NM_003183), and its gene is located on chromosome 2p25. ADAM-17 is widely expressed in various tissues including the brain, heart, kidney, and skeletal muscle and its expression changes during embryonic development and adult life (Black et al., 1997). ADAM-17 is a multi-domain protein starting with a signal sequence (1–17 aa), followed by a prodomain (18–214 aa), a metalloenzyme or catalytic domain (215–473 aa) with the typical HEXXHXXGXXH (X being any amino acid residue) sequence, a disintegrin domain (474–572 aa), an a cysteine-rich domain (603–671 aa), followed by a transmembrane domain (672–694 aa) and a cytoplasmic tail (695–824 aa). ADAM-17 has very little sequence similarities with other ADAMs (see phylogenic review in (Edwards et al., 2008)); its closest relative is ADAM-10 (NM_001110.2); however, their protein sequence homology is less than 30% according to NCBI Blast.
FUNCTION OF ADAM-17
Protein ectodomain shedding and its functional consequences
The most well-known function of catalytically active ADAMs, including ADAM-17, is to cleave ectodomains of various transmembrane proteins. The proteolysis usually occurs at the membrane-adjacent part of the molecule. Proteins with different function can be processed by ectodomain shedding: EGFR ligands, proinflammatory cytokines like TNFα and its receptor TNFRI, adhesion molecules and the amyloid precursor protein (Black et al., 1997, Garton et al., 2003, Lammich et al., 1999, Moss et al., 1997b, Peschon et al., 1998, Reddy et al., 2000). ADAM-17 has preferences for certain proteins as discussed below but catalytically active ADAMs can have overlapping substrate “specificity” (for lists of known ADAM substrates, see recent review (Edwards et al., 2008)). After cleavage, the molecules can bind to their receptor on the same cell (autocrine effect), or can bind to receptors on neighboring cells. Alternatively, they can reach more distant cells in the same tissue (juxtacrine and paracrine effect) and even enter the bloodstream (endocrine effect) (Wiley et al., 1998, Borrell-Pages et al., 2003). Usually, the Golgi apparatus provides a reserve for the shedded ligand, and ligand shedding can mobilize ligand movement to the membrane (Wang et al., 2003).
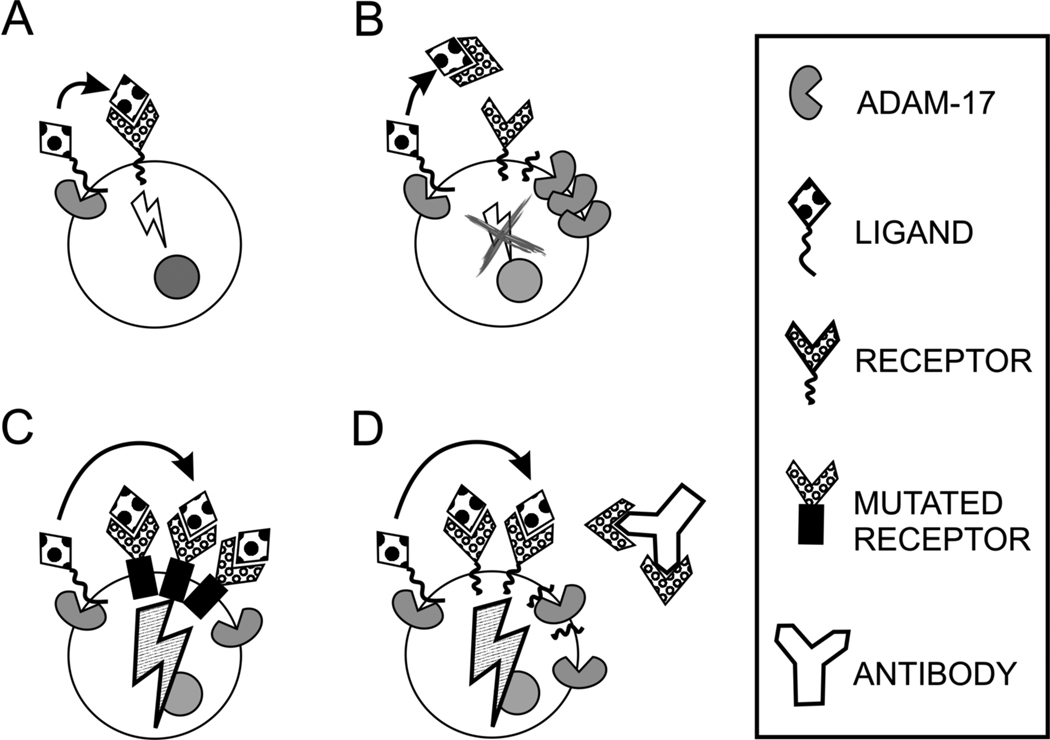
Protein cleavage can regulate cellular signaling and affect cell behavior, but the result always depends on the cellular context. Because both substrate and receptor can be cleaved, several scenarios are possible. The cleaved substrate can bind to its receptor and then the activated receptor initiates downstream signaling events: ADAM-17 can cleave HB-EGF which, in turn, activates EGFR and initiates cell proliferation (Gooz et al., 2006) (Fig. 2A). However, the receptor can be cleaved from the cell surface; thus, ectodomain shedding can actually stop the ligand-initiated signaling: ADAM-17-induced macrophage colony-stimulating factor receptor cleavage downregulates activated macrophages (Rovida et al., 2001) (Fig. 2B). Failure in receptor downregulation and normal turnover can have serious consequences as observed in TNF-receptor associated periodic febrile syndrome (TRAPS) (McDermott et al., 1999). A mutation in the cleavage site of the TNF receptor I can lead to receptor accumulation on the cell surface due to inefficient shedding. Receptor abundance increases cell susceptibility to TNFα and TRAPS patients have increased inflammatory responses with concomitant fever (Fig. 2C). If the receptor shedding occurs before the ligand binding, the receptor can serve as a decoy and inhibit the ligand binding to cell surface receptors. For example, breast cancer cells that overexpress Her-2 escaped anti-Her-2 antibody-mediated inhibition of cell proliferation by shedding large amounts of soluble Her-2, which captured the antibody prior to it reaching cells (Brodowicz et al., 1997) (Fig. 2D).
Fig 2.
Interestingly, activation of membrane-bound receptors (for example the EGFR) by EGFR ligand binding can result in either cell proliferation or apoptosis depending on which other signaling pathways are activated or “connected” to the host-receptors pathway (Jackson et al., 2003, Blobel, 2005). Therefore, inhibiting or knocking down a receptor which activates inhibitory signaling pathways can actually activate the previously inhibited processes. An example of this mechanism is the contribution of EGFR activation to normal heart development via the Smad/BMP signaling pathway. EGFR activates Smad signaling, thereby inhibiting hyperproliferation of cells in developing heart valves (Kretzschmar et al., 1997). Also, Smad6−/− mice were confirmed to have enlarged heart valves (Galvin et al., 2000) and that EGFR knockout mice and HB-EGF−/− mice develop hyperplasic heart valves, too (Chen et al., 2000, Jackson et al., 2003). Insufficient or inactive EGFR during heart valve development therefore hinders activation of an inhibitory mechanism (Smad) and leads to activation of a proliferative pathway.
Regulated intramembrane proteolysis (RIP)
What then happens with the cleaved intramembrane/cytoplasmic part of the proteins is presently unclear. However, the transmembrane module of certain proteins has been shown to be further processed by regulated intramembrane proteolysis or RIP (Schroeter et al., 1998, De Strooper et al., 1999, Wolfe et al., 1999). Signaling through Notch, a protein involved in cell-fate decisions, involves cleavage of the Notch receptor first on its ectodomain by ADAM-10 or ADAM-17 at a so-called α-position (Brou et al., 2000), which is followed by a second intramembrane cleavage by multi-protein protease complex γ-secretase, which includes enzymes like presenilin (Six et al., 2003, Hartmann et al., 2001). The intracellular domain of Notch is transferred to the nucleus where it regulates transcription of various genes (Lewis, 1998). Another example is the processing of the β-amyloid precursor protein (APP), which is responsible for amyloid plaque formation in Alzheimer’s disease (Masters et al., 1985). APP is cleaved first by ADAM-17 or ADAM-10 and, similar to Notch, the remaining molecule is processed by presenilin (Buxbaum et al., 1998). The intracellular fragment induces intracellular Ca2+ signaling (Leissring et al., 2002). This process results in a soluble extracellular fragment that does not contribute to amyloid plaque formation (Anderson et al., 1992, Kojro and Fahrenholz, 2005). In contrast, decreased activity or lack of α-secretases allows β-secretase to cleave the extracellular module of APP at a more distal region which will result in formation of the amyloidogenic β peptide (Yan et al., 1999). Furthermore, recent investigations provided evidence that after ectodomain shedding the EGFR ligands HB-EGF and neuregulin are proteolytically cleaved on their C-terminal domain and signal to the nucleus (Nanba et al., 2003, Bao et al., 2003). Therefore, other than soluble factor-induced events, backwards signaling could provide cellular feedback and could mobilize transport of additional factors to the cell surface. The consequences of this “backwards signaling” require further investigations. Also, it would be interesting to know whether all transmembrane proteins for which the ectodomain is processed undergo transmembrane proteolysis.
Inter-receptor crosstalk
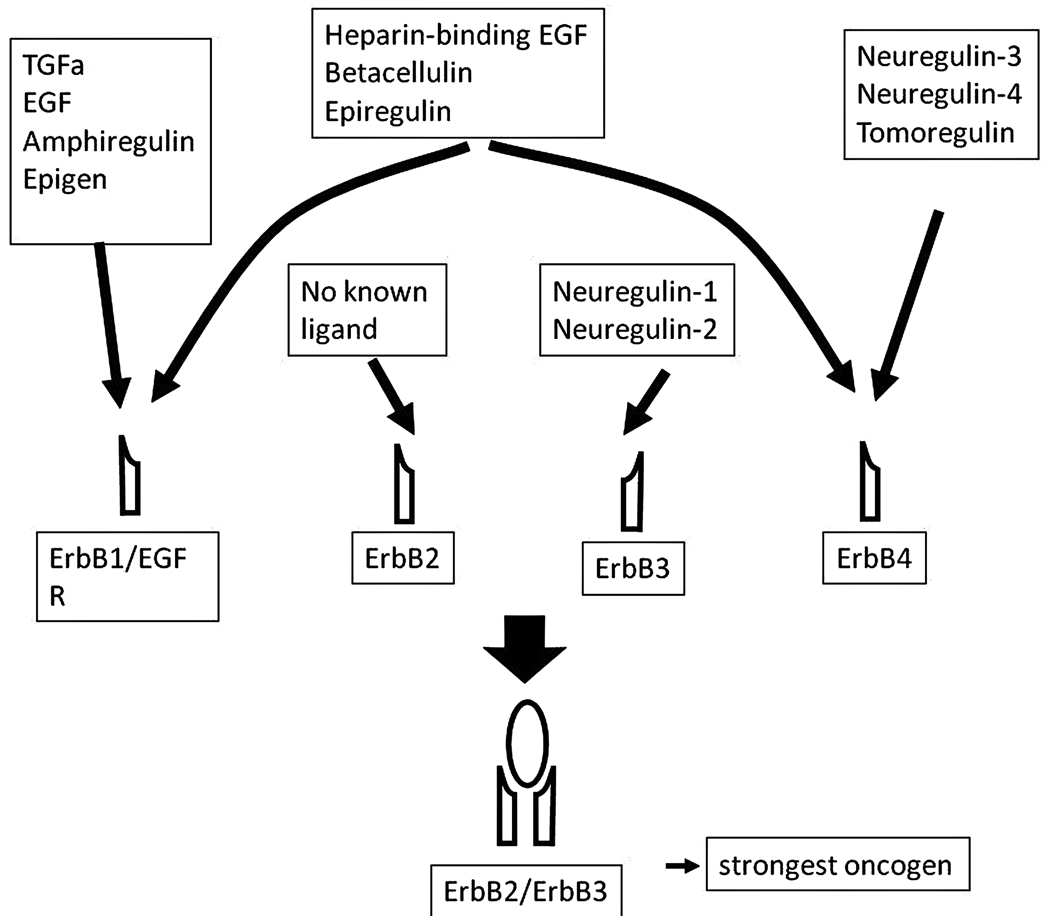
Ectodomain shedding is an important element of communication among different types of cell surface receptors. Crosstalk between G protein-coupled receptors (GPCRs) and receptor tyrosine kinases (RTKs) was first reported in 1999 (Prenzel et al., 1999). The mechanism which involved GPCR-induced activation of a matrix metalloenzyme and subsequent shedding of the EGFR ligand heparin-binding EGF (HB-EGF) was the archetype of “triple-membrane spanning signaling”. Prenzel’s showed that activation of several distinct GPCRs can induce phosphorylation of the EGFR through the same ligand: HB-EGF. Since then, several other EGFR ligands have been identified in inter-receptor crosstalk including EGF, amphiregulin, neuregulin 1–4, epiregulin, epigen, TGFα, betacellulin. ADAM-10, -12, -15 and -17 were documented as growth factor shedding enzymes (for recent review see (Edwards et al., 2008). GPCR agonists implicated in inter-receptor crosstalk that utilizes ADAM-17 include angiotensin-II (Lautrette et al., 2005), ATP (Yin and Yu, 2009), lysophosphatidic acid (LPA) and thrombin (Gschwind et al., 2003), membrane type bile acid receptor (M-BAR)/TGR5 (Yasuda et al., 2007), endothelin-1 (Sanderson et al., 2006), carbachol (Alfa Cisse et al., 2007), and serotonin (Gooz et al., 2006). It is important to note that the same GPCR agonists can induce cleavage of various substrates depending on the cell type (and probably the cellular context). Thus, angiotensin-II is implicated in ADAM-17-mediated TGFα shedding of kidney cells (Lautrette et al., 2005), in HB-EGF shedding of ACHN kidney carcinoma cells (Schafer et al., 2004) and in HB-EGF shedding of vascular smooth muscle cells (Ohtsu et al., 2006). In contrast, the same ligand can be involved in different pathophysiological processes mediated by ADAM-17. Thus, the enzyme can shed amphiregulin both in lung (Lemjabbar et al., 2003), and in colon cancer cells (Merchant et al., 2005). Another regulatory checkpoint of the GPCR-RTK crosstalk which can add variability to the signaling process is that, upon activation, EGFR monomers can form not only homodimers but also heterodimers. This process involves engagement of other ErbB family members like the ErbB4 receptor, which can add additional variability to the outcome of the signaling process as heterodimers have different substrate preferences than the EGFR homodimer (Murali et al., 1996, Citri et al., 2003, Li et al., 2007b) (Fig. 3).
Fig 3.
REGULATION OF ADAM-17
ADAM-17 domains and their role in regulating ADAM-17 enzymatic activity
The prodomain of ADAM-17 serves as the initial inhibitor of the enzyme during its translation (Gonzales et al., 2004, Li et al., 2009). Interestingly, there is evidence that its function is enhanced by the presence of the disintegrin domain of the enzyme (Gonzales et al., 2004). The prodomain of ADAM-17 is cleaved by furin, a pro-protein convertase, in the trans-Golgi network (Schlondorff et al., 2000) at the last 4 amino acids (RVKR) preceding the catalytic domain. Interestingly, in contrast to other MMPs, the cysteine switch mechanism—the binding of Cys184 to the Zn2+ ion in the catalytic domain’s active center—does not appear to be the mechanism of auto-inhibition in the case of ADAM-17. Prodomain variants containing a Cys184A mutation are still capable of inhibiting the enzyme activity (Leonard et al., 2005). Recently, the key inhibitory sequence was identified as Phe72 and the Asp-Asp-Val-Ile137 motif (Gonzales et al., 2008). In contrast, others have described a 19-amino acid, leucine-rich domain, corresponding to ADAM-17 amino acids 30–48 (Buckley et al., 2005) which inhibited ADAM-17-mediated TNF receptor 2 (TNFR2) cleavage independent of the cysteine switch mechanism. Cleaved pro-domains can form a complex with the mature form of the ADAM enzyme in some instances (Fahrenholz et al., 2000, Wewer et al., 2006, Moss et al., 2007); however, for ADAM-17, we have no reports concerning the possible interaction between mature ADAM-17 and its processed prodomain. Immunohistochemical studies suggest that most of the active form of ADAM-17 is localized in the cellular perinuclear region, with a small amount present in the plasma membrane (Schlondorff et al., 2000). It was also shown that removal of the prodomain is not necessary for ADAM-17 transport to the cell surface, as during pro-protein convertase inhibition immature form of ADAM-17 was present in the plasma membrane (Peiretti et al., 2003a). There is evidence that ADAM-17 is “packaged” into lipid rafts during its transport and maturation through the Golgi apparatus, and that this spatial distribution also contributes to ADAM-17 activity regulation by keeping the enzyme separated from its substrates (Tellier et al., 2006).
The catalytic domain or metalloenzyme domain of ADAM-17 contains the classical Zn2+ chelating sequence HEXXHXXGXXH (X being any amino acid residue) and is responsible for processing membrane bound proteins such as TNFα. ADAM-17 and its closest relative, ADAM-10, have overlapping substrate specificity as well as substrates unique to each. A recent investigation underscored the differences in substrate recognition of these enzymes using peptide library screening and systematic analysis of amino acids next to the cleavage site, and these experiments showed that ADAM-17 prefers smaller aliphatic residues at the P’1 position and that ADAM-10 prefers larger residues such as leucine (Caescu et al., 2009). The active center of the enzymes were also compared in mutational studies, and the residues responsible for differences in substrate recognition were found to be present in the S’1 pocket of the enzymes. Also, Val440 was reported to be sufficient and necessary for ADAM-17 substrate specificity (Caescu et al., 2009). The classical cleavage sequence in TNFα was identified as Ala 76/ Val 77 (-Arg-Ser-Ser-Ser) (Kriegler et al., 1988). Analysis of growth hormone receptor (GHR) proteolysis by ADAM-17 revealed that the cleavage site is eight residues from the cell membrane in the extracellular domain of GHR (Wang et al., 2002). It was suggested that the sequence of the substrate cleavage site is less important than its position; the so called “stalk region” between the transmembrane, and the first globular part of the molecule determines whether ADAM-17 can cleave the protein (Schlondorff and Blobel, 1999, Wang et al., 2002, Hinkle et al., 2004). However, a recent thorough analysis of an ADAM-17 peptide substrate library showed that the substrate sequence is indeed important (Lambert et al., 2005).
Tissue inhibitors of metalloproteinases-3 (TIMP-3), a native inhibitor of ADAM-17, binds to the ADAM-17 catalytic domain (Amour et al., 1998). However, the effectiveness of this inhibition is greatly influenced by the C-terminal modules of ADAM-17 (disintegrin, and cysteine-rich domains), which significantly weaken the inhibitory activity of TIMP-3 (Lee et al., 2002). Studies of the crystal structure of the TIMP-3/ADAM-17 catalytic domain complex revealed that during interaction, TIMP-3 extends into a unique hydrophobic pocket of the ADAM-17 surface, which together with two leucine residues, allows effective and unique binding of TIMP-3 to the enzyme (Wisniewska et al., 2008). Mutational analyses (M435I, C225Y, C600Y) identified amino acids indispensible for the catalytic activity of ADAM-17 both in the metalloproteinase domain and also in the disintegrin domain (Li and Fan, 2004). The importance of methionine 435 (the so-called met-turn, which gives the metzincin enzyme family its name) in the active site of ADAM-17 was further investigated using M435I, M435L, and M435S mutational constructs (Perez et al., 2007). In contrast to another metzincin enzyme MMP-2, M453 seems to be necessary for ADAM-17 cleavage of transmembrane growth factors (Perez et al., 2007). The important role of distal ADAM-17 modules in regulating the catalytic domain activity was further confirmed by elegant stopped-flow x-ray spectroscopy methods and transient kinetic analyses which showed that the Zn2+ ion in the catalytic domain undergoes charge transitions before the substrate binds to the metal ion (Solomon et al., 2007). This phenomenon was possibly due to interaction of the substrate with ADAM-17 modules other than the catalytic domain, which underwent conformational changes, triggering change in the catalytic domain. The redox state of two highly conserved cysteinyl sulfhydryl groups (cystein-X-X-cystein) in the disintegrin domain of ADAM-17 also have important roles in regulating ADAM-17 activity as evidenced by their role in L-selectin secretion in macrophages (Wang et al., 2009b).
Integrins are heterodimeric (αβ) transmembrane glycoproteins best known as adhesion receptors mediating cell-cell and cell-matrix interactions (for review see (Giancotti and Ruoslahti, 1999)). At present, there are 18 α and 8 β integrin subunits identified, which can assemble in 24 different ways. The subunit assembly determines the integrin’s substrate specificity; although, several integrins can bind to the same type of matrix molecules (Giancotti and Ruoslahti, 1999). Integrins are involved in other cellular processes than adhesion: they participate in outside-in and inside out signaling, cell cytoskeleton re-organization, cell adhesion, migration, cell survival, and proliferation (Giancotti and Ruoslahti, 1999, Huveneers et al., 2007). The disintegrin domain of ADAM-17, like other ADAMs, was always regarded as an integrin-binding module, based on its similarity to snake venom disintegrins which are known for competitively inhibit integrin function. Disintegrins are small molecules (20–100 amino acid) which originate from Class II–IV snake hemorrhagic metalloproteinases (SVMP) by proteolytic or autoproteolytic cleavage (Kini and Evans, 1992). Class I SVMPs have only MMP domains, and consequently have the weakest anticoagulant activity among the snake venom metalloproteinases. Disintegrins were first discovered in viper venom based on their ability to inhibit platelet aggregation by interfering with platelet integrin αIIbβ3 receptor binding to fibrinogen (Huang et al., 1987). Later, snake venom disintegrins were actually found to interact with a wide range of integrins (α2β1, α4β1, α4β7, α9β1, αvβ3, αvβ5 and α5β1) (Zhou et al., 2000, Cominetti et al., 2003, Tselepis et al., 1997, Danen et al., 1998, Marcinkiewicz et al., 1999), and to thereby affect the cellular function of not only platelets but also endothelial cells (Olfa et al., 2005), rat mesangial cells (Tsai et al., 1995), fibroblasts (Yeh et al., 2001), osteoclasts (Mercer et al., 1998), and tumor cells such as human melanoma cells (Trikha et al., 1994), lung tumor cells (Galan et al., 2008), and gliomas (Schmitmeier et al., 2003). The integrin-binding affinity of snake venom disintegrins greatly depends on their active-loop sequence for which position is defined by the specific alignment of cysteines (reviewed in detail in (Calvete et al., 2003)). Most of the Class II SVMPs belong to the RGD-containing disintegrins and these, together with “modified RGD”-containing disintegrins (KDG, MVD, MGD, KGD, WGD), are under intense investigation for therapeutic use in circulation diseases, rheumatoid arthritis, cancer treatment, osteoporosis, and Alzheimer’s disease by targeting not only αIIbβ3 and αvβ3, but also the fibronectin receptor α5β1 integrin (Mercer et al., 1998, Chung et al., 2003). The other group of disintegrins includes members of the non-RGD containing or MLD-, and KTS disintegrins which bind mainly to α4β1, α4β7, α9β1 integrins and α1β1 integrin, respectively (reviewed in (Marcinkiewicz, 2005, Calvete, 2005)). These disintegrins are a recent discovery, so further studies are warranted to understand their function.
The disintegrin or disintegrin-like domain of ADAMs resembles the class III SVMPs. However, only ADAM-15 has an RGD sequence in its substrate recognition loop. ADAM-9 has a KTS sequence and many ADAMs have the XCD (usually ECD) motif in their loop sequence. Overall, ADAM disintegrin domains are much larger and more degenerate than snake venom disintegrins (Bridges and Bowditch, 2005). ADAM-17 has an ECD sequence, but it has a tyrosine (Y) in the 5th position whereas all other ADAMs have a cysteine residue. Nonetheless, there is evidence that ADAMs do interact with integrins even if the exact binding loops have not been well characterized. For example ADAM-15 was shown to interact with both αvβ3 and α5β1 (Nath et al., 1999), and ADAM-2, 3, 7, 12, 28 and 33 are capable of both α4β1 and α9β1 binding (reviewed in (Arribas et al., 2006)). However, at this time, the only integrin which was shown to associate with ADAM-17 is integrin α5β1 (Bax et al., 2004) and there is evidence that ADAM-17 inhibits α5β1 integrin-mediated cell migration. It was also shown that ADAM-17 does not influence α4β1 integrin-mediated cell migration, possibly because it cannot bind to this integrin (Huang et al., 2005).
It is important to emphasize that, to date, ADAM/integrin interactions have been studied mostly in the context of cell-cell interactions and cell-matrix interactions or adhesion (Kawaguchi et al., 2003, Eto et al., 2000) and very often with recombinant disintegrin domains “glued” to plastic surfaces, which is essentially studying the molecule out of context. Thus, these investigations provide limited understanding about in vivo ADAM/integrin binding. Even if recombinant ADAM-17 was reported to bind purified α5β1 integrin, and the two proteins co-localized in migrating HeLa cells (Bax et al., 2004), the physiological significance of these interactions is unlcear. Most importantly, we have no data to show whether α5β1 integrin binding affects ADAM-17 activity and, consequently, ADAM-17-mediated signaling pathways, and we do not fully understand the meaning of ADAM-integrin interactions. For catalytically inactive ADAMs, disintegrin binding to cell surface integrins can mediate cellular adhesion both of the same cell and of the neighboring cells. Disintegrin-integrin engagement can have other consequences as well: ligation of integrins can initiate important signaling cascades through activation of growth factor receptors, GPCRs, ion channels, and cytokine receptors (Giancotti and Ruoslahti, 1999). Intracellular molecules can cluster at and phosphorylate integrins and influence cell shape and cell behavior, such as migration, growth, and apoptosis. This could be true for the catalytically active ADAMs, too. By binding to integrins on neighboring cells, ADAM enzymes can initiate the above-mentioned signaling cascade and simultaneously influence cell attachment. ADAMs binding to integrins on the same cell can influence host cell attachment and initiate signaling cascades. In contrast, disintegrin binding to integrin can signal “backwards” and may influence ADAM activity as disintegrin domains have regulatory role on the catalytic module (Li and Fan, 2004, Smith et al., 2002). Thus integrin binding could modulate disintegrin-catalytic domain interaction. Theoretically, integrin-ADAM binding can have both inhibitory and stimulatory effect on ADAM catalytic activity through inducing conformational change or by regulating spatial, intramembrane distribution of the enzyme and its substrate. Through conformational change, integrins can sequester ADAM, so it cannot access its substrate. Also, integrins can bind ADAM substrate as was shown for HB-EGF and α3β1 integrin (Nakamura et al., 1995) and could make it available for the enzyme on the same cell or on neighboring cells, thus promoting ADAM enzyme function. Interestingly, some evidence suggests that integrins can use different activation states when binding to ADAMs versus extracellular matrix molecules (Chen et al., 1999).
The cytoplasmic domain of ADAM-17 is a proline rich, short, 130 amino acid module. It contains a potential tyrosine phosphorylation site, DKKLDKQYESL, promixal to the transmembrane domain and a Src-homology 3 domain (SH3) binding site (731–738) (UniProtKB/SwissProt), PAPQTPGR (Black et al., 1997). The cytoplasmic domain was suggested to have important signaling function and its potential role in ADAM-17 trafficking was proposed (Soond et al., 2005). However, our data on ADAM-17 trafficking are sparse. Overexpression studies using truncated forms of ADAM-17 showed that the enzyme must be expressed with its transmembrane domain to effectively cleave TNFα when the cells are stimulated with phorbol ester (Reddy et al., 2000). There is evidence that the cytoplasmic domain was not necessary for phorbol ester-induced ADAM-17 shedding activity (Doedens et al., 2003, Reddy et al., 2000). Phorbol esters are strong inducers of metalloenzyme activity and are useful tools for studying ADAM activity changes. But the question still remains whether more physiological stimuli would require the cytoplasmic module for a complete cellular response. It was shown recently that ADAM-10 undergoes regulated intramembrane proteolysis by presenilin after its ectodomain was shed by ADAM-9 or -15 (Tousseyn et al., 2009). The cytoplasmic domain of ADAM-10 then translocates to the nucleus to gene loci undergoing transcription. Because ADAM-10 is the closest relative of ADAM-17, there is a high possibility (and it is tempting to speculate) that the cytoplasmic domain of ADAM-17 itself can undergo RIP and participate in gene transcription regulation.
Post-translational modification of the enzyme
One of the most important post-translational modifications of ADAM-17 is its pro-domain removal in the trans-Golgi network as described above, a process which results in the mature form of the enzyme. Other important post-translational modifications include glycosylation and phosphorylation of the enzyme. There are six potential N-linked glycosylation sites on the mature form of ADAM-17: three are located on the metalloenzyme modul, two are located on the disintegrin modul, and one is present on the cysteine-rich domain. There are three further potential N-glycosylation sites on the prodomain which suggests that glycosylation can be an important regulator of ADAM-1 7 activity. Indeed, ADAM-17 has been shown to be N-glycosylated (Peiretti et al., 2003a) and it is usually referred to as the 130 kDa form of the enzyme. However, we have no data to show how glycosylation of ADAM-17 affects the activity and/or transport of the enzyme.
Another important post-translational modification of the enzyme is its phosphorylation. The cytoplasmic domain, especially phosphorylation of its serine and theronine residues was studied extensively. However, the data so far are very contradictory. Phorbol ester (PMA) and EGF treatments induced ADAM-17 phosphorylation on threonine 735 by mitogen activated protein kinase ERK in transfected CHO cells and HEK293 cells with concomitant increases in cleavage of the transmembrane TrkA neurotrophin receptor (Diaz-Rodriguez et al., 2002). In contrast, others showed that PMA and EGF induced not threonine (or tyrosine) but serine 819 phosphorylation of ADAM-17 by ERK (Fan et al., 2003). The authors also showed that ADAM-17 undergoes Ser 791 dephosphorylation during growth factor stimulation. However, there are few data regarding non-PMA-mediated phosphorylation of ADAM-17. Current, Zhang and colleagues have shown that the GPCR agonist, gastrin releasing peptide, induces activation of Src-PI3K (phosphoinositid 3-kinase)-PDK1 (phosphoinositide-dependent kinase) in cancer cells (Zhang et al., 2006). They identified PDK1 as the enzyme that directly phosphorylates ADAM-17 both on threonine and serine residues. However, the exact phosphorylation sites of or the role of the cytoplasmic tail were not studied. Another GPCR agonist carbachol induced threonine 735 and tyrosine 702 phosphorylation of ADAM-17 in HEK 293 cells acting on muscarinic M1 and M3 receptors. Wounding, ATP- or LPA-treatment of corneal epithelial cells induced ERK-dependent ADAM-17 serine phosphorylation through GPCR-activation (Yin and Yu, 2009). The contradictory data presented by these papers can be due to the fact that investigators used artificial systems: transformed or transfected cells with overexpressed ADAM-17 and mainly PMA to study ADAM-17 phosphorylation. But it is also possible that ADAM-17 gets phosphorylated on different residues depending on the cell type and stimulus. To better understand this phenomenon, more investigation is needed to study ADAM-17 post-translational modification in primary, untransfected cells and using more physiologically relevant stimuli instead of PMA.
ADAM-17 binding partners
Besides TIMP-3, which is the naturally occurring inhibitor of ADAM-17 binding to the enzyme active center and integrin α5β1 which binds to the disintegrin/cystein rich domain (Bax et al., 2004), various substrates of the enzyme (discussed below), and several other molecules were identified as binding partners and potential regulatory proteins by yeast two-hybrid screening and co-immunoprecipitation studies. Mitotic arrest deficient 2 (MAD 2) was shown to interact with ADAM-17 cytoplasmic domain (Nelson et al., 1999). The PDZ-domain (Post synaptic density protein [PSD95]/Drosophila disc large tumor suppressor (DlgA), and Zonula occludens-1 protein [zo-1]) of protein tyrosine phosphatase-1H was shown to bind also to ADAM-17 cytoplasmic domain (Zheng et al., 2002). Yeast two-hybrid assay and co-localization studies showed that SAP9 (synapse associated protein-9) interacts through its PDZ3 domain with the extreme C-terminal of ADAM-17, and overexpression of SAP9 inhibited substrate cleavage (TNFRI, TNFR2 and TNFα) of the enzyme (Peiretti et al., 2003b). Interestingly, others showed that SAP9 can interact with the cytoplasmic domain of both ADAM-17 and its ligand TGFα, and in this way regulate substrate availability of the enzyme (Surena et al., 2009). However, this mechanism would suggest that overexpression of SAP9 could actually enhance and not inhibit the effect of ADAM-17. Another study identified a so far unknown protein with SH3-domains as a novel ADAM binding partner using a yeast two-hybrid screening assay. The protein, which they called Eve-1, co-precipitated with ADAM-17 and several other ADAMs in cell lysates (Tanaka et al., 2004). The down-regulation of Eve-1 inhibited shedding of several EGF receptor ligands. ADAM-17 was also shown to interact with the Four and Half LIM domain 2 protein (FHL2) between the amino acid sequences 721–739. This protein provides a bridge between the actin cytoskeleton and the enzyme (Canault et al., 2006b). Nardilysin (N-arginine dibasic convertase), a metalloendopeptidase of the M16 family was shown to bind both HB-EGF and ADAM-17, and enhance ADAM-17 activity during phorbol ester stimulation (Nishi et al., 2006). The exact site of the interaction was not studied in this case. In another study, the cytoplasmic tail of ADAM-17 together with the cytoplasmic tail of ACE-2 (angiotensin converting enzyme-2) was shown to be necessary for severe acute respiratory syndrome (SARS) coronavirus entry into host cells (Haga et al., 2008). The number of identified binding partners is increasing, but there is much to learn about the exact mechanisms and consequences of these interactions on ADAM-17 activity. Also, we have no data regarding how the binding partners’ binding is regulated. For example, there is currently no evidence in the literature about whether phosphorylation of ADAM-17 induces changes in the binding of ADAM-17 to regulatory molecules.
Role of trafficking in ADAM-17 activity regulation
Besides post-translational modification and the regulatory role of binding partners, trafficking is the process that can also determine ADAM-17 sheddase activity. Recent data showed that after and during furin cleavage, active ADAM-17 is localized separately from its substrate in cholesterol rich membrane domains, lipid rafts, which can thus spatially restrict its activity (Tellier et al., 2006). Furthermore, there is evidence that ERK-dependent threonine 735 phosphorylation is necessary for ADAM-17 to reach the secretory pathway during maturation (Soond et al., 2005). Doedens’ group showed that PMA but not GPCR stimulation decreased ADAM-17 cell-surface immunostaining. In this paper, the authors showed internalization and degradation of ADAM-17 molecule (Doedens and Black, 2000). In contrast, another group found that PMA stimulation did not cause ADAM-17 redistribution in the cells; however, the treatment decreased total cellular ADAM-17 content (Diaz-Rodriguez et al., 2002). Despite these observations, we lack a mechanistic understanding about how ADAM-17 trafficking is regulated. It would be interesting to see whether binding partners affect ADAM-17 transport. Furthermore, because most of the studies on ADAM-17 fate were carried out in transfected and PMA stimulated cells, we do not know how a physiological stimulus like GPCR activation affects ADAM-17 trafficking. As it was pointed out by a group who analyzed ADAM-17 trafficking, transfected ADAM-17 is processed differently than its endogenous counterpart, and is perhaps not the best model for studying the fate of the endogenously expressed protein (Borroto et al., 2003).
Structural studies on ADAM-17: how to best target the enzyme activity
In 1998 Maskos et al. published the crystal structure of the ADAM-17 catalytic or metalloenzyme domain (Maskos et al., 1998). This opened up the possibility of designing ADAM-17 inhibitors based on structural similarities to TNFα binding. At the same time, there was intense work to identify and develop selective ADAM-17 inhibitors (reviewed in (Newton and Decicco, 1999) either by changing the structure of broad spectrum MMP inhibitors or by designing new compounds (Xue et al., 2001, Rabinowitz et al., 2001, Duan et al., 2002). Improvement of the structure (Ingram et al., 2006) and description of the crystal structure of the ADAM-17 catalytic domain in complex with its naturally occurring inhibitor tissue inhibitor of metalloenzymes-3 (TIMP-3) (Wisniewska et al., 2008) was useful for the recent development of several highly selective, potent ADAM-17 inhibitors targeted against the active center (Gilmore et al., 2006, Gilmore et al., 2007, Huang et al., 2007, Venkatesan et al., 2004). Orally active, selective ADAM inhibitors were developed and tested in combination with ErbB2 receptor antibodies for the treatment of breast cancer (Fridman et al., 2007). The newest inhibitors were promising in that they had no musculoskeletal side effects or liver toxicity often observed with previously developed MMP and ADAM inhibitors (for detailed description of structure and effectiveness of ADAM-17 inhibitors, and discussion of clinical trials see (Moss et al., 2008, DasGupta et al., 2009). It is noteworthy that the inhibitors presently developed target only the enzyme active center and other molecular areas were not explored. Because ADAM-17 is a promiscuous enzyme, we must discover how it is regulated and what proteins it binds to in certain cells to develop potential combinational therapy.
ADAM-17 IN DEVELOPMENT
What we learned from knockout studies
The first data on the role of ADAM-17 in development were generated by Peshon’s laboratory (Peschon et al., 1998) who found that ADAM-17 knockout mice are not viable and have severe epithelial abnormalities. After creating catalytic domain inactive transgenic animals which lacked the Zn2+−binding site of the metalloenzyme domain (ADAM-17 ΔZn/ΔZn) they showed that lack of ADAM-17 activity leads to perinatal lethality (between embryonic day 17.5 and birth), likely caused by defects in the placenta and in the fetal lung. Dysregulation of epithelial development lead to defects in development of skin, hair, and cornea, and several organs (thyroid, parathyroid, stomach and intestine). Recent investigations also showed that ADAM-17 is important in energy homeostasis, possibly increasing sympathetic outflow; mice with catalytically inactive ADAM-17 have hypermetabolism without increased appetite which renders the animals extremely lean (Gelling et al., 2008).
The pulmonary hypoplasia seen in ADAM-17 knockout animals were later investigated by Zhao’s group (Zhao et al., 2001) who found that the lungs in ADAM-17-null animals at embryonic day 16.5 had impaired branching morphogenesis and delays in vascularization. Epithelial cell proliferation and differentiation was also delayed as evidenced by repressed gene expression of surfactant-C and aquaporin-5. Newborn ADAM-17-null mice had thicker mesenchyme and fewer peripheral epithelial sacs. Shi and colleagues investigated cardiac development of ADAM-17 ΔZn/ΔZn animals and described enlarged heart (atria and ventricles) at embryonic age 18.5 with hypertrophic, loosely packed cardiac myocytes (Shi et al., 2003). Later Jackson’s group reported that newborn ADAM-17 knockout animals have enlarged heart valves similar to HB-EGF-null (Jackson et al., 2003) and EGFR-null mice (Miettinen et al., 1995, Sibilia and Wagner, 1995, Threadgill et al., 1995), and showed that lack of these proteins separately lead to loss of inhibition on bone morphogenic protein (BMP) signaling which is responsible for abnormal valve thickening (Jackson et al., 2003). Similarities in the phenotype of ADAM-17, EGFR, TGFα (Luetteke et al., 1993, Mann et al., 1993), HB-EGF and amphiregulin knockout mice (Luetteke et al., 1999) is discussed in a recent review (Blobel, 2005).
The role of ADAM-17 sheddase activity was also studied in bone development and in animals with catalytically inactive ADAM-17, osteoclasts do not move to the diaphysis of metatarsals and therefore do not contribute to normal marrow cavity formation of long bones (Boissy et al., 2003). It seems that a factor (or factors) which are only processed by ADAM-17 is necessary for normal bone marrow cavity formation, but further investigations are needed to identify these molecules. To study the role of ADAM-17 in hematopoiesis, normal mice were irradiated and transplanted with bone marrow of mice with inactive ADAM-17 (Li et al., 2007a). Li and co-workers reported that the animals had partially blocked T cell development, and they noted that ADAM-17 is also necessary for peripheral B cell development. The first data with ADAM-17 conditional knockout mouse were published in 2007 by Blobel’s group, who inactivated ADAM-17 in myeloid cells and prevented endotoxin-induced lethality of the animals by inhibiting TNFα shedding and preventing increased serum TNFα (Horiuchi et al., 2007). In another study, to characterize the role of ADAM-17 in non-hemopoietic cells, ADAM-17 was conditionally inactivated in mice using Cre recombinase gene expression under the control of the Sox9 transcription factor’s promoter, which participates in skeletal development. This maneuver resulted in shortened bones with an osteoporosis-like phenotype characterized by prominent bone loss, and bone marrow hypercellularity and extramedullary hematopoiesis in the liver and spleen (Horiuchi et al., 2009a). These data suggest that conditional knockout animals are useful models for studying cell and tissue-specific roles of ADAM-17 during development and in adult animals.
ADAM-17 IN HUMAN DISEASES
ADAM-17 in inflammation
Because ADAM-17 was first identified as the TNF-α cleaving enzyme, it was expected that ADAM-17 enzymatic activity would be increased in diseases with increased circulating or tissue TNFα such as rheumatoid arthritis and inflammatory bowel disease (Feldmann et al., 1996, Papadakis and Targan, 2000). Indeed, increased TNFα converting enzyme was found in whole cartilage (Patel et al., 1998) and chondrocytes of patients with osteoarthritis (Amin, 1999). Elevated ADAM-17 activity was also reported in synovial tissue of rheumatoid arthritis patients (Ohta et al., 2001). In an experimental model of rheumatoid arthritis, low oxygen and TNFα treatment induced ADAM-17 mRNA expression in synovial cells with a concomitant increase in ADAM-17 activity and TNFα shedding rate (Charbonneau et al., 2007). Upregulated ADAM-17 in monocytes of patients with early systemic sclerosis was reported (Bohgaki et al., 2005). However, there is evidence in the literature which suggests that the role of ADAM-17 in inflammation is far beyond its effect on TNFα shedding. Chemoattractants in inflammed tissue attract leukocytes from blood which attach and roll on the vascular endothelium near the inflammation site. This attachment is mediated by the transmembrane protein selectins (Spertini et al., 1992). After adhering to the endothelium, leukocytes migrate through the basal membrane, which is promoted by leukocyte (L)-selectin cleavage (Faveeuw et al., 2001). ADAM-17 was identified as the sheddase cleaving L-selectin (Peschon et al., 1998) from macrophages (Wang et al., 2009b, Condon et al., 2001). It was shown that L-selectin co-clustering and cleavage by ADAM-17 is indeed promoted by leukocyte attachment to endothelial or E-selectins (Schaff et al., 2008). There is additional evidence that ADAM-17 cleaves other molecules than L-selectin which participate in leukocyte activation, such as the CX3CL1/Fractalkine (Garton et al., 2001, Hundhausen et al., 2003). ADAM-17 also serves as a sheddase for V-CAM (or vascular cell adhesion molecule), the ligand of the leukocyte very late antigen 4 (VLA-4 or α4β1 integrin) which mediates leukocyte adhesion to the vascular endothelium (Garton et al., 2003). ADAM-17 also cleaves intercellular adhesion molecule-1 (ICAM-1), which participates in leukocytes adhesion to integrins (Tsakadze et al., 2006). Recently ADAM-17 was implicated in the shedding of the tight junction molecule junctional adhesion molecule-1 (JAM-1) which serves as a zipper between endothelial cells (Koenen et al., 2009). Shedding of JAM-1 can contribute to leukocyte diapedesis through the endothelial layer and the shedded molecule can serve as a biomarker of inflammation. Thus, ADAM-17 was shown to shed several factors contributing to successful recruitment of leukocytes to the inflammation site. Recent studies showed that TIMP-3 knockout animals have increased TNF-dependent systemic immunresponse to both antigen (methylated bovine serum albumin) and LPS-induced inflammation (Mahmoodi et al., 2005, Smookler et al., 2006). Increased degradation of collagen and aggrecan, similar to human osteoarthritis, was also observed in the joints of these animals (Sahebjam et al., 2007). Furthermore, increased soluble VCAM-release was observed from cytokine stimulated TIMP-3 −/− endothelial cells and aortic explants compared to control (Singh et al., 2005). Synthetic metalloenzyme inhibition (Smookler et al., 2006) or ADAM-17 gene silencing (Singh et al., 2005) rescued the phenotype, suggesting that TIMP-3 is important in balancing ADAM-17 activity during the inflammatory response.
Interestingly, there is some evidence for the anti-inflammatory role of ADAM-17. The enzyme was reported to cleave colony stimulating factor-1 (CSF-1) from surface of activated macrophages, thereby downregulating their activation (Rovida et al., 2001). Furthermore, ADAM-17 was identified recently as the major sheddase for the IL-15 receptor α (Budagian et al., 2004), the recombinant soluble form of which was previously shown to inhibit collagen-induced arthritis (Ruchatz et al., 1998) and cardiac allograph rejection. Also, through its role in cleaving IL-6 receptor α ADAM-17 was implicated in IL-6 trans-signaling, a process which contributes to the decline of neutrophil infiltration and promotes monocyte recruitment allowing resolution of the inflammation (Marin et al., 2002). However, these data are somewhat contradictory as elevated levels of IL-6 and IL-6-soluble IL6 receptor complex was found in systemic juvenile rheumatoid arthritis (De Benedetti et al., 1994) and Crohn’s disease (see below), and IL-6 was implicated in the pathogenesis of these autoimmune diseases. These data suggest that further studies are needed to better characterize the net effect of ADAM-17 in various inflammatory processes.
Because many people are affected by arthritis and inflammation-mediated bone and joint disease, ADAM-17 as a potential therapeutic target is under intense investigation. Metalloenzymes and ADAMs as a target of multilevel therapy (molecular polypharmacy) in osteoarthritis has been recently reviewed (Burrage and Brinckerhoff, 2007) and a recent paper summarizes present knowledge and the advancements made in anti-ADAM-17 therapy in inflammatory disease (Moss et al., 2008).
Endotoxic shock, sepsis
In a murine model of sepsis, LPA-induced morbidity was inhibited by anti-TNFα antibody which also inhibited development of arthritis in TNFα transgenic animals (Tsuji et al., 2002). In Neisseria meningitidis or LPS-stimulated mononuclear cells, TNFα rapidly decreased from the cell surfaces after stimulation which supports the role of ADAM-17 in sepsis (Robertshaw and Brennan, 2005). Elevated ADAM-17 was found in blood and peritoneal fluid in patients with peritonitis, and their polymorphonuclear neutrophils had decreased TNFα and L-selectin on their surfaces (Kermarrec et al., 2005). Furthermore, in a recent study, ADAM-17 inactivation in murine myeloid cells prevented endotoxin shock-induced lethality in mice by inhibiting an increase in circulating TNFα, thus confirming the role of ADAM-17 in this disease (Horiuchi et al., 2007).
During endotoxin shock, growth hormone receptor is shedded from liver cells of LPS-treated nude mice, decreasing cell surface receptor availability, desensitizing the cells for growth hormone, and inhibiting the hormones’ anabolic effects (Wang et al., 2008). This mechanism seemed to be ADAM-17–independent: in LPS-treated animals, ADAM-17 was not activated. In another study, investigators showed that selenium treatment was beneficial in endotoxin shock, possibly due to metalloenzyme-dependent L-selectin shedding of monocytes resulting in decreased monocyte rolling (Ahrens et al., 2008). L-selectin cleavage was shown previously to actually promote leukocyte transmigration through the vascular endothelium (see above (Faveeuw et al., 2001). However, it is possible that if L-selectin is cleaved before leukocyte attachment, the cells rolling can be seriously affected which would inhibit their transmigration. Sepsis was also shown to inhibit anticoagulatory pathways by downregulating the endothelial protein C receptor (EPCR) through activation of the NFκB pathway (Song et al., 2009). It was shown previously that shedding of EPCR is mediated by ADAM-17 activation, which suggests that the enzyme can contribute to sepsis-induced coagulation (Qu et al., 2007).
Inflammatory bowel disease
Recently, upregulated ADAM-17 expression in intestinal epithelial cells during the active phase of Crohn’s disease was reported (Cesaro et al., 2009). It was also shown that T cells in chronically inflamed intestines of patients with Crohn’s disease and colitis are resistant to apoptosis because through their g130 protein they bind an IL-6-soluble IL-6 receptor complex (IL-6-sIL6R) even if they do not have transmembrane IL-6 receptors (Atreya et al., 2000). There is evidence that this signaling, which is called IL-6 trans-signaling, is driven by ADAM-17-mediated shedding of the IL-6 receptor (Briso et al., 2008). Interestingly, this is the same mechanism which was reported to initiate “normal” immune response and resolve inflammation by other investigators (Marin et al., 2002).
Psoriasis
The beneficial effect of topical MMP/ADAM inhibitors for the treatment of psoriasis was shown in phorbol ester-induced epidermal hyperplasia, a murine model of the disease (Moriyama et al., 2004). Immunohistological study of psoriatic lesion from patients showed that a wide range of cells including keratinocytes and blood vessels—but mainly inflammatory cells such as mast cells—expressed high levels of ADAM-17 (Kawaguchi et al., 2005). Peripheral blood mononuclear cells had elevated ADAM-17 activity and this correlated positively with both plasma soluble TNF-receptor 1 levels and disease severity, which resolved to almost normal levels after patients were treated with narrowband ultraviolet B light (Serwin et al., 2007). It was shown recently that Jun protein controls TNFα shedding in the epidermis by regulating TIMP-3 expression and thus ADAM-17 activation (Guinea-Viniegra et al., 2009). This mechanism can provide an additional therapeutic target for the treatment of psoriasis.
Pulmonary inflammation
ADAM-17 is expressed in bronchial epithelial cells, vascular smooth muscle cells, and macrophages in the lung (Ermert et al., 2003). Epithelial ADAM-17 was implicated in airway inflammation through cytokine activation during bacterial infection because ADAM-17 co-localized with the IL-6 receptor and initiated its shedding (Gomez et al., 2005). Furthermore, alveolar macrophages produce TNFα, which was shown to activate IL-8 production of lung epithelial cells through amphiregulin and EGFR activation (Chokki et al., 2006). In an animal model of chronic obstructive pulmonary disease, elevated tissue levels of ADAM-17, TNFα, and ErbB3 were observed with increased bronchoalveolar and serum TNF (Ju et al., 2007). ADAM-17 contributed to EGFR-dependent acidic mammalian chitinase secretion of lung epithelial cells which in turn increased chemokine production of the cells (Hartl et al., 2008). Inhibitors of ADAM-17/MMPs were tested in models of airway inflammation (intranasal LPS challenge) and decreased influx of neutrophils and lymphocytes was found (Trifilieff et al., 2002). Similarly, in a model of acute allergic lung inflammation ADAM-17/MMP inhibitors reduced invasion of neutrophils and eosinophils (Trifilieff et al., 2002). Inhibitors of ADAM-17 also inhibited post-transplantation lung injury in animal models (Goto et al., 2004). However, it was shown that inflammatory mediators such as neutrophil elastase upregulated the MUC1 gene in airway epithelia in vitro in an ADAM-17 dependent way and served as negative regulator of airway inflammation (Kuwahara et al., 2007). Furthermore, in TIMP-3 knockout animals, septic lung stress increased lung compliance and severity of disease (Martin et al., 2003) . These data again confirm that ADAM-17 can be both a negative and positive regulator of inflammatory processes.
ADAM-17 in the CNS
ADAM-17 in ischemic stroke and brain repair
ADAM-10 and -17 were shown to be present at various regions of the rodent brain by Northern blot analysis (Karkkainen et al., 2000). Immunohistochemical and in situ hybridization studies showed that ADAM-17 is expressed in endothelial cells and astrocytes (Goddard et al., 2001). Both ADAM enzymes were implicated in the development of the nervous system through activating neural cell adhesion and neurite outgrow by cleaving L1 (Maretzky et al., 2005) and neuronal cell adhesion molecule, NCAM (Kalus et al., 2006). In pathophysiological states ADAM-17/MMP inhibitors were shown to be beneficial as adjuvant therapy in bacterial meningitis (Meli et al., 2004), possibly due to their inhibition of TNFα–induced leukocyte invasion and extracellular matrix degradation (Leib et al., 2001). ADAM-17 inhibition was effective even in Toll-like receptor 2 (TLR2)-deficient mice and could compensate for the loss of the important anti-inflammatory action of TLR2 in pneumococcal meningitis (Echchannaoui et al., 2007). In oxygen-glucose deprivation, an in vitro model of brain ischemia/reperfusion injury, ADAM-17 was increased in rat forebrain slices with a concomitant increase in inducible nitric oxide synthase (iNOS) (Hurtado et al., 2001). A similar mechanism was observed in long-term and short-term immobilization-induced stress. In this case, glutamate receptor activation induced TNFα shedding and consequent iNOS upregulation through NF-κB signaling which was inhibited by an ADAM-17 inhibitor, suggesting a neuroprotective effect of ADAM-17 inhibition during stress (Madrigal et al., 2002). Another group, however, showed that ADAM-17-dependent TNFα cleavage actually inhibited apoptosis of neurons through NFκB activation in rat mixed cortical cultures (Hurtado et al., 2002). Also, in ischemic preconditioning, ADAM-17 was increased, protecting the animal brains during a subsequent longer ischemic period by reducing the size of infarction (Cardenas et al., 2002). This mechanism involved upregulation of glutamate receptors (Romera et al., 2004) and TNFR1 (Pradillo et al., 2005). Preconditioning with normobaric hyperoxia seemed to have similar effect: it also upregulated ADAM-17 and activated NFκB signaling (Bigdeli and Khoshbaten, 2008). ADAM-17 was also shown to be beneficial in a rodent stroke model in which ADAM-17 mediated neurogenesis in the subventricular zone and thus promoted recovery of the animals (Katakowski et al., 2007). Others showed that ADAM-17 can contribute to brain repair by inducing neuronal stem cell proliferation and migration in interaction with the endocannabioid system (Rubio-Araiz et al., 2008). Furthermore, niacin induced arteriogenesis after middle cerebral artery occlusion, a rat stroke model, was mediated by ADAM-17 activation and Notch cleavage (Chen et al., 2009).
In contrast to the above-mentioned findings, in the same middle cerebral artery occlusion model model of ischemic stroke, inhibition of ADAM-17 activity decreased the infarction size and the authors observed no change in apoptotic cell number upon ADAM-17 inhibition (Wang et al., 2004). Also, ADAM-17 was shown to worsen outcomes of cerebral ischemia during immobilization stress (Caso et al., 2006). Furthermore, in TIMP-3 knockout animals cerebral ischemia induced less neuronal death and inflammation in the hippocampus than in wild-type animals (Walker and Rosenberg, 2009). Because there is evidence for both neuroprotective and neurodegenerative effects of ADAM-17, further investigations are needed to characterize the net result of TNFα (and thus ADAM-17)-mediated signaling mechanisms and their consequences during brain injury, and to determine under which circumstances ADAM-17 inhibition is beneficial or harmful. The therapeutic potential of ADAM-17 inhibition in stroke has been discussed in a recent review (Lovering and Zhang, 2005).
Role of ADAM-17 in memory
The importance of ADAM-17 in learning and memory was also implicated by thorough experiments which showed that the enzyme participates in glutamate receptor 1/5-induced long-term depression (LTD), a cellular model of synaptic plasticity (Cho et al., 2008). ADAM-17 was shown to cleave neuronal pentraxin (NPR) which then binds to AMPA-type glutamate receptors and regulates their endocytosis, a process necessary for LTD in hippocampal and cerebellar synapses. Another protein, the cell adhesion molecule RA175/SynCAM1, which also participates in synaptic connection formation and plasticity, is also processed by ADAM-17-like enzymes on neuronal dendrits (Tanabe et al., 2008). Thus, ADAM-17 has an important role in synaptic formation. Interestingly, it was also shown that TIMP-3 knockout animals have impaired cognitive functions compared to wild-type animals, which may be due to increased MMP activity in the hippocampus (Baba et al., 2009).
Alzheimer’s Disease
ADAM-17 has an important role in Alzheimer’s disease (AD). As detailed above, together with ADAM-10, they act as α-secretases to cleave amyloid precursor protein (APP) and produce a soluble, non-amyloidogenic fragment, APPsα (Allinson et al., 2003). If APP is cleaved by β-secretase, the resulting product is the amyloidogenic Aβ. Amyloid Aβ peptide can induce the inflammation that is observed in the AD brain, which manifests in microglia and astrocyte activation around amyloid plaques and increased inflammatory mediators (Sastre et al., 2008). Interestingly, ADAM-17 was shown to localize in brain areas which contain amyloid plaques (Skovronsky et al., 2001). Recent work showed that FHL2 (or DRAL), a previously described ADAM-17 cytoplasmic domain partner which influences ADAM-17 activity (Canault et al., 2006b), is necessary for APPsα formation (Tanahashi and Yoshioka, 2008). Other regulators were also reported: the pro-inflammatory cytokine interleukin-1 is capable of enhancing ADAM-17 activity and soluble APP formation in astrocytes, and at the same time decreases Aβ production (Tachida et al., 2008). Interestingly, interleukin-1 can also stimulate translation of APP in cells (Bandyopadhyay et al., 2007). Increased APPsα production is not always followed by a decrease in Aβ production. In CHO cells expressing APP, ADAM-17 inhibition decreased soluble APP without increasing production of amyloid β peptide (Kim et al., 2008). Other factors which influence APPsα shedding, possibly through ADAM-17 (or ADAM-10) activation, include the acetylcholinesterase inhibitor huperzine A (Peng et al., 2007), the metalloendopeptidase nardilysin (Hiraoka et al., 2007); and the GPCR agonists neuropeptide PACAP and P2Y2R in human astrocytomas (Camden et al., 2005, Kojro et al., 2006). A better understanding of ADAM-17 and ADAM-10 regulation will enhance our knowledge of APP processing regulation and will enable us to design drugs for the treatment of disease.
ADAM-17 and Multiple Sclerosis
Based on its role in inflammation, ADAM-17 was implicated in neuroinflammatory disorders including multiple sclerosis (MS) (Moss et al., 1997a). This is supported by data that reflect increased ADAM-17 in peripheral mononuclear cells in patients with active MS (Seifert et al., 2002). Furthermore, ADAM-17 was observed together with TNFR2 in active MS plaques and invading T-lymphocytes in postmortem brain tissue of MS patients using immunohistochemistry (Kieseier et al., 2003). Also, increased amounts of ADAM-17 and soluble TNFR2 was observed in cerebrospinal fluid of MS patients in the same study. Expression of ADAM-17 was thoroughly analyzed in various types of MS, and investigators found that ADAM-17 was highest in secondary progressive and remitting-relapsing MS in relapse (Comabella et al., 2006). ADAM-17 expression was shown in active MS lesions at the time of myeloid breakdown in endothelial cells, microglia, and astrocytes (Plumb et al., 2006). One possible way that ADAM-17 can contribute to plaque formation is to alter growth factor signaling and prolong lesion activity by shedding the tyrosine kinases Axl and Mer decoy receptors in MS (Weinger et al., 2009). However, this possibility warrants further investigation.
ADAM-17 in malignancies
ADAM-17 was implicated in carcinogenesis because the enzyme sheds growth factors necessary for tumor progression and growth, and because it contributes to inflammation often observed in tumors. In accordance with this finding, increased shedding of epidermal growth factor ligands in tissues was shown to contribute to the development of a malignant phenotype (Katakowski et al., 2009), and elevated expression of ADAMs, including ADAM-17, are usually correlated with poor disease progression (reviewed recently (Duffy et al., 2009). Even if ADAM-17 has a role in various malignancies (Arribas et al., 2006), at present, the role of ADAM-17 is best studied in breast cancer. ADAM-17 overexpression in breast cancer correlated with TGFα expression (Borrell-Pages et al., 2003), tumor progression, and metastasis (McGowan et al., 2007). Furthermore, increased ADAM-17 was a predictor for shorter survival in breast cancer patients (McGowan et al., 2008). A breast cancer serum marker, nectin-4 which is shed by ADAM-17, was detected in patients with metastatic breast cancer (Fabre-Lafay et al., 2005). In vitro ADAM-17 induced proliferation, migration, and tube formation of breast cancer cells through activation of the EGFR-PI3K-AKT pathway (Zheng et al., 2009) and the important pathophysiological role of the enzyme was further confirmed in experiments in which a malignant phenotype of a breast cancer cell line was reverted to normal using small interfering RNAs against ADAM-17 (Kenny and Bissell, 2007).
In ovarian cancer tissues, upregulation of HB-EGF and ADAM-17 was observed, and a positive correlation between HB-EGF and ADAM-17 was found (Tanaka et al., 2005). The authors suggested that the high level of lysophosphatidic acid usually seen in patients with ovarian cancer possibly contributes to cancer progression through EGFR ligand shedding and ADAM-17 activation. The importance of ADAM-17 in cancer invasion also was shown in oral squamous cell carcinoma (Takamune et al., 2008). In this case, TNFα induced NFkB activation which, in turn, contributed to ADAM-17 maturation and induced cancer cell migration. ADAM-17 was increased in primary colon carcinoma (Blanchot-Jossic et al., 2005), in polarized colorectal cancer cells (Merchant et al., 2008), and in Helicobacter pylori-positive gastric carcinoma (Yoshimura et al., 2002). Expression of ADAM-17 was higher in nodular hepatocellular carcinoma (HCC) than in solitary large or small HCC (Ding et al., 2004). In hepatocellular carcinoma of nude mice, ADAM-17 silencing resulted in significant decrease in tumorigenesis, invasion, and angiogenesis (Tsai et al., 2009) suggesting that ADAM-17 inhibition can be beneficial in this type of cancer. In various lung cancer cell lines, ADAM-17 was identified as a possible novel lung cancer biomarker together with soluble TNFRI (Planque et al., 2009). Increased expression of ADAM-17 was observed in skin malignancies, specifically in invading cells of basal cell carcinoma (Oh et al., 2009). In another study, ADAM-17 expression was observed in 30% of benign prostatic hyperplasia biopsies, and all tumor samples and investigated prostatic tumor cell lines expressed ADAM-17 (Karan et al., 2003). Studying the role of ADAM-17 in brain tumor development, investigators found that overexpression of ADAM-17 in normal cortical astrocytes mediated their conversion into a malignant phenotype, resulting in non-adherent growth, increased cell proliferation, and production of angiogenic factors (Katakowski et al., 2009). ADAM-17 was also implicated in hematopoietic malignancies through shedding of the tyrosine kinase FLT3 ligand (Horiuchi et al., 2009b). Based on these observations, ADAM-17 is a potentially attractive therapeutic target for the treatment of several malignancies.
ADAM-17 in angiogenesis
Angiogenesis or new vessel development is not only an important developmental process but also it is indispensible for heart tissue recovery after ischemic episodes, from proliferative retinopathy, or from tumor growth and invasion. Angiogenesis is initiated by vascular endothelial growth factor (VEGF) which stimulates endothelial cells and their progenitors to differentiate and proliferate. The subsequent events include migration of endothelial cells into the surrounding extracellular matrix where they form cord-like structures (sprouts) which later develop lumens and interconnect. These events are followed by recruitment of accessory cells. Metalloproteinases also have important roles in regulating angiogenesis (Rundhaug, 2005). The significance of ADAM-17 in angiogenesis was first shown in transgenic mice expressing only catalytically inactive ADAM-17. These animals had pulmonary hypovascularization and heart-valve deformities (Zhao et al., 2001, Shi et al., 2003). In a murine model of retinal neovascularization, TNFα was upregulated in the retina during the hypoxia-induced neovascularization period (Majka et al., 2002). Moreover, VEGF induced ADAM-17 activation in retinal microvascular endothelial cells (Majka et al., 2002) and in human umbilical vascular endothelial cells (HUVECs) through ERK activation (Swendeman et al., 2008). A thorough analysis of the role of ADAM-17 in angiogenesis using ADAM-17 silencing revealed that a lack of the enzyme affects invasion and proliferation of the endothelial cells and contributes to destabilization of the developing network of HUVECS (Gooz et al., 2009). Furthermore, ADAM-17 was required for VEGF-induced invasion of HUVECs into a 3D matrix and VEGF-induced expression and activation of MMP-2. The role of ADAM-17 in HUVEC sprouting was confirmed by others (Kwak et al., 2009), and it was also shown that a chemical inhibitor of ADAM-17 inhibited both VEGF-induced tube formation of HUVECs and ischemia-induced retinal neovascularization in mice (Chikaraishi et al., 2009). Similarly, increased angiogenic activity and abnormal choroidal vascularization was observed in the eye of TIMP-3 −/− mice which was attenuated with VEGFR2 inhibition (Janssen et al., 2008). Increased MMP activity was also observed in the eye of the animals suggesting an important role of TIMP-3 in regulating choroidal vascularization. Further, in the ischemic brain increased arteriogenesis after stroke was initiated by niacin-induced ADAM-17 upregulation (Chen et al., 2009) as described above. These novel findings emphasize the role of ADAM-17 in angiogenesis both during development and during pathophysiological states such as proliferative retinopathy and stroke. Because the enzyme’s expression and activity is upregulated in various cancers, it is possible that other than initiating tumor cell proliferation, ADAM-17 also contributes to tumor neovascularization. Therefore, successful and specific ADAM-17 inhibition could attack tumor proliferation, inflammation and vasculogenesis.
ADAM-17 in heart diseases and in atherosclerosis
ADAM-17 has an important role in heart development, regulating valvulogenesis and centricular modeling (Shi et al., 2003) but several lines of evidence suggest that it contributes to various heart diseases. ADAM-17 was upregulated together with TNFα in myocarditis and correlated negatively with left ventricular systolic function in patients (Satoh et al., 2000). Increased expression of ADAM-17 was noted in peripheral mononuclear cells (PMNC) of patients with advanced congestive heart failure (Satoh et al., 2004). In patients with acute myocardial infarction ADAM-17 in PMNCs was higher in those subjects who had complications such as malignant recurrent ventricular arrhythmia or pump failure (Shimoda et al., 2005).
In an animal model of concentrated cardiac hypertrophy, ADAM-17 inhibition attenuated development of left ventricle dilatation, perhaps by preserving TNFα on myocyte surfaces because the phenotype was similar to mice expressing non-cleavable TNFα on their myocytes (Dibbs et al., 2003). Dilated cardiomyopathy with increased TNFα activity is also observed in TIMP-3 deficient animals, which further supports the role of ADAM-17 in cardiac remodeling (Fedak et al., 2004). Ablation of TNFα attenuated left ventricular dilatation in these animals, and together, with a metalloenzyme inhibitor, rescued the phenotype to normal (Kassiri et al., 2005). Recent data confirmed the role of ADAM-17 in cardiac remodeling both in spontaneous hypertensive rat and in animals treated with angiotensin-II; ADAM-17 knockdown with small interfering RNA inhibited development of both agonist-induced and hypertension-induced cardiac hypertrophy and fibrosis (Wang et al., 2009a). Similarly, in TIPM-3 knockout animals L-NAME raised blood pressure less than that observed in wild-type animals, and the hypertension-induced fibrotic changes in vascular walls were also attenuated in the TIMP-3 −/− animals (Higuchi et al., 2007).
Aging of coronary arteries is associated with transition to an inflammatory phenotype and is characterized by an upregulation of TNFα activity which promotes endothelial cell apoptosis (Csiszar et al., 2004). In another study, investigators showed increased ADAM-17 immunostaining in atherosclerotic plaques of the aortic arch and sinus in apolipoprotein E-deficient mice and in humans (Canault et al., 2006a). At the same time, elevated circulating plasma TNFR I and 2 was also observed. There is also evidence of increased ADAM-17 and TNFα expression in areas of ruptured coronary plaques in patients with myocardial infarction (Satoh et al., 2008). These findings strongly suggest that inhibiting ADAM-17 activity can be beneficial for preventing complications of acute myocardial infarction and cardiac remodeling in hypertension.
ADAM-17 and diabetes
In an animal model of nonobese, insulin-resistant diabetes (hypertensive fructose-fed rats) peritoneally applied ADAM-17 inhibitor restored insulin sensitivity of the animals to normal, which suggests that TNFα inhibition through ADAM-17 could be an attractive treatment for nonobese insulin-resistant diabetes (Togashi et al., 2002). In another publication, heterozygous ADAM-17 (−/+) animals fed a high-fat diet were less prone to obesity and insulin resistance than their wild type littermates (Serino et al., 2007). In diabetes, cardiovascular complications are attributed to low levels of inflammation. In a recent investigation, aldose reductase was shown to be involved in high-glucose-induced TNFα shedding and ADAM-17 activation in rat aortic smooth muscle cells (Reddy et al., 2009). Inhibition of aldose reductase by sorbitol decreased shedded TNFαboth in cell culture and in rats with streptozotocin-induced diabetes, suggesting that the enzyme could be a therapeutic target for treating diabetes-induced vascular inflammation. Similarly, decreased TIMP-3 was implicated in inflammation and insulin resistance; and hepatic steatosis, liver and fat tissue inflammation was observed in TIMP-3 knockout animals fed a high-fat diet (Menghini et al., 2009). This suggests that increased ADAM-17 could be important in obesity-associated fatty liver disease.
Interestingly, ADAM-17 as α-secretase was also shown to cleave Klotho, an anti-aging protein expressed mainly in the kidney and brain (Bloch et al., 2009). Klotho also regulates glucose metabolism: Klotho knockdown animals have low blood insulin levels and are hypersensitive to exogenous insulin (Utsugi et al., 2000). Regulating Klotho could provide another control point for ADAM-17 to influence glucose homeostasis.
ADAM-17 in kidney disease
Increased activation of the EGFR through TGFα or HB-EGF was shown in several kidney diseases including polycystic kidney disease (Richards et al., 1998). In bpk mice, a model of autosomal recessive polycystic kidney disease (PKD), increased TGFα immunostaining was found in the proximal tubules of cystic kidneys (Dell et al., 2001). ADAM-17 inhibitor treatment significantly decreased cyst formation and improved kidney function. Another group showed that TGFα knockout bpk mice also developed PKD, suggesting that other ADAM-17 substrates can be responsible for the phenotype (Nemo et al., 2005). Indeed, EGF, amphiregulin, HB-EGF, EGFR and ErbbB4 are overexpressed in polycystic kidney disease, which all could be substrates of ADAM-17. Chronic kidney disease (CKD) which is characterized by extracellular matrix deposition and fibrotic transformation of kidney tissue can develop from renal diseases of different origins. Because angiotensin converting enzyme inhibitors slowed disease progression, the role of angiotensin-II in CKD was studied intensively. Data are accumulating to suggest that increased EGFR activation can contribute to the fibrotic process, and it was shown that the GPCR agonist angiotensin-II induces EGFR transactivation by ADAM-17 activation and TGFα shedding (Lautrette et al., 2005). Chemical inhibition of ADAM-17 attenuated chronic angiotensin-II-induced glomerular fibrosis suggesting that ADAM-17 inhibition can be a valuable therapeutic tool in treating fibrotic kidney disease. In TGFα knockout mice, chronic angiotensin-II infusion-induced proteinuria, tubular atrophy, and glomerulosclerosis were greatly attenuated compared to wild type, angiotensin-II treated animals (Shah and Catt, 2006). In TIMP-3 −/− animals unilateral ureteral obstruction, an experimental model of interstitial kidney fibrosis, led to increased renal injury compared to wild-type animals (Kassiri et al., 2009). The increased fibrosis and inflammation that was accompanied with elevated ADAM-17 activity was attenuated with additional TNFα ablation. In an in vitro model of glomerular disease, angiotensin-II induced HB-EGF shedding from mesangial cells (Uchiyama-Tanaka et al., 2001). In another study, the pro-fibrotic GPCR agonist, serotonin, was shown to stimulate EGFR activation and proliferation of mesangial cells through ADAM-17-mediated HB-EGF shedding (Gooz et al., 2006). Downregulating ADAM-17 by silencing RNA completely inhibited the effect of serotonin. In mesangial cells, the chemokine CXCL16 is shed by ADAM-17 and ADAM-10 (Schramme et al., 2008). Inhibiting metalloenzyme activity in mesangial cells inhibited T-Jurkat cell chemotaxis, and silencing either CXCL16 or ADAM-17 expression attenuated proliferation of mesangial cells. These data show that ADAM-17 can be an exciting novel target for the treatment of kidney diseases. Because many people are affected by chronic kidney disease—1 in 10 Americans, based on the National Kidney Foundation http://www.nkf.org–and that presently ACE inhibitors are the only therapy available to slow CKD progression, therapeutic targeting of ADAM-17 in kidney diseases should be considered and investigated more intensively.
CONCLUDING REMARKS
ADAM-17 is expressed by most mammalian cells and may have a role in controlling many physiological and pathophysiological processes. It is indispensible for normal mammalian development and through its contribution as an important sheddase, it is indispensible for adult life. The number of identified ADAM-17 substrates is increasing and evidence is mounting that ADAM-17 has a role in almost every cellular function. Because of the ubiquitous nature of the enzyme, mechanisms for controlling ADAM-17 activity are worth investigating. For example, how to specifically target ADAM-17 in cancer or in kidney disease without targeting the function of the enzyme in other tissues is a concern. Even with specific inhibitors that inhibit only ADAM-17 and no other metalloenzymes, the side effects would be enormous. Also, the dual nature of ADAM-17 (it is both pro-proliferative and inhibitory) is problematic because some effects may be beneficial for some tissues or diseases and harmful in others. Because ADAM-17 activity must be regarded in its full cellular context, molecules binding to the enzymes and signaling pathways interconnected with the ADAM-17 signaling pathway all have unique cell- and tissue-specific effects which influence the outcome of ADAM-17 activation. We are only beginning to understand the effect of post-translational modifications on the enzyme activity and its role in ADAM-17 interactions and trafficking. Also, we have data regarding phorbol ester-induced activation of the enzyme, and these experiments, while providing valuable data—such as a list of molecules cleaved by the enzyme—do not tell us specifically which molecules regulate ADAM-17 activity. Understanding ADAM-17activity regulation in vivo is crucial for the development of adequate patient therapy.
ACKNOWLEDGEMENTS
I am thankful for Dr. Pal Gooz for critical reading of the manuscript and for Dr. Jennifer Schnellmann at the Office of Scientific Editing and Publications at the Medical University of South Carolina for critical reading and editing the manuscript.
DECLARATION OF INTEREST
This work was supported by grants from the National Institutes of Health (NIH/NIDDK K01 DK070054 and NIH/NIDDK ARRA supplement 3K01DK070054-05S1) and from the Paul Teschan Research Fund of the Dialysis Clinic, Inc.
REFERENCES
- Ahrens I, Ellwanger C, Smith BK, Bassler N, Chen YC, Neudorfer I, Ludwig A, Bode C, Peter K. J Leukoc Biol. 2008;83:1388–1395. doi: 10.1189/jlb.0707497. [DOI] [PubMed] [Google Scholar]
- Alfa Cisse M, Sunyach C, Slack BE, Fisher A, Vincent B, Checler F. J Neurosci. 2007;27:4083–4092. doi: 10.1523/JNEUROSCI.5293-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allinson TM, Parkin ET, Turner AJ, Hooper NM. J Neurosci Res. 2003;74:342–352. doi: 10.1002/jnr.10737. [DOI] [PubMed] [Google Scholar]
- Amin AR. Osteoarthritis Cartilage. 1999;7:392–394. doi: 10.1053/joca.1998.0221. [DOI] [PubMed] [Google Scholar]
- Amour A, Slocombe PM, Webster A, Butler M, Knight CG, Smith BJ, Stephens PE, Shelley C, Hutton M, Knauper V, Docherty AJ, Murphy G. FEBS Lett. 1998;435:39–44. doi: 10.1016/s0014-5793(98)01031-x. [DOI] [PubMed] [Google Scholar]
- Anderson JP, Chen Y, Kim KS, Robakis NK. J Neurochem. 1992;59:2328–2331. doi: 10.1111/j.1471-4159.1992.tb10128.x. [DOI] [PubMed] [Google Scholar]
- Arribas J, Bech-Serra JJ, Santiago-Josefat B. Cancer Metastasis Rev. 2006;25:57–68. doi: 10.1007/s10555-006-7889-6. [DOI] [PubMed] [Google Scholar]
- Atreya R, Mudter J, Finotto S, Mullberg J, Jostock T, Wirtz S, Schutz M, Bartsch B, Holtmann M, Becker C, Strand D, Czaja J, Schlaak JF, Lehr HA, Autschbach F, Schurmann G, Nishimoto N, Yoshizaki K, Ito H, Kishimoto T, Galle PR, Rose-John S, Neurath MF. Nat Med. 2000;6:583–588. doi: 10.1038/75068. [DOI] [PubMed] [Google Scholar]
- Baba Y, Yasuda O, Takemura Y, Ishikawa Y, Ohishi M, Iwanami J, Mogi M, Doe N, Horiuchi M, Maeda N, Fukuo K, Rakugi H. Lab Invest. 2009;89:1340–1347. doi: 10.1038/labinvest.2009.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay S, Goldstein LE, Lahiri DK, Rogers JT. Curr Med Chem. 2007;14:2848–2864. doi: 10.2174/092986707782360060. [DOI] [PubMed] [Google Scholar]
- Bao J, Wolpowitz D, Role LW, Talmage DA. J Cell Biol. 2003;161:1133–1141. doi: 10.1083/jcb.200212085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bax DV, Messent AJ, Tart J, van Hoang M, Kott J, Maciewicz RA, Humphries MJ. J Biol Chem. 2004;279:22377–22386. doi: 10.1074/jbc.M400180200. [DOI] [PubMed] [Google Scholar]
- Bigdeli MR, Khoshbaten A. Neuroscience. 2008;153:671–678. doi: 10.1016/j.neuroscience.2008.02.064. [DOI] [PubMed] [Google Scholar]
- Black RA, Rauch CT, Kozlosky CJ, Peschon JJ, Slack JL, Wolfson MF, Castner BJ, Stocking KL, Reddy P, Srinivasan S, Nelson N, Boiani N, Schooley KA, Gerhart M, Davis R, Fitzner JN, Johnson RS, Paxton RJ, March CJ, Cerretti DP. Nature. 1997;385:729–733. doi: 10.1038/385729a0. [DOI] [PubMed] [Google Scholar]
- Blanchot-Jossic F, Jarry A, Masson D, Bach-Ngohou K, Paineau J, Denis MG, Laboisse CL, Mosnier JF. J Pathol. 2005;207:156–163. doi: 10.1002/path.1814. [DOI] [PubMed] [Google Scholar]
- Blobel CP. Nat Rev Mol Cell Biol. 2005;6:32–43. doi: 10.1038/nrm1548. [DOI] [PubMed] [Google Scholar]
- Bloch L, Sineshchekova O, Reichenbach D, Reiss K, Saftig P, Kuro-o M, Kaether C. FEBS Lett. 2009;583:3221–3224. doi: 10.1016/j.febslet.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohgaki T, Amasaki Y, Nishimura N, Bohgaki M, Yamashita Y, Nishio M, Sawada KI, Jodo S, Atsumi T, Koike T. Ann Rheum Dis. 2005;64:1165–1173. doi: 10.1136/ard.2004.030338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boissy P, Lenhard TR, Kirkegaard T, Peschon JJ, Black RA, Delaisse JM, del Carmen Ovejero M. FEBS Lett. 2003;553:257–261. doi: 10.1016/s0014-5793(03)01022-6. [DOI] [PubMed] [Google Scholar]
- Borrell-Pages M, Rojo F, Albanell J, Baselga J, Arribas J. EMBO J. 2003;22:1114–1124. doi: 10.1093/emboj/cdg111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borroto A, Ruiz-Paz S, de la Torre TV, Borrell-Pages M, Merlos-Suarez A, Pandiella A, Blobel CP, Baselga J, Arribas J. J Biol Chem. 2003;278:25933–25939. doi: 10.1074/jbc.M301673200. [DOI] [PubMed] [Google Scholar]
- Bridges LC, Bowditch RD. Curr Pharm Des. 2005;11:837–847. doi: 10.2174/1381612053381747. [DOI] [PubMed] [Google Scholar]
- Briso EM, Dienz O, Rincon M. J Immunol. 2008;180:7102–7106. doi: 10.4049/jimmunol.180.11.7102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodowicz T, Wiltschke C, Budinsky AC, Krainer M, Steger GG, Zielinski CC. Int J Cancer. 1997;73:875–879. doi: 10.1002/(sici)1097-0215(19971210)73:6<875::aid-ijc19>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Brou C, Logeat F, Gupta N, Bessia C, LeBail O, Doedens JR, Cumano A, Roux P, Black RA, Israel A. Mol Cell. 2000;5:207–216. doi: 10.1016/s1097-2765(00)80417-7. [DOI] [PubMed] [Google Scholar]
- Buckley CA, Rouhani FN, Kaler M, Adamik B, Hawari FI, Levine SJ. Am J Physiol Lung Cell Mol Physiol. 2005;288:L1132–L1138. doi: 10.1152/ajplung.00429.2004. [DOI] [PubMed] [Google Scholar]
- Budagian V, Bulanova E, Orinska Z, Ludwig A, Rose-John S, Saftig P, Borden EC, Bulfone-Paus S. J Biol Chem. 2004;279:40368–40375. doi: 10.1074/jbc.M404125200. [DOI] [PubMed] [Google Scholar]
- Burrage PS, Brinckerhoff CE. Curr Drug Targets. 2007;8:293–303. doi: 10.2174/138945007779940098. [DOI] [PubMed] [Google Scholar]
- Buxbaum JD, Liu KN, Luo Y, Slack JL, Stocking KL, Peschon JJ, Johnson RS, Castner BJ, Cerretti DP, Black RA. J Biol Chem. 1998;273:27765–27767. doi: 10.1074/jbc.273.43.27765. [DOI] [PubMed] [Google Scholar]
- Caescu CI, Jeschke GR, Turk BE. Biochem J. 2009;424:79–88. doi: 10.1042/BJ20090549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvete JJ. Curr Pharm Des. 2005;11:829–835. doi: 10.2174/1381612053381783. [DOI] [PubMed] [Google Scholar]
- Calvete JJ, Marcinkiewicz C, Sanz L. Curr Pharm Des. 2007;13:2853–2859. doi: 10.2174/138161207782023766. [DOI] [PubMed] [Google Scholar]
- Calvete JJ, Moreno-Murciano MP, Theakston RD, Kisiel DG, Marcinkiewicz C. Biochem J. 2003;372:725–734. doi: 10.1042/BJ20021739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camden JM, Schrader AM, Camden RE, Gonzalez FA, Erb L, Seye CI, Weisman GA. J Biol Chem. 2005;280:18696–18702. doi: 10.1074/jbc.M500219200. [DOI] [PubMed] [Google Scholar]
- Canault M, Peiretti F, Kopp F, Bonardo B, Bonzi MF, Coudeyre JC, Alessi MC, Juhan-Vague I, Nalbone G. Atherosclerosis. 2006a;187:82–91. doi: 10.1016/j.atherosclerosis.2005.08.031. [DOI] [PubMed] [Google Scholar]
- Canault M, Tellier E, Bonardo B, Mas E, Aumailley M, Juhan-Vague I, Nalbone G, Peiretti F. J Cell Physiol. 2006b;208:363–372. doi: 10.1002/jcp.20671. [DOI] [PubMed] [Google Scholar]
- Cardenas A, Moro MA, Leza JC, O'Shea E, Davalos A, Castillo J, Lorenzo P, Lizasoain I. J Cereb Blood Flow Metab. 2002;22:1297–1302. doi: 10.1097/01.WCB.0000033968.83623.D0. [DOI] [PubMed] [Google Scholar]
- Caso JR, Lizasoain I, Lorenzo P, Moro MA, Leza JC. Neuroscience. 2006;142:59–69. doi: 10.1016/j.neuroscience.2006.06.009. [DOI] [PubMed] [Google Scholar]
- Cesaro A, Abakar-Mahamat A, Brest P, Lassalle S, Selva E, Filippi J, Hebuterne X, Hugot JP, Doglio A, Galland F, Naquet P, Vouret-Craviari V, Mograbi B, Hofman PM. Am J Physiol Gastrointest Liver Physiol. 2009;296:G1332–G1343. doi: 10.1152/ajpgi.90641.2008. [DOI] [PubMed] [Google Scholar]
- Charbonneau M, Harper K, Grondin F, Pelmus M, McDonald PP, Dubois CM. J Biol Chem. 2007;282:33714–33724. doi: 10.1074/jbc.M704041200. [DOI] [PubMed] [Google Scholar]
- Charrier-Hisamuddin L, Laboisse CL, Merlin D. FASEB J. 2008;22:641–653. doi: 10.1096/fj.07-8876rev. [DOI] [PubMed] [Google Scholar]
- Chen B, Bronson RT, Klaman LD, Hampton TG, Wang JF, Green PJ, Magnuson T, Douglas PS, Morgan JP, Neel BG. Nat Genet. 2000;24:296–299. doi: 10.1038/73528. [DOI] [PubMed] [Google Scholar]
- Chen J, Cui X, Zacharek A, Ding GL, Shehadah A, Jiang Q, Lu M, Chopp M. J Cereb Blood Flow Metab. 2009;29:911–920. doi: 10.1038/jcbfm.2009.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen MS, Almeida EA, Huovila AP, Takahashi Y, Shaw LM, Mercurio AM, White JM. J Cell Biol. 1999;144:549–561. doi: 10.1083/jcb.144.3.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikaraishi Y, Shimazawa M, Yokota K, Yoshino K, Hara H. Curr Neurovasc Res. 2009;6:140–147. doi: 10.2174/156720209788970072. [DOI] [PubMed] [Google Scholar]
- Cho RW, Park JM, Wolff SB, Xu D, Hopf C, Kim JA, Reddy RC, Petralia RS, Perin MS, Linden DJ, Worley PF. Neuron. 2008;57:858–871. doi: 10.1016/j.neuron.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chokki M, Mitsuhashi H, Kamimura T. Life Sci. 2006;78:3051–3057. doi: 10.1016/j.lfs.2005.12.023. [DOI] [PubMed] [Google Scholar]
- Chung KH, Kim SH, Han KY, Sohn YD, Chang SI, Baek KH, Jang Y, Kim DS, Kang IC. J Pharm Pharmacol. 2003;55:1577–1582. doi: 10.1211/0022357022160. [DOI] [PubMed] [Google Scholar]
- Citri A, Skaria KB, Yarden Y. Exp Cell Res. 2003;284:54–65. doi: 10.1016/s0014-4827(02)00101-5. [DOI] [PubMed] [Google Scholar]
- Comabella M, Romera C, Camina M, Perkal H, Moro MA, Leza JC, Lizasoain I, Castillo M, Montalban X. J Neurol. 2006;253:701–706. doi: 10.1007/s00415-006-0090-6. [DOI] [PubMed] [Google Scholar]
- Cominetti MR, Ribeiro JU, Fox JW, Selistre-de-Araujo HS. Arch Biochem Biophys. 2003;416:171–179. doi: 10.1016/s0003-9861(03)00298-4. [DOI] [PubMed] [Google Scholar]
- Condon TP, Flournoy S, Sawyer GJ, Baker BF, Kishimoto TK, Bennett CF. Antisense Nucleic Acid Drug Dev. 2001;11:107–116. doi: 10.1089/108729001750171353. [DOI] [PubMed] [Google Scholar]
- Csiszar A, Ungvari Z, Koller A, Edwards JG, Kaley G. Physiol Genomics. 2004;17:21–30. doi: 10.1152/physiolgenomics.00136.2003. [DOI] [PubMed] [Google Scholar]
- Danen EH, Marcinkiewicz C, Cornelissen IM, van Kraats AA, Pachter JA, Ruiter DJ, Niewiarowski S, van Muijen GN. Exp Cell Res. 1998;238:188–196. doi: 10.1006/excr.1997.3821. [DOI] [PubMed] [Google Scholar]
- DasGupta S, Murumkar PR, Giridhar R, Yadav MR. Bioorg Med Chem. 2009;17:444–459. doi: 10.1016/j.bmc.2008.11.067. [DOI] [PubMed] [Google Scholar]
- De Benedetti F, Massa M, Pignatti P, Albani S, Novick D, Martini A. J Clin Invest. 1994;93:2114–2119. doi: 10.1172/JCI117206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Strooper B, Annaert W, Cupers P, Saftig P, Craessaerts K, Mumm JS, Schroeter EH, Schrijvers V, Wolfe MS, Ray WJ, Goate A, Kopan R. Nature. 1999;398:518–522. doi: 10.1038/19083. [DOI] [PubMed] [Google Scholar]
- Dell KM, Nemo R, Sweeney WE, Jr, Levin JI, Frost P, Avner ED. Kidney Int. 2001;60:1240–1248. doi: 10.1046/j.1523-1755.2001.00963.x. [DOI] [PubMed] [Google Scholar]
- Diaz-Rodriguez E, Montero JC, Esparis-Ogando A, Yuste L, Pandiella A. Mol Biol Cell. 2002;13:2031–2044. doi: 10.1091/mbc.01-11-0561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibbs ZI, Diwan A, Nemoto S, DeFreitas G, Abdellatif M, Carabello BA, Spinale FG, Feuerstein G, Sivasubramanian N, Mann DL. Circulation. 2003;108:1002–1008. doi: 10.1161/01.CIR.0000085203.46621.F4. [DOI] [PubMed] [Google Scholar]
- Ding X, Yang LY, Huang GW, Wang W, Lu WQ. World J Gastroenterol. 2004;10:2735–2739. doi: 10.3748/wjg.v10.i18.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doedens JR, Black RA. J Biol Chem. 2000;275:14598–14607. doi: 10.1074/jbc.275.19.14598. [DOI] [PubMed] [Google Scholar]
- Doedens JR, Mahimkar RM, Black RA. Biochem Biophys Res Commun. 2003;308:331–338. doi: 10.1016/s0006-291x(03)01381-0. [DOI] [PubMed] [Google Scholar]
- Duan JJ, Chen L, Wasserman ZR, Lu Z, Liu RQ, Covington MB, Qian M, Hardman KD, Magolda RL, Newton RC, Christ DD, Wexler RR, Decicco CP. J Med Chem. 2002;45:4954–4957. doi: 10.1021/jm0255670. [DOI] [PubMed] [Google Scholar]
- Duffy MJ, McKiernan E, O'Donovan N, McGowan PM. Clin Cancer Res. 2009;15:1140–1144. doi: 10.1158/1078-0432.CCR-08-1585. [DOI] [PubMed] [Google Scholar]
- Echchannaoui H, Leib SL, Neumann U, Landmann RM. BMC Infect Dis. 2007;7:25. doi: 10.1186/1471-2334-7-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards DR, Handsley MM, Pennington CJ. Mol Aspects Med. 2008;29:258–289. doi: 10.1016/j.mam.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermert M, Pantazis C, Duncker HR, Grimminger F, Seeger W, Ermert L. Cytokine. 2003;22:89–100. doi: 10.1016/s1043-4666(03)00117-0. [DOI] [PubMed] [Google Scholar]
- Eto K, Puzon-McLaughlin W, Sheppard D, Sehara-Fujisawa A, Zhang XP, Takada Y. J Biol Chem. 2000;275:34922–34930. doi: 10.1074/jbc.M001953200. [DOI] [PubMed] [Google Scholar]
- Fabre-Lafay S, Garrido-Urbani S, Reymond N, Goncalves A, Dubreuil P, Lopez M. J Biol Chem. 2005;280:19543–19550. doi: 10.1074/jbc.M410943200. [DOI] [PubMed] [Google Scholar]
- Fahrenholz F, Gilbert S, Kojro E, Lammich S, Postina R. Ann N Y Acad Sci. 2000;920:215–222. doi: 10.1111/j.1749-6632.2000.tb06925.x. [DOI] [PubMed] [Google Scholar]
- Fan H, Turck CW, Derynck R. J Biol Chem. 2003;278:18617–18627. doi: 10.1074/jbc.M300331200. [DOI] [PubMed] [Google Scholar]
- Faveeuw C, Preece G, Ager A. Blood. 2001;98:688–695. doi: 10.1182/blood.v98.3.688. [DOI] [PubMed] [Google Scholar]
- Fedak PW, Smookler DS, Kassiri Z, Ohno N, Leco KJ, Verma S, Mickle DA, Watson KL, Hojilla CV, Cruz W, Weisel RD, Li RK, Khokha R. Circulation. 2004;110:2401–2409. doi: 10.1161/01.CIR.0000134959.83967.2D. [DOI] [PubMed] [Google Scholar]
- Feldmann M, Brennan FM, Elliott MJ, Williams RO, Maini RN. Ann N Y Acad Sci. 1995;766:272–278. doi: 10.1111/j.1749-6632.1995.tb26675.x. [DOI] [PubMed] [Google Scholar]
- Feldmann M, Brennan FM, Maini RN. Annu Rev Immunol. 1996;14:397–440. doi: 10.1146/annurev.immunol.14.1.397. [DOI] [PubMed] [Google Scholar]
- Fridman JS, Caulder E, Hansbury M, Liu X, Yang G, Wang Q, Lo Y, Zhou BB, Pan M, Thomas SM, Grandis JR, Zhuo J, Yao W, Newton RC, Friedman SM, Scherle PA, Vaddi K. Clin Cancer Res. 2007;13:1892–1902. doi: 10.1158/1078-0432.CCR-06-2116. [DOI] [PubMed] [Google Scholar]
- Galan JA, Sanchez EE, Rodriguez-Acosta A, Soto JG, Bashir S, McLane MA, Paquette-Straub C, Perez JC. Toxicon. 2008;51:1186–1196. doi: 10.1016/j.toxicon.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvin KM, Donovan MJ, Lynch CA, Meyer RI, Paul RJ, Lorenz JN, Fairchild-Huntress V, Dixon KL, Dunmore JH, Gimbrone MA, Jr, Falb D, Huszar D. Nat Genet. 2000;24:171–174. doi: 10.1038/72835. [DOI] [PubMed] [Google Scholar]
- Garton KJ, Gough PJ, Blobel CP, Murphy G, Greaves DR, Dempsey PJ, Raines EW. J Biol Chem. 2001;276:37993–38001. doi: 10.1074/jbc.M106434200. [DOI] [PubMed] [Google Scholar]
- Garton KJ, Gough PJ, Philalay J, Wille PT, Blobel CP, Whitehead RH, Dempsey PJ, Raines EW. J Biol Chem. 2003;278:37459–37464. doi: 10.1074/jbc.M305877200. [DOI] [PubMed] [Google Scholar]
- Gelling RW, Yan W, Al-Noori S, Pardini A, Morton GJ, Ogimoto K, Schwartz MW, Dempsey PJ. Endocrinology. 2008;149:6053–6064. doi: 10.1210/en.2008-0775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giancotti FG, Ruoslahti E. Science. 1999;285:1028–1032. doi: 10.1126/science.285.5430.1028. [DOI] [PubMed] [Google Scholar]
- Gilmore JL, King BW, Asakawa N, Harrison K, Tebben A, Sheppeck JE, 2nd, Liu RQ, Covington M, Duan JJ. Bioorg Med Chem Lett. 2007;17:4678–4682. doi: 10.1016/j.bmcl.2007.05.100. [DOI] [PubMed] [Google Scholar]
- Gilmore JL, King BW, Harris C, Maduskuie T, Mercer SE, Liu RQ, Covington MB, Qian M, Ribadeneria MD, Vaddi K, Trzaskos JM, Newton RC, Decicco CP, Duan JJ. Bioorg Med Chem Lett. 2006;16:2699–2704. doi: 10.1016/j.bmcl.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Goddard DR, Bunning RA, Woodroofe MN. Glia. 2001;34:267–271. doi: 10.1002/glia.1060. [DOI] [PubMed] [Google Scholar]
- Gomez MI, Sokol SH, Muir AB, Soong G, Bastien J, Prince AS. J Immunol. 2005;175:1930–1936. doi: 10.4049/jimmunol.175.3.1930. [DOI] [PubMed] [Google Scholar]
- Gonzales PE, Galli JD, Milla ME. Biochemistry. 2008;47:9911–9919. doi: 10.1021/bi801049v. [DOI] [PubMed] [Google Scholar]
- Gonzales PE, Solomon A, Miller AB, Leesnitzer MA, Sagi I, Milla ME. J Biol Chem. 2004;279:31638–31645. doi: 10.1074/jbc.M401311200. [DOI] [PubMed] [Google Scholar]
- Gooz M, Gooz P, Luttrell LM, Raymond JR. J Biol Chem. 2006;281:21004–21012. doi: 10.1074/jbc.M512096200. [DOI] [PubMed] [Google Scholar]
- Gooz P, Gooz M, Baldys A, Hoffman S. Biochem Biophys Res Commun. 2009;380:33–38. doi: 10.1016/j.bbrc.2009.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto T, Ishizaka A, Kobayashi F, Kohno M, Sawafuji M, Tasaka S, Ikeda E, Okada Y, Maruyama I, Kobayashi K. Am J Respir Crit Care Med. 2004;170:1239–1246. doi: 10.1164/rccm.200402-146OC. [DOI] [PubMed] [Google Scholar]
- Gschwind A, Hart S, Fischer OM, Ullrich A. EMBO J. 2003;22:2411–2421. doi: 10.1093/emboj/cdg231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guinea-Viniegra J, Zenz R, Scheuch H, Hnisz D, Holcmann M, Bakiri L, Schonthaler HB, Sibilia M, Wagner EF. Genes Dev. 2009;23:2663–2674. doi: 10.1101/gad.543109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haga S, Yamamoto N, Nakai-Murakami C, Osawa Y, Tokunaga K, Sata T, Sasazuki T, Ishizaka Y. Proc Natl Acad Sci U S A. 2008;105:7809–7814. doi: 10.1073/pnas.0711241105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl D, He CH, Koller B, Da Silva CA, Homer R, Lee CG, Elias JA. J Biol Chem. 2008;283:33472–33482. doi: 10.1074/jbc.M805574200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann D, Tournoy J, Saftig P, Annaert W, De Strooper B. J Mol Neurosci. 2001;17:171–181. doi: 10.1385/JMN:17:2:171. [DOI] [PubMed] [Google Scholar]
- Higashiyama S, Nanba D. Biochim Biophys Acta. 2005;1751:110–117. doi: 10.1016/j.bbapap.2004.11.009. [DOI] [PubMed] [Google Scholar]
- Higuchi M, Yasuda O, Kawamoto H, Yotsui T, Baba Y, Ozaki T, Maeda N, Fukuo K, Rakugi H, Ogihara T. Hypertens Res. 2007;30:563–571. doi: 10.1291/hypres.30.563. [DOI] [PubMed] [Google Scholar]
- Hinkle CL, Sunnarborg SW, Loiselle D, Parker CE, Stevenson M, Russell WE, Lee DC. J Biol Chem. 2004;279:24179–24188. doi: 10.1074/jbc.M312141200. [DOI] [PubMed] [Google Scholar]
- Hiraoka Y, Ohno M, Yoshida K, Okawa K, Tomimoto H, Kita T, Nishi E. J Neurochem. 2007;102:1595–1605. doi: 10.1111/j.1471-4159.2007.04685.x. [DOI] [PubMed] [Google Scholar]
- Horiuchi K, Kimura T, Miyamoto T, Miyamoto K, Akiyama H, Takaishi H, Morioka H, Nakamura T, Okada Y, Blobel CP, Toyama Y. J Immunol. 2009a;182:2093–2101. doi: 10.4049/jimmunol.0802491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi K, Kimura T, Miyamoto T, Takaishi H, Okada Y, Toyama Y, Blobel CP. J Immunol. 2007;179:2686–2689. doi: 10.4049/jimmunol.179.5.2686. [DOI] [PubMed] [Google Scholar]
- Horiuchi K, Morioka H, Takaishi H, Akiyama H, Blobel CP, Toyama Y. J Immunol. 2009b;182:7408–7414. doi: 10.4049/jimmunol.0801931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang A, Joseph-McCarthy D, Lovering F, Sun L, Wang W, Xu W, Zhu Y, Cui J, Zhang Y, Levin JI. Bioorg Med Chem. 2007;15:6170–6181. doi: 10.1016/j.bmc.2007.06.031. [DOI] [PubMed] [Google Scholar]
- Huang J, Bridges LC, White JM. Mol Biol Cell. 2005;16:4982–4991. doi: 10.1091/mbc.E05-03-0258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang TF, Holt JC, Lukasiewicz H, Niewiarowski S. J Biol Chem. 1987;262:16157–16163. [PubMed] [Google Scholar]
- Hundhausen C, Misztela D, Berkhout TA, Broadway N, Saftig P, Reiss K, Hartmann D, Fahrenholz F, Postina R, Matthews V, Kallen KJ, Rose-John S, Ludwig A. Blood. 2003;102:1186–1195. doi: 10.1182/blood-2002-12-3775. [DOI] [PubMed] [Google Scholar]
- Huovila AP, Turner AJ, Pelto-Huikko M, Karkkainen I, Ortiz RM. Trends Biochem Sci. 2005;30:413–422. doi: 10.1016/j.tibs.2005.05.006. [DOI] [PubMed] [Google Scholar]
- Hurtado O, Cardenas A, Lizasoain I, Bosca L, Leza JC, Lorenzo P, Moro MA. Neuropharmacology. 2001;40:1094–1102. doi: 10.1016/s0028-3908(01)00035-1. [DOI] [PubMed] [Google Scholar]
- Hurtado O, Lizasoain I, Fernandez-Tome P, Alvarez-Barrientos A, Leza JC, Lorenzo P, Moro MA. J Cereb Blood Flow Metab. 2002;22:576–585. doi: 10.1097/00004647-200205000-00009. [DOI] [PubMed] [Google Scholar]
- Huveneers S, Truong H, Danen HJ. Int J Radiat Biol. 2007;83:743–751. doi: 10.1080/09553000701481808. [DOI] [PubMed] [Google Scholar]
- Huxley-Jones J, Clarke TK, Beck C, Toubaris G, Robertson DL, Boot-Handford RP. BMC Evol Biol. 2007;7:63. doi: 10.1186/1471-2148-7-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram RN, Orth P, Strickland CL, Le HV, Madison V, Beyer BM. Protein Eng Des Sel. 2006;19:155–161. doi: 10.1093/protein/gzj014. [DOI] [PubMed] [Google Scholar]
- Jackson LF, Qiu TH, Sunnarborg SW, Chang A, Zhang C, Patterson C, Lee DC. Embo J. 2003;22:2704–2716. doi: 10.1093/emboj/cdg264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen A, Hoellenriegel J, Fogarasi M, Schrewe H, Seeliger M, Tamm E, Ohlmann A, May CA, Weber BH, Stohr H. Invest Ophthalmol Vis Sci. 2008;49:2812–2822. doi: 10.1167/iovs.07-1444. [DOI] [PubMed] [Google Scholar]
- Ju CR, Xia XZ, Chen RC. Chin Med J (Engl) 2007;120:1505–1510. [PubMed] [Google Scholar]
- Kalus I, Bormann U, Mzoughi M, Schachner M, Kleene R. J Neurochem. 2006;98:78–88. doi: 10.1111/j.1471-4159.2006.03847.x. [DOI] [PubMed] [Google Scholar]
- Karan D, Lin FC, Bryan M, Ringel J, Moniaux N, Lin MF, Batra SK. Int J Oncol. 2003;23:1365–1371. [PubMed] [Google Scholar]
- Karkkainen I, Rybnikova E, Pelto-Huikko M, Huovila AP. Mol Cell Neurosci. 2000;15:547–560. doi: 10.1006/mcne.2000.0848. [DOI] [PubMed] [Google Scholar]
- Kassiri Z, Oudit GY, Kandalam V, Awad A, Wang X, Ziou X, Maeda N, Herzenberg AM, Scholey JW. J Am Soc Nephrol. 2009;20:1223–1235. doi: 10.1681/ASN.2008050492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassiri Z, Oudit GY, Sanchez O, Dawood F, Mohammed FF, Nuttall RK, Edwards DR, Liu PP, Backx PH, Khokha R. Circ Res. 2005;97:380–390. doi: 10.1161/01.RES.0000178789.16929.cf. [DOI] [PubMed] [Google Scholar]
- Katakowski M, Chen J, Zhang ZG, Santra M, Wang Y, Chopp M. J Cereb Blood Flow Metab. 2007;27:669–678. doi: 10.1038/sj.jcbfm.9600390. [DOI] [PubMed] [Google Scholar]
- Katakowski M, Jiang F, Zheng X, Gutierrez JA, Szalad A, Chopp M. Cancer Sci. 2009;100:1597–1604. doi: 10.1111/j.1349-7006.2009.01221.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi M, Mitsuhashi Y, Kondo S. Br J Dermatol. 2005;152:915–919. doi: 10.1111/j.1365-2133.2005.06440.x. [DOI] [PubMed] [Google Scholar]
- Kawaguchi N, Sundberg C, Kveiborg M, Moghadaszadeh B, Asmar M, Dietrich N, Thodeti CK, Nielsen FC, Moller P, Mercurio AM, Albrechtsen R, Wewer UM. J Cell Sci. 2003;116:3893–3904. doi: 10.1242/jcs.00699. [DOI] [PubMed] [Google Scholar]
- Kenny PA, Bissell MJ. J Clin Invest. 2007;117:337–345. doi: 10.1172/JCI29518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kermarrec N, Selloum S, Plantefeve G, Chosidow D, Paoletti X, Lopez A, Mantz J, Desmonts JM, Gougerot-Pocidalo MA, Chollet-Martin S. Crit Care Med. 2005;33:1359–1364. doi: 10.1097/01.ccm.0000166359.47577.57. [DOI] [PubMed] [Google Scholar]
- Kieseier BC, Pischel H, Neuen-Jacob E, Tourtellotte WW, Hartung HP. Glia. 2003;42:398–405. doi: 10.1002/glia.10226. [DOI] [PubMed] [Google Scholar]
- Kim ML, Zhang B, Mills IP, Milla ME, Brunden KR, Lee VM. J Neurosci. 2008;28:12052–12061. doi: 10.1523/JNEUROSCI.2913-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kini RM, Evans HJ. Toxicon. 1992;30:265–293. doi: 10.1016/0041-0101(92)90869-7. [DOI] [PubMed] [Google Scholar]
- Koenen RR, Pruessmeyer J, Soehnlein O, Fraemohs L, Zernecke A, Schwarz N, Reiss K, Sarabi A, Lindbom L, Hackeng TM, Weber C, Ludwig A. Blood. 2009;113:4799–4809. doi: 10.1182/blood-2008-04-152330. [DOI] [PubMed] [Google Scholar]
- Kojro E, Fahrenholz F. Subcell Biochem. 2005;38:105–127. doi: 10.1007/0-387-23226-5_5. [DOI] [PubMed] [Google Scholar]
- Kojro E, Postina R, Buro C, Meiringer C, Gehrig-Burger K, Fahrenholz F. Faseb J. 2006;20:512–514. doi: 10.1096/fj.05-4812fje. [DOI] [PubMed] [Google Scholar]
- Kretzschmar M, Doody J, Massague J. Nature. 1997;389:618–622. doi: 10.1038/39348. [DOI] [PubMed] [Google Scholar]
- Kriegler M, Perez C, DeFay K, Albert I, Lu SD. Cell. 1988;53:45–53. doi: 10.1016/0092-8674(88)90486-2. [DOI] [PubMed] [Google Scholar]
- Kuwahara I, Lillehoj EP, Koga T, Isohama Y, Miyata T, Kim KC. Am J Respir Cell Mol Biol. 2007;37:691–698. doi: 10.1165/rcmb.2007-0072OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak HI, Mendoza EA, Bayless KJ. Matrix Biol. 2009;28:470–479. doi: 10.1016/j.matbio.2009.07.007. [DOI] [PubMed] [Google Scholar]
- Lambert MH, Blackburn RK, Seaton TD, Kassel DB, Kinder DS, Leesnitzer MA, Bickett DM, Warner JR, Andersen MW, Badiang JG, Cowan DJ, Gaul MD, Petrov KG, Rabinowitz MH, Wiethe RW, Becherer JD, McDougald DL, Musso DL, Andrews RC, Moss ML. Comb Chem High Throughput Screen. 2005;8:327–339. doi: 10.2174/1386207054020840. [DOI] [PubMed] [Google Scholar]
- Lammich S, Kojro E, Postina R, Gilbert S, Pfeiffer R, Jasionowski M, Haass C, Fahrenholz F. Proc Natl Acad Sci U S A. 1999;96:3922–3927. doi: 10.1073/pnas.96.7.3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lautrette A, Li S, Alili R, Sunnarborg SW, Burtin M, Lee DC, Friedlander G, Terzi F. Nat Med. 2005;11:867–874. doi: 10.1038/nm1275. [DOI] [PubMed] [Google Scholar]
- Lee MH, Verma V, Maskos K, Becherer JD, Knauper V, Dodds P, Amour A, Murphy G. FEBS Lett. 2002;520:102–106. doi: 10.1016/s0014-5793(02)02776-x. [DOI] [PubMed] [Google Scholar]
- Leib SL, Clements JM, Lindberg RL, Heimgartner C, Loeffler JM, Pfister LA, Tauber MG, Leppert D. Brain. 2001;124:1734–1742. doi: 10.1093/brain/124.9.1734. [DOI] [PubMed] [Google Scholar]
- Leissring MA, Murphy MP, Mead TR, Akbari Y, Sugarman MC, Jannatipour M, Anliker B, Muller U, Saftig P, De Strooper B, Wolfe MS, Golde TE, LaFerla FM. Proc Natl Acad Sci U S A. 2002;99:4697–4702. doi: 10.1073/pnas.072033799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemjabbar H, Li D, Gallup M, Sidhu S, Drori E, Basbaum C. J Biol Chem. 2003;278:26202–26207. doi: 10.1074/jbc.M207018200. [DOI] [PubMed] [Google Scholar]
- Leonard JD, Lin F, Milla ME. Biochem J. 2005;387:797–805. doi: 10.1042/BJ20041727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis J. Nature. 1998;393:304–305. doi: 10.1038/30597. [DOI] [PubMed] [Google Scholar]
- Li N, Boyd K, Dempsey PJ, Vignali DA. J Immunol. 2007a;178:4214–4221. doi: 10.4049/jimmunol.178.7.4214. [DOI] [PubMed] [Google Scholar]
- Li X, Fan H. J Biol Chem. 2004;279:27365–27375. doi: 10.1074/jbc.M401690200. [DOI] [PubMed] [Google Scholar]
- Li X, Yan Y, Huang W, Yang Y, Wang H, Chang L. Mol Biol Rep. 2009;36:641–651. doi: 10.1007/s11033-008-9224-5. [DOI] [PubMed] [Google Scholar]
- Li Z, Mei Y, Liu X, Zhou M. Cell Signal. 2007b;19:466–471. doi: 10.1016/j.cellsig.2006.07.020. [DOI] [PubMed] [Google Scholar]
- Lovering F, Zhang Y. Curr Drug Targets CNS Neurol Disord. 2005;4:161–168. doi: 10.2174/1568007053544147. [DOI] [PubMed] [Google Scholar]
- Luetteke NC, Qiu TH, Fenton SE, Troyer KL, Riedel RF, Chang A, Lee DC. Development. 1999;126:2739–2750. doi: 10.1242/dev.126.12.2739. [DOI] [PubMed] [Google Scholar]
- Luetteke NC, Qiu TH, Peiffer RL, Oliver P, Smithies O, Lee DC. Cell. 1993;73:263–278. doi: 10.1016/0092-8674(93)90228-i. [DOI] [PubMed] [Google Scholar]
- Madrigal JL, Hurtado O, Moro MA, Lizasoain I, Lorenzo P, Castrillo A, Bosca L, Leza JC. Neuropsychopharmacology. 2002;26:155–163. doi: 10.1016/S0893-133X(01)00292-5. [DOI] [PubMed] [Google Scholar]
- Mahmoodi M, Sahebjam S, Smookler D, Khokha R, Mort JS. Am J Pathol. 2005;166:1733–1740. doi: 10.1016/S0002-9440(10)62483-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majka S, McGuire PG, Das A. Invest Ophthalmol Vis Sci. 2002;43:260–266. [PubMed] [Google Scholar]
- Mann GB, Fowler KJ, Gabriel A, Nice EC, Williams RL, Dunn AR. Cell. 1993;73:249–261. doi: 10.1016/0092-8674(93)90227-h. [DOI] [PubMed] [Google Scholar]
- Marcinkiewicz C. Curr Pharm Des. 2005;11:815–827. doi: 10.2174/1381612053381765. [DOI] [PubMed] [Google Scholar]
- Marcinkiewicz C, Calvete JJ, Vijay-Kumar S, Marcinkiewicz MM, Raida M, Schick P, Lobb RR, Niewiarowski S. Biochemistry. 1999;38:13302–13309. doi: 10.1021/bi9906930. [DOI] [PubMed] [Google Scholar]
- Maretzky T, Schulte M, Ludwig A, Rose-John S, Blobel C, Hartmann D, Altevogt P, Saftig P, Reiss K. Mol Cell Biol. 2005;25:9040–9053. doi: 10.1128/MCB.25.20.9040-9053.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin V, Montero-Julian F, Gres S, Bongrand P, Farnarier C, Kaplanski G. Eur J Immunol. 2002;32:2965–2970. doi: 10.1002/1521-4141(2002010)32:10<2965::AID-IMMU2965>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- Martin EL, Moyer BZ, Pape MC, Starcher B, Leco KJ, Veldhuizen RA. Am J Physiol Lung Cell Mol Physiol. 2003;285:L1222–L1232. doi: 10.1152/ajplung.00141.2003. [DOI] [PubMed] [Google Scholar]
- Maskos K, Fernandez-Catalan C, Huber R, Bourenkov GP, Bartunik H, Ellestad GA, Reddy P, Wolfson MF, Rauch CT, Castner BJ, Davis R, Clarke HR, Petersen M, Fitzner JN, Cerretti DP, March CJ, Paxton RJ, Black RA, Bode W. Proc Natl Acad Sci U S A. 1998;95:3408–3412. doi: 10.1073/pnas.95.7.3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters CL, Simms G, Weinman NA, Multhaup G, McDonald BL, Beyreuther K. Proc Natl Acad Sci U S A. 1985;82:4245–4249. doi: 10.1073/pnas.82.12.4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott MF, Aksentijevich I, Galon J, McDermott EM, Ogunkolade BW, Centola M, Mansfield E, Gadina M, Karenko L, Pettersson T, McCarthy J, Frucht DM, Aringer M, Torosyan Y, Teppo AM, Wilson M, Karaarslan HM, Wan Y, Todd I, Wood G, Schlimgen R, Kumarajeewa TR, Cooper SM, Vella JP, Amos CI, Mulley J, Quane KA, Molloy MG, Ranki A, Powell RJ, Hitman GA, O'Shea JJ, Kastner DL. Cell. 1999;97:133–144. doi: 10.1016/s0092-8674(00)80721-7. [DOI] [PubMed] [Google Scholar]
- McGeehan GM, Becherer JD, Bast RC, Jr, Boyer CM, Champion B, Connolly KM, Conway JG, Furdon P, Karp S, Kidao S, et al. Nature. 1994;370:558–561. doi: 10.1038/370558a0. [DOI] [PubMed] [Google Scholar]
- McGowan PM, McKiernan E, Bolster F, Ryan BM, Hill AD, McDermott EW, Evoy D, O'Higgins N, Crown J, Duffy MJ. Ann Oncol. 2008;19:1075–1081. doi: 10.1093/annonc/mdm609. [DOI] [PubMed] [Google Scholar]
- McGowan PM, Ryan BM, Hill AD, McDermott E, O'Higgins N, Duffy MJ. Clin Cancer Res. 2007;13:2335–2343. doi: 10.1158/1078-0432.CCR-06-2092. [DOI] [PubMed] [Google Scholar]
- Meli DN, Loeffler JM, Baumann P, Neumann U, Buhl T, Leppert D, Leib SL. J Neuroimmunol. 2004;151:6–11. doi: 10.1016/j.jneuroim.2004.01.026. [DOI] [PubMed] [Google Scholar]
- Menghini R, Menini S, Amoruso R, Fiorentino L, Casagrande V, Marzano V, Tornei F, Bertucci P, Iacobini C, Serino M, Porzio O, Hribal ML, Folli F, Khokha R, Urbani A, Lauro R, Pugliese G, Federici M. Gastroenterology. 2009;136:663–672. doi: 10.1053/j.gastro.2008.10.079. e4. [DOI] [PubMed] [Google Scholar]
- Mercer B, Markland F, Minkin C. J Bone Miner Res. 1998;13:409–414. doi: 10.1359/jbmr.1998.13.3.409. [DOI] [PubMed] [Google Scholar]
- Merchant NB, Rogers CM, Trivedi B, Morrow J, Coffey RJ. Surgery. 2005;138:415–421. doi: 10.1016/j.surg.2005.06.030. [DOI] [PubMed] [Google Scholar]
- Merchant NB, Voskresensky I, Rogers CM, Lafleur B, Dempsey PJ, Graves-Deal R, Revetta F, Foutch AC, Rothenberg ML, Washington MK, Coffey RJ. Clin Cancer Res. 2008;14:1182–1191. doi: 10.1158/1078-0432.CCR-07-1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miettinen PJ, Berger JE, Meneses J, Phung Y, Pedersen RA, Werb Z, Derynck R. Nature. 1995;376:337–341. doi: 10.1038/376337a0. [DOI] [PubMed] [Google Scholar]
- Mohler KM, Sleath PR, Fitzner JN, Cerretti DP, Alderson M, Kerwar SS, Torrance DS, Otten-Evans C, Greenstreet T, Weerawarna K, et al. Nature. 1994;370:218–220. doi: 10.1038/370218a0. [DOI] [PubMed] [Google Scholar]
- Mohler KM, Torrance DS, Smith CA, Goodwin RG, Stremler KE, Fung VP, Madani H, Widmer MB. J Immunol. 1993;151:1548–1561. [PubMed] [Google Scholar]
- Moriyama H, Tsukida T, Inoue Y, Yokota K, Yoshino K, Kondo H, Miura N, Nishimura S. J Med Chem. 2004;47:1930–1938. doi: 10.1021/jm0304313. [DOI] [PubMed] [Google Scholar]
- Moss ML, Bomar M, Liu Q, Sage H, Dempsey P, Lenhart PM, Gillispie PA, Stoeck A, Wildeboer D, Bartsch JW, Palmisano R, Zhou P. J Biol Chem. 2007;282:35712–35721. doi: 10.1074/jbc.M703231200. [DOI] [PubMed] [Google Scholar]
- Moss ML, Jin SL, Becherer JD, Bickett DM, Burkhart W, Chen WJ, Hassler D, Leesnitzer MT, McGeehan G, Milla M, Moyer M, Rocque W, Seaton T, Schoenen F, Warner J, Willard D. J Neuroimmunol. 1997a;72:127–129. doi: 10.1016/s0165-5728(96)00180-4. [DOI] [PubMed] [Google Scholar]
- Moss ML, Jin SL, Milla ME, Bickett DM, Burkhart W, Carter HL, Chen WJ, Clay WC, Didsbury JR, Hassler D, Hoffman CR, Kost TA, Lambert MH, Leesnitzer MA, McCauley P, McGeehan G, Mitchell J, Moyer M, Pahel G, Rocque W, Overton LK, Schoenen F, Seaton T, Su JL, Becherer JD, et al. Nature. 1997b;385:733–736. doi: 10.1038/385733a0. [DOI] [PubMed] [Google Scholar]
- Moss ML, Sklair-Tavron L, Nudelman R. Nat Clin Pract Rheumatol. 2008;4:300–309. doi: 10.1038/ncprheum0797. [DOI] [PubMed] [Google Scholar]
- Murali R, Brennan PJ, Kieber-Emmons T, Greene MI. Proc Natl Acad Sci U S A. 1996;93:6252–6257. doi: 10.1073/pnas.93.13.6252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K, Iwamoto R, Mekada E. J Cell Biol. 1995;129:1691–1705. doi: 10.1083/jcb.129.6.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Abe H, Hirata A, Shimoda C. Eukaryot Cell. 2004;3:27–39. doi: 10.1128/EC.3.1.27-39.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanba D, Mammoto A, Hashimoto K, Higashiyama S. J Cell Biol. 2003;163:489–502. doi: 10.1083/jcb.200303017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath D, Slocombe PM, Stephens PE, Warn A, Hutchinson GR, Yamada KM, Docherty AJ, Murphy G. J Cell Sci. 1999;112(Pt 4):579–587. doi: 10.1242/jcs.112.4.579. [DOI] [PubMed] [Google Scholar]
- Nelson KK, Schlondorff J, Blobel CP. Biochem J. 1999;343(Pt 3):673–680. [PMC free article] [PubMed] [Google Scholar]
- Nemo R, Murcia N, Dell KM. Pediatr Res. 2005;57:732–737. doi: 10.1203/01.PDR.0000159513.51898.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton RC, Decicco CP. J Med Chem. 1999;42:2295–2314. doi: 10.1021/jm980541n. [DOI] [PubMed] [Google Scholar]
- Nishi E, Hiraoka Y, Yoshida K, Okawa K, Kita T. J Biol Chem. 2006;281:31164–31172. doi: 10.1074/jbc.M601316200. [DOI] [PubMed] [Google Scholar]
- Oh ST, Schramme A, Stark A, Tilgen W, Gutwein P, Reichrath J. J Cutan Pathol. 2009;36:395–401. doi: 10.1111/j.1600-0560.2008.01082.x. [DOI] [PubMed] [Google Scholar]
- Ohta S, Harigai M, Tanaka M, Kawaguchi Y, Sugiura T, Takagi K, Fukasawa C, Hara M, Kamatani N. J Rheumatol. 2001;28:1756–1763. [PubMed] [Google Scholar]
- Ohtsu H, Dempsey PJ, Frank GD, Brailoiu E, Higuchi S, Suzuki H, Nakashima H, Eguchi K, Eguchi S. Arterioscler Thromb Vasc Biol. 2006;26:e133–e137. doi: 10.1161/01.ATV.0000236203.90331.d0. [DOI] [PubMed] [Google Scholar]
- Olfa KZ, Jose L, Salma D, Amine B, Najet SA, Nicolas A, Maxime L, Raoudha Z, Kamel M, Jacques M, Jean-Marc S, Mohamed el A, Naziha M. Lab Invest. 2005;85:1507–1516. doi: 10.1038/labinvest.3700350. [DOI] [PubMed] [Google Scholar]
- Papadakis KA, Targan SR. Annu Rev Med. 2000;51:289–298. doi: 10.1146/annurev.med.51.1.289. [DOI] [PubMed] [Google Scholar]
- Patel IR, Attur MG, Patel RN, Stuchin SA, Abagyan RA, Abramson SB, Amin AR. J Immunol. 1998;160:4570–4579. [PubMed] [Google Scholar]
- Peiretti F, Canault M, Deprez-Beauclair P, Berthet V, Bonardo B, Juhan-Vague I, Nalbone G. Exp Cell Res. 2003a;285:278–285. doi: 10.1016/s0014-4827(03)00052-1. [DOI] [PubMed] [Google Scholar]
- Peiretti F, Deprez-Beauclair P, Bonardo B, Aubert H, Juhan-Vague I, Nalbone G. J Cell Sci. 2003b;116:1949–1957. doi: 10.1242/jcs.00415. [DOI] [PubMed] [Google Scholar]
- Peng Y, Lee DY, Jiang L, Ma Z, Schachter SC, Lemere CA. Neuroscience. 2007;150:386–395. doi: 10.1016/j.neuroscience.2007.09.022. [DOI] [PubMed] [Google Scholar]
- Perez L, Kerrigan JE, Li X, Fan H. Biochem Cell Biol. 2007;85:141–149. doi: 10.1139/o06-179. [DOI] [PubMed] [Google Scholar]
- Peschon JJ, Slack JL, Reddy P, Stocking KL, Sunnarborg SW, Lee DC, Russell WE, Castner BJ, Johnson RS, Fitzner JN, Boyce RW, Nelson N, Kozlosky CJ, Wolfson MF, Rauch CT, Cerretti DP, Paxton RJ, March CJ, Black RA. Science. 1998;282:1281–1284. doi: 10.1126/science.282.5392.1281. [DOI] [PubMed] [Google Scholar]
- Planque C, Kulasingan V, Smith CR, Reckamp K, Goodglick L, Diamandis EP. Mol Cell Proteomics. 2009 doi: 10.1074/mcp.M900134-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plumb J, McQuaid S, Cross AK, Surr J, Haddock G, Bunning RA, Woodroofe MN. Mult Scler. 2006;12:375–385. doi: 10.1191/135248506ms1276oa. [DOI] [PubMed] [Google Scholar]
- Pradillo JM, Romera C, Hurtado O, Cardenas A, Moro MA, Leza JC, Davalos A, Castillo J, Lorenzo P, Lizasoain I. J Cereb Blood Flow Metab. 2005;25:193–203. doi: 10.1038/sj.jcbfm.9600019. [DOI] [PubMed] [Google Scholar]
- Prenzel N, Zwick E, Daub H, Leserer M, Abraham R, Wallasch C, Ullrich A. Nature. 1999;402:884–888. doi: 10.1038/47260. [DOI] [PubMed] [Google Scholar]
- Qu D, Wang Y, Esmon NL, Esmon CT. J Thromb Haemost. 2007;5:395–402. doi: 10.1111/j.1538-7836.2007.02347.x. [DOI] [PubMed] [Google Scholar]
- Rabinowitz MH, Andrews RC, Becherer JD, Bickett DM, Bubacz DG, Conway JG, Cowan DJ, Gaul M, Glennon K, Lambert MH, Leesnitzer MA, McDougald DL, Moss ML, Musso DL, Rizzolio MC. J Med Chem. 2001;44:4252–4267. doi: 10.1021/jm0102654. [DOI] [PubMed] [Google Scholar]
- Reddy AB, Ramana KV, Srivastava S, Bhatnagar A, Srivastava SK. Endocrinology. 2009;150:63–74. doi: 10.1210/en.2008-0677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy P, Slack JL, Davis R, Cerretti DP, Kozlosky CJ, Blanton RA, Shows D, Peschon JJ, Black RA. J Biol Chem. 2000;275:14608–14614. doi: 10.1074/jbc.275.19.14608. [DOI] [PubMed] [Google Scholar]
- Reiss K, Saftig P. Semin Cell Dev Biol. 2009;20:126–137. doi: 10.1016/j.semcdb.2008.11.002. [DOI] [PubMed] [Google Scholar]
- Richards WG, Sweeney WE, Yoder BK, Wilkinson JE, Woychik RP, Avner ED. J Clin Invest. 1998;101:935–939. doi: 10.1172/JCI2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertshaw HJ, Brennan FM. Br J Anaesth. 2005;94:222–228. doi: 10.1093/bja/aei021. [DOI] [PubMed] [Google Scholar]
- Romera C, Hurtado O, Botella SH, Lizasoain I, Cardenas A, Fernandez-Tome P, Leza JC, Lorenzo P, Moro MA. J Neurosci. 2004;24:1350–1357. doi: 10.1523/JNEUROSCI.1596-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovida E, Paccagnini A, Del Rosso M, Peschon J, Dello Sbarba P. J Immunol. 2001;166:1583–1589. doi: 10.4049/jimmunol.166.3.1583. [DOI] [PubMed] [Google Scholar]
- Rubio-Araiz A, Arevalo-Martin A, Gomez-Torres O, Navarro-Galve B, Garcia-Ovejero D, Suetterlin P, Sanchez-Heras E, Molina-Holgado E, Molina-Holgado F. Mol Cell Neurosci. 2008;38:374–380. doi: 10.1016/j.mcn.2008.03.010. [DOI] [PubMed] [Google Scholar]
- Ruchatz H, Leung BP, Wei XQ, McInnes IB, Liew FY. J Immunol. 1998;160:5654–5660. [PubMed] [Google Scholar]
- Rundhaug JE. J Cell Mol Med. 2005;9:267–285. doi: 10.1111/j.1582-4934.2005.tb00355.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahebjam S, Khokha R, Mort JS. Arthritis Rheum. 2007;56:905–909. doi: 10.1002/art.22427. [DOI] [PubMed] [Google Scholar]
- Sanderson MP, Abbott CA, Tada H, Seno M, Dempsey PJ, Dunbar AJ. J Cell Biochem. 2006;99:609–623. doi: 10.1002/jcb.20968. [DOI] [PubMed] [Google Scholar]
- Sastre M, Walter J, Gentleman SM. J Neuroinflammation. 2008;5:25. doi: 10.1186/1742-2094-5-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh M, Ishikawa Y, Itoh T, Minami Y, Takahashi Y, Nakamura M. Eur J Clin Invest. 2008;38:97–105. doi: 10.1111/j.1365-2362.2007.01912.x. [DOI] [PubMed] [Google Scholar]
- Satoh M, Iwasaka J, Nakamura M, Akatsu T, Shimoda Y, Hiramori K. Eur J Heart Fail. 2004;6:869–875. doi: 10.1016/j.ejheart.2004.02.007. [DOI] [PubMed] [Google Scholar]
- Satoh M, Nakamura M, Satoh H, Saitoh H, Segawa I, Hiramori K. J Am Coll Cardiol. 2000;36:1288–1294. doi: 10.1016/s0735-1097(00)00827-5. [DOI] [PubMed] [Google Scholar]
- Schafer B, Marg B, Gschwind A, Ullrich A. J Biol Chem. 2004;279:47929–47938. doi: 10.1074/jbc.M400129200. [DOI] [PubMed] [Google Scholar]
- Schaff U, Mattila PE, Simon SI, Walcheck B. J Leukoc Biol. 2008;83:99–105. doi: 10.1189/jlb.0507304. [DOI] [PubMed] [Google Scholar]
- Schlondorff J, Becherer JD, Blobel CP. Biochem J. 2000;347(Pt 1):131–138. [PMC free article] [PubMed] [Google Scholar]
- Schlondorff J, Blobel CP. J Cell Sci. 1999;112(Pt 21):3603–3617. doi: 10.1242/jcs.112.21.3603. [DOI] [PubMed] [Google Scholar]
- Schmitmeier S, Markland FS, Ritter MR, Sawcer DE, Chen TC. Cell Commun Adhes. 2003;10:1–16. doi: 10.1080/15419060302062. [DOI] [PubMed] [Google Scholar]
- Schramme A, Abdel-Bakky MS, Kampfer-Kolb N, Pfeilschifter J, Gutwein P. Biochem Biophys Res Commun. 2008;370:311–316. doi: 10.1016/j.bbrc.2008.03.088. [DOI] [PubMed] [Google Scholar]
- Schroeter EH, Kisslinger JA, Kopan R. Nature. 1998;393:382–386. doi: 10.1038/30756. [DOI] [PubMed] [Google Scholar]
- Seifert T, Kieseier BC, Ropele S, Strasser-Fuchs S, Quehenberger F, Fazekas F, Hartung HP. Mult Scler. 2002;8:447–451. doi: 10.1191/1352458502ms830oa. [DOI] [PubMed] [Google Scholar]
- Serino M, Menghini R, Fiorentino L, Amoruso R, Mauriello A, Lauro D, Sbraccia P, Hribal ML, Lauro R, Federici M. Diabetes. 2007;56:2541–2546. doi: 10.2337/db07-0360. [DOI] [PubMed] [Google Scholar]
- Serwin AB, Sokolowska M, Chodynicka B. Photodermatol Photoimmunol Photomed. 2007;23:130–134. doi: 10.1111/j.1600-0781.2007.00296.x. [DOI] [PubMed] [Google Scholar]
- Shah BH, Catt KJ. Trends Pharmacol Sci. 2006;27:235–237. doi: 10.1016/j.tips.2006.03.010. [DOI] [PubMed] [Google Scholar]
- Shi W, Chen H, Sun J, Buckley S, Zhao J, Anderson KD, Williams RG, Warburton D. Dev Biol. 2003;261:371–380. doi: 10.1016/s0012-1606(03)00315-4. [DOI] [PubMed] [Google Scholar]
- Shimoda Y, Satoh M, Nakamura M, Akatsu T, Hiramori K. Clin Sci (Lond) 2005;108:339–347. doi: 10.1042/CS20040229. [DOI] [PubMed] [Google Scholar]
- Sibilia M, Wagner EF. Science. 1995;269:234–238. doi: 10.1126/science.7618085. [DOI] [PubMed] [Google Scholar]
- Singh RJ, Mason JC, Lidington EA, Edwards DR, Nuttall RK, Khokha R, Knauper V, Murphy G, Gavrilovic J. Cardiovasc Res. 2005;67:39–49. doi: 10.1016/j.cardiores.2005.02.020. [DOI] [PubMed] [Google Scholar]
- Six E, Ndiaye D, Laabi Y, Brou C, Gupta-Rossi N, Israel A, Logeat F. Proc Natl Acad Sci U S A. 2003;100:7638–7643. doi: 10.1073/pnas.1230693100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skovronsky DM, Fath S, Lee VM, Milla ME. J Neurobiol. 2001;49:40–46. doi: 10.1002/neu.1064. [DOI] [PubMed] [Google Scholar]
- Smith KM, Gaultier A, Cousin H, Alfandari D, White JM, DeSimone DW. J Cell Biol. 2002;159:893–902. doi: 10.1083/jcb.200206023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smookler DS, Mohammed FF, Kassiri Z, Duncan GS, Mak TW, Khokha R. J Immunol. 2006;176:721–725. doi: 10.4049/jimmunol.176.2.721. [DOI] [PubMed] [Google Scholar]
- Solomon A, Akabayov B, Frenkel A, Milla ME, Sagi I. Proc Natl Acad Sci U S A. 2007;104:4931–4936. doi: 10.1073/pnas.0700066104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song D, Ye X, Xu H, Liu SF. Blood. 2009;114:2521–2529. doi: 10.1182/blood-2009-02-205914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soond SM, Everson B, Riches DW, Murphy G. J Cell Sci. 2005;118:2371–2380. doi: 10.1242/jcs.02357. [DOI] [PubMed] [Google Scholar]
- Spertini O, Luscinskas FW, Gimbrone MA, Jr, Tedder TF. J Exp Med. 1992;175:1789–1792. doi: 10.1084/jem.175.6.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surena AL, de Faria GP, Studler JM, Peiretti F, Pidoux M, Camonis J, Chneiweiss H, Formstecher E, Junier MP. Biochim Biophys Acta. 2009;1793:264–272. doi: 10.1016/j.bbamcr.2008.09.005. [DOI] [PubMed] [Google Scholar]
- Swendeman S, Mendelson K, Weskamp G, Horiuchi K, Deutsch U, Scherle P, Hooper A, Rafii S, Blobel CP. Circ Res. 2008;103:916–918. doi: 10.1161/CIRCRESAHA.108.184416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachida Y, Nakagawa K, Saito T, Saido TC, Honda T, Saito Y, Murayama S, Endo T, Sakaguchi G, Kato A, Kitazume S, Hashimoto Y. J Neurochem. 2008;104:1387–1393. doi: 10.1111/j.1471-4159.2007.05127.x. [DOI] [PubMed] [Google Scholar]
- Takamune Y, Ikebe T, Nagano O, Shinohara M. Biochem Biophys Res Commun. 2008;365:393–398. doi: 10.1016/j.bbrc.2007.11.010. [DOI] [PubMed] [Google Scholar]
- Tanabe Y, Kasahara T, Momoi T, Fujita E. Neurosci Lett. 2008;444:16–21. doi: 10.1016/j.neulet.2008.08.023. [DOI] [PubMed] [Google Scholar]
- Tanahashi H, Yoshioka K. Neurosci Lett. 2008;439:293–297. doi: 10.1016/j.neulet.2008.05.039. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Nanba D, Mori S, Shiba F, Ishiguro H, Yoshino K, Matsuura N, Higashiyama S. J Biol Chem. 2004;279:41950–41959. doi: 10.1074/jbc.M400086200. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Miyamoto S, Suzuki SO, Oki E, Yagi H, Sonoda K, Yamazaki A, Mizushima H, Maehara Y, Mekada E, Nakano H. Clin Cancer Res. 2005;11:4783–4792. doi: 10.1158/1078-0432.CCR-04-1426. [DOI] [PubMed] [Google Scholar]
- Tellier E, Canault M, Rebsomen L, Bonardo B, Juhan-Vague I, Nalbone G, Peiretti F. Exp Cell Res. 2006;312:3969–3980. doi: 10.1016/j.yexcr.2006.08.027. [DOI] [PubMed] [Google Scholar]
- Threadgill DW, Dlugosz AA, Hansen LA, Tennenbaum T, Lichti U, Yee D, LaMantia C, Mourton T, Herrup K, Harris RC, et al. Science. 1995;269:230–234. doi: 10.1126/science.7618084. [DOI] [PubMed] [Google Scholar]
- Togashi N, Ura N, Higashiura K, Murakami H, Shimamoto K. Hypertension. 2002;39:578–580. doi: 10.1161/hy0202.103290. [DOI] [PubMed] [Google Scholar]
- Tousseyn T, Thathiah A, Jorissen E, Raemaekers T, Konietzko U, Reiss K, Maes E, Snellinx A, Serneels L, Nyabi O, Annaert W, Saftig P, Hartmann D, De Strooper B. J Biol Chem. 2009;284:11738–11747. doi: 10.1074/jbc.M805894200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifilieff A, Walker C, Keller T, Kottirsch G, Neumann U. Br J Pharmacol. 2002;135:1655–1664. doi: 10.1038/sj.bjp.0704616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trikha M, De Clerck YA, Markland FS. Cancer Res. 1994;54:4993–4998. [PubMed] [Google Scholar]
- Tsai TJ, Sheu JR, Chen YM, Yen CJ, Chen CF, Huang TF. Nephron. 1995;70:83–90. doi: 10.1159/000188549. [DOI] [PubMed] [Google Scholar]
- Tsai WC, Hsu PW, Lai TC, Chau GY, Lin CW, Chen CM, Lin CD, Liao YL, Wang JL, Chau YP, Hsu MT, Hsiao M, Huang HD, Tsou AP. Hepatology. 2009;49:1571–1582. doi: 10.1002/hep.22806. [DOI] [PubMed] [Google Scholar]
- Tsakadze NL, Sithu SD, Sen U, English WR, Murphy G, D'Souza SE. J Biol Chem. 2006;281:3157–3164. doi: 10.1074/jbc.M510797200. [DOI] [PubMed] [Google Scholar]
- Tselepis VH, Green LJ, Humphries MJ. J Biol Chem. 1997;272:21341–21348. doi: 10.1074/jbc.272.34.21341. [DOI] [PubMed] [Google Scholar]
- Tsuji F, Oki K, Okahara A, Suhara H, Yamanouchi T, Sasano M, Mita S, Horiuchi M. Cytokine. 2002;17:294–300. doi: 10.1006/cyto.2002.1015. [DOI] [PubMed] [Google Scholar]
- Uchiyama-Tanaka Y, Matsubara H, Nozawa Y, Murasawa S, Mori Y, Kosaki A, Maruyama K, Masaki H, Shibasaki Y, Fujiyama S, Nose A, Iba O, Hasagawa T, Tateishi E, Higashiyama S, Iwasaka T. Kidney Int. 2001;60:2153–2163. doi: 10.1046/j.1523-1755.2001.00067.x. [DOI] [PubMed] [Google Scholar]
- Utsugi T, Ohno T, Ohyama Y, Uchiyama T, Saito Y, Matsumura Y, Aizawa H, Itoh H, Kurabayashi M, Kawazu S, Tomono S, Oka Y, Suga T, Kuro-o M, Nabeshima Y, Nagai R. Metabolism. 2000;49:1118–1123. doi: 10.1053/meta.2000.8606. [DOI] [PubMed] [Google Scholar]
- Venkatesan AM, Davis JM, Grosu GT, Baker J, Zask A, Levin JI, Ellingboe J, Skotnicki JS, Dijoseph JF, Sung A, Jin G, Xu W, McCarthy DJ, Barone D. J Med Chem. 2004;47:6255–6269. doi: 10.1021/jm040086x. [DOI] [PubMed] [Google Scholar]
- Walker EJ, Rosenberg GA. Exp Neurol. 2009;216:122–131. doi: 10.1016/j.expneurol.2008.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Al-Lamki RS, Zhang H, Kirkiles-Smith N, Gaeta ML, Thiru S, Pober JS, Bradley JR. J Biol Chem. 2003;278:21751–21760. doi: 10.1074/jbc.M212662200. [DOI] [PubMed] [Google Scholar]
- Wang X, Feuerstein GZ, Xu L, Wang H, Schumacher WA, Ogletree ML, Taub R, Duan JJ, Decicco CP, Liu RQ. Mol Pharmacol. 2004;65:890–896. doi: 10.1124/mol.65.4.890. [DOI] [PubMed] [Google Scholar]
- Wang X, He K, Gerhart M, Huang Y, Jiang J, Paxton RJ, Yang S, Lu C, Menon RK, Black RA, Baumann G, Frank SJ. J Biol Chem. 2002;277:50510–50519. doi: 10.1074/jbc.M208738200. [DOI] [PubMed] [Google Scholar]
- Wang X, Jiang J, Warram J, Baumann G, Gan Y, Menon RK, Denson LA, Zinn KR, Frank SJ. Mol Endocrinol. 2008;22:1427–1437. doi: 10.1210/me.2007-0561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Oka T, Chow FL, Cooper SB, Odenbach J, Lopaschuk GD, Kassiri Z, Fernandez-Patron C. Hypertension. 2009a;54:575–582. doi: 10.1161/HYPERTENSIONAHA.108.127670. [DOI] [PubMed] [Google Scholar]
- Wang Y, Herrera AH, Li Y, Belani KK, Walcheck B. J Immunol. 2009b;182:2449–2457. doi: 10.4049/jimmunol.0802770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinger JG, Omari KM, Marsden K, Raine CS, Shafit-Zagardo B. Am J Pathol. 2009;175:283–293. doi: 10.2353/ajpath.2009.080807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wewer UM, Morgelin M, Holck P, Jacobsen J, Lydolph MC, Johnsen AH, Kveiborg M, Albrechtsen R. J Biol Chem. 2006;281:9418–9422. doi: 10.1074/jbc.M513580200. [DOI] [PubMed] [Google Scholar]
- Wiley HS, Woolf MF, Opresko LK, Burke PM, Will B, Morgan JR, Lauffenburger DA. J Cell Biol. 1998;143:1317–1328. doi: 10.1083/jcb.143.5.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewska M, Goettig P, Maskos K, Belouski E, Winters D, Hecht R, Black R, Bode W. J Mol Biol. 2008;381:1307–1319. doi: 10.1016/j.jmb.2008.06.088. [DOI] [PubMed] [Google Scholar]
- Wolfe MS, De Los Angeles J, Miller DD, Xia W, Selkoe DJ. Biochemistry. 1999;38:11223–11230. doi: 10.1021/bi991080q. [DOI] [PubMed] [Google Scholar]
- Xue CB, Voss ME, Nelson DJ, Duan JJ, Cherney RJ, Jacobson IC, He X, Roderick J, Chen L, Corbett RL, Wang L, Meyer DT, Kennedy K, DeGradodagger WF, Hardman KD, Teleha CA, Jaffee BD, Liu RQ, Copeland RA, Covington MB, Christ DD, Trzaskos JM, Newton RC, Magolda RL, Wexler RR, Decicco CP. J Med Chem. 2001;44:2636–2660. doi: 10.1021/jm010127e. [DOI] [PubMed] [Google Scholar]
- Yan R, Bienkowski MJ, Shuck ME, Miao H, Tory MC, Pauley AM, Brashier JR, Stratman NC, Mathews WR, Buhl AE, Carter DB, Tomasselli AG, Parodi LA, Heinrikson RL, Gurney ME. Nature. 1999;402:533–537. doi: 10.1038/990107. [DOI] [PubMed] [Google Scholar]
- Yasuda H, Hirata S, Inoue K, Mashima H, Ohnishi H, Yoshiba M. Biochem Biophys Res Commun. 2007;354:154–159. doi: 10.1016/j.bbrc.2006.12.168. [DOI] [PubMed] [Google Scholar]
- Yeh CH, Peng HC, Yang RS, Huang TF. Mol Pharmacol. 2001;59:1333–1342. doi: 10.1124/mol.59.5.1333. [DOI] [PubMed] [Google Scholar]
- Yin J, Yu FS. Invest Ophthalmol Vis Sci. 2009;50:132–139. doi: 10.1167/iovs.08-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura T, Tomita T, Dixon MF, Axon AT, Robinson PA, Crabtree JE. J Infect Dis. 2002;185:332–340. doi: 10.1086/338191. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Thomas SM, Lui VW, Xi S, Siegfried JM, Fan H, Smithgall TE, Mills GB, Grandis JR. Proc Natl Acad Sci U S A. 2006;103:6901–6906. doi: 10.1073/pnas.0509719103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Chen H, Peschon JJ, Shi W, Zhang Y, Frank SJ, Warburton D. Dev Biol. 2001;232:204–218. doi: 10.1006/dbio.2001.0176. [DOI] [PubMed] [Google Scholar]
- Zheng X, Jiang F, Katakowski M, Zhang ZG, Lu QE, Chopp M. Cancer Biol Ther. 2009;8:1045–1054. doi: 10.4161/cbt.8.11.8539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Schlondorff J, Blobel CP. J Biol Chem. 2002;277:42463–42470. doi: 10.1074/jbc.M207459200. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Nakada MT, Brooks PC, Swenson SD, Ritter MR, Argounova S, Arnold C, Markland FS. Biochem Biophys Res Commun. 2000;267:350–355. doi: 10.1006/bbrc.1999.1965. [DOI] [PubMed] [Google Scholar]