Abstract
The conformation of a transmembrane peptide, sMTM7, encompassing the cytoplasmic hemi-channel domain of the seventh transmembrane section of subunit a from V-ATPase from Saccharomyces cerevisiae solubilized in SDS solutions was studied by circular dichroism (CD) spectroscopy and fluorescence spectroscopy of the single tryptophan residue of this peptide. The results show that the peptide adopts an α-helical conformation or aggregated β-sheet depending on the peptide-to-SDS ratio used. The results are compared with published data about a longer version of the peptide (i.e., MTM7). It is concluded that the bulky, positively charged arginine residue located in the center of both peptides has a destabilizing effect on the helical conformation of the SDS-solubilized peptides, leading to β-sheet formation and subsequent aggregation.
Keywords: V-ATPase, SDS micelles, Secondary structure, Aggregation, Membrane-mimicking solvent, Spectroscopy
Introduction
For the structure determination of transmembrane peptides and proteins by liquid-state high-resolution NMR spectroscopy in aqueous solutions, detergents are often used to solubilize the hydrophobic polypeptide molecules in micellar-like complexes (Fernandez et al. 2001; Nielsen et al. 2003; Papavoine et al. 1995, 1998; Vinogradova et al. 1998). It is supposed that in these complexes the detergent micelle covers the hydrophobic region of the polypeptide, while possible hydrophilic termini are embedded in the water phase (Arora et al. 2001). Several studies have shown that transmembrane polypeptides can retain and exhibit their natural conformation when solubilized in SDS solutions at concentrations well above the critical micelle concentration (cmc) (Arora et al. 2001; Henry and Sykes 1994; Lazarova et al. 2004; Papavoine et al. 1994; Underhaug et al. 2006). Due to its physicochemical characteristics, SDS is considered to be an ideal solubilizing detergent for hydrophobic membrane polypeptides; as in biological membranes, it has a negative net charge and can provide a good interface for the folding of surface-active peptides (Mammi and Peggion 1990).
Recently conformational studies of transmembrane peptides located in the cytoplasmic hemi-channel region of TM7 of V-ATPase subunit a have been published, based on two synthesized peptides: MTM7 and sMTM7 (see Fig. 1) (Duarte et al. 2007a, b; Vos et al. 2007). These putative transmembrane peptides include the arginine residue (R735), which is known to be part of the proton translocation channel, interacting with the glutamic acids from the subunits c, c′, and c″ (Nishi and Forgac 2002). This work sheds light on the low- and high-resolution structure of this domain of V-ATPase that is essential for proton translocation. MTM7 is a 37-residue peptide that was studied in organic solvents and SDS micellar solutions. In organic solvents (TFE and DMSO) and SDS micelles, this peptide adopted an α-helical conformation, however, the increase in the molecular weight of the peptide/SDS micelles hampered the use of NMR spectroscopy to obtain a high-resolution structure. Peptide sMTM7 is a smaller version of peptide MTM7 containing 25 residues (see Fig. 1). This peptide was also studied in organic solvents (Duarte et al. 2007a) and in SDS micellar solutions (Vos et al. 2007). The relevance of the use of such membrane-mimicking solvents to study these peptides has been discussed previously (Duarte et al. 2008). When solubilized in DMSO, peptide sMTM7 exhibits two α-helical regions and a fraying C-terminal, as determined by high-resolution NMR spectroscopy. In SDS micelles, the overall conformation of the peptide was determined by circular dichroism to be α-helical. Similar to peptide MTM7, the study of peptide sMTM7 in SDS micelles by NMR was impossible due to the strong broadening and low signal-to-noise ratio of the NMR spectra.
Fig. 1.

Primary structure of peptides MTM7 and sMTM7. The coloring of the residues is representative of their hydrophobicity (Larkin et al. 2007), ranging from high hydrophilicity (blue) to high hydrophobicity (red). The numbering of the residues of the peptides is according to the numbering of the entire V-ATPase subunit a (Nishi and Forgac 2002)
In this paper, we report the conformational behavior of peptide sMTM7 in different SDS micellar solutions. Circular dichroism spectroscopy is applied to study the secondary structure and thermal stability of the peptide in the micelles. Taking advantage of the presence of a single tryptophan at position 737 (see Fig. 1), the fluorescence of peptide sMTM7 was measured enabling us to characterize the hydrophobicity of the environment that solvates the peptide. Our results indicate that at SDS concentrations above the cmc, peptide sMTM7 can adopt an α-helical or β-sheet conformation depending on the number of SDS molecules available in solution. In comparing these data with the solubilizing data obtained for the longer peptide MTM7, we conclude that the bulky, positively charged arginine residue located in the center of the peptide has a destabilizing effect on the helical conformation of the SDS-solubilized peptides, leading to β-sheet formation and subsequent aggregation.
Materials and methods
Peptide design and synthesis
Peptide sMTM7 is composed of 25 residues (Fig. 1) and was designed based on the putative localization of transmembrane segment TM7 located in the cytoplasmic hemi-channel region of V-ATPase subunit a (Duarte et al. 2007a). The peptide is a smaller version of the 37-residue peptide MTM7 (Fig. 1) that was used in previous work (Duarte et al. 2007a). Throughout this paper the sequential numbering of amino acid residues of peptide sMTM7 is identical to the one used for subunit a in V-ATPase (Fig. 1). Peptide sMTM7 was produced on solid support using continuous flow chemistry by Pepceuticals, Leicester, UK. Its purity was tested by mass spectrometry and found to be larger than 90%.
Solvents
Peptide sMTM7 is a highly hydrophobic peptide with a poor solubility in DPC micelles, TFE:water mixtures, and aqueous systems (Duarte et al. 2007a). To perform conformational studies, two membrane-mimicking solvents commonly used in the literature were selected: SDS micelles (SDS from Merck, Darmstadt, Germany), and TFE (Acros Organics, Geel, Belgium).
Sample preparation
To solubilize peptide sMTM7 in aqueous solutions of SDS, the peptide was initially dissolved in TFA and dried under a stream of nitrogen. A 2-mM peptide solution was prepared in TFE resulting in a clear solution. This peptide solution was then mixed with a freshly made SDS solution at the desired concentration to achieve each of three different peptide-to-SDS molar ratios of 1:250, 1:100, and 1:70 (the final concentrations of SDS were 500, 200 and 140 mM, respectively). The obtained final solutions were clear. Deionized water was added to yield a ratio of 16:1 (vwater/vSDS). The samples were mixed for 15 min and rapidly frozen followed by drying under vacuum at −70°C. The dry samples were rehydrated with deionized water (pH 6.0) resulting in clear solutions. Samples of peptide sMTM7 in TFE were prepared by dissolving the peptide in pure TFE. The final peptide concentration was 0.19 mM for all samples. In all prepared SDS samples the concentration of SDS was well above the cmc, which is 8 mM.
CD measurements
For the CD measurements, the sample preparation of peptide sMTM7 in SDS micelles was performed as described above. For each peptide-to-SDS ratio a freshly prepared sample was used. The final concentration of peptide sMTM7 was determined spectrophotometrically by measuring the UV absorbance at 280 nm, using an extinction coefficient of 7,210 l mol−1 cm−1 (Gill and von Hippel 1989). For all the experiments, the optical density at 280 nm was below 0.5.
All CD experiments were performed on a Jasco Spectropolarimeter J-715 (Jasco, Tokyo, Japan), equipped with a Peltier thermal-controlled cuvette holder. The spectra were recorded using a 0.1-cm path length cuvette from 260 to 200 nm with a 1.0-nm step resolution and a response time of 0.25 s. The spectra were collected and averaged over 25 scans. To analyze the thermal stability of peptide sMTM7 dissolved in the different peptide-to-SDS ratios, CD spectra were collected at 20, 30, 40 and 50°C. After reaching the desired temperature the sample was allowed to stabilize for 10 min after which the spectrum was measured. After the measurement at 50°C, the temperature was lowered to 20°C and a spectrum was acquired to check for reversibility of the conformation. CD spectra of TFE and SDS solutions without the peptide with a SDS concentration equivalent to each peptide-to-SDS ratio were measured as background and subtracted for each spectrum.
The secondary structure content of peptide sMTM7 was calculated using the CD spectrum deconvolution software CDNN (Bohm et al. 1992). This software calculates the secondary structure of the peptide by comparison with a CD database of known protein structures.
Fluorescence spectroscopy
For the fluorescence measurements, the sample preparation of peptide sMTM7 in TFE and SDS micelles was performed as described above. Fluorescence spectra were recorded on a Fluorolog 3.22 (Jobin Yvon-Spex, Longjumeau, France) in quartz cuvettes with path lengths of 10 and 4 mm in the excitation and emission direction, respectively. Fluorescence spectra were recorded between 300 and 400 nm, using an excitation wavelength of 280 nm and using excitation and emission slits giving a 2-nm bandpass. Background fluorescence from a sample without peptide was subtracted from each spectrum.
Results
CD spectroscopy
Figure 2 shows the CD spectrum of peptide sMTM7 dissolved in TFE. The CD spectrum exhibits two negative bands located at 208 and 222 nm combined with a positive band near 200 nm, suggesting the presence of an α-helical conformation. This is confirmed by secondary structure analysis using CDNN (Table 1) that suggests that 50% of the peptide populates an α-helical conformation, whereas the remaining peptide remains unstructured in TFE.
Fig. 2.
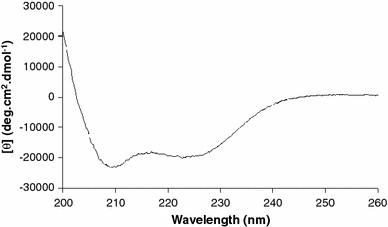
Far-UV CD spectrum of peptide sMTM7 in TFE. The final peptide concentration was 0.19 mM. The temperature was 20°C
Table 1.
Secondary structure content of peptide sMTM7 in TFE and SDS micelles at 20°C determined by the CDNN software (Bohm et al. 1992)
| Sample conditions | Secondary structure calculation (%) | ||
|---|---|---|---|
| α-Helix | β-Sheet | Unfolded | |
| TFE | 50 | 0 | 50 |
| Peptide-to-SDS ratio | |||
| 1:70 | 0 | 60 | 40 |
| 1:100 | 12 | 49 | 39 |
| 1:250 | 30 | 0 | 70 |
The standard error in the percentage is approximately ±4%
The CD spectra of peptide sMTM7 solubilized in the different SDS micellar solutions are presented in Fig. 3. From the line shapes one can observe that the peptide adopts different conformations for each of the three different peptide-to-SDS ratios used. When peptide sMTM7 is dissolved in the highest concentration of SDS (47.5 mM of SDS corresponding to the 1:250 ratio), the CD spectrum shows two minima at 208 and 226 nm, suggesting that peptide sMTM7 adopts an α-helix conformation. Indeed, spectral analysis obtained from CDNN (Table 1) indicates that 30% of the peptide populates the α-helix conformation and the remaining 70% is unstructured.
Fig. 3.
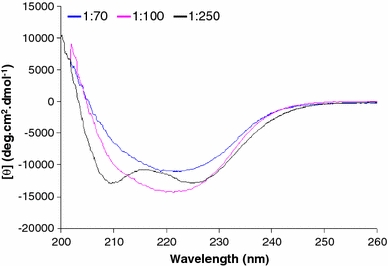
Far-UV CD spectra of peptide sMTM7 solubilized at peptide-to-SDS ratios of 1:250, 1:100, and 1:70. The final peptide concentration for all samples was 0.19 mM. The temperature was 20°C
By decreasing the amount of SDS molecules available to solvate the peptide to an SDS concentration of 19 mM (i.e., a peptide-to-SDS ratio of 1:100), the CD spectrum of peptide sMTM7 exhibits one strong minima at 222 nm and a less pronounced minima at 210 nm. CDNN analysis of this spectrum indicates that it arises from a linear combination of CD signals originating from both α-helix and β-sheet conformations (Table 1). In the SDS solution where the concentration of detergent is further lowered to 13.3 mM of SDS (corresponding to the 1:70 ratio), the CD spectrum of peptide sMTM7 shows one minimum at 222 nm similar to the one identified for the 1:100 ratio, however, with a lower intensity. CDNN analysis (Table 1) indicates that 60% of peptide sMTM7 populates the β-sheet conformation and the remaining 40% remains unstructured.
To analyze the thermal stability of the peptide at different peptide-to-SDS ratios, the CD spectra were recorded at various temperatures. At the peptide-to-SDS ratios used, the ellipticity changes with temperature in a linear way, but no defined unfolding transition is observed in the temperature range studied (spectra not shown). Upon cooling of peptide sMTM7 back to 20°C, the ellipticity returns to its original value for all the three peptide-to-SDS ratios studied. Thus, the conformational exchange between the α-helical or β-sheet and unfolded conformations of peptide sMTM7 is fully reversible. The plot in Fig. 4 shows that increasing the temperature leads to a more pronounced decrease in the CD signal of the samples in which an α-helix conformation was found (i.e., peptide-to-SDS ratio 1:100 and 1:250). However, for the sample in which only β-sheet conformation was found (i.e., peptide-to-SDS ratio 1:70), the effect of temperature is reduced.
Fig. 4.
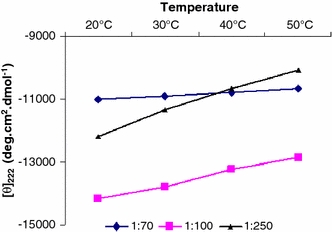
Thermal stability experiments of peptide sMTM7 solubilized at peptide-to-SDS ratios of 1:250, 1:100, and 1:70. The effect of temperature was monitored by measuring the ellipticity at 222 nm
Fluorescence experiments
Peptide sMTM7 contains a single tryptophan residue at position 737 (W737). This opens the possibility to record the fluorescence spectra to provide information about the polarity of the tryptophan environment (Lakowicz 2006). The fluorescence spectrum of peptide sMTM7 dissolved at three peptide-to-SDS ratios using an excitation wavelength of 280 nm exhibits a fluorescence maximum for W737 at 330 nm (Fig. 5). By comparing the fluorescence spectra with different excitation wavelengths, the sharp peak at 311 nm can be assigned to a Raman scattering peak. The maximum of fluorescence of peptide sMTM7 dissolved in TFE is red-shifted to 340 nm. This suggests that when peptide sMTM7 is solubilized in SDS micelles, W737 is located in a more hydrophobic environment (Lakowicz 2006).
Fig. 5.
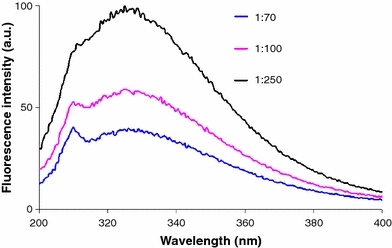
Fluorescence spectra of peptide sMTM7 dissolved at peptide-to-SDS ratios of 1:250, 1:100, and 1:70 at an excitation wavelength of 280 nm. The emission peak at 311 nm is due to Raman scattering. The final peptide concentration for all samples was 0.19 mM. The temperature was 20°C
The intensities of the fluorescence maximum of the spectra in Fig. 5 show that an increase in the number of SDS molecules available to the peptide (from 1:70 to 1:250 ratios) leads to an increase in fluorescence intensity. This increase in the signal intensity and the fact that the maximum is located at the same wavelength lead to the conclusion that the peptide is subject to self-quenching. This effect could be due to the formation of aggregated peptide sMTM7.
Discussion
Recently, several papers have been published about the intrinsic conformational properties of a transmembrane segment (TM7) located in the cytoplasmic hemi-channel region of V-ATPase subunit a (Duarte et al. 2007a, b; Hesselink et al. 2005; Vos et al. 2006, 2007). These papers provided detailed information about the low- and high-resolution structure of this domain of V-ATPase that is essential for proton translocation. A large part of the data was obtained for peptides MTM7 and sMTM7 (Fig. 1) representing the transmembrane region of TM7 dissolved in DMSO as a membrane-mimicking organic solvent (Duarte et al. 2007a, b).
In spite of the high-quality structural data obtained in DMSO by 2D-NMR spectroscopy, it is still challenging to understand the structural behavior of peptides MTM7 and sMTM7 in detergent micelles. When dissolved at peptide-to-SDS ratios of 1:100 and 1:250, peptide MTM7 adopts a helical conformation (Duarte et al. 2007b). Meanwhile, peptide sMTM7 is also known to adopt a helical conformation in SDS micelles (Vos et al. 2007). However, for the last, only the peptide-to-SDS ratio of 1:1,600 was used and no further information about the physicochemical behavior of the peptide in the micelles was obtained. The present paper focuses on a combined CD and fluorescence study to describe the solubilization and secondary structure characterization of peptides MTM7 and sMTM7 in SDS micelles. For peptide MTM7, we will rely on data from previously published work, whereas here we will present new experimental results for peptide sMTM7.
The CD data of peptide sMTM7 dissolved at three peptide-to-SDS ratios (Table 1) indicate that the overall conformation of the peptide is strongly influenced by the detergent concentration, even above the detergent cmc. Indeed, a decrease in the number of SDS molecules (from 1:250 to 1:100 to 1:70) available to solubilize peptide sMTM7 leads to a conformational change α-helix → β-sheet (Fig. 6). However, when the peptide is solubilized at a peptide-to-SDS ratio of 1:250, the percentage of α-helical conformation is similar to the one detected previously for a peptide-to-SDS ratio of 1:1,600 (Vos et al. 2007). The analysis of the CD spectra at a peptide-to-SDS ratio of 1:100 shows a mixture of α-helix and β-sheet conformations (Table 1; Fig. 6). It is not evident from the CD results whether different peptides exhibiting α-helix or β-sheet conformations at this ratio are located in different micelles (center illustration of Fig. 7), or whether they can exchange conformations.
Fig. 6.
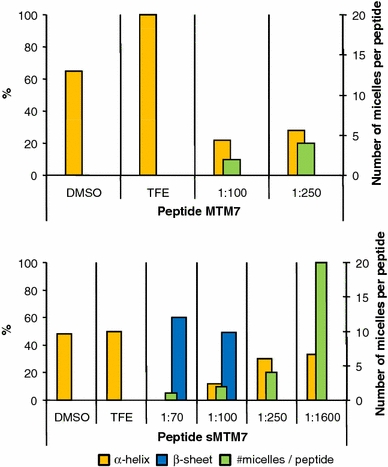
Top Graphical representation of the secondary structure content (α-helix, orange columns) of peptide MTM7 as determined by Duarte et al. (2007b). The number of available SDS micelles in solution per peptide is also indicated [green columns; considering that one SDS micelle is formed by 60 SDS molecules (Yoshii et al. 2006)]. Bottom Graphical representation of the helical (orange columns) and β-sheet (blue columns) conformers detected for peptide sMTM7 dissolved in SDS micelles [peptide-to-SDS ratios 1:70, 1:100, and 1:250 determined in the present work; peptide-to-SDS 1:1,600 determined by Vos et al. (2007)], TFE (determined in the present work), and DMSO (determined from the conformational ensemble published previously (Duarte et al. 2007a)
Fig. 7.
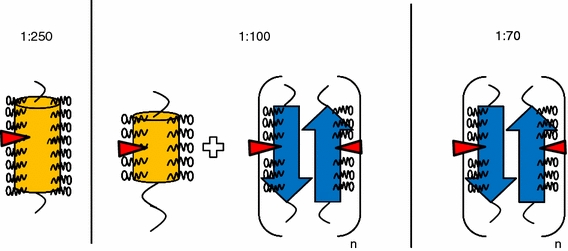
Schematic illustration of possible structures of peptide sMTM7 dissolved at different peptide-to-SDS ratios. The orange cylinders represent peptide sMTM7 in an α-helical conformation, the size of the cylinder is relative to the amount of detected α-helix. The blue arrows represent peptide sMTM7 in a β-sheet conformation. The red triangles represent arginine residue R735
The temperature scans of the samples in which an α-helix conformation is present (1:100, 1:250) show a reversible change in CD intensity, suggesting a conformational exchange between α-helix and unfolded conformations (Fig. 4). Alternatively, the sample in which a β-sheet conformation is detected (1:70) is shown to be quite stable as a function of temperature (Fig. 4), indicating that it is not involved in an exchange with α-helical or unfolded conformations. From these results, we might conclude that at a peptide-to-SDS ratio of 1:100, 12% of peptide sMTM7 populating an α-helix conformation can exchange with unstructured peptide, while 49% of peptide populating the β-sheet is not involved with conformational exchange.
The position of the fluorescence maximum of tryptophan residue W737 of peptide sMTM7 remains the same at the three peptide-to-SDS ratios studied (Fig. 5). This indicates that this residue stays located in a region with the same polarity. The change in fluorescence intensity in Fig. 5 indicates that at increasing peptide-to-SDS ratios, the peptide tends to aggregate. This is concomitant with a β-sheet formation (Table 1), suggesting that the peptide forms β-sheet aggregates.
The relevant conformational data for peptides MTM7 and sMTM7 are summarized in Fig. 6. In this figure, the number of micelles per peptide represents the theoretical number of SDS micelles per peptide assuming that an SDS micelle is composed of 60 SDS molecules. At peptide-to-SDS ratios of 1:70 and 1:100, the number of available SDS micelles (1 or about 2, respectively) for both peptides is sufficient to stabilize the peptides in solution, however, the number of micelles is not high enough to prevent aggregation and β-sheet formation as is observed for peptide sMTM7. In the case of a peptide-to-SDS ratio of 1:250, the number of micelles becomes high enough (≈4) to allow both peptides to adopt an α-helical conformation. Upon increasing the SDS concentration to allow a peptide-to-SDS ratio of 1:1,600, the α-helical content does not increase, whereas the number of micelles per peptide is increased fivefold.
From this analysis it is reasonable to assume that for peptide-to-SDS ratios below 1:250, the peptide is in a monomeric state with a stable α-helical conformation surrounded by SDS molecules forming a micellar-like structure. A schematic illustration of such a peptide-SDS complex is shown in Fig. 7. Considering that tryptophan W737 is in an SDS environment when it adopts an α-helical conformation, its polarity independence (Fig. 5) indicates a similar environment when it aggregates adopting a β-sheet conformation. In this view we propose that monolayers of β-sheet aggregates are surrounded by SDS micelles in such a way that W737 is always in contact with the SDS acyl chains (Fig. 7).
A possible explanation for the SDS-dependent aggregational and conformational change of peptide sMTM7 could be due to the limited way SDS molecules can stabilize the α-helix conformation. The primary sequence of peptide sMTM7 contains several hydrophobic residues (Duarte et al. 2007a), which could lead to strong hydrophobic interactions with the SDS acyl chains. However, the peptide contains a positively charged bulky arginine residue at position 735 (R735) and a fraying C-terminal domain with an unfolded conformation (Duarte et al. 2007a) that could reduce the interaction with SDS molecules. Only with a sufficiently high number of SDS molecules, well above the cmc will an energetically stable peptide-SDS complex be formed with the peptide in a monomeric α-helical conformation. This conformation can be assumed to be the native conformation of peptide sMTM7 (Duarte et al. 2007a; Vos et al. 2007). If these conditions are not fulfilled already at the sample preparation stage, the peptide will aggregate, the β-sheet being then the favorable conformation, i.e., similar to that found for M13 coat protein (Spruijt and Hemminga 1991) and amyloid peptides (Bisaglia et al. 2006).
Based on the present information and published data (Duarte et al. 2007b), it is interesting to carry out a comparative study between peptides sMTM7 and MTM7 (Fig. 1) dissolved and solubilized under different conditions (Fig. 6). Peptide MTM7 is a 37-residue peptide that contains 35% of hydrophobic residues compared to the 28% of peptide sMTM7. The conformational data of peptide MTM7 in SDS micelles and DMSO indicate that this peptide has a tendency to adopt an α-helical conformation in membrane-mimicking environments. The relatively low percentage of helical content was explained previously (Duarte et al. 2007b) to be due to (1) the presence of R735 in the center of the peptide sequence, which might interfere with the peptide solubilization in the micelles, and (2) the fact that CD spectroscopy detected a low ellipticity due to possible breaks in the helical segments, inducing a calculation of lower helical content.
In comparing the conformational results of peptide MTM7 with those of peptide sMTM7, it is interesting to note that, at an intermediate peptide-to-SDS ratio of 1:100, peptide MTM7 exhibits an α-helical conformation, while peptide sMTM7 shows the presence of α-helical and β-sheet conformations. This difference should primarily be due to the removal of residues from the termini of peptide MTM7. However, the decrease in length of the polypeptide chain does not justify per se these conformational changes. One effect that might play a role is the presence of R735 in the middle of both peptides, which can prevent the formation of a well-fitting SDS coat, hampering a correct solubilization of the peptide. This effect would also explain the more pronounced instability of the short sMTM7 peptide as compared to the longer MTM7 peptide. The higher stabilization of the latter might be due to the higher number of amino acid residues that permit the formation of an α-helical conformation.
In conclusion, both peptides MTM7 and sMTM7 are highly hydrophobic, thus a high number of SDS molecules is required to solubilize them in aqueous solution. The presence of arginine residue R735 in the center of the peptides, due to its positively charged long side chain, imposes an extra condition on the binding of SDS molecules. When the available number of SDS molecules decreases, the bulky positively charged arginine residue will tend to be solubilized in the aqueous environment, while the SDS molecules will cover the remaining peptide domains. This effect is more prominent in the shorter sMTM7 peptide due to the lower number of residues that would permit the formation of two α-helices (before and after residue R735). We hypothesize that this decrease in the number of SDS molecules will destabilize the helical conformation of the peptides and lead to β-sheet formation and subsequent aggregation.
Acknowledgments
This work was supported by contract no. QLG-CT-2000-01801 of the European Commission (MIVase—New Therapeutic Approaches to Osteoporosis: targeting the osteoclast V-ATPase).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Abbreviations
- CD
Circular dichroism
- CMC
Critical micellar concentration
- DMSO
Dimethylsulfoxide
- DPC
Dodecylphophocholine
- MTM7
Peptide mimicking the cytoplasmic side of the seventh transmembrane segment of subunit a from V-ATPase (from residue 716 to 753)
- sMTM7
Peptide mimicking the cytoplasmic side of the seventh transmembrane segment of subunit a from V-ATPase (from residue 719 to 745)
- SDS
Sodium dodecyl sulfate
- TFE
Trifluoroethanol
- V-ATPase
Vacuolar proton-translocating adenosine triphosphatase
Footnotes
The more you see: Spectroscopy in molecular biophysics.
References
- Arora A, Abildgaard F, Bushweller JH, Tamm LK. Structure of outer membrane protein A transmembrane domain by NMR spectroscopy. Nat Struct Mol Biol. 2001;8:334–338. doi: 10.1038/86214. [DOI] [PubMed] [Google Scholar]
- Bisaglia M, Trolio A, Bellanda M, Bergantino E, Bubacco L, Mammi S. Structure and topology of the non-amyloid-β component fragment of human α-synuclein bound to micelles: implications for the aggregation process. Protein Sci. 2006;15:1408–1416. doi: 10.1110/ps.052048706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohm G, Muhr R, Jaenicke R. CDNN: quantitative analysis of protein far UV circular dichroism spectra by neural networks. Protein Eng. 1992;5:191–195. doi: 10.1093/protein/5.3.191. [DOI] [PubMed] [Google Scholar]
- Duarte AMS, De Jong ER, Wechselberger R, Van Mierlo CPM, Hemminga MA. Segment TM7 from the cytoplasmic hemi-channel from VO-H+-V-ATPase includes a flexible region that has a potential role in proton translocation. Biochim Biophys Acta. 2007;1768:2263–2270. doi: 10.1016/j.bbamem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Duarte AMS, Wolfs CJAM, Van Nuland NAJ, Harrison MA, Findlay JBC, Van Mierlo CPM, Hemminga MA. Structure and localization of an essential transmembrane segment of the proton translocation channel of yeast H+-V-ATPase. Biochim Biophys Acta. 2007;1768:218–227. doi: 10.1016/j.bbamem.2006.07.014. [DOI] [PubMed] [Google Scholar]
- Duarte AMS, Van Mierlo CPM, Hemminga MA. Molecular dynamics study of the solvation of an α-helical transmembrane peptide by DMSO. J Phys Chem B. 2008;112:8664–8671. doi: 10.1021/jp076678j. [DOI] [PubMed] [Google Scholar]
- Fernandez C, Hilty C, Bonjour S, Adeishvili K, Pervushin K, Wuthrich K. Solution NMR studies of the integral membrane proteins OmpX and OmpA from Escherichia coli. FEBS Lett. 2001;504:173–178. doi: 10.1016/S0014-5793(01)02742-9. [DOI] [PubMed] [Google Scholar]
- Gill SC, von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- Henry GD, Sykes BD. Methods to study membrane protein structure in solution. Methods Enzymol. 1994;239:515–535. doi: 10.1016/S0076-6879(94)39020-7. [DOI] [PubMed] [Google Scholar]
- Hesselink RW, Koehorst RBM, Nazarov PV, Hemminga MA. Membrane-bound peptides mimicking transmembrane Vph1p helix 7 of yeast V-ATPase: a spectroscopic and polarity mismatch study. Biochim Biophys Acta. 2005;1716:137–145. doi: 10.1016/j.bbamem.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Lakowicz JR. Principles of fluorescence spectroscopy. 3. New York: Springer; 2006. [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. ClustalW2 and ClustalX version 2. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lazarova T, Brewin KA, Stoeber K, Robinson CR. Characterization of peptides corresponding to the seven transmembrane domains of human adenosine A2a receptor. Biochemistry. 2004;43:12945–12954. doi: 10.1021/bi0492051. [DOI] [PubMed] [Google Scholar]
- Mammi S, Peggion E. Conformational studies of human [15–2-aminohexanoic acid]little gastrin in sodium dodecyl sulfate micelles by 1H NMR. Biochemistry. 1990;29:5265–5269. doi: 10.1021/bi00474a007. [DOI] [PubMed] [Google Scholar]
- Nielsen G, Malmendal A, Meissner A, Moller JV, Nielsen NC. NMR studies of the fifth transmembrane segment of sarcoplasmic reticulum Ca2+-ATPase reveals a hinge close to the Ca2+-ligating residues. FEBS Lett. 2003;544:50–56. doi: 10.1016/S0014-5793(03)00448-4. [DOI] [PubMed] [Google Scholar]
- Nishi T, Forgac M. The vacuolar (H+)-ATPases—nature’s most versatile proton pumps. Nat Rev Mol Cell Biol. 2002;3:94–103. doi: 10.1038/nrm729. [DOI] [PubMed] [Google Scholar]
- Papavoine CHM, Konings RNH, Hilbers CW, Van de Ven FJM. Location of M13 coat protein in sodium dodecyl sulfate micelles as determined by NMR. Biochemistry. 1994;33:12990–12997. doi: 10.1021/bi00248a007. [DOI] [PubMed] [Google Scholar]
- Papavoine CHM, Aelen JMA, Konings RNH, Hilbers CW, Van de Ven FJM. NMR studies of the major coat protein of bacteriophage M13. Structural information of gVIIIp in dodecylphosphocholine micelles. Eur J Biochem. 1995;232:490–500. doi: 10.1111/j.1432-1033.1995.490zz.x. [DOI] [PubMed] [Google Scholar]
- Papavoine CHM, Christiaans BEC, Folmer RHA, Konings RNH, Hilbers CW. Solution structure of the M13 major coat protein in detergent micelles: a basis for a model of phage assembly involving specific residues. J Mol Biol. 1998;282:401–419. doi: 10.1006/jmbi.1998.1860. [DOI] [PubMed] [Google Scholar]
- Spruijt RB, Hemminga MA. The in situ aggregational and conformational state of the major coat protein of bacteriophage M13 in phospholipid bilayers mimicking the inner membrane of host Escherichia coli. Biochemistry. 1991;30:11147–11154. doi: 10.1021/bi00110a018. [DOI] [PubMed] [Google Scholar]
- Underhaug J, Jakobsen LO, Esmann M, Malmendal A, Nielsen NC. NMR studies of the fifth transmembrane segment of Na+, K+-ATPase reveals a non-helical ion-binding region. FEBS Lett. 2006;580:4777–4783. doi: 10.1016/j.febslet.2006.07.063. [DOI] [PubMed] [Google Scholar]
- Vinogradova O, Sonnichsen F, Sanders CR. On choosing a detergent for solution NMR studies of membrane proteins. J Biomol NMR. 1998;11:381–386. doi: 10.1023/A:1008289624496. [DOI] [PubMed] [Google Scholar]
- Vos WL, Vermeer LS, Wolfs CJAM, Spruijt RB, Hemminga MA. Decomposition of ESR spectra using MALDI-TOF mass spectrometry. Anal Chem. 2006;78:5296–5301. doi: 10.1021/ac0521448. [DOI] [PubMed] [Google Scholar]
- Vos WL, Vermeer LS, Hemminga MA. Conformation of a peptide encompassing the proton translocation channel of vacuolar H+-ATPase. Biophys J. 2007;92:138–146. doi: 10.1529/biophysj.106.089854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshii N, Iwahashi K, Okazaki S. A molecular dynamics study of free energy of micelle formation for sodium dodecyl sulfate in water and its size distribution. J Chem Phys. 2006;124:184901–184906. doi: 10.1063/1.2179074. [DOI] [PubMed] [Google Scholar]


