Abstract
Conventional electron paramagnetic resonance (EPR) spectra of lipids that are spin-labelled close to the terminal methyl end of the acyl chains are able to resolve the lipids directly contacting the protein from those in the fluid bilayer regions of the membrane. This allows determination of both the stoichiometry of lipid–protein interaction (i.e., number of lipid sites at the protein perimeter) and the selectivity of the protein for different lipid species (i.e., association constants relative to the background lipid). Spin-label EPR data are summarised for 20 or more different transmembrane peptides and proteins, and 7 distinct species of lipids. Lineshape simulations of the two-component conventional spin-label EPR spectra allow estimation of the rate at which protein-associated lipids exchange with those in the bulk fluid regions of the membrane. For lipids that do not display a selectivity for the protein, the intrinsic off-rates for exchange are in the region of 10 MHz: less than 10× slower than the rates of diffusive exchange in fluid lipid membranes. Lipids with an affinity for the protein, relative to the background lipid, have off-rates for leaving the protein that are correspondingly slower. Non-linear EPR, which depends on saturation of the spectrum at high radiation intensities, is optimally sensitive to dynamics on the timescale of spin-lattice relaxation, i.e., the microsecond regime. Both progressive saturation and saturation transfer EPR experiments provide definitive evidence that lipids at the protein interface are exchanging on this timescale. The sensitivity of non-linear EPR to low frequencies of spin exchange also allows the location of spin-labelled membrane protein residues relative to those of spin-labelled lipids, in double-labelling experiments.
Keywords: Spin label, Electron paramagnetic resonance (EPR), Electron spin resonance (ESR), Non-linear EPR, Saturation transfer EPR, Relaxation enhancements, Lipid–protein interactions
Introduction
Since the original observation of motionally restricted boundary lipid in progressively delipidated preparations of cytochrome oxidase by Griffith and colleagues (Jost et al. 1973), electron paramagnetic resonance (EPR) of spin-labelled lipids has developed into an extremely versatile method for studying lipid–protein interactions in biological membranes (Marsh et al. 1982, 2002a; Marsh 1983, 1987, 1990, 1995, 1996, 2008b; Marsh and Powell 1988; Knowles and Marsh 1991; Marsh and Horváth 1998; Marsh and Páli 2004; Esmann and Marsh 2006).
The spin-label EPR method is based primarily on the spectral resolution of lipids directly in contact with the intramembranous sector of the protein from those exhibiting the characteristic chain fluidity gradient in the bulk membrane (cf. Schorn and Marsh 1996a, b). The fraction, f b, of spin-labelled lipids contacting the protein is related directly to the stoichiometry and selectivity of interaction with the protein (Marsh 1985; Brotherus et al. 1981):
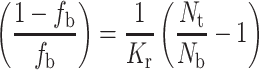 |
1 |
where N t is the total lipid/protein mole ratio. In Eq. 1, N b is the number of lipid association sites on the protein and K r is the association constant of spin-labelled lipid with the protein, relative to that of the unlabelled background host lipid, according to the lipid exchange reaction:
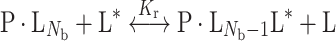 |
where P is protein, L is lipid and the asterisk (*) indicates spin label. Material balance relates the fraction of protein-associated spin-labelled lipids to the kinetics of lipid exchange in the above reaction:
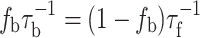 |
2 |
where τ −1f is the on-rate and τ −1b is the off-rate for spin-labelled lipids L* interacting with the protein. These dynamic parameters governing lipid exchange also can be determined by spin-label EPR.
In addition to the systematic dependences on lipid–protein ratio and lipid selectivity that are observed, and invariance with respect to different spin-label configurations (Esmann et al. 1988a), there are also spectroscopic verifications of the method. These include multi-frequency EPR (Horváth et al. 1994) and saturation transfer EPR (Marsh 2007; Horváth et al. 1993b). The latter techniques demonstrate unambiguously that the two-component nature of the spin-label spectra is preserved under quite different conditions of g-shift anisotropy or dynamic window of observation (see also Watts et al. 1981).
Stoichiometry and selectivity of lipid–protein interaction
Amongst the various available techniques, spin-label EPR remains unique in yielding directly the stoichiometry of the protein-associated lipid, a quantity that is related to the intramembranous structure and assembly of integral proteins (see Fig. 1; Marsh 1997; Páli et al. 2006) and can be used to detect oligomer formation (Arora et al. 2003; Ryba et al. 1993; Kóta et al. 2008; Marsh 2009). Representative data on the numbers of lipids, N b, that contact the intramembranous perimeter of different membrane proteins are presented in Table 1.
Fig. 1.
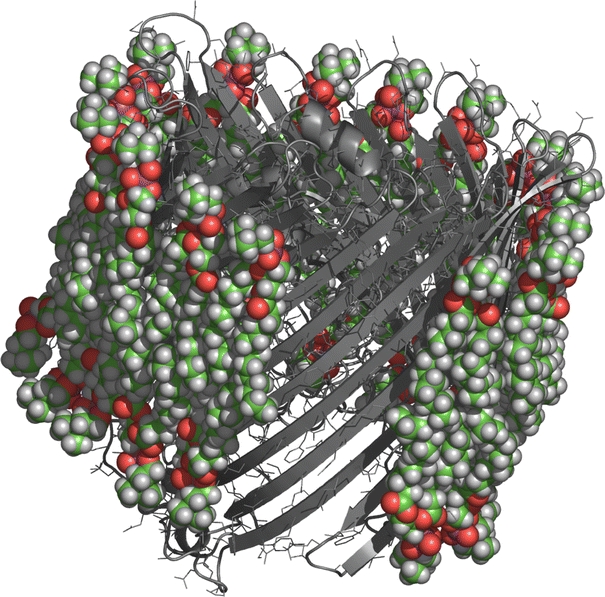
First shell of lipids surrounding the crystal structure of Escherichia coli outer membrane protein FhuA (PDB code 2FCP) (Ferguson et al. 1998). Part of the shell of energy-minimised diC14:0PtdCho lipids (space-filling representation) is cut away to show the protein in ribbon and wire-frame representation. In total, 34 lipids contact the intramembrane perimeter of the protein (Páli et al. 2006). The stoichiometry of motionally restricted lipids observed by EPR is N b = 32 (Ramakrishnan et al. 2004)
Table 1.
Stoichiometries, N b of first-shell lipids surrounding integral proteins and peptides from spin-label EPR, model building with X-ray protein structures (Páli et al. 2006), and geometrical predictions for α-helical bundles and β-sheets (Marsh 1997)
| Protein | N b (mol/mol) | Referencesa | ||
|---|---|---|---|---|
| EPR | X-ray | Geometry | ||
| α-helical: | ||||
| Rhodopsin | 22–25 | 27 ± 2 | 24.5 | 1–5 |
| ADP-ATP carrier | 25 | 28 ± 2 | 22.5 | 6 |
| Ca-ATPase | 22–24 | 28 ± 4 | 31 | 7, 8 |
| Na, K-ATPase | 32–33 | – | 33 | 9–11 |
| Cytochrome reductase | 38 ± 3 | 46 ± 4 | 37 | 12 |
| Nicotinic acetylcholine receptor | 41 ± 7 | 52 ± 4 | 52 | 13, 14 |
| Cytochrome oxidase | 56 ± 5 | 57 ± 4 | 68.5 | 15 |
| 43 ± 4b | 63b | 16 | ||
| Phospholamban A36,41,46 | 11.3c | 10 | 17 | |
| 4.0 ± 0.1d | 4e | 17 | ||
| M13 coat protein | 4–5 | 4f | 18 | |
| Myelin proteolipid | 10 ± 2 | 10f | 19 | |
| 16-kDa proteolipid | 5–6 | 10f | 20 | |
| β-barrel/sheet: | ||||
| OmpA | 11 | 11 | 21 | |
| OmpG | 20 ± 1 | 19 | 22 | |
| FomA | 23 | 23 | ||
| FhuA | 32 | 30 | 21 | |
| M13 phage coat proteing | 4 | 18 | ||
| K27, K-channel peptideh | 2.2 | 24 | ||
| K27-Δ2 | 2.5 | 25 | ||
| β 6.3-helix: | ||||
| Gramicidin A | 3.6 ± 0.3 | 26 | ||
a1. Watts et al. (1979), 2. Pates and Marsh (1987), 3. Ryba et al. (1987), 4. Ryba and Marsh (1992), 5. Pates et al. (1985), 6. Horváth et al. (1990c), 7. Silvius et al. (1984), 8. Thomas et al. (1982), 9. Brotherus et al. (1981), 10. Esmann et al. (1985), 11. Esmann et al. (1988a), 12. Powell et al. (submitted), 13. Mantipragada et al. (2003), 14. Ellena et al. (1983), 15. Knowles et al. (1979), 16. Kleinschmidt et al. (1998), 17. Arora et al. (2003), 18. Peelen et al. (1992), 19. Brophy et al. (1984), 20. Páli et al. (1995), 21. Ramakrishnan et al. (2004), 22. Anbazhagan et al. (2008a), 23. Anbazhagan et al. (2008b), 24. Horváth et al. (1997), 25. Horváth et al. (1995), 26. Kóta et al. (2004), 27. Páli et al. (2006)
bValue per monomer deduced from structure of dimer (27)
cIn diC14:0PtdGro
dIn C16:0C18:1cPtdCho
ePredictions for pentamer: n agg = 5
fPredictions for hexamer: n agg = 6
gβ-sheet form of M13 coat protein
hSequence: KLEALYILMVLGFFGFFTLGIMLSYIR. Both K27 and K27-Δ2 are in the β-sheet form
An extension of this method is to investigate the partial penetration of surface-associated proteins and peptides into the membrane (Görrissen et al. 1986; Rietveld et al. 1985, 1986; Jordi et al. 1989; Snel and Marsh 1994; Sankaram et al. 1989a, b, c; Keller et al. 1995, 1996; Snel et al. 1995; Montich and Marsh 1995; Montich et al. 1995; Ramakrishnan et al. 2001; Swamy et al. 2002). In these cases of partial penetration, the lipid stoichiometries are relatively low and the degree of motional restriction of the lipid chains is less than that for truly transmembrane proteins.
A wide variety of spin-labelled species is available (see Marsh 2008a) that can be used together with the EPR method to determine the selectivity patterns of interaction with different transmembrane proteins (see Fig. 2). Representative values of the mean relative association constants for different lipid species are listed for a variety of transmembrane proteins in Table 2. The free energy of association, ΔG ass(L*), that characterises the lipid selectivity is related to the relative association constant by:
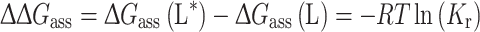 |
3 |
where T is the absolute temperature and R is the ideal gas constant. Selectivities are observed for certain negatively charged lipids that are partly, but not exclusively, of electrostatic origin (see, e.g., Fig. 3). Hydration and hydrogen bonding probably play an additional role in lipid selectivity (cf. Cevc et al. 1980).
Fig. 2.

Spin-labelled lipids used for investigating lipid–protein interactions. The spin-label nitroxyl ring is rigidly attached to the C-n atom of the lipid hydrocarbon chain (n = 14 in the figure), or to the steroid nucleus. For transmembrane proteins, two-component spectra are detected with lipids spin-labelled at the C14-position of the hydrocarbon chain (14-PXSL and 14-SASL). Two-component spectra are also resolved with the spin-labelled steroids, cholestane and androstanol (CSL and ASL, respectively). n-SASL, stearic acid; n-PCSL, -PESL, -PGSL, -PSSL, -PISL, and -PASL: phosphatidylcholine, phosphatidylethanolamine, phosphatidylglycerol, phosphatidylserine, phosphatidylinositol and phosphatidic acid, respectively. n-CLSL, cardiolipin. n-SMSL, sphingomyelin; n-GM1SL, monosialoganglioside GM1; n-GM2SL, monosialoganglioside GM2; n-GM3SL, monosialoganglioside GM3; n-GD1bSL, disialoganglioside GD1b; n-MGDGSL, monogalactosyl diglyceride; CSL, cholestane; and ASL, androstanol
Table 2.
Relative association constants K r, referred to phosphatidylcholine or phosphatidylglycerol (K r,o ≈ 1), for various spin-labelled phospholipid species interacting with different transmembrane proteins or peptides
| Protein | K r/K r,o | Referencesa | ||||||
|---|---|---|---|---|---|---|---|---|
| CL | Ptd | St | PtdSer | PtdGro | PtdEtn | PtdCho | ||
| α-helix: | ||||||||
| PLP | – | 10.4 | 6.5 | 2.2 | 1.8 | – | 1.0 | 1 |
| DM-20 | – | 2.3 | 2.0 | 1.1 | 1.0 | 1.0 | 1 | |
| Myelin proteolipidb | 1.5c | 2.9c | 7.0c | 1.4c | 1.1c | 0.5c | 1.0 | 2 |
| 3.0d | 2.4d | 2.9d | 1.4d | 2.0d | 1.7d | 1.0 | 3 | |
| Na, K-ATPase | 3.8 | 1.5 | 1.7 | 1.7 | 0.9 | 0.9 | 1.0 | 4, 5 |
| Na, K-ATPase–trypsinised | – | – | 1.5, 2.8e | 1.9 | – | – | 1.0 | 6 |
| Cytochrome c oxidase | 5.4 | 1.9 | – | 1.0 | 1.0 | 1.0 | 1.0 | 7 |
| ADP-ATP-carrier | 3.8 | 4.3 | 4.1 | 2.4 | 0.8 | – | 1.0 | 8 |
| Nicotinic acetylcholine receptor | 5.1 | – | 4.9 | 2.7 | 1.7 | 0.5 | 1.0 | 9 |
| M13 phage coat protein | – | 1.6f | 1.2f | 1.2f | 1.1f | 1.0f | 1.0 | 10 |
| 16-kDa proteolipid | – | – | 2.4 | 1.5 | 1.4 | – | 1.0 | 11 |
| Phospholamban–A36,41,46 | – | 1.3 | 2.8 | 1.0 | 1.0 | 0.9 | 1.0 | 12 |
| Phospholamban-Δ1–25A36,41,46 | – | 1.1 | 1.8 | 1.0 | 1.0 | 0.8 | 1.0 | 12 |
| GalP | – | 1.0, 2.9e | 1.9, 4.3e | 1.0 | 0.9 | 1.0 | 1.0 | 13 |
| Rhodopsin | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 14, 15 |
| β-barrel/sheet: | ||||||||
| OmpAg | – | 2.5 | ~0.2 | 0.6 | 1.0 | 0.5 | 0.6 | 16 |
| OmpG | – | 1.2 | 0.7 | – | 1.1 | 0.9 | 1.0 | 17 |
| FomA | – | 1.2 | 4.1 | 1.6 | 1.1 | 1.1 | 1.0 | 18 |
| FhuAg | – | 1.5 | 3.0 | 1.4 | 1.0 | 0.6 | 1.1 | 19 |
| M13 phage coat protein | 4.2 h | 4.2h | 2.3h | 2.1h | 1.6h | 0.9h | 1.0 | 20 |
| K27, K-channel peptidei | – | 3.3 | 2.0 | 2.0 | 1.1 | - | 1.1 | 21 |
| K27-Δ2 | – | 4.3 | 4.3 | 2.5 | 1.0 | - | 1.0 | 22 |
| β 6.3-helix: | ||||||||
| Gramicidin A | – | 1.2 | 0.7, 1.4e | 1.1 | 0.8 | 0.8 | 1.0 | 23 |
a1. Horváth et al. (1990b), 2. Brophy et al. (1984), 3. Sankaram et al. (1991), 4. Esmann et al. (1985), 5. Esmann and Marsh 1985), 6. Arora et al. (1998), 7. Knowles et al. (1981), 8. Horváth et al. (1990c), 9. Mantipragada et al. (2003), 10. Peelen et al. (1992), 11. Páli et al. (1995), 12. Arora et al. (2003), 13. Hubert et al. (2003), 14. Marsh et al. (1982), 15. Watts et al. (1979), 16. Ramakrishnan et al. (2004), 17. Anbazhagan et al. (2008a), 18. Anbazhagan et al. (2008b), 19. Ramakrishnan et al. (2004), 20. Datema et al. (1987), 21. Horváth et al. (1997), 22. Horváth et al. (1995), 23. Kóta et al. (2004)
CL, cardiolipin (Ptd2Gro); Ptd, phosphatidic acid; St, stearic acid; PtdSer, phosphatidylserine; PtdGro, phosphatidylglycerol; PtdEtn, phosphatidylethanolamine; PtdCho, phosphatidylcholine; PLP, myelin proteolipid protein
bNatural mixture of the proteolipid protein (PLP) and the DM-20 isoform (PLP-Δ116–150)
cIn diC14:0PtdCho
dIn diC14:0PtdGro
eValues for protonated and charged forms, respectively
fα-helical (partly) form of the protein in diC14:0PtdCho
gRelative to PtdGro
hβ-sheet form of the protein in diC14:0PtdCho/diC14:0PtdGro (80:20 mol/mol)
iSequence: KLEALYILMVLGFFGFFTLGIMLSYIR. β-sheet forms of K27 and K27-Δ2
Fig. 3.

Clusters of basic amino-acids that give rise to the pronounced selectivity of FhuA for anionic lipids (phosphatidic acid, phosphatidylserine and stearic acid). Electrostatic surfaces are coloured with blue positive, red negative and white neutral (Ramakrishnan et al. 2004)
Selectivity studies have also been performed with spin-labelled glycolipids (Li et al. 1989a, b, 1990), gangliosides (Schwarzmann et al. 1983; Esmann et al. 1988b; Mantipragada et al. 2003), and a variety of lyso and acyl derivatives of CL (Powell et al. 1987; Abramovitch et al. 1990; Esmann et al. 1988c). In addition to the selective interaction with lipids, the method may also be extended to spin-labelled drugs: for example, local anaesthetics (Miyazaki et al. 1992; Horváth et al. 1990a), and inhibitors of the osteoclast V-ATPase (Dixon et al. 2004, 2008).
Lipid exchange dynamics at the protein interface
Whereas the great power of the spin-label method lies in the determination of structural parameters (i.e., stoichiometries) and thermodynamic parameters (i.e., selectivities), the dynamic parameters that can be determined by EPR spectroscopy are equally of interest. Of especial relevance is the rate at which lipids in direct contact with the protein exchange with those in the bulk fluid bilayer regions of the membrane.
Resolution of two distinct components in the conventional EPR spectra of spin-labelled lipids in lipid–protein systems implies that exchange between the two lipid populations is slower than the difference in their spectral frequencies (Horváth et al. 1994). The critical rate is ~5 × 108 s−1 (see, e.g., Marsh et al. 2002b), which exceeds that for translational diffusion of free lipids in fluid bilayer membranes (Sachse et al. 1987; King et al. 1987). Thus, even if spin-labelled lipids at the protein interface exchange at rates comparable to those of translation in the bulk, they are resolved as a separate component of slower rotational mobility. The exchange is slow on the conventional spin-label EPR timescale, but is still sufficiently rapid to affect transverse relaxation, i.e., linewidths.
The rate of exchange of spin-labelled lipids at the intramembranous surface of the protein can be estimated by simulating the two-component EPR lineshapes with the Bloch Equations that incorporate two-site exchange:
 |
where subscripts ‘b’ and ‘f’ refer to protein-associated and bilayer-associated environments, respectively, of the lipid spin label. The steady-state rate equations for the complex transverse magnetisations: M b = u b + iv b and M f = u f + iv f in the rotating frame, can be written as (Horváth et al. 1988a, 1994):
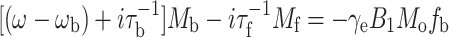 |
4 |
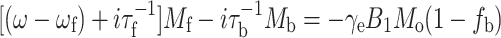 |
5 |
where ω b = ω o,b − iT −12,b and ω f = ω o,f − iT −12,f are the complex angular resonance frequencies of the protein-associated and bilayer-associated spin-label components, respectively, and T 2,b and T 2,f are the corresponding transverse relaxation times. In Eqs. 4 and 5, γe is the electron gyromagnetic ratio, B 1 is the microwave magnetic field intensity and M o is the equilibrium z-magnetisation. The slow-exchange solution for the EPR absorption lineshape is then given by:
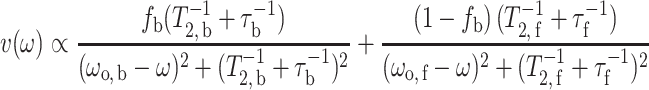 |
6 |
when τ −1b, τ −1f << (ω o,b − ω o,f). The two rate constants are related by Eq. 2, where τ −1b is the intrinsic exchange rate that depends on the affinity of L* for the protein (i.e., on K r), and is independent of the lipid–protein ratio. The on-rate, on the other hand, is diffusion-controlled and depends on the size of the free pool of lipid according to Eq. 2.
Data on the intrinsic off-rate constants (τ −1b,0) for exchange of phosphatidylcholine (a lipid that does not express selectivity for the protein) at the interface with different transmembrane proteins are listed in Table 3. These are obtained from simulation of the spectral lineshapes by using the exchange-coupled Bloch Equations. For comparison, the intrinsic diffusional lipid–lipid exchange rates in fluid phosphatidylcholine bilayers are τ −1diff ≈ 8 × 107 s−1 (Sachse et al. 1987; King et al. 1987). The rates of exchange at the protein interface are such that 2H-labelled lipids would be in fast exchange on the quadrupolar NMR timescale, explaining why only single-component lipid spectra are observed in lipid–protein systems by solid-state NMR (Meier et al. 1987).
Table 3.
Off-rate constants (τ −1b,o) and activation energies (E a,o) for exchange of spin-labelled phosphatidylcholine (14-PCSL) at the intramembranous surface of different transmembrane proteins
| Protein/lipid | T (°C) | τ −1b,o (s−1) | E a,o (kJ/mol) | Referencesa |
|---|---|---|---|---|
| Myelin proteolipid protein/diC14:0PtdCho | 30 | 1.6 × 107 | 20 | 1, 2 |
| Myelin DM-20 protein/diC14:0PtdCho | 30 | 1.5 × 107 | – | 3 |
| ADP-ATP carrier/egg PtdCho | 10 | 1.4 × 107 | – | 4 |
| M13 coat protein(α-helix)/diC18:1cPtdCho | 24 | 3.0 × 107 | – | 5 |
| M13 coat protein(α-helix)/diC14:0PtdCho | 30 | 2.3 × 107 | – | 5 |
| M13 coat protein(β-sheet)/diC14:0PtdCho | 30 | 5.3 × 106 | – | 6 |
| Rhodopsin/diC14:0PtdCho | 30 | 1.6 × 107 | 20 | 7 |
Because the on-rate is diffusion-controlled for all lipids, the off-rates must reflect the relative affinities of the different lipids for the protein. Combining Eqs. 1 and 2 for any fixed total lipid/protein ratio, one sees that the intrinsic off-rates of lipids A and B depend inversely on their relative association constants (Horváth et al. 1988a):
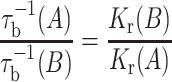 |
7 |
This reciprocal relation is found to hold for most systems investigated (Horváth et al. 1988b, 1990b, c; Wolfs et al., 1989; Peelen et al. 1992), with few exceptions that must reflect the existence of highly specific sites (Horváth et al. 1990c).
Sensitivity of spin-lattice relaxation to exchange of spin-labelled lipids
Slow-exchange simulations, which depend on transverse relaxation rates, lie at the limits of dynamic sensitivity for conventional spin-label EPR spectroscopy. More sensitive determination of the exchange rate comes from EPR saturation experiments because these depend on longitudinal (i.e., spin-lattice) relaxation rates, which are optimally matched in timescale to lipid exchange processes (Páli et al. 1999, 2000; Arora and Marsh 1998; Arora et al. 1999).
The effect of two-site exchange on spin-lattice relaxation can be determined from the rate equations for the population differences (n) between the up and down spin orientations (see Fig. 4). These population differences determine the longitudinal z-magnetisation. The steady-state rate equations when spin transitions are induced at rate 2W by B 1-irradiation of the protein-associated component b are (Horváth et al. 1993a):
 |
8 |
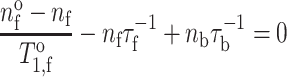 |
9 |
where n ob and n of are the spin population differences at Boltzmann equilibrium, and T o1,b and T o1,f are the spin-lattice relaxation times in the absence of exchange, for protein-associated and bilayer-associated spin-labelled lipids, respectively, in both cases. Solution of these rate equations, together with Eq. 2 (which also holds for population differences), yields the standard expression for saturation of the spin system at the protein-associated site ‘b’ (see, e.g., Slichter 1978):
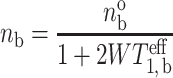 |
10 |
where T eff1,b is the effective spin-lattice relaxation time in the presence of exchange. This is given by (Horváth et al. 1993a):
 |
11 |
from which the intrinsic off-rate, τ −1b, for exchange is obtained.
Fig. 4.

Energy levels, spin populations (N ±b, N ±f), and transitions for two spin-label sites, ‘b’ and ‘f’. The spin population difference is given by: n b = N −b − N +b. The transition rate for spin-lattice relaxation is: 2W e = 1/T o1. The rate of exchange between the two sites is: N b τ −1b = N f τ −1f, for both spin populations and population differences. The rate of Heisenberg exchange between spins ‘b’ and ‘f’ is: 2K x N ±b N ∓f (Marsh 1993)
Figure 5 shows progressive saturation curves for a spin-labelled lipid in lipid bilayers alone, associated with a delipidated protein (PLP), and in lipid membranes containing PLP. Data are given for temperatures both below and above the chain-melting transition of the lipid. In both cases, spin-labelled lipids associated with the delipidated protein saturate more readily than those in membranes of the lipid alone, indicating a pronounced difference in the lipid chain dynamics at the protein interface (Livshits et al. 1998, 2003). In the fluid phase, this mobility difference is clear from the two-component lineshapes, i.e., occurs on the timescale of T 2-relaxation. The saturation properties show that this distinction extends also to the gel phase, where differences in mobility are found on the longer timescale of T 1-relaxation.
Fig. 5.

Progressive saturation curves for the integrated intensity of the conventional EPR spectra of spin-labelled phosphatidylcholine 14-PCSL in: diC14:0PtdCho membranes (solid circles), delipidated myelin proteolipid protein PLP (open circles), and PLP/diC14:0PtdCho membranes of lipid/protein ratio 24:1 mol/mol (solid squares). Left in the lipid gel phase (4°C), right in the lipid fluid phase (30°C). Solid lines are fits of Eq. 12 for saturation of the single components (f b = 0 or 1), and dotted lines are predictions for saturation of the lipid–protein membranes assuming no exchange between the two components (f b = 0.40) (Horváth et al. 1993a)
The saturation behaviour in the protein-containing membranes is intermediate between that in the lipid-alone and protein-alone environments. However, in the fluid phase, the saturation curve is strongly biassed towards that from the lipid alone. The dependence of the double-integrated intensity of a two-component spectrum on the microwave field strength, B 1, is given by (Snel and Marsh 1994; Páli et al. 1993):
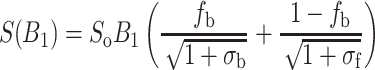 |
12 |
where the saturation parameter for the protein-associated lipids is σ b = γ 2e B 21 T 1,b T 2,b and similarly for σf (γe = 1.76086 × 1011 rad s−1 T−1). (Note that Eq. 12 illustrates the double advantage of using the integrated EPR intensity, instead of spectral lineheights, for constructing saturation curves: contributions from different components in multi-component systems are strictly additive, and the saturation curves do not depend on the degree of inhomogeneous broadening of any of the components.) The dotted saturation curves in Fig. 5 correspond to predictions from Eq. 12 by using the saturation parameters determined from the corresponding single-component systems. In the gel phase, where the lipid mobility is low, the spectrum from the lipid–protein membrane saturates exactly as predicted for a linear combination of the two isolated environments. In the fluid phase, on the other hand, the spectrum saturates less readily than predicted from the isolated components. Alleviation of saturation arises from lipid exchange between the two environments that must take place on the timescale of spin-lattice relaxation (i.e., in the microsecond regime), in fluid membranes. Exchange at these rates (cf. Table 3), although still relatively rapid, confirms that the two-component lineshapes observed by conventional EPR correspond to slow exchange.
Of the various non-linear EPR methods (Marsh 1993), out-of-phase detection (as in saturation transfer EPR) has proved to be an especially powerful tool for probing lipid exchange in protein-containing membranes. In the absence of exchange between components on the T
1-timescale, the first integral I
ST of the 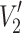 -STEPR spectrum (second harmonic detected in phase quadrature) is additive between the different components (Horváth and Marsh 1983):
-STEPR spectrum (second harmonic detected in phase quadrature) is additive between the different components (Horváth and Marsh 1983):
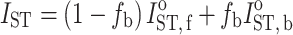 |
13 |
where I
oST,b and I
oST,f are the values of I
ST for components ‘b’ and ‘f’, respectively, and f
b is the fraction of component ‘b’ in the two-component mixture. Figure 6 gives the integrated 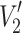 -STEPR intensity as a function of the fraction of spin-labelled lipid that is associated with the PLP protein in reconstituted membranes. Samples all have the same total lipid/protein ratio; f
b is varied by using spin-labelled lipid species with differing affinities for PLP. The value of f
b is determined by spectral subtraction with the two-component conventional V
1-EPR spectra, as for the data in Tables 1 and 2. Below the lipid chain-melting temperature (T
t), I
ST depends linearly on f
b, as predicted by Eq. 13, showing that in the gel phase any lipid exchange is extremely slow, relative to the microsecond timescale. In the fluid lipid phase, above T
t, however, the dependence of I
ST on f
b lies below the straight (dashed) line expected for no exchange. Saturation is partially alleviated by exchange between sites on and off the protein at rates comparable to the spin-lattice relaxation time. This result, which is based on differential selectivities of various lipid species for PLP (cf. Table 2), complements that from the progressive saturation experiments given in the right panel of Fig. 5.
-STEPR intensity as a function of the fraction of spin-labelled lipid that is associated with the PLP protein in reconstituted membranes. Samples all have the same total lipid/protein ratio; f
b is varied by using spin-labelled lipid species with differing affinities for PLP. The value of f
b is determined by spectral subtraction with the two-component conventional V
1-EPR spectra, as for the data in Tables 1 and 2. Below the lipid chain-melting temperature (T
t), I
ST depends linearly on f
b, as predicted by Eq. 13, showing that in the gel phase any lipid exchange is extremely slow, relative to the microsecond timescale. In the fluid lipid phase, above T
t, however, the dependence of I
ST on f
b lies below the straight (dashed) line expected for no exchange. Saturation is partially alleviated by exchange between sites on and off the protein at rates comparable to the spin-lattice relaxation time. This result, which is based on differential selectivities of various lipid species for PLP (cf. Table 2), complements that from the progressive saturation experiments given in the right panel of Fig. 5.
Fig. 6.

Dependence of the integrated V′2 saturation transfer EPR intensity, I ST, from different spin-labelled lipids on the fraction, f b, of each lipid species associated with the myelin proteolipid protein in diC14:0PtdCho membranes at fixed lipid/protein ratio (37:1 mol/mol). Measurements correspond to the gel-phase (T < T t) and the fluid phase (T > T t) at 4 and 30°C, respectively. Straight lines are predictions for zero exchange rate (τ −1b = 0) at the two temperatures, and the curved line is a non-linear least-squares fit of Eq. 14 with Eq. 11 (and equivalent) giving a constant lipid on-rate of T o1,bτ−1f = 2.9 in the fluid phase at 30°C (Horváth et al. 1993a)
Both theoretical simulations and experiment indicate that the normalised saturation transfer intensity, I ST, is approximately proportional to T 1, over a considerable range of spin-lattice relaxation times (Páli et al. 1996). The net ST-EPR intensity in the presence of exchange between components ‘b’ and ‘f’ is thus given by (cf. Marsh and Horváth 1992; Páli et al. 1992):
 |
14 |
where T 1,b and T 1,f are the spin-lattice relaxation times at lipid locations, respectively, on and off the protein, and T o1,b, T o1,f are the corresponding values in the absence of exchange. Combining Eq. 14 with Eq. 11 and its equivalent for T 1,f/T o1,f gives the predicted dependence of I ST on f b in the presence of exchange. The non-linear least-squares fit to the data shown for the fluid phase in Fig. 6 yields a normalised on-rate constant for lipid exchange of T o1,b τ −1f = 2.9 (at fixed lipid/protein ratio of 37:1 mol/mol and T = 30°C). With this on-rate, the off-rates τ −1b for the lipids with different affinities for PLP are determined from the values of f b by material balance, i.e., Eq. 2 (see Table 4).
Table 4.
Normalised off-rates for exchange of different spin-labelled lipid species at the protein interface in myelin proteolipid (PLP)/diC14:0PtdCho membranes at 30°C (Horváth et al. 1993a)
| Lipid | ΔI ST × 102 | T o1,b τ −1b |
|---|---|---|
| Stearic acid | −0.19 | 0.67 |
| Phosphatidic acid | −0.27 | 0.86 |
| Phosphatidylserine | −0.20 | 2.45 |
| Phosphatidylglycerol | −0.13 | 4.9 |
| Phosphatidylcholine | −0.14 | 4.9 |
Intrinsic off-rates, τ −1b, are normalised with respect to the intrinsic spin-lattice relaxation time of the protein-associated lipids, T o1,b ~ 1 μs. ΔI ST is the difference in normalised ST-EPR intensity from that predicted by linear additivity
Accessibility of penetrant proteins to spin exchange with labelled lipids
The extent of membrane penetration by a spin-labelled segment of a surface-associated or transmembrane protein can be assessed from spin–spin interactions with spin labels located at different positions in a lipid molecule (Snel and Marsh 1994; Páli et al. 1999). Spin exchange in the double-labelled system is analogous to molecular exchange at the lipid–protein interface (Marsh 1992a, b; and see Fig. 4). In the presence of Heisenberg spin-exchange between the two distinguishable spin-labelled species, the spin-lattice relaxation time of the spin-labelled protein is given by (Snel and Marsh 1994):
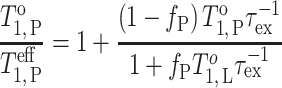 |
15 |
where subscripts P and L refer to spin-labelled protein and lipid, respectively, and f P is the fraction of the total spin-label intensity that is associated with the protein. The mutual spin exchange frequency τ −1ex is proportional to the total spin label concentration and is a measure of the collision frequency between the two spin-labelled species.
Table 5 gives the spin exchange frequencies between a spin label on Cys14 or Cys17 in the N-terminal region of apocytochrome c bound to phosphatidylglycerol membranes and spin labels in the polar group (T-PASL) or at different positions in the acyl chains (5-PGSL and 14-PGSL) of the membrane lipids. (Note that Cys14 and Cys17 are the sites of covalent attachment of the haem group in the holoprotein and, at labelling levels of 1:1 mol/mol, either one or other of these residues in the apoprotein bears the spin label.) The maximum exchange frequency is found with a spin label on the C-14 atom of the lipid chain. This indicates that the N-terminal section of apocytochrome c penetrates deeply into the membrane, consistent with lipid–protein interactions facilitating the translocation of this haem-less precursor into the mitochondrion (Görrissen et al. 1986; Snel et al. 1994). In contrast, for the mature holocytochrome c protein spin labelled on Cys102 close to the C-terminal, the largest exchange frequency is found with a lipid spin labelled in the polar headgroup, as expected for a native peripheral protein bound at the outer surface of the inner mitochondrial membrane (Kostrzewa et al. 2000).
Table 5.
Normalised spin exchange frequencies between lipids spin-labelled at different positions and spin-labelled proteins (cytochrome c and apocytochrome c) bound to phosphatidylglycerol bilayer membranes (Snel and Marsh 1994)
| Spin-labelled lipid | Δ(1/T 1 T 2)×10−13 (s−2) | T o1,P τ −1ex |
|---|---|---|
| Apocytochrome c/diC14:0PtdGro: | ||
| T-PASL | 0 | 0 |
| 5-PGSL | 0.3 | 0.2 |
| 14-PGSL | 8.5 | ≥5 |
| Apocytochrome c/diC18:1cPtdGro: | ||
| T-PASL | 5.6 | ≥5 |
| 5-PGSL | 0 | 0 |
| 14-PGSL | 5.4 | 23 |
| Cytochrome c/diC18:1cPtdGro: | ||
| T-PASL | 1.6 | 0.8 |
| 5-PGSL | 0.2 | 0.1 |
| 14-PGSL | 0 | 0 |
Spin exchange frequencies, τ −1ex, are normalised with respect to the intrinsic spin-lattice relaxation time of the spin-labelled protein, T o1,P ~ 1 μs. Δ(1/T 1 T 2) is the difference in 1/T 1 T 2 of the double-labelled system from that predicted by Eq. 12 with saturation parameters from the single spin-labelled systems. T-PASL is phosphatidic acid spin-labelled on the phosphate headgroup
The vertical depth of the DCCD-reactive glutamate that is essential for proton translocation by the 16-kDa proteolipid subunit of the vacuolar ATPase (Vma3p in yeast) was mapped in a similar way by using different spin-labelled lipids and a nitroxyl analogue of DCCD (Páli et al. 1999). Most importantly, the STEPR intensities of the double-labelled system demonstrate that the essential glutamate residue is exposed to lipid, as is required by the rotary mechanism for proton transport by stepwise interaction with the Vph1p subunit of the yeast V-ATPase.
Conclusion
Spin-label EPR has provided a wealth of data on both stoichiometry and selectivity of lipid–protein interactions in biological membranes. The experiments are, however, mostly restricted to spin-labelled lipids at probe concentrations, which means that a small number of high-affinity sites are not distinguished from a smaller generalised selectivity for all lipid sites at the intramembranous perimeter of the protein. In one instance, competition with exogenous cardiolipin failed to yield evidence for a very high-affinity site on cytochrome oxidase (Powell et al. 1985), where endogenous cardiolipin had been replaced by phosphatidylcholine (Watts et al. 1978). On the other hand, both X-ray crystallography and fluorescence experiments have located non-annular lipid at intersubunit sites. Therefore, this remains a future challenge for the EPR spectroscopist. More data are also needed on the dynamics of lipid exchange: the methods are in place but the current database is of relatively modest size. Nevertheless, a specific site for cardiolipin on the ADP-ATP translocator is already indicated by comparison of exchange rates and relative association constants for this lipid (Horváth et al. 1990c).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
The more you see: spectroscopy in molecular biophysics.
References
- Abramovitch DA, Marsh D, Powell GL. Activation of beef heart cytochrome oxidase by cardiolipin and analogues of cardiolipin. Biochim Biophys Acta. 1990;1020:34–42. doi: 10.1016/0005-2728(90)90090-q. [DOI] [PubMed] [Google Scholar]
- Anbazhagan V, Qu J, Kleinschmidt JH, Marsh D. Incorporation of outer membrane protein OmpG in lipid membranes. Protein–lipid interactions and β-barrel orientation. Biochemistry. 2008;47:6189–6198. doi: 10.1021/bi800203g. [DOI] [PubMed] [Google Scholar]
- Anbazhagan V, Vijay N, Kleinschmidt JH, Marsh D. Protein–lipid interactions with Fusobacterium nucleatum major membrane protein FomA: spin-label EPR and polarised infrared spectroscopy. Biochemistry. 2008;47:8414–8423. doi: 10.1021/bi800750s. [DOI] [PubMed] [Google Scholar]
- Arora A, Marsh D. Protein-induced vertical lipid dislocation in a model membrane system: spin-label relaxation studies on avidin–biotinylphosphatidylethanolamine interactions. Biophys J. 1998;75:2915–2922. doi: 10.1016/S0006-3495(98)77733-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora A, Esmann M, Marsh D. Selectivity of lipid–protein interactions with trypsinized Na, K-ATPase studied by spin-label EPR. Biochim Biophys Acta. 1998;1371:163–167. doi: 10.1016/s0005-2736(98)00030-3. [DOI] [PubMed] [Google Scholar]
- Arora A, Esmann M, Marsh D. Microsecond motions of the lipids associated with trypsinized Na,K-ATPase membranes. Progressive saturation spin-label electron spin resonance studies. Biochemistry. 1999;38:10084–10091. doi: 10.1021/bi9826916. [DOI] [PubMed] [Google Scholar]
- Arora A, Williamson IM, Lee AG, Marsh D. Lipid–protein interactions with cardiac phospholamban studied by spin-label electron spin resonance. Biochemistry. 2003;42:5151–5158. doi: 10.1021/bi020663o. [DOI] [PubMed] [Google Scholar]
- Brophy PJ, Horváth LI, Marsh D. Stoichiometry and specificity of lipid–protein interaction with myelin proteolipid protein studied by spin-label electron spin resonance. Biochemistry. 1984;23:860–865. doi: 10.1021/bi00300a011. [DOI] [PubMed] [Google Scholar]
- Brotherus JR, Griffith OH, Brotherus MO, Jost PC, Silvius JR, Hokin LE. Lipid–protein multiple binding equilibria in membranes. Biochemistry. 1981;20:5261–5267. doi: 10.1021/bi00521a026. [DOI] [PubMed] [Google Scholar]
- Cevc G, Watts A, Marsh D. Non-electrostatic contribution to the titration of the ordered-fluid phase transition of phosphatidylglycerol bilayers. FEBS Lett. 1980;120:267–270. doi: 10.1016/0014-5793(80)80313-9. [DOI] [PubMed] [Google Scholar]
- Datema KP, Wolfs CJAM, Marsh D, Watts A, Hemminga MA. Spin-label electron spin resonance study of bacteriophage M13 coat protein incorporation into mixed lipid bilayers. Biochemistry. 1987;26:7571–7574. doi: 10.1021/bi00398a006. [DOI] [PubMed] [Google Scholar]
- Dixon N, Páli T, Kee TP, Marsh D. Spin-labelled vacuolar-ATPase inhibitors in lipid membranes. Biochim Biophys Acta. 2004;1665:177–183. doi: 10.1016/j.bbamem.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Dixon N, Páli T, Kee TP, Ball S, Harrison MA, Findlay JBC, Nyman J, Väänänen HK, Finbow ME, Marsh D. Interaction of spin-labelled inhibitors of the vacuolar H+-ATPase with the transmembrane V0-sector. Biophys J. 2008;94:506–514. doi: 10.1529/biophysj.107.111781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellena JF, Blazing MA, McNamee MG. Lipid–protein interactions in reconstituted membranes containing acetylcholine receptor. Biochemistry. 1983;22:5523–5535. doi: 10.1021/bi00293a012. [DOI] [PubMed] [Google Scholar]
- Esmann M, Marsh D. Spin-label studies on the origin of the specificity of lipid–protein interactions in Na+,K+-ATPase membranes from Squalus acanthias. Biochemistry. 1985;24:3572–3578. doi: 10.1021/bi00335a027. [DOI] [PubMed] [Google Scholar]
- Esmann M, Marsh D. Lipid–protein interactions with the Na,K-ATPase. Chem Phys Lipids. 2006;141:94–104. doi: 10.1016/j.chemphyslip.2006.02.018. [DOI] [PubMed] [Google Scholar]
- Esmann M, Watts A, Marsh D. Spin-label studies of lipid–protein interactions in (Na+,K+)-ATPase membranes from rectal glands of Squalus acanthias. Biochemistry. 1985;24:1386–1393. doi: 10.1021/bi00327a016. [DOI] [PubMed] [Google Scholar]
- Esmann M, Hideg K, Marsh D. Novel spin-labels for the study of lipid–protein interactions. Application to (Na+,K+)-ATPase membranes. Biochemistry. 1988;27:3913–3917. doi: 10.1021/bi00411a004. [DOI] [PubMed] [Google Scholar]
- Esmann M, Marsh D, Schwarzmann G, Sandhoff K. Ganglioside–protein interactions: spin-label electron spin resonance studies with (Na+, K+)-ATPase membranes. Biochemistry. 1988;27:2398–2403. doi: 10.1021/bi00407a022. [DOI] [PubMed] [Google Scholar]
- Esmann M, Powell GL, Marsh D. Spin label studies on the selectivity of lipid–protein interaction of cardiolipin analogues with the Na+/K+-ATPase. Biochim Biophys Acta. 1988;941:287–292. doi: 10.1016/0005-2736(88)90190-3. [DOI] [PubMed] [Google Scholar]
- Ferguson AD, Hofmann E, Coulton JW, Diederichs K, Welte W. Siderophore-mediated iron transport: crystal structure of FhuA with bound lipopolysaccharide. Science. 1998;282:2215–2220. doi: 10.1126/science.282.5397.2215. [DOI] [PubMed] [Google Scholar]
- Görrissen H, Marsh D, Rietveld A, De Kruijff B. Apocytochrome c binding to negatively charged lipid dispersions studied by spin-label electron spin resonance. Biochemistry. 1986;25:2904–2910. doi: 10.1021/bi00358a025. [DOI] [PubMed] [Google Scholar]
- Horváth LI, Marsh D. Analysis of multicomponent saturation transfer ESR spectra using the integral method: application to membrane systems. J Magn Reson. 1983;54:363–373. [Google Scholar]
- Horváth LI, Brophy PJ, Marsh D. Exchange rates at the lipid–protein interface of myelin proteolipid protein studied by spin-label electron spin resonance. Biochemistry. 1988;27:46–52. doi: 10.1021/bi00401a009. [DOI] [PubMed] [Google Scholar]
- Horváth LI, Brophy PJ, Marsh D. Influence of lipid headgroup on the specificity and exchange dynamics in lipid–protein interactions. A spin label study of myelin proteolipid apoprotein–phospholipid complexes. Biochemistry. 1988;27:5296–5304. doi: 10.1021/bi00414a052. [DOI] [PubMed] [Google Scholar]
- Horváth LI, Arias HR, Hankovszky HO, Hideg K, Barrantes FJ, Marsh D. Association of spin-labeled local anaesthetics at the hydrophobic surface of acetylcholine receptor in native membranes from Torpedo marmorata. Biochemistry. 1990;29:8707–8713. doi: 10.1021/bi00489a029. [DOI] [PubMed] [Google Scholar]
- Horváth LI, Brophy PJ, Marsh D. Influence of polar residue deletions on lipid–protein interactions with the myelin proteolipid protein. Spin-label ESR studies with DM-20/lipid recombinants. Biochemistry. 1990;29:2635–2638. doi: 10.1021/bi00463a002. [DOI] [PubMed] [Google Scholar]
- Horváth LI, Drees M, Beyer K, Klingenberg M, Marsh D. Lipid–protein interactions in ADP-ATP carrier/egg phosphatidylcholine recombinants studied by spin-label ESR spectroscopy. Biochemistry. 1990;29:10664–10669. doi: 10.1021/bi00499a013. [DOI] [PubMed] [Google Scholar]
- Horváth LI, Brophy PJ, Marsh D. Exchange rates at the lipid–protein interface of the myelin proteolipid protein determined by saturation transfer electron spin resonance and continuous wave saturation studies. Biophys J. 1993;64:622–631. doi: 10.1016/S0006-3495(93)81421-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horváth LI, Brophy PJ, Marsh D. Spin label saturation transfer EPR determinations of the stoichiometry and selectivity of lipid–protein interactions in the gel phase. Biochim Biophys Acta. 1993;1147:277–280. doi: 10.1016/0005-2736(93)90014-q. [DOI] [PubMed] [Google Scholar]
- Horváth LI, Brophy PJ, Marsh D. Microwave frequency dependence of ESR spectra from spin labels undergoing two-site exchange in myelin proteolipid membranes. J Magn Reson. 1994;B105:120–128. doi: 10.1006/jmrb.1994.1111. [DOI] [PubMed] [Google Scholar]
- Horváth LI, Heimburg T, Kovachev P, Findlay JBC, Hideg K, Marsh D. Integration of a K+ channel-associated peptide in a lipid bilayer: conformation, lipid–protein interactions, and rotational diffusion. Biochemistry. 1995;34:3893–3898. doi: 10.1021/bi00012a004. [DOI] [PubMed] [Google Scholar]
- Horváth LI, Knowles PF, Kovachev P, Findlay JBC, Marsh D. A single-residue deletion alters the lipid selectivity of a K+ channel-associated peptide in the β-conformation: spin label electron spin resonance studies. Biophys J. 1997;73:2588–2594. doi: 10.1016/S0006-3495(97)78288-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubert A, Henderson PJF, Marsh D. Lipid–protein interactions in Escherichia coli membranes overexpressing the sugar-H+ symporter, GalP. EPR of spin-labelled lipids. Biochim Biophys Acta. 2003;1611:243–248. doi: 10.1016/s0005-2736(03)00081-6. [DOI] [PubMed] [Google Scholar]
- Jordi W, De Kruijff B, Marsh D. Specificity of the interaction of amino- and carboxy-terminal fragments of the mitochondrial precursor protein apocytochrome c with negatively charged phospholipids. A spin-label electron spin resonance study. Biochemistry. 1989;28:8998–9005. doi: 10.1021/bi00449a007. [DOI] [PubMed] [Google Scholar]
- Jost PC, Griffith OH, Capaldi RA, Vanderkooi G. Evidence for boundary lipid in membranes. Proc Natl Acad Sci USA. 1973;70:480–484. doi: 10.1073/pnas.70.2.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller RCA, Snel MME, De Kruijff B, Marsh D. SecA restricts, in a nucleotide-dependent manner, acyl chain mobility up to the center of a phospholipid bilayer. FEBS Lett. 1995;358:251–254. doi: 10.1016/0014-5793(94)01439-8. [DOI] [PubMed] [Google Scholar]
- Keller RCA, ten Berge D, Nouwen N, Snel MME, Tommassen J, Marsh D, De Kruijff B. Mode of insertion of the signal sequence of a bacterial precursor protein into phospholipid bilayers as revealed by cysteine-based site-directed spectroscopy. Biochemistry. 1996;35:3063–3071. doi: 10.1021/bi951870+. [DOI] [PubMed] [Google Scholar]
- King MD, Sachse J-H, Marsh D. Unconstrained optimization method for interpreting the concentration and temperature dependence of the linewidths of interacting nitroxide spin labels. Application to the measurement of translational diffusion coefficients of spin-labeled phospholipids in membranes. J Magn Reson. 1987;72:257–267. [Google Scholar]
- Kleinschmidt JH, Powell GL, Marsh D. Cytochrome c-induced increase of motionally restricted lipid in reconstituted cytochrome c oxidase membranes, revealed by spin-label ESR spectroscopy. Biochemistry. 1998;37:11579–11585. doi: 10.1021/bi980235b. [DOI] [PubMed] [Google Scholar]
- Knowles PF, Marsh D. Magnetic resonance of membranes. Biochem J. 1991;274:625–641. doi: 10.1042/bj2740625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles PF, Watts A, Marsh D. Spin label studies of lipid immobilization in dimyristoylphosphatidylcholine-substituted cytochrome oxidase. Biochemistry. 1979;18:4480–4487. doi: 10.1021/bi00588a005. [DOI] [PubMed] [Google Scholar]
- Knowles PF, Watts A, Marsh D. Spin label studies of headgroup specificity in the interaction of phospholipids with yeast cytochrome oxidase. Biochemistry. 1981;20:5888–5894. doi: 10.1021/bi00523a036. [DOI] [PubMed] [Google Scholar]
- Kostrzewa A, Páli T, Froncisz W, Marsh D. Membrane location of spin-labeled cytochrome c determined by paramagnetic relaxation agents. Biochemistry. 2000;39:6066–6074. doi: 10.1021/bi992559l. [DOI] [PubMed] [Google Scholar]
- Kóta Z, Páli T, Marsh D. Orientation and lipid–peptide interactions of gramicidin A in lipid membranes: polarized ATR infrared spectroscopy and spin-label electron spin resonance. Biophys J. 2004;86:1521–1531. doi: 10.1016/S0006-3495(04)74220-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kóta Z, Páli T, Dixon N, Kee TP, Harrison M, Findlay JBC, Finbow ME, Marsh D. Incorporation of transmembrane peptides from the vacuolar H+-ATPase in phospholipid membranes: spin-label electron paramagnetic resonance and polarized infrared spectroscopy. Biochemistry. 2008;47:3937–3949. doi: 10.1021/bi7025112. [DOI] [PubMed] [Google Scholar]
- Li G, Horváth LI, Knowles PF, Murphy DJ, Marsh D. Spin label saturation transfer ESR studies of protein–lipid interactions in photosystem II-enriched membranes. Biochim Biophys Acta. 1989;987:187–192. [Google Scholar]
- Li G, Knowles PF, Murphy DJ, Nishida I, Marsh D. Spin-label ESR studies of lipid–protein interactions in thylakoid membranes. Biochemistry. 1989;28:7446–7452. doi: 10.1021/bi00444a044. [DOI] [PubMed] [Google Scholar]
- Li G, Knowles PF, Murphy DJ, Marsh D. Lipid–protein interactions in thylakoid membranes of chilling-resistant and -sensitive plants studied by spin label electron spin resonance spectroscopy. J Biol Chem. 1990;265:16867–16872. [PubMed] [Google Scholar]
- Livshits VA, Páli T, Marsh D. Relaxation time determinations by progressive saturation EPR: Effects of molecular motion and Zeeman modulation for spin labels. J Magn Reson. 1998;133:79–91. doi: 10.1006/jmre.1998.1434. [DOI] [PubMed] [Google Scholar]
- Livshits VA, Dzikovski BG, Marsh D. Anisotropic motion effects in CW non-linear EPR spectra: relaxation enhancement of lipid spin labels. J Magn Reson. 2003;162:429–442. doi: 10.1016/s1090-7807(03)00076-4. [DOI] [PubMed] [Google Scholar]
- Mantipragada S, Horváth LI, Arias HR, Schwarzmann G, Sandhoff K, Barrantes FJ, Marsh D. Lipid–protein interactions and effect of local anesthetics in acetylcholine receptor-rich membranes from Torpedo marmorata electric organ. Biochemistry. 2003;42:9167–9175. doi: 10.1021/bi034485q. [DOI] [PubMed] [Google Scholar]
- Marsh D. Spin label answers to lipid–protein interactions. Trends Biochem Sci. 1983;8:330–333. [Google Scholar]
- Marsh D. ESR spin label studies of lipid–protein interactions. In: Watts A, de Pont JJHHM, editors. Progress in protein–lipid interactions. Amsterdam: Elsevier; 1985. pp. 143–172. [Google Scholar]
- Marsh D. Selectivity of lipid–protein interactions. J Bioenerg Biomembr. 1987;19:677–689. doi: 10.1007/BF00762302. [DOI] [PubMed] [Google Scholar]
- Marsh D. Lipid–protein interactions in membranes. FEBS Lett. 1990;268:371–375. doi: 10.1016/0014-5793(90)81288-y. [DOI] [PubMed] [Google Scholar]
- Marsh D. Exchange and dipolar spin–spin interactions and rotational diffusion in saturation transfer EPR spectroscopy. Appl Magn Reson. 1992;3:53–65. [Google Scholar]
- Marsh D. Influence of nuclear relaxation on the measurement of exchange frequencies in CW saturation EPR studies. J Magn Reson. 1992;99:332–337. [Google Scholar]
- Marsh D. Progressive saturation and saturation transfer ESR for measuring exchange processes of spin-labelled lipids and proteins in membranes. Chem Soc Rev. 1993;22:329–335. [Google Scholar]
- Marsh D. Lipid–protein interactions and heterogeneous lipid distribution in membranes. Mol Membr Biol. 1995;12:59–64. doi: 10.3109/09687689509038496. [DOI] [PubMed] [Google Scholar]
- Marsh D. Peptide models for membrane channels. Biochem J. 1996;315:345–361. doi: 10.1042/bj3150345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh D. Stoichiometry of lipid–protein interaction and integral membrane protein structure. Eur Biophys J. 1997;26:203–208. [Google Scholar]
- Marsh D. Saturation transfer EPR studies of slow rotational motion in membranes. Appl Magn Reson. 2007;31:387–410. [Google Scholar]
- Marsh D. Electron spin resonance in membrane research: protein–lipid interactions. Methods. 2008;46:83–96. doi: 10.1016/j.ymeth.2008.07.001. [DOI] [PubMed] [Google Scholar]
- Marsh D. Protein modulation of lipids, and vice-versa, in membranes. Biochim Biophys Acta. 2008;1778:1545–1575. doi: 10.1016/j.bbamem.2008.01.015. [DOI] [PubMed] [Google Scholar]
- Marsh D. Orientation and peptide–lipid interactions of alamethicin incorporated in lipid membranes: polarized infrared and spin-label EPR spectroscopy. Biochemistry. 2009;48:729–737. doi: 10.1021/bi801279n. [DOI] [PubMed] [Google Scholar]
- Marsh D, Horváth LI. Influence of Heisenberg spin exchange on conventional and phase-quadrature EPR lineshapes and intensities under saturation. J Magn Reson. 1992;97:13–26. [Google Scholar]
- Marsh D, Horváth LI. Structure, dynamics and composition of the lipid–protein interface. Perspectives from spin-labelling. Biochim Biophys Acta. 1998;1376:267–296. doi: 10.1016/s0304-4157(98)00009-4. [DOI] [PubMed] [Google Scholar]
- Marsh D, Páli T. The protein–lipid interface: perspectives from magnetic resonance and crystal structures. Biochim Biophys Acta. 2004;1666:118–141. doi: 10.1016/j.bbamem.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Marsh D, Powell GL. Properties of cardiolipin and functional implications for cytochrome oxidase activity. Bioelectrochem Bioenerg. 1988;20:73–82. [Google Scholar]
- Marsh D, Watts A, Pates RD, Uhl R, Knowles PF, Esmann M. ESR spin label studies of lipid–protein interactions in membranes. Biophys J. 1982;37:265–274. doi: 10.1016/S0006-3495(82)84675-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh D, Horváth LI, Swamy MJ, Mantripragada S, Kleinschmidt JH. Interaction of membrane-spanning proteins with peripheral and lipid-anchored membrane proteins. Perspectives from protein–lipid interactions. Mol Membr Biol. 2002;19:247–255. doi: 10.1080/09687680210162419. [DOI] [PubMed] [Google Scholar]
- Marsh D, Kurad D, Livshits VA. High-field electron spin resonance of spin labels in membranes. Chem Phys Lipids. 2002;116:93–114. doi: 10.1016/s0009-3084(02)00022-1. [DOI] [PubMed] [Google Scholar]
- Meier P, Sachse J-H, Brophy PJ, Marsh D, Kothe G. Integral membrane proteins significantly decrease the molecular motion in lipid bilayers: a deuteron NMR relaxation study of membranes containing myelin proteolipid apoprotein. Proc Natl Acad Sci USA. 1987;84:3704–3708. doi: 10.1073/pnas.84.11.3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki J, Hideg K, Marsh D. Interfacial ionization and partitioning of membrane-bound local anaesthetics. Biochim Biophys Acta. 1992;1103:62–68. doi: 10.1016/0005-2736(92)90057-s. [DOI] [PubMed] [Google Scholar]
- Montich GG, Marsh D. Interaction of α-lactalbumin with phosphatidylglycerol. Influence of protein binding on the lipid phase transition and lipid acyl chain mobility. Biochemistry. 1995;34:13139–13145. doi: 10.1021/bi00040a027. [DOI] [PubMed] [Google Scholar]
- Montich GG, Montecucco C, Papini E, Marsh D. Insertion of diphtheria toxin in lipid bilayers studied by spin label ESR. Biochemistry. 1995;34:11561–11567. doi: 10.1021/bi00036a032. [DOI] [PubMed] [Google Scholar]
- Páli T, Bartucci R, Horváth LI, Marsh D. Distance measurements using paramagnetic ion-induced relaxation in the saturation transfer electron spin resonance of spin-labeled biomolecules. Application to phospholipid bilayers and interdigitated gel phases. Biophys J. 1992;61:1595–1602. doi: 10.1016/S0006-3495(92)81963-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Páli T, Horváth LI, Marsh D. Continuous-wave saturation of two-component, inhomogeneously broadened, anisotropic EPR spectra. J Magn Reson. 1993;A101:215–219. [Google Scholar]
- Páli T, Finbow ME, Holzenburg A, Findlay JBC, Marsh D. Lipid–protein interactions and assembly of the 16-kDa channel polypeptide from Nephrops norvegicus. Studies with spin-label electron spin resonance spectroscopy and electron microscopy. Biochemistry. 1995;34:9211–9218. doi: 10.1021/bi00028a034. [DOI] [PubMed] [Google Scholar]
- Páli T, Livshits VA, Marsh D. Dependence of saturation-transfer EPR intensities on spin-lattice relaxation. J Magn Reson B. 1996;113:151–159. doi: 10.1006/jmrb.1996.0168. [DOI] [PubMed] [Google Scholar]
- Páli T, Finbow ME, Marsh D. Membrane assembly of the 16-kDa proteolipid channel from Nephrops norvegicus studied by relaxation enhancements in spin-label ESR. Biochemistry. 1999;38:14311–14319. doi: 10.1021/bi991459c. [DOI] [PubMed] [Google Scholar]
- Páli T, Kleinschmidt JH, Powell GL, Marsh D. Nonlinear electron paramagnetic resonance studies of the interaction of cytochrome c oxidase with spin-labeled lipids in gel-phase membranes. Biochemistry. 2000;39:2355–2361. doi: 10.1021/bi992478p. [DOI] [PubMed] [Google Scholar]
- Páli T, Bashtovyy D, Marsh D. Stoichiometry of lipid interaction with transmembrane proteins, deduced from the 3-D structures. Prot Sci. 2006;15:1153–1161. doi: 10.1110/ps.052021406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pates RD, Marsh D. Lipid mobility and order in bovine rod outer segment disk membranes. A spin-label study of lipid–protein interactions. Biochemistry. 1987;26:29–39. doi: 10.1021/bi00375a005. [DOI] [PubMed] [Google Scholar]
- Pates RD, Watts A, Uhl R, Marsh D. Lipid–protein interactions in frog rod outer segment disc membranes. Characterization by spin labels. Biochim Biophys Acta. 1985;814:389–397. doi: 10.1016/0005-2736(85)90460-2. [DOI] [PubMed] [Google Scholar]
- Peelen SJCJ, Sanders JC, Hemminga MA, Marsh D. Stoichiometry, selectivity, and exchange dynamics of lipid–protein interaction with bacteriophage M13 coat protein studied by spin label electron spin resonance. Effects of protein secondary structure. Biochemistry. 1992;31:2670–2677. doi: 10.1021/bi00125a006. [DOI] [PubMed] [Google Scholar]
- Powell GL, Knowles PF, Marsh D. Association of spin-labelled cardiolipin with dimyristoylphosphatidylcholine-substituted bovine heart cytochrome c oxidase. A generalized specificity increase rather than highly specific binding sites. Biochim Biophys Acta. 1985;816:191–194. doi: 10.1016/0005-2736(85)90409-2. [DOI] [PubMed] [Google Scholar]
- Powell GL, Knowles PF, Marsh D. Spin label studies on the specificity of interaction of cardiolipin with beef heart cytochrome oxidase. Biochemistry. 1987;26:8138–8145. doi: 10.1021/bi00399a018. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan M, Anbazhagan V, Pratap TV, Marsh D, Swamy MJ. Membrane insertion and lipid–protein interactions of bovine seminal plasma protein PDC-109 investigated by spin-label electron spin resonance spectroscopy. Biophys J. 2001;81:2215–2225. doi: 10.1016/S0006-3495(01)75869-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan M, Pocanschi CL, Kleinschmidt JH, Marsh D. Association of spin-labelled lipids with β-barrel proteins from the outer membrane of Escherichia coli. Biochemistry. 2004;43:11630–11636. doi: 10.1021/bi048858e. [DOI] [PubMed] [Google Scholar]
- Rietveld A, Ponjee GAE, Schiffers P, Jordi W, Van de Coolwijk PJFM, Demel RA, Marsh D, De Kruijff B. Investigations on the insertion of the mitochondrial precursor protein apocytochrome c into model membranes. Biochim Biophys Acta. 1985;818:398–409. doi: 10.1016/0005-2736(85)90015-x. [DOI] [PubMed] [Google Scholar]
- Rietveld A, Berkhout TA, Roenhorst A, Marsh D, De Kruijff B. Preferential association of apocytochrome c with negatively charged phospholipids in mixed model membranes. Biochim Biophys Acta. 1986;858:38–46. doi: 10.1016/0005-2736(86)90289-0. [DOI] [PubMed] [Google Scholar]
- Ryba NJP, Marsh D. Protein rotational diffusion and lipid/protein interactions in recombinants of bovine rhodopsin with saturated diacylphosphatidylcholines of different chain lengths studied by conventional and saturation transfer electron spin resonance. Biochemistry. 1992;31:7511–7518. doi: 10.1021/bi00148a011. [DOI] [PubMed] [Google Scholar]
- Ryba NJP, Horváth LI, Watts A, Marsh D. Molecular exchange at the lipid–rhodopsin interface: spin-label electron spin resonance studies of rhodopsin-dimyristoyl phosphatidylcholine recombinants. Biochemistry. 1987;26:3234–3240. doi: 10.1021/bi00385a045. [DOI] [PubMed] [Google Scholar]
- Ryba NJP, Hoon MA, Findlay JBC, Saibil HR, Wilkinson JR, Heimburg T, Marsh D. Rhodopsin mobility, structure, and lipid–protein interaction in squid photoreceptor membranes. Biochemistry. 1993;32:3298–3305. doi: 10.1021/bi00064a012. [DOI] [PubMed] [Google Scholar]
- Sachse J-H, King MD, Marsh D. ESR determination of lipid diffusion coefficients at low spin-label concentrations in biological membranes, using exchange broadening, exchange narrowing, and dipole–dipole interactions. J Magn Reson. 1987;71:385–404. [Google Scholar]
- Sankaram MB, Brophy PJ, Marsh D. Interaction of two complementary fragments of the bovine spinal cord myelin basic protein with phospholipid bilayers. An ESR spin label study. Biochemistry. 1989;28:9692–9698. doi: 10.1021/bi00451a023. [DOI] [PubMed] [Google Scholar]
- Sankaram MB, Brophy PJ, Marsh D. Selectivity of interaction of phospholipids with bovine spinal cord myelin basic protein studied by spin-label electron spin resonance. Biochemistry. 1989;28:9699–9707. doi: 10.1021/bi00451a024. [DOI] [PubMed] [Google Scholar]
- Sankaram MB, Brophy PJ, Marsh D. Spin label ESR studies on the interaction of bovine spinal cord myelin basic protein with dimyristoylphosphatidylglycerol dispersions. Biochemistry. 1989;28:9685–9691. doi: 10.1021/bi00451a022. [DOI] [PubMed] [Google Scholar]
- Sankaram MB, Brophy PJ, Marsh D. Lipid–protein and protein–protein interactions in double recombinants of myelin proteolipid apoprotein and myelin basic protein with dimyristoylphosphatidylglycerol. Biochemistry. 1991;30:5866–5873. doi: 10.1021/bi00238a009. [DOI] [PubMed] [Google Scholar]
- Schorn K, Marsh D. Lipid chain dynamics and molecular location of diacylglycerol in hydrated binary mixtures with phosphatidylcholine: spin label ESR studies. Biochemistry. 1996;35:3831–3836. doi: 10.1021/bi952688b. [DOI] [PubMed] [Google Scholar]
- Schorn K, Marsh D. Lipid chain dynamics in diacylglycerol–phosphatidylcholine mixtures studied by slow-motional simulations of spin label ESR spectra. Chem Phys Lipids. 1996;82:7–14. [Google Scholar]
- Schwarzmann G, Hoffmann-Bleihauer P, Schubert J, Sandhoff K, Marsh D. Incorporation of ganglioside analogues into fibroblast cell membranes. A spin-label study. Biochemistry. 1983;22:5041–5048. doi: 10.1021/bi00290a025. [DOI] [PubMed] [Google Scholar]
- Silvius JR, McMillen DA, Saley ND, Jost PC, Griffith OH. Competition between cholesterol and phosphatidylcholine for the hydrophobic surface of sarcoplasmic reticulum Ca2+-ATPase. Biochemistry. 1984;23:538–547. doi: 10.1021/bi00298a022. [DOI] [PubMed] [Google Scholar]
- Slichter CP. Principles of magnetic resonance. Berlin: Springer; 1978. [Google Scholar]
- Snel MME, Marsh D. Membrane location of apocytochrome c and cytochrome c determined from lipid–protein spin exchange interactions by continuous wave saturation electron spin resonance. Biophys J. 1994;67:737–745. doi: 10.1016/S0006-3495(94)80534-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snel MME, De Kruijff B, Marsh D. Interaction of spin-labeled apocytochrome c and spin-labeled cytochrome c with negatively charged lipids studied by electron spin resonance. Biochemistry. 1994;33:7146–7156. doi: 10.1021/bi00189a018. [DOI] [PubMed] [Google Scholar]
- Snel MME, de Kroon AIPM, Marsh D. Mitochondrial presequence inserts differently into membranes containing cardiolipin and phosphatidylglycerol. Biochemistry. 1995;34:3605–3613. doi: 10.1021/bi00011a015. [DOI] [PubMed] [Google Scholar]
- Swamy MJ, Marsh D, Anbazhagan V, Ramakrishnan M. Effect of cholesterol on the interaction of seminal plasma protein, PDC-109 with phosphatidylcholine membranes. FEBS Lett. 2002;528:230–234. doi: 10.1016/s0014-5793(02)03316-1. [DOI] [PubMed] [Google Scholar]
- Thomas DD, Bigelow DJ, Squier TJ, Hidalgo C. Rotational dynamics of protein and boundary lipid in sarcoplasmic reticulum membrane. Biophys J. 1982;37:217–225. doi: 10.1016/S0006-3495(82)84671-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts A, Marsh D, Knowles PF. Lipid-substituted cytochrome oxidase: no absolute requirement of cardiolipin for activity. Biochem Biophys Res Commun. 1978;81:397–402. doi: 10.1016/0006-291x(78)91547-4. [DOI] [PubMed] [Google Scholar]
- Watts A, Volotovski ID, Marsh D. Rhodopsin–lipid associations in bovine rod outer segment membranes. Identification of immobilized lipid by spin labels. Biochemistry. 1979;18:5006–5013. doi: 10.1021/bi00589a031. [DOI] [PubMed] [Google Scholar]
- Watts A, Davoust J, Marsh D, Devaux PF. Distinct states of lipid mobility in bovine rod outer segment membranes. Resolution of spin label results. Biochim Biophys Acta. 1981;643:673–676. doi: 10.1016/0005-2736(81)90365-5. [DOI] [PubMed] [Google Scholar]
- Wolfs CJAM, Horváth LI, Marsh D, Watts A, Hemminga MA. Spin-label ESR of bacteriophage M13 coat protein in mixed lipid bilayers. Characterization of molecular selectivity of charged phospholipids for the bacteriophage coat protein in lipid bilayers. Biochemistry. 1989;28:9995–10001. doi: 10.1021/bi00452a018. [DOI] [PubMed] [Google Scholar]


