Abstract
Transient structures in unfolded proteins are important in elucidating the molecular details of initiation of protein folding. Recently, native and non-native secondary structure have been discovered in unfolded A. vinelandii flavodoxin. These structured elements transiently interact and subsequently form the ordered core of an off-pathway folding intermediate, which is extensively formed during folding of this α–β parallel protein. Here, site-directed spin-labelling and paramagnetic relaxation enhancement are used to investigate long-range interactions in unfolded apoflavodoxin. For this purpose, glutamine-48, which resides in a non-native α-helix of unfolded apoflavodoxin, is replaced by cysteine. This replacement enables covalent attachment of nitroxide spin-labels MTSL and CMTSL. Substitution of Gln-48 by Cys-48 destabilises native apoflavodoxin and reduces flexibility of the ordered regions in unfolded apoflavodoxin in 3.4 M GuHCl, because of increased hydrophobic interactions in the unfolded protein. Here, we report that in the study of the conformational and dynamic properties of unfolded proteins interpretation of spin-label data can be complicated. The covalently attached spin-label to Cys-48 (or Cys-69 of wild-type apoflavodoxin) perturbs the unfolded protein, because hydrophobic interactions occur between the label and hydrophobic patches of unfolded apoflavodoxin. Concomitant hydrophobic free energy changes of the unfolded protein (and possibly of the off-pathway intermediate) reduce the stability of native spin-labelled protein against unfolding. In addition, attachment of MTSL or CMTSL to Cys-48 induces the presence of distinct states in unfolded apoflavodoxin. Despite these difficulties, the spin-label data obtained here show that non-native contacts exist between transiently ordered structured elements in unfolded apoflavodoxin.
Keywords: Unfolded protein, Flavodoxin, α–β Parallel protein, Paramagnetic relaxation enhancement, MTSL, CMTSL
Introduction
Understanding the molecular mechanisms of protein folding is one of the fundamental challenges of structural biology. Proteins initially fold from disordered unfolded states. Transient structures in unfolded proteins are important in elucidation of the molecular details of initiation of protein folding. Early observations of residual structure in unfolded proteins prompted the suggestion that native-like structure is present in the denatured state (Gillespie and Shortle 1997; Yi et al. 2000; Lietzow et al. 2002) and that this residual structure biases the subsequent conformational search toward the native conformation (Yi et al. 2000; Daggett and Fersht 2003). However, evidence of non-native structure has been found in the unfolded states of few proteins (Kristjansdottir et al. 2005; Platt et al. 2005; Reed et al. 2006; Marsh et al. 2007).
Recently, non-native secondary structure and non-native hydrophobic interactions have been observed in the unfolded state of a 179-residue flavodoxin from Azotobacter vinelandii (Nabuurs et al. 2008), which is the protein of interest in the study presented here. Remarkably, structure formation in unfolded flavodoxin does not direct folding to the native state, but instead causes formation of a misfolded off-pathway intermediate (Nabuurs et al. 2009a).
Flavodoxins are monomeric proteins involved in electron transport and contain a non-covalently bound flavin mononucleotide (FMN) cofactor. The proteins consist of a single structural domain and adopt the flavodoxin-like or α–β parallel topology (Fig. 1), which is widely prevalent in nature.
Fig. 1.
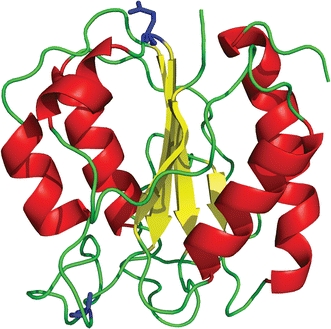
Schematic diagram of native C69A flavodoxin from A. vinelandii (pdb ID 1YOB (Alagaratnam et al. 2005)). The protein contains a parallel β-sheet surrounded by α-helices at either side of the sheet. Residues Ala-69 and Gln-48 are shown in ball-and-stick representation (coloured blue). The FMN cofactor is not shown
Both denaturant-induced equilibrium and kinetic (un)folding of flavodoxin and apoflavodoxin (i.e., flavodoxin without FMN) have been characterized using guanidine hydrochloride (GuHCl) as denaturant (van Mierlo et al. 1998; van Mierlo and Steensma 2000; Bollen et al. 2004, 2005, 2006; Bollen and van Mierlo 2005). The folding data show that apoflavodoxin autonomously folds to its native state, which is structurally identical with that of flavodoxin except that residues in the flavin-binding region of the apo protein have considerable dynamics (Steensma et al. 1998; Steensma and van Mierlo 1998). Binding of FMN to native apoflavodoxin is the last step in flavodoxin folding.
Apoflavodoxin kinetic folding involves an energy landscape with two folding intermediates and is described by: Ioff ↔ unfolded apoflavodoxin ↔ Ion ↔ native apoflavodoxin (Bollen et al. 2004). Intermediate Ion lies on the productive route from unfolded to native protein, is highly unstable and is, therefore, not observed during denaturant-induced equilibrium unfolding of apoflavodoxin. Consequently, GuHCl-induced equilibrium unfolding of apoflavodoxin is described by: Ioff ↔ unfolded apoflavodoxin ↔ native apoflavodoxin (Bollen et al. 2004). Approximately 90% of folding molecules fold via molten globule-like off-pathway intermediate Ioff, which is a relatively stable species that needs to unfold to produce native protein and thus acts as a trap (Bollen et al. 2004). Elevated protein concentrations (van Mierlo et al. 2000) and molecular crowding (Engel et al. 2008) cause severe aggregation of this species. The formation of an off-pathway species is typical for proteins with a flavodoxin-like topology (Bollen and van Mierlo 2005). An off-pathway intermediate is experimentally observed for all other α–β parallel proteins of which the kinetic folding has been investigated, i.e., apoflavodoxin from Anabaena (Fernandez-Recio et al. 2001), CheY (Kathuria et al. 2008), cutinase (Otzen et al. 2007), and UMP/CMP kinase (Lorenz and Reinstein 2008).
To better understand why the off-pathway intermediate is formed during flavodoxin folding, GuHCl-unfolded apoflavodoxin has been characterized at the residue-level using heteronuclear NMR spectroscopy (Nabuurs et al. 2008, 2009a). Secondary shifts analysis and investigation of 1H–15N relaxation rates reveal four structured elements that transiently exist in unfolded apoflavodoxin. These transiently ordered regions have restricted flexibility on the (sub)nanosecond timescale and comprise residues Ala-41–Gly-53, Glu-72–Gly-83, Gln-99–Ala-122, and Thr-160–Gly-176 (Nabuurs et al. 2008). These regions match with regions of large average area buried upon folding (AABUF), which correlates with hydrophobicity (Rose and Roy 1980) and corresponds to sequence-dependent dynamic variations due to hydrophobic interactions in unfolded proteins (Schwarzinger et al. 2002; Le Duff et al. 2006).
Restricted flexibility in unfolded apoflavodoxin is due to transient helix formation and local and non-local hydrophobic interactions (Nabuurs et al. 2008, 2009a). On reducing the denaturant concentration, the four structured elements in unfolded apoflavodoxin transiently interact and subsequently form the ordered core of the molten globule (Nabuurs et al. 2008, 2009a). As a consequence, the molten globule has a totally different topology compared with native apoflavodoxin: it is helical and contains no β-sheet (Nabuurs et al. 2009b). Structure formation within virtually all parts of unfolded apoflavodoxin precedes folding to the molten globule state. This folding transition is non-cooperative and involves a series of distinct transitions (Nabuurs et al. 2009a). Part of Ioff remains random coil down to a GuHCl concentration of 1.58 M (i.e., residues Lys-13 to Val-36).
To probe long-range interactions in unfolded apoflavodoxin and to consolidate the findings of non-native interactions that lead to formation of the off-pathway intermediate, in the study presented here use is made of site-directed spin-labelling. Paramagnetic relaxation agents are often used to investigate long-distance interactions in unfolded proteins (Dyson and Wright 2005). Site-specific labelling with thiol-specific nitroxide electron spin-labels, for example MTSL ((1-oxy-,2,2,5,5-tetramethyl-D-pyrroline-3-methyl)-methanethiosulfonate) (Berliner et al. 1982) or CMTSL ((1-oxy-,2,2,5,5-tetramethylpyrroline-3-yl)-carbamidoethyl methanethiosulfonate) (Card et al. 2005) (Fig. 2), enables determination of long-range contacts in unfolded proteins (Mittag and Forman-Kay 2007). MTSL (or CMTSL) can exist in two electronic states, an oxidized paramagnetic state and a reduced diamagnetic state. In the oxidized state an unpaired electron in MTSL (or CMTSL) reduces the intensity of NMR signals of atoms that are within approximately 20 to 25 Å of the spin-label, making it a workable probe of distances in the range from 10 to 20 Å in folded proteins (Gillespie and Shortle 1997). This phenomenon of signal intensity reduction is called paramagnetic relaxation enhancement (PRE). Regions of a polypeptide chain that form tertiary interactions with the spin-labelled region will exhibit strong PRE effects. Conversely, regions that remain distant at all times from the spin-labelled region should exhibit weak PRE effects.
Fig. 2.
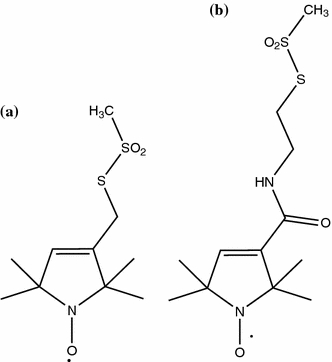
Schematic representations of the chemical structures of the hydrophobic compounds MTSL (a) and CMTSL (b). Because of the presence of an amide group in the linker between protein and the proxyl ring in CMTSL this spin-label is more polar than MTSL
Wild-type flavodoxin contains a single cysteine at position 69. This cysteine is poorly accessible to solvent in the holo form of the protein, because of the presence of FMN. However, in apoflavodoxin it is solvent accessible and can be used to attach a spin-label to the protein. Another variant of apoflavodoxin with a single cysteine was made for the purpose of the study presented here. In the latter variant, Gln-48, which resides in the non-native α-helix that is formed in unfolded apoflavodoxin (Nabuurs et al. 2008), is replaced by cysteine. In native apoflavodoxin Gln-48 and Cys-69 are positioned at opposite sites of the protein (Fig. 1).
Here, we show that attachment of a hydrophobic spin-label leads to hydrophobic interactions between the spin-label and various residues of an unfolded protein. Such perturbation of an unfolded protein has not been reported previously, despite the common use of spin-labels to characterise unfolded proteins. Still, even though this perturbation exists, valid information about residual structure in unfolded apoflavodoxin is obtained.
Materials and methods
Sample preparation
Site-directed mutagenesis was used to replace the single cysteine in wild-type A. vinelandii (strain ATCC 478) flavodoxin II (i.e., WT flavodoxin) by alanine (i.e., C69A flavodoxin, which is largely similar to WT flavodoxin (Steensma et al. 1996; van Mierlo et al. 1998)). In addition, glutamine at position 48 was replaced by cysteine, resulting in the double mutant C69A-Q48C flavodoxin, hereafter named Q48C flavodoxin.
Uniformly 15N-labelled Q48C and C69A flavodoxins were obtained from transformed E. coli cells grown on 15N-labelled algae medium (Silantes, Germany). Uniformly 15N-labelled WT flavodoxin was obtained from transformed E. coli cells grown on 15N-labelled minimal medium. All protein variants were purified as described elsewhere (van Mierlo et al. 1998).
Unfolded apoflavodoxin was obtained by denaturing flavodoxin in 6 M GuHCl. Subsequently, FMN was removed via gel filtration in 5 M GuHCl. WT and Q48C protein variants were labelled with MTSL or CMTSL (Toronto Research Chemicals, Toronto, Canada) by adding a 3:1 molar ratio of spin-label to unfolded protein at room temperature. After 2 h, free spin-label was separated from labelled protein by gel filtration. Spin-labelled WT and Q48C apoflavodoxin are referred to as WT(C)MTSL and Q48C(C)MTSL apoflavodoxin, respectively.
All NMR samples contained about 0.3–0.5 mM apoflavodoxin, 10% D2O, and 2,2-dimethyl-2-silapentane-5-sulfonic acid (DSS) as internal chemical shift reference. To avoid covalent dimerisation of protein molecules that are not spin-labelled, a sufficient amount of DTT was present in samples of WT and Q48C apoflavodoxin.
The buffer used in all experiments was 100 mM potassium pyrophosphate (KPPi), pH 6.0, and refractometry was used to verify GuHCl concentration (Nozaki 1972).
NMR experiments
Gradient enhanced 1H–15N HSQC spectra were recorded on a Bruker Avance 700 MHz machine. Sample temperature was kept at 25°C. In the 1H dimension of the 1H–15N HSQC experiments, 2048 complex data points were acquired, whereas in the indirect 15N dimension 360 complex data points were collected. Spectral widths were 6010 and 1750 Hz in t 2 and t 1, respectively, and the number of scans was 16. All NMR experiments performed with spin-labelled protein in the paramagnetic state were repeated using spin-labelled protein in the diamagnetic state. Diamagnetic protein was obtained by adding 5 μl concentrated stock solution of ascorbic acid to the NMR samples containing paramagnetic protein (the resulting dilution of the NMR sample was less than 1%). This addition resulted in a threefold molar excess of ascorbic acid compared with protein.
Fluorescence spectroscopy
Thermal unfolding of C69A apoflavodoxin, Q48C apoflavodoxin, and Q48CMTSL apoflavodoxin was followed by fluorescence emission. Protein unfolding was achieved by increasing the temperature in a 1.5-ml stirred quartz cuvette (path-length 0.4 cm) from 20 to 70°C at a rate of 0.5°/min. Temperature was measured in the cuvette by using an internal probe. The excitation wavelength used was 280 nm, and fluorescence emission was recorded at 340 nm. Excitation and emission slits were set at 5 nm. In thermal unfolding experiments, protein was in 100 mM KPPi, pH 6.0, and protein concentration ranged between 4 and 6 μM.
Data analysis
NMR data
All NMR data were processed using NMRPipe (Delaglio et al. 1995) and analysed using NMRViewJ (Johnson and Blevins 1994). Paramagnetic relaxation enhancements by nitroxide spin-labels in unfolded apoflavodoxin variants were measured as ratios of maximal cross peak intensities, I, between 1H–15N HSQC spectra of unfolded WT(C)MTSL or Q48C(C)MTSL apoflavodoxin in the paramagnetic and diamagnetic state, respectively:
 |
1 |
Thermally induced equilibrium unfolding data
The change in free energy for thermally induced protein unfolding, ΔG(T), is described by the modified Gibbs–Helmholtz equation:
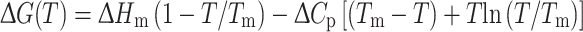 |
2 |
where ΔH m is the enthalpy change for unfolding measured at T m, T is the absolute temperature, T m, is the temperature at the midpoint of the (un)folding transition, and ΔC p is the difference in heat capacity between the unfolded and folded states (Pace and Laurents 1989). Under the assumption that ΔC p is temperature-independent (Privalov and Khechinashvili 1974; Becktel and Schellman 1987), a two-state mechanism of unfolding can be fitted to individual thermal unfolding curves:
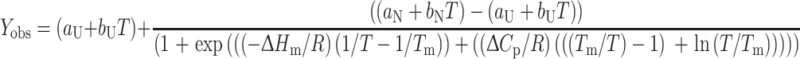 |
3 |
where Y obs is the measured fluorescence signal, R is the gas constant, and a and b are the intercepts and slopes, respectively, of the pre-unfolding and post-unfolding baselines.
Results and discussion
Introducing a cysteine at position 48 and subsequent labelling with MTSL both reduce the thermal midpoint of unfolding of apoflavodoxin
Thermal-induced unfolding experiments show that replacement of Gln-48 by Cys-48 destabilizes native apoflavodoxin in 100 mM KPPi at pH 6.0. Fluorescence emission enables monitoring of unfolding of native protein molecules, because both folding intermediate and unfolded apoflavodoxin have comparable fluorescence signals. Using fluorescence emission, the thermal midpoint of unfolding (T m) of Q48C apoflavodoxin is determined to be 40.4 ± 0.2°C, whereas C69A apoflavodoxin has a T m of 48.2 ± 0.1°C (Fig. 3). Attachment of the MTSL spin-label to Cys-48 further reduces T m to 33.0 ± 1.6°C.
Fig. 3.
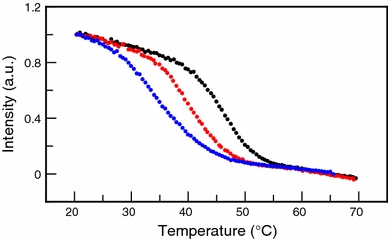
Thermal unfolding of apoflavodoxin shows that replacement of Gln-48 by Cys-48 and subsequent attachment of an MTSL spin-label to Cys-48 both destabilize the protein against thermal unfolding. Through detection of changes in fluorescence emission at 340 nm thermal unfolding is measured for C69A apoflavodoxin (black dots), Q48C apoflavodoxin (red dots), and Q48CMTSL apoflavodoxin (blue dots). Midpoints of unfolding are 48.2 ± 0.1°C for C69A apoflavodoxin, 40.4 ± 0.2°C for Q48C apoflavodoxin, and 33.0 ± 1.6°C for Q48CMTSL apoflavodoxin
Introducing a cysteine at position 48 changes the dynamic features of unfolded apoflavodoxin
Comparison of chemical shifts of cross peaks in 1H–15N HSQC spectra of Q48C and C69A apoflavodoxin in 3.4 M GuHCl previously showed that replacement of a glutamine by a cysteine at position 48 leads to chemical shift changes in unfolded apoflavodoxin. These chemical shift changes indicate long-range non-native interactions between transiently formed helices in unfolded apoflavodoxin (Nabuurs et al. 2008).
In 3.4 M GuHCl, several cross peak intensities of unfolded Q48C apoflavodoxin are reduced compared with the cross peak intensities of the backbone amides of unfolded C69A apoflavodoxin, because of broadening of cross peaks. This broadening is caused by reduced flexibility of backbone amides. Fig. 4 shows that four distinct regions in unfolded Q48C apoflavodoxin have reduced flexibility of their backbone amides compared with the corresponding amides of unfolded C69A apoflavodoxin. These distinct regions coincide with the four regions with restricted flexibility detected in unfolded C69A apoflavodoxin (Nabuurs et al. 2008), which are highlighted by grey bars in Fig. 4. Consequently, upon replacing Gln-48 by Cys-48 the four regions with restricted flexibility in unfolded apoflavodoxin become even less flexible, probably because of non-native hydrophobic interactions between Cys-48 and the four ordered regions identified.
Fig. 4.
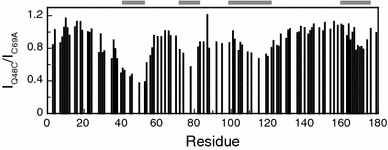
Upon replacing Gln-48 by Cys-48 the four regions with restricted flexibility in unfolded apoflavodoxin in 3.4 M GuHCl become less flexible. Shown are the ratios between cross peak intensities of backbone amides of unfolded Q48C apoflavodoxin (I Q48C) and unfolded C69A apoflavodoxin (I C69A). The ratio I Q48C/I C69A of residue 179 is set to 1. Horizontal grey bars highlight the four regions of unfolded C69A apoflavodoxin with restricted flexibility on the (sub)nanosecond timescale (Nabuurs et al. 2008)
Unfolded Q48C apoflavodoxin in 6.0 M GuHCl behaves as a random coil; subsequent attachment of MTSL causes the protein to become more ordered
Far-UV CD data and transverse relaxation rates (Nabuurs et al. 2008) show that C69A apoflavodoxin in 6.0 M GuHCl behaves as a random coil. The Q48C variant of apoflavodoxin is also a random coil in 6.0 M GuHCl. This random coil behaviour is concluded because the 1H–15N HSQC spectra of both Q48C and C69A apoflavodoxin at this concentration denaturant are very similar regarding both cross peak positions and cross peak intensities (data not shown).
The ratios between cross peak intensities of reduced Q48CMTSL and Q48C apoflavodoxin, both unfolded in 6.0 M GuHCl, are shown in Fig. 5. The direct sequential neighbours of Cys-48 are either not visible or could not be assigned in the 1H–15N HSQC spectrum of Q48CMTSL apoflavodoxin. In 6.0 M GuHCl, the backbone amides of Ser-40, Ala-41, Glu-42, Ile-51, Leu-52, and Gly-53 of reduced Q48CMTSL apoflavodoxin have reduced intensities compared with the corresponding intensities in the HSQC spectrum of Q48C apoflavodoxin (Fig. 5). Restricted motions of these residues because of interactions with MTSL most likely cause this decrease.
Fig. 5.
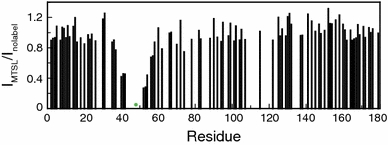
Attachment of MTSL to unfolded Q48C apoflavodoxin in 6.0 M GuHCl causes the protein to behave in a more ordered fashion than random coil apoflavodoxin. Shown are the ratios between cross peak intensities of backbone amides of unfolded Q48CMTSL apoflavodoxin in the diamagnetic state (I MTSL) and unfolded Q48C apoflavodoxin (I nolabel), which are both in 6.0 M GuHCl. The ratio I MTSL/I nolabel of residue 179 is set to 1. The green dot indicates the position of the spin-label
In conclusion, attachment of MTSL to unfolded Q48C apoflavodoxin in 6.0 M GuHCl causes the protein to behave in a more ordered fashion than random coil apoflavodoxin.
Use of PRE experiments
Magnetic interaction between an unpaired electron in a paramagnetic nitroxide spin-label and a nearby proton causes broadening of the corresponding 1H NMR signal because of the increased transverse relaxation rate of the proton involved (Gillespie and Shortle 1997). This relaxation rate has an r −6-dependence on electron–proton distance and thus enables detection of long-range interactions in proteins. Consequently, ratios of cross peak intensities (I para/I dia) extracted from two redox state-dependent 1H–15N HSQC spectra of the spin-labelled protein enable estimation of distances between the spin-label and affected protons in the protein (Teilum et al. 2002). Because of the ubiquitous backbone fluctuations in unfolded apoflavodoxin (Nabuurs et al. 2008), no attempt is made to extract quantitative distances from the data. Rather, a trend in PRE as a function of primary structure can be observed, as shown in the following discussion.
PRE shows that WTMTSL apoflavodoxin in 6.0 M GuHCl does not behave as a random coil, because of hydrophobic interactions between MTSL and unfolded protein
1H–15N HSQC spectra were acquired of MTSL-labelled WT apoflavodoxin and Q48C apoflavodoxin both unfolded in 6.0 M GuHCl, with the spin-label either in the paramagnetic or the diamagnetic state. Subsequently, the ratios of the intensities of cross peak maxima (I para/I dia) of backbone amides were determined. Fig. 6 shows these ratios and demonstrates that for both protein variants resonances of residues that are sequential neighbours of the cysteine to which a nitroxide radical is attached are broadened beyond detection. In addition, for WTMTSL apoflavodoxin the nitroxide radical also reduces the backbone amide cross peak intensities of residues Ser-40–Leu-62. PRE data of WTMTSL apoflavodoxin thus indicate that Gln-48 is in the vicinity of the spin-label that is attached to Cys-69. Our observations show that in 6.0 M GuHCl WTMTSL apoflavodoxin does not behave as a random coil.
Fig. 6.
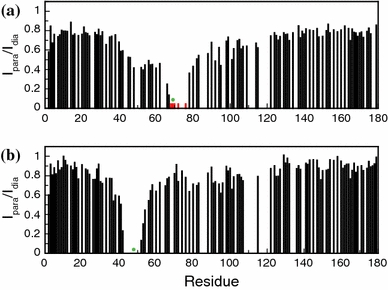
PRE of amide protons of WTMTSL apoflavodoxin and Q48CMTSL apoflavodoxin, which are both unfolded in 6.0 M GuHCl. 1H–15N HSQC spectra of both protein variants with the spin-label either in the paramagnetic or diamagnetic state were recorded at 25°C. Subsequently, the ratio of the intensities of cross peak maxima (I para/I dia) of backbone amides was determined. Shown are I para/I dia of a WTMTSL apoflavodoxin and b Q48CMTSL apoflavodoxin. Green dots highlight the positions of the spin-label. Red bars in (a) highlight residues for which no cross peaks are visible in the HSQC spectrum of the protein with paramagnetic spin-label, but for which cross peaks are observed when the spin-label is diamagnetic
In contrast with the above observation, PRE data of Q48CMTSL apoflavodoxin (Fig. 6b) show no evidence of an interaction between Cys-48 and Ala-69. Note however, as discussed in a previous section, that attachment of MTSL to unfolded Q48C apoflavodoxin in 6.0 M GuHCl causes the protein to behave more ordered than random coil apoflavodoxin. The nitroxide radical of MTSL attached to Cys-48 does not broaden resonances of residues in the vicinity of Ala-69 (the cross peak of the backbone amide of Ala-69 suffers from severe overlap and is thus not assigned). The interaction between the MTSL spin-label attached to Cys-69 and region Ser-40–Leu-62 of unfolded apoflavodoxin in 6.0 M GuHCl is thus due to hydrophobic interactions of the label with residues in this region. Similar hydrophobic interactions with MTSL spin-label were also observed in dimerization studies of ARNT PAS-B (Card et al. 2005).
In both WTCMTSL and Q48CCMTSL apoflavodoxin unfolded in 3.4 M GuHCl the spin-label interacts with four transiently structured regions
To reduce hydrophobic interactions in unfolded apoflavodoxin due to MTSL, another spin-label, i.e., CMTSL (Card et al. 2005), which is more hydrophilic than MTSL, was chosen as paramagnetic relaxation agent. Compared with MTSL, CMTSL contains an additional amide group in the linker between protein and the proxyl ring (Fig. 2), thereby introducing a degree of polarity into an otherwise hydrophobic compound.
Figure 7a and b show the ratios of the intensities of HSQC cross peak maxima (I para/I dia) of backbone amides of the paramagnetic and diamagnetic states of WTCMTSL apoflavodoxin and Q48CCMTSL apoflavodoxin, respectively, which are both unfolded in 6.0 M GuHCl. The pattern of interactions of the CMTSL spin-label in both unfolded proteins is similar to the pattern of interactions observed using MTSL as spin-label. Consequently, also with CMTSL attached to unfolded apoflavodoxin in 6.0 M GuHCl, hydrophobic interactions exist between the spin-label at position 69 and the hydrophobic region comprising residues 40–60 of unfolded apoflavodoxin.
Fig. 7.

PRE of amide protons of WTCMTSL and Q48CCMTSL apoflavodoxin unfolded in GuHCl at 25°C. The ratio of the intensities of HSQC cross peak maxima (I para/I dia) of backbone amides was determined as described in the legend of Fig. 6. Shown are I para/I dia of a WTMTSL and b Q48CMTSL apoflavodoxin, both in 6.0 M GuHCl, and I para/I dia of c WTMTSL and d Q48CMTSL, both in 3.4 M GuHCl. Green dots highlight the positions of the CMTSL spin-label. Residues Ser-40, Ala-41, Glu-42, Gln-46, Phe-49, Leu-50, Leu-52, Gly-53, Thr-54, and Leu-78 of Q48CCMTSL apoflavodoxin in 3.4 M GuHCl each give rise to two backbone amide cross peaks (a few examples are shown in Fig. 8) and are indicated with red bars in (d)
In Fig. 7c and d PRE data of WTCMTSL and Q48CCMTSL apoflavodoxin in 3.4 M GuHCl are shown. In both WTCMTSL and Q48CCMTSL apoflavodoxin the CMTSL spin-label interacts with various regions of the unfolded protein. In particular, interactions are observed between the spin-label and the four transiently structured regions in unfolded apoflavodoxin in 3.4 M GuHCl, strengthening our previous findings (Nabuurs et al. 2008) that non-native contacts do, indeed, exist between transiently ordered structured elements in unfolded apoflavodoxin.
In Q48CCMTSL apoflavodoxin unfolded in 3.4 M GuHCl, the spin-label interacts with residues spread over the entire sequence of the protein. Residues 40 to 54 and Leu-78 stand out, as the corresponding backbone amide resonances are broadened beyond detection in the paramagnetic state of the unfolded protein. These residues will be discussed in the next section. In addition, resonances of residues 82 to 129 are more severely broadened than the resonances of most of the residues of unfolded Q48CCMTSL apoflavodoxin.
The attached spin-label induces the presence of two distinct states in unfolded apoflavodoxin
We notice that in the HSQC spectrum of reduced Q48CMTSL and Q48CCMTSL apoflavodoxin in 3.4 M GuHCl multiple cross peaks are observed for several backbone amides. We focus here on Q48CCMTSL apoflavodoxin for which two cross peaks are observed for each individual backbone amide of residues Ser-40, Ala-41, Glu-42, Gln-46, Phe-49, Leu-50, Leu-52, Gly-53, Thr-54, and Leu-78. In Fig. 8 examples of this cross peak doubling are shown. For each doubled cross peak it is observed that upon bringing the spin-label into the paramagnetic state one of these two cross peaks broadens beyond detection (this disappearing cross peak is used to calculate I para/I dia of the corresponding residues in Fig. 7c,d). Doubling of the cross peaks of the residues mentioned is not because of the presence of non-labelled protein molecules. In the latter case, additional cross peaks would appear at different positions in the HSQC spectrum, as the HSQC spectrum of Q48C apoflavodoxin in 3.4 M GuHCl shows (Fig. 8).
Fig. 8.
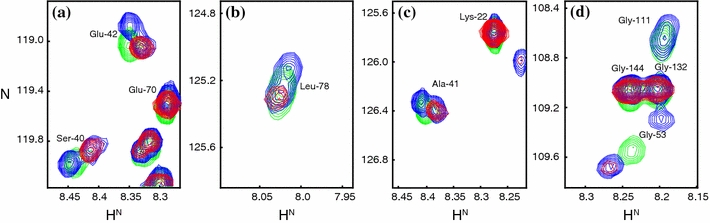
A backbone amide that is in close vicinity to the CMTSL spin-label of Q48CCMTSL apoflavodoxin in 3.4 M GuHCl can give rise to two cross peaks in the corresponding 1H–15N HSQC spectrum. Blue contour levels highlight cross peaks arising from diamagnetic Q48CCMTSL apoflavodoxin, red contour levels highlight cross peaks arising from paramagnetic Q48CCMTSL apoflavodoxin, and green contour levels highlight cross peaks arising from Q48C apoflavodoxin with no spin-label attached to it. A set of two cross peaks is shown for a Ser-40 and Glu-42, b Leu-78, c Ala-41 and for d Gly-53
A plausible explanation of the observed doubling of cross peaks is that the attached spin-label induces the presence of two distinct states in unfolded apoflavodoxin. In one of these states the spin-label is in the proximity of the above-mentioned residues whereas in the other state it is not. Both folding states are in slow exchange with one another on the NMR chemical shift time scale, because two separate, sharp cross peaks are observed per backbone amide of the residues discussed.
Leu-78 is the only residue that is not sequentially close to the CMTSL-label at position 48, but nevertheless gives rise to two backbone amide cross peaks. Indeed, chemical shift deviations upon replacing residue 48 show that interactions between Cys-48 and Leu-78 must exist in unfolded apoflavodoxin (Nabuurs et al. 2008). Residual structure that is neither an α-helix nor a β-sheet is found in the region Glu-72–Gly-83 of unfolded apoflavodoxin (Nabuurs et al. 2008). These observations are further support for the existence of persistent hydrophobic interactions between Leu-78 and the CMTSL spin-label in one of the two distinct states within unfolded Q48CCMTSL apoflavodoxin. The tertiary interaction between residues 48 and 78 in unfolded apoflavodoxin, as revealed by PRE experiments, must be non-native. The latter is concluded because complete disappearance of the backbone amide cross peak of Leu-78 implies that the distance between residues 48 and 78 must be much shorter than the 17.28 Å distance between the corresponding Cα atoms in native flavodoxin (calculated using the X-ray structure of A. vinelandii flavodoxin (Alagaratnam et al. 2005)).
Introducing cysteine residues in (unfolded) proteins to enable labelling with probes is used frequently (Fanucci and Cafiso 2006; Clore et al. 2007). To detect long-range interactions within (natively) unfolded proteins, for example apomyoglobin (Lietzow et al. 2002), α-synuclein (Bertoncini et al. 2005), ACBP (Teilum et al. 2002), and N-PGK (Cliff et al. 2009), MTSL is widely used as a nitroxide spin-label. However, the results presented here show that in an unfolded protein covalent attachment of a spin-label to cysteine can introduce hydrophobic interactions between the hydrophobic spin-label and various amino acid residues. These interactions alter the hydrophobic free energy (Baldwin 2005) of the unfolded state (and possibly of the off-pathway intermediate) and, as a consequence, the free energy difference between native apoflavodoxin and non-native protein molecules diminishes. Indeed, covalent attachment of a MTSL-label can alter the stability of the protein involved, as is shown here for native apoflavodoxin and as is reported for ACBP variants (Teilum et al. 2002). Remarkably, in contrast with unfolded apoflavodoxin, no hydrophobic interactions, either short-range or long-range, with the MTSL-label have been reported for the unfolded proteins mentioned. Neither has doubling of cross peaks of these spin-labelled unfolded proteins been reported.
Native apoflavodoxin is an α–β parallel protein and contains in its core many hydrophobic residues that are shielded from the solvent. It is possible that in unfolded protein these residues are susceptible to hydrophobic interactions with the hydrophobic nitroxide spin-labels used. These interactions can give rise to the existence of distinct states in the unfolded protein, as this work shows. Despite the difficulties mentioned, the spin-label data obtained here show that non-native contacts exist between transiently ordered structured elements in unfolded apoflavodoxin.
Acknowledgments
The Netherlands Organization for Scientific Research supported this work. NMR spectra were recorded at the Utrecht Facility for High-Resolution NMR, The Netherlands.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
The more you see: spectroscopy in molecular biophysics.
References
- Alagaratnam S, van Pouderoyen G, Pijning T, Dijkstra BW, Cavazzini D, Rossi GL, Van Dongen WM, van Mierlo CP, van Berkel WJ, Canters GW. A crystallographic study of Cys69Ala flavodoxin II from Azotobacter vinelandii: structural determinants of redox potential. Protein Sci. 2005;14:2284–2295. doi: 10.1110/ps.051582605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin RL. Weak interactions in protein folding: hydrophobic free energy, van der Waals interactions, peptide hydrogen bonds, and peptide solvation. In: Buchner J, Kiefhaber T, editors. Protein folding handbook. Washington: Wiley-VCH; 2005. pp. 127–162. [Google Scholar]
- Becktel WJ, Schellman JA. Protein stability curves. Biopolymers. 1987;26:1859–1877. doi: 10.1002/bip.360261104. [DOI] [PubMed] [Google Scholar]
- Berliner LJ, Grunwald J, Hankovszky HO, Hideg K. A novel reversible thiol-specific spin label: papain active site labeling and inhibition. Anal Biochem. 1982;119:450–455. doi: 10.1016/0003-2697(82)90612-1. [DOI] [PubMed] [Google Scholar]
- Bertoncini CW, Jung YS, Fernandez CO, Hoyer W, Griesinger C, Jovin TM, Zweckstetter M. Release of long-range tertiary interactions potentiates aggregation of natively unstructured alpha-synuclein. Proc Natl Acad Sci U S A. 2005;102:1430–1435. doi: 10.1073/pnas.0407146102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollen YJ, van Mierlo CP. Protein topology affects the appearance of intermediates during the folding of proteins with a flavodoxin-like fold. Biophys Chem. 2005;114:181–189. doi: 10.1016/j.bpc.2004.12.005. [DOI] [PubMed] [Google Scholar]
- Bollen YJ, Sanchez IE, van Mierlo CP. Formation of on- and off-pathway intermediates in the folding kinetics of Azotobacter vinelandii apoflavodoxin. Biochemistry. 2004;43:10475–10489. doi: 10.1021/bi049545m. [DOI] [PubMed] [Google Scholar]
- Bollen YJ, Nabuurs SM, van Berkel WJ, van Mierlo CP. Last in, first out: the role of cofactor binding in flavodoxin folding. J Biol Chem. 2005;280:7836–7844. doi: 10.1074/jbc.M412871200. [DOI] [PubMed] [Google Scholar]
- Bollen YJ, Kamphuis MB, van Mierlo CP. The folding energy landscape of apoflavodoxin is rugged: hydrogen exchange reveals nonproductive misfolded intermediates. Proc Natl Acad Sci U S A. 2006;103:4095–4100. doi: 10.1073/pnas.0509133103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Card PB, Erbel PJ, Gardner KH. Structural basis of ARNT PAS-B dimerization: use of a common beta-sheet interface for hetero- and homodimerization. J Mol Biol. 2005;353:664–677. doi: 10.1016/j.jmb.2005.08.043. [DOI] [PubMed] [Google Scholar]
- Cliff MJ, Craven CJ, Marston JP, Hounslow AM, Clarke AR, Waltho JP. The denatured state of N-PGK is compact and predominantly disordered. J Mol Biol. 2009;385:266–277. doi: 10.1016/j.jmb.2008.10.004. [DOI] [PubMed] [Google Scholar]
- Clore GM, Tang C, Iwahara J. Elucidating transient macromolecular interactions using paramagnetic relaxation enhancement. Curr Opin Struct Biol. 2007;17:603–616. doi: 10.1016/j.sbi.2007.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daggett V, Fersht A. The present view of the mechanism of protein folding. Nat Rev Mol Cell Biol. 2003;4:497–502. doi: 10.1038/nrm1126. [DOI] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe–a multidimensional spectral processing system based on unix pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Dyson HJ, Wright PE. Elucidation of the protein folding landscape by NMR. Methods Enzymol. 2005;394:299–321. doi: 10.1016/S0076-6879(05)94011-1. [DOI] [PubMed] [Google Scholar]
- Engel R, Westphal AH, Huberts DH, Nabuurs SM, Lindhoud S, Visser AJ, van Mierlo CP. Macromolecular crowding compacts unfolded apoflavodoxin and causes severe aggregation of the off-pathway intermediate during apoflavodoxin folding. J Biol Chem. 2008;283:27383–27394. doi: 10.1074/jbc.M802393200. [DOI] [PubMed] [Google Scholar]
- Fanucci GE, Cafiso DS. Recent advances and applications of site-directed spin labeling. Curr Opin Struct Biol. 2006;16:644–653. doi: 10.1016/j.sbi.2006.08.008. [DOI] [PubMed] [Google Scholar]
- Fernandez-Recio J, Genzor CG, Sancho J. Apoflavodoxin folding mechanism: an alpha/beta protein with an essentially off-pathway intermediate. Biochemistry. 2001;40:15234–15245. doi: 10.1021/bi010216t. [DOI] [PubMed] [Google Scholar]
- Gillespie JR, Shortle D. Characterization of long-range structure in the denatured state of staphylococcal nuclease.I. Paramagnetic relaxation enhancement by nitroxide spin labels. J Mol Biol. 1997;268:158–169. doi: 10.1006/jmbi.1997.0954. [DOI] [PubMed] [Google Scholar]
- Johnson BA, Blevins RA. NMR view–a computer-program for the visualization and analysis of NMR data. J Biomol NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- Kathuria SV, Day IJ, Wallace LA, Matthews CR. Kinetic traps in the folding of beta alpha-repeat proteins: CheY initially misfolds before accessing the native conformation. J Mol Biol. 2008;382:467–484. doi: 10.1016/j.jmb.2008.06.054. [DOI] [PubMed] [Google Scholar]
- Kristjansdottir S, Lindorff-Larsen K, Fieber W, Dobson CM, Vendruscolo M, Poulsen FM. Formation of native and non-native interactions in ensembles of denatured ACBP molecules from paramagnetic relaxation enhancement studies. J Mol Biol. 2005;347:1053–1062. doi: 10.1016/j.jmb.2005.01.009. [DOI] [PubMed] [Google Scholar]
- Le Duff CS, Whittaker SBM, Radford SE, Moore GR. Characterisation of the conformational properties of urea-unfolded Im7: implications for the early stages of protein folding. J Mol Biol. 2006;364:824–835. doi: 10.1016/j.jmb.2006.09.037. [DOI] [PubMed] [Google Scholar]
- Lietzow MA, Jamin M, Jane Dyson HJ, Wright PE. Mapping long-range contacts in a highly unfolded protein. J Mol Biol. 2002;322:655–662. doi: 10.1016/S0022-2836(02)00847-1. [DOI] [PubMed] [Google Scholar]
- Lorenz T, Reinstein J. The influence of proline isomerization and off-pathway intermediates on the folding mechanism of eukaryotic UMP/CMP Kinase. J Mol Biol. 2008;381:443–455. doi: 10.1016/j.jmb.2008.06.001. [DOI] [PubMed] [Google Scholar]
- Marsh JA, Neale C, Jack FE, Choy WY, Lee AY, Crowhurst KA, Forman-Kay JD. Improved structural characterizations of the drkN SH3 domain unfolded state suggest a compact ensemble with native-like and non-native structure. J Mol Biol. 2007;367:1494–1510. doi: 10.1016/j.jmb.2007.01.038. [DOI] [PubMed] [Google Scholar]
- Mittag T, Forman-Kay JD. Atomic-level characterization of disordered protein ensembles. Curr Opin Struct Biol. 2007;17:3–14. doi: 10.1016/j.sbi.2007.01.009. [DOI] [PubMed] [Google Scholar]
- Nabuurs SM, Westphal AH, van Mierlo CP. Extensive formation of off-pathway species during folding of an alpha-beta parallel protein is due to docking of (non)native structure elements in unfolded molecules. J Am Chem Soc. 2008;130:16914–16920. doi: 10.1021/ja803841n. [DOI] [PubMed] [Google Scholar]
- Nabuurs SM, Westphal AH, van Mierlo CPM. Noncooperative formation of the off-pathway molten globule during folding of the α–β parallel protein apoflavodoxin. J Am Chem Soc. 2009;131:2739–2746. doi: 10.1021/ja8089476. [DOI] [PubMed] [Google Scholar]
- Nabuurs SM, Westphal AH, aan den Toorn M, Lindhoud S, van Mierlo CP. Topological switching between an alpha-beta parallel protein and a remarkably helical molten globule. J Am Chem Soc. 2009;131:8290–8295. doi: 10.1021/ja9014309. [DOI] [PubMed] [Google Scholar]
- Nozaki Y. The preparation of guanidine hydrochloride. In: Hirs CHW, Timasheff SN, editors. Methods in enzymology. New York: Academic Press; 1972. pp. 43–50. [DOI] [PubMed] [Google Scholar]
- Otzen DE, Giehm L, Baptista RP, Kristensen SR, Melo EP, Pedersen S. Aggregation as the basis for complex behaviour of cutinase in different denaturants. Biochim Biophys Acta. 2007;1774:323–333. doi: 10.1016/j.bbapap.2006.11.012. [DOI] [PubMed] [Google Scholar]
- Pace CN, Laurents DV. A new method for determining the heat capacity change for protein folding. Biochemistry. 1989;28:2520–2525. doi: 10.1021/bi00432a026. [DOI] [PubMed] [Google Scholar]
- Platt GW, McParland VJ, Kalverda AP, Homans SW, Radford SE. Dynamics in the unfolded state of beta2-microglobulin studied by NMR. J Mol Biol. 2005;346:279–294. doi: 10.1016/j.jmb.2004.11.035. [DOI] [PubMed] [Google Scholar]
- Privalov PL, Khechinashvili NN. A thermodynamic approach to the problem of stabilization of globular protein structure: a calorimetric study. J Mol Biol. 1974;86:665–684. doi: 10.1016/0022-2836(74)90188-0. [DOI] [PubMed] [Google Scholar]
- Reed MA, Jelinska C, Syson K, Cliff MJ, Splevins A, Alizadeh T, Hounslow AM, Staniforth RA, Clarke AR, Craven CJ, Waltho JP. The denatured state under native conditions: a non-native-like collapsed state of N-PGK. J Mol Biol. 2006;357:365–372. doi: 10.1016/j.jmb.2005.12.080. [DOI] [PubMed] [Google Scholar]
- Rose GD, Roy S. Hydrophobic basis of packing in globular-proteins. Proc Natl Acad Sci USA. 1980;77:4643–4647. doi: 10.1073/pnas.77.8.4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzinger S, Wright PE, Dyson HJ. Molecular hinges in protein folding: The urea-denatured state of apomyoglobin. Biochemistry. 2002;41:12681–12686. doi: 10.1021/bi020381o. [DOI] [PubMed] [Google Scholar]
- Steensma E, van Mierlo CP. Structural characterisation of apoflavodoxin shows that the location of the stable nucleus differs among proteins with a flavodoxin-like topology. J Mol Biol. 1998;282:653–666. doi: 10.1006/jmbi.1998.2045. [DOI] [PubMed] [Google Scholar]
- Steensma E, Heering HA, Hagen WR, Van Mierlo CP. Redox properties of wild-type, Cys69Ala, and Cys69Ser Azotobacter vinelandii flavodoxin II as measured by cyclic voltammetry and EPR spectroscopy. Eur J Biochem. 1996;235:167–172. doi: 10.1111/j.1432-1033.1996.00167.x. [DOI] [PubMed] [Google Scholar]
- Steensma E, Nijman MJ, Bollen YJ, de Jager PA, van den Berg WA, van Dongen WM, van Mierlo CP. Apparent local stability of the secondary structure of Azotobacter vinelandii holoflavodoxin II as probed by hydrogen exchange: implications for redox potential regulation and flavodoxin folding. Protein Sci. 1998;7:306–317. doi: 10.1002/pro.5560070210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teilum K, Kragelund BB, Poulsen FM. Transient structure formation in unfolded acyl-coenzyme A-binding protein observed by site-directed spin labelling. J Mol Biol. 2002;324:349–357. doi: 10.1016/S0022-2836(02)01039-2. [DOI] [PubMed] [Google Scholar]
- van Mierlo CP, Steensma E. Protein folding and stability investigated by fluorescence, circular dichroism (CD), and nuclear magnetic resonance (NMR) spectroscopy: the flavodoxin story. J Biotechnol. 2000;79:281–298. doi: 10.1016/S0168-1656(00)00244-3. [DOI] [PubMed] [Google Scholar]
- van Mierlo CP, van Dongen WM, Vergeldt F, van Berkel WJ, Steensma E. The equilibrium unfolding of Azotobacter vinelandii apoflavodoxin II occurs via a relatively stable folding intermediate. Protein Sci. 1998;7:2331–2344. doi: 10.1002/pro.5560071110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Mierlo CP, van den Oever JM, Steensma E. Apoflavodoxin (un)folding followed at the residue level by NMR. Protein Sci. 2000;9:145–157. doi: 10.1110/ps.9.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi Q, Scalley-Kim ML, Alm EJ, Baker D. NMR characterization of residual structure in the denatured state of protein L. J Mol Biol. 2000;299:1341–1351. doi: 10.1006/jmbi.2000.3816. [DOI] [PubMed] [Google Scholar]


