Abstract
Until recently, phosphotyrosine signaling was thought to be restricted to multicellular animals. Surprisingly, the unicellular choanoflagellate Monosiga brevicollis contains a number and diversity of tyrosine kinases that exceeds that of any metazoan, including humans. Many of these M. brevicollis tyrosine kinases possess combinations of signaling domains that do not occur in metazoans. One such kinase, the Src-like protein MbSrc4, contains a lipid-binding C2 domain N-terminal to the conserved SH3-SH2-kinase domains. Here, we report that the enzyme is highly active as a tyrosine kinase and that the targeting functions of the C2, SH3, and SH2 domains are similar to the mammalian counterparts. The membrane-binding activity of the C2 domain is functionally equivalent to the myristoylation signal of c-Src, suggesting that it is an example of convergent evolution. When expressed in mammalian cells, full-length MbSrc4 displays low activity toward endogenous proteins, and it cannot functionally substitute for mammalian c-Src in a reporter gene assay. Removal of the MbSrc4 C2 domain leads to increased phosphorylation of cellular proteins. Thus, in contrast to the related M. brevicollis Src-like kinase MbSrc1, MbSrc4 is not targeted properly to mammalian Src substrates, suggesting that the C2 domain plays a specific role in M. brevicollis signaling.
Choanoflagellates are believed to be the closest living unicellular relatives of metazoans (1–3). The sequencing of the genome of the choanoflagellate Monosiga brevicollis has highlighted the presence of a number of signaling molecules that were previously thought to be unique to multicellular animals (2). In particular, the machinery necessary for phosphotyrosine-based signal transduction (tyrosine kinases, tyrosine phosphatases, and SH21 domains) is present in M. brevicollis. This suggests that the evolution of tyrosine kinases predated their widespread use in cellular signaling in metazoans. A comparison of M. brevicollis and metazoan tyrosine kinases can therefore reveal the structural and functional features that arose more recently in the metazoan lineage.
A striking feature of the M. brevicollis genome is the large number of receptor and nonreceptor tyrosine kinases as compared with metazoans (4, 5). M. brevicollis contains orthologues of the mammalian Src, Csk, Abl, and Tec kinases, but most of the tyrosine kinases have no obvious mammalian orthologues (2, 4, 5). Many of the kinases contain combinations of domains that are not seen in any metazoan. For example, 10 of the 15 HMTKs (HM-motif tyrosine kinases) contain one or more PTB domains, FYTK contains an inositol lipid-binding FYVE domain, and Src4 contains a lipid-binding C2 domain (5); none of these domains is found in combination with a tyrosine kinase catalytic domain in metazoans. These variations in the domain architecture highlight the role of domain shuffling in the evolution of signal transduction pathways.
Several M. brevicollis tyrosine kinases show possible examples of convergent evolution (5). There are four identifiable Src-family nonreceptor tyrosine kinases (Src1–4) in the M. brevicollis genome. Each kinase contains the SH3, SH2, and kinase catalytic domains found in metazoan Src family kinases. Three of the four (Src1–3) contain an N-terminal myristoylation consensus sequence, similar to mammalian Src-family kinases (SFKs). For mammalian c-Src, myristoylation is critical for proper membrane targeting of the enzyme. Mutation of the myristoylation sequence yields a form of c-Src that is enzymatically active, yet unable to transform cells (6, 7). In contrast, Src4 possesses a C2 domain, which in other signaling molecules functions as a lipid targeting module (8). Thus, Src4 may possess an alternative mechanism for membrane localization. Similarly, the PTB domains in several HMTK kinases may play analogous functions to the SH2 domains found in many families of nonreceptor tyrosine kinases; for example, the PTB domains may be involved in targeting the HMTK kinase domains to cellular proteins for phosphorylation.
We previously cloned, expressed, and purified the nonreceptor tyrosine kinase MbSrc1 from M. brevicollis (9). The kinase has the same domain arrangement as mammalian Src kinases, and the individual SH3, SH2, and catalytic domains have similar functions to their mammalian counterparts. We also cloned and expressed the M. brevicollis homologue of c-Src C-terminal kinase (MbCsk) and showed that it phosphorylates the C-terminus of MbSrc1, yet this phosphorylation does not inhibit MbSrc to the same degree seen in the mammalian Src/Csk pair. Our results suggested that the targeting function of the SH3 and SH2 domains in SFKs may have evolved earlier than the autoinhibitory roles of these domains. Here, we have carried out a biochemical characterization of the C2-containing MbSrc4. Our studies have revealed that the targeting functions of the C2, SH2, and SH3 domains are intact in Src4 and similar to their mammalian counterparts. Nonetheless, MbSrc4 cannot functionally substitute for mammalian c-Src.
MATERIALS AND METHODS
cDNA Cloning
The sequence of MbSrc4 was identified within the kinase.com database (http://kinase.com/kinbase). This sequence differs from the Joint Genome Institute (JGI) predicted gene model (NCBI accession number = XP_001749643.1), in that the JGI model contains an extra 120 residues at the N-terminus. RT-PCR of the longer JGI form of MbSrc4 was unsuccessful (Susan Young, personal communication). The extra 120 amino acids in the JGI model lack a myristoylation signal but contain two long stretches of hydrophobic amino acids that could target the protein to the membrane. The C2, SH3, SH2, and kinase domains are identical in the kinase.com and JGI models.
The kinase.com form of MbSrc4 was amplified by PCR from a M. brevicollis cDNA library (10) using the5′ primer GGAATTCAATGGCAGACTCCTCCACGCCCAGCCGCAAGGGC and the 3′ primer GCTCTAGATCAATGTGCTGGCATCGGCATATTGAGCCTCGCC. The predicted translation product for MbSrc4 contains 606 amino acids, but we were unable to amplify a cDNA using primers to the extreme C-terminus. The PCR product encodes a protein that ends at Leu599; the last seven amino acids are not predicted to be in a conserved domain or regulatory region (Figure 1). For baculovirus expression, MbSrc4 DNA (encoding residues 1–599) was cloned into the EcoRI site of pFastBac-Htc (Invitrogen). Flag-tagged MbSrc4 was expressed in mammalian cells by cloning into the EcoRI site of p3XFLAG-CMV (Sigma). The MbSrc4 N-terminus (residues 1–136) was expressed in Escherichia coli NB42 cells by cloning into the BamHI and XbaI sites of pProEX-Htb (Invitrogen). The MbSrc4-32K construct (residues 154–599, lacking the C2 domain) was cloned into the p3XFLAG-CMV vector. All constructs were confirmed by DNA sequencing.
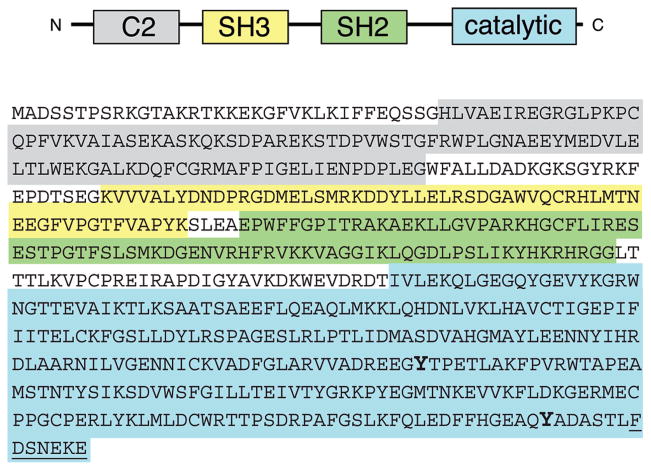
Figure 1.
Domain arrangement of MbSrc4. Tyrosines 483 and 593, which are homologous to Y416 and Y527 of c-Src, respectively, are shown in boldface. The C-terminal seven amino acids (underlined) were not included in the construct expressed in Sf9 cells.
Protein Expression and Purification
His-tagged MbSrc4 was expressed in Sf9 insect cells using the Bac-to-Bac system (Invitrogen), as described previously (9). Sf9 cells (600 mL) were infected with recombinant MbSrc4 baculovirus at a multiplicity of infection of 10 plaque-forming units/cell for a period of 72 h. MbSrc4 was purified on a 5 mL column of nickel-nitrilotriacetic acid resin (Qiagen), as described previously (9, 11). Purified MbSrc4 was concentrated in an Ultrafree-10 concentrator (Millipore) and stored in 40% glycerol at −20 °C. The MbSrc4 N-terminus was expressed as a His-tagged protein in E. coli and purified using similar methods. In some experiments, the His tag was removed by overnight treatment with tobacco etch virus at 4 °C.
Tyrosine Kinase Assays
Kinase assays were performed with [γ-32P]ATP using the phosphocellulose paper binding assay (12,13). Reaction mixtures contained 20 mM Tris-HCl (pH 7.4), 10 mM MgCl2, 0.1 mM Na3VO4, 0.5 mM DTT, 0.25 mM ATP, varying concentrations of peptide substrate, and [γ-32P]ATP (200–400 cpm/pmol). The following substrates were used: Src optimal substrate, AEEEIYGEFEAKKKKG; SH3-binding substrate, AEEEIYGEFGGRGAAPPPPPVPRGRG; SH3 control substrate, AEEEIYGEFGGRGAAAAAAAVARGRG; SH2-binding substrate, RRLEDAIYAAGGGGGEPPQpYEEIG; SH2 control substrate, RRLEDAIYAAGGGGGEPPQFEEIG; MbSTAT, KKKASGYVMADIA; RTKB2-1, SEEVYGAVVDKKK; RTKB2-2, AEEVYEAIADKKK.
Cell Transfection and Western Blotting
COS-7 or Src/Yes/Fyn-deficient (SYF) cells (14) were cultured in Dulbecco’s modified Eagle’s medium plus 10% fetal bovine serum at 37 °C in 5% CO2. Cells were transfected at 50% confluency using TransIT polyamine transfection reagent (Mirus) at a ratio of 1:3.5 (DNA: TransIT). For Western blotting, cells were lysed 48 h after transfection in buffer containing 10 mM Tris-HCl, pH 7.4, 50 mM NaCl, 5 mM EDTA, 1% Triton X-100, 50 mM NaF, 2mMNa3VO4, 1 mM PMSF, 1 mg/mL aprotinin, and 1 mg/mL leupeptin at 4 °C for 1 h. The cell lysates were centrifuged at 14000g for 10 min at 4 °C, and protein concentrations were determined using Bio-Rad protein detection reagent. Lysates (25 μg) were analyzed by 7.5% SDS–PAGE, and proteins in the lysates were transferred onto PVDF membrane. Immunodetection was carried out with anti-phosphotyrosine (4G10; Upstate) and anti-Flag (M2; Sigma) antibodies.
Luciferase Reporter Assay
The SYF cells were seeded in 12 well plates (5 × 104 cells/well) and transfected in triplicate with protein expression plasmids, GAS luciferase reporter plasmid, or a promoterless renilla luciferase plasmid (pRL-null) as a transfection control, as described previously (60). Transfected cells were harvested after 48 h. Preparation of cell lysates and measurement of light units were carried out using the dual-luciferase reporter assay system (Promega) following the manufacturer’s instructions. Relative light units were calculated as (firefly luciferase activity/renilla luciferase activity).
Fluorescence Labeling
Proteins were thawed and dialyzed for 45 min against 500 mL of 20 mM Hepes (pH 7.4) and 150 mM NaCl buffer. Afterward, the proteins were labeled on ice with the thiol-reactive probe 7-diethylamino-3-(4′-maleimidylphenyl)-4-methylcoumarin (CPM; Invitrogen). Unreacted CPM is not fluorescent, but the probe becomes fluorescent once it covalently attaches to a thiol group. After 1 h, the reaction was stopped by adding DTT to a final concentration of 1 mM. The labeled proteins were separated from unreacted probe by dialysis against 20 mM Hepes (pH 7.4), 150 mM NaCl, and 1 mM DTT, 3 times for 30 min. The final molar ratios of probe:protein were 1:1, as determined by absorption to determine the probe concentration and BCA analysis to determine the protein concentration (15, 16).
Membrane Binding Experiments
Fluorescence measurements were performed on an ISS spectrofluorometer (Champaign, IL) using 3 mm quartz cuvettes. The emission spectrum of CPM-labeled protein was measured from 415 to 530 nm (λex = 384 nm). Background signal from unlabeled protein accounted for less than 0.1% of the emission and was not considered. The lipids 1-palmitoyl-2-oleoylphosphatidylethanolamine (POPE), 1-palmitoyl-2-oleoylphosphatidylserine (POPS), and 1-palmitoyl-2-oleoylphosphatidylcholine (POPC) were purchased from Avanti Polar Lipids, Inc. (Alabaster, AL).
Membrane binding was carried out by titrating large, unilamellar vesicles (prepared by manual extrusion through a 0.1 μm polycarbonate filter) into a 100 nM solution of labeled protein and by measuring the change in the integrated area of the sample as compared to control samples that substituted buffer for labeled protein. After correcting for dilution and background, the change in fluorescence intensity was plotted as a function of lipid concentration and fit to a hyperbolic curve using SigmaPlot (Jandel, Inc.) to obtain the apparent partition coefficient (Kp), which corresponds to the lipid concentration at which 50% of the protein or peptide is bound. Measurements in the presence of calcium ions were carried out by incrementally adding calcium carbonate to a final concentration of 500 nM.
Subcellular Fractionation
SYF cells were transfected with the appropriate Flag-tagged constructs. After 44–48 h, the cells were harvested in ice-cold PBS and centrifuged at 500g for 5 min. The cells were immediately resuspended in buffer A (10 mM HEPES, pH 7.0, 5 mM MgCl2, 25 mM KCl, 150 mM PMSF, 1mM Na3VO4, 10 μg/mL leupeptin, and 10 μg/mL aprotinin). Cells were gently homogenized by forcing them 10 times through 23-gauge needles. The cell homogenates were immediately mixed with buffer B (1:1 v/v) (10 mM HEPES, pH 7.0, 5 mM MgCl2, 25 mM KCl, 1 M sucrose, 150 mM PMSF, 1 mM Na3VO4, 10 μg/mL leupeptin, and 10 μg/mL aprotinin). Nuclei, unbroken cells, mitochontria, peroxisomes, and microsomes were pelleted by centrifugation at 16000g for 15 min. The supernatants were further centrifuged at 100000g for 1 h to separate the membrane fraction and clear soluble cytosolic fraction. The membrane fraction was washed once and then dissolved in lysis buffer. The lysates were separated by SDS–PAGE, and the proteins were visualized by anti-Flag Western blotting.
RESULTS
Cloning M. brevicollis MbSrc4
A cDNA encoding MbSrc4 (residues 1–599) was amplified by PCR from an M. brevicollis cDNA library (10). The domain structure of MbSrc4 is shown in Figure 1. The enzyme contains the conserved SH3-SH2-kinase domain structure found in all Src family kinases. The N-terminal region of MbSrc4 contains a C2 domain, which in other signaling proteins is a lipid-binding module. No metazoan tyrosine kinase with a C2 domain has been described. The overall amino acid identity between MbSrc4 and human c-Src is 37.5%, and within the SH3-SH2-kinase regions, the identity rises to 45.7%. By comparison, the overall amino acid identity between MbSrc1 and human c-Src is 57%. Metazoan Src family kinases possess two tyrosine residues that are important for enzyme regulation: Y416 (chicken Src numbering), which is a site for autophosphorylation and activation, and Y527, which is an inhibitory site when phosphorylated (7, 17, 18). MbSrc4 possesses tyrosines at homologous positions (Y483 and Y593, respectively).
Expression and Purification of MbSrc4 from Insect Cells
To test whether MbSrc4 is an enzymatically active tyrosine kinase, we cloned the MbSrc4 DNA into a baculovirus expression vector and expressed the His-tagged enzyme in Spodoptera frugiperda (Sf9) insect cells. MbSrc4 was purified in a single step from Sf9 cell lysates by chromatography on nickel-nitrilotriacetic acid resin (Figure 2A).
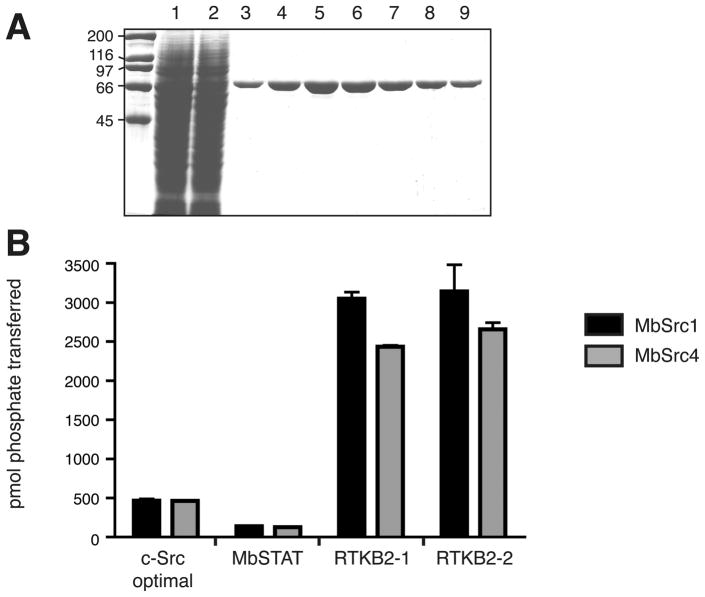
Figure 2.
(A) Purification of MbSrc4 followed by SDS–PAGE with Coomassie blue staining. Lanes: 1, Sf9 whole cell lysate; 2, unbound fraction after Ni-NTA chromatography; 3–9, Ni-NTA column fractions. (B) Enzymatic activity of MbSrc4 toward synthetic peptide substrates. Kinase activity was measured using the phosphocellulose paper assay. The peptides used were as follows: c-Src optimal, AEEEIYGEFEAKKKG; MbSTAT, KKKASGYVMADIA; RTKB2-1, SEEVYGAVVDKKK; RTKB2-2, AEEVYEAIADKKK. The peptides were tested at concentrations of 750 μM. Reactions proceeded for 5 min at 30 °C.
We tested the enzymatic activity of MbSrc4 toward a series of synthetic peptide substrates. Peptide AEEEIYGEFEAKKKKG incorporated the EEEIYGEF motif identified as the optimal c-Src phosphorylation sequence in peptide library studies (19). We also tested two peptides derived from the intracellular domains of M. brevicollis receptor tyrosine kinases (RTKB2-1 and -2), along with a peptide derived from a predicted M. brevicollis STAT molecule (5). We compared the activity of MbSrc4 with a comparable preparation of MbSrc1, a non-C2 domain containing Src kinase from M. brevicollis (Figure 2B). Consistent with our previous data (5), MbSrc1 preferred the RTKB2-derived peptides from this group of substrates and showed ≈6-fold lower activity with the c-Src optimal sequence. The activity of MbSrc4 was slightly lower than that of MbSrc1 toward this panel of substrates, but the specificity of the two enzymes was similar (Figure 2B). Kinetic analysis of the MbSrc4 reaction with peptide RTKB2-2 yielded a kcat/Km value of 2.9 × 105 min−1 M−1, comparable to the catalytic efficiency of a mammalian Src family kinase (Hck) with its optimal substrate (4.5 × 105 min−1 M−1)(20). Thus, MbSrc4 is a catalytically competent tyrosine kinase, and its substrate specificity appears to be more closely related to that of M. brevicollis MbSrc1 than that of mammalian c-Src. The sequences of the P + 1 loops (regions responsible for interacting with protein substrates) are well conserved between c-Src and MbSrc4, suggesting that additional regions or conformational differences account for the differences in substrate specificity.
Autophosphorylation of MbSrc4
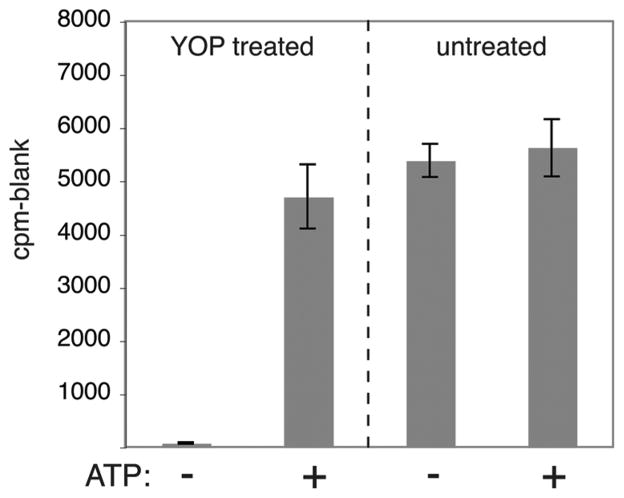
Mammalian SFKs undergo autophosphorylation at Y416, which lies in the activation loop, a flexible segment between the N- and C-terminal lobes of the catalytic domain. Phosphorylation of Y416 substantially increases tyrosine kinase activity (21, 22). We incubated purified MbSrc4 with ATP under conditions that have previously been shown to promote Y416 phosphorylation (9, 22) and then measured MbSrc4 activity toward the synthetic peptide. In addition, to test the possibility that MbSrc4 purified from insect cells might already be phosphorylated, we pretreated the enzyme with the Yersinia YOP tyrosine phosphatase. YOP-treated MbSrc4 had barely detectable levels of kinase activity, and incubation with ATP strongly activated the enzyme (Figure 3). In contrast, MbSrc4 that was not treated with YOP showed high activity in the presence or absence of ATP (Figure 3). Similar results were previously obtained for MbSrc1 (9). These experiments suggest that MbSrc4 is controlled by autophosphorylation in a similar manner to other SFKs.
Figure 3.
Effect of autophosphorylation on MbSrc4 activity. MbSrc4 (5 μL of 68 μM) was incubated with immobilized GST-YOP in 50 mM Tris (pH 7.5) and 50 mM NaCl (200 μL final volume). The reaction was mixed at room temperature for 30 min. After removal of GST-YOP by centrifugation, MbSrc4 was assayed directly or after incubation with 0.5 mM ATP for 30 min at 30 °C.
Substrate Targeting by the SH3 and SH2 Domains of MbSrc4
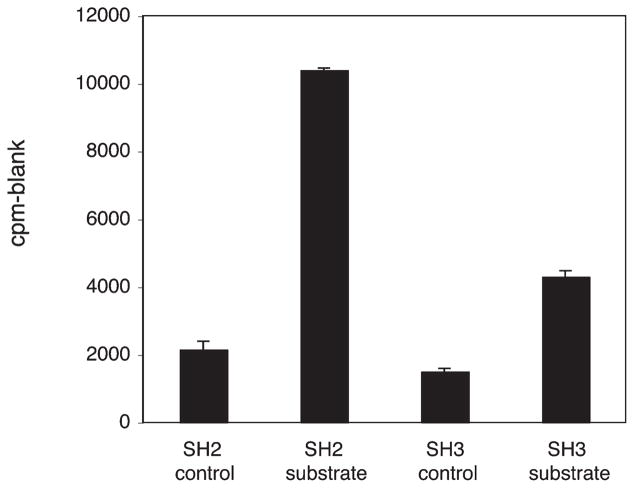
In mammalian Src family kinases, the SH3 and SH2 domains play an important role in the recognition of protein substrates (7, 23). SH3 domains bind to short proline-rich sequences, and SH2 domains bind to sequences containing phosphotyrosine. Many cellular substrates for SFKs contain ligands for one or both of these domains, and binding leads to phosphorylation by the Src catalytic domain. Substrate targeting can be measured in vitro using synthetic peptides that contain SH3 or SH2 ligand sequences in addition to a SFK phosphorylation site (24, 25). Mammalian SFKs preferentially phosphorylate such peptides, as compared to control sequences in which the SH3/SH2 ligand sequences are altered. M. brevicollis MbSrc1 displays SH3-mediated substrate targeting but not SH2-mediated targeting (9). We tested the ability of purified MbSrc4 to phosphorylate peptides containing an SH3 or SH2 ligand. Phosphorylation of the SH3- and SH2-containing substrates by MbSrc4 was higher than controls (Figure 4), and the increase in activity (5-fold for the SH2 substrate, 2.5–3-fold for the SH3 substrate) is comparable to that observed for other SFKs. Thus, the SH3 and SH2 domains of MbSrc4 are functional and retain their ability to target the kinase domain to potential substrates.
Figure 4.
Substrate targeting by MbSrc4. MbSrc4 (400 nM) was assayed with 70 μM concentrations of the SH2-binding substrate, SH2 control substrate, SH3-binding substrate, or SH3 control substrate (sequences given in the Materials and Methods section). Kinase activity was measured using the phosphocellulose paper assay; reactions were carried out for 5 min at 30 °C.
Membrane Binding by MbSrc4
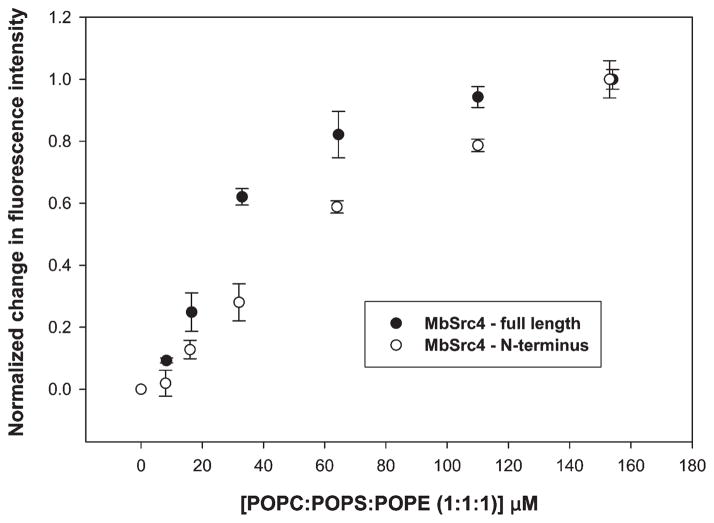
To test for membrane binding of MbSrc4, we prepared a fluorescently labeled version of the protein and measured binding to large unilamellar vesicles (LUVs) composed of POPC:POPS:POPE (1:1:1). This substrate is often used as a model for the plasma membrane since the large size of LUVs allows one to measure protein binding to a flat, uniform surface and the unsaturated oleoyl hydrocarbon chains insure that the lipid will be in the fluid phase. The plasma membrane compositions of different cell types vary (www.lipidmaps.org). Most cells contain ~30% negatively charged lipids along with a high concentration of phosphaditylcholine and phosphatidylethanolamine lipids (26, 27). Phosphatidylserine is often used in binding measurements because C2 domains prefer negatively charged lipids. The binding of various C2 domains to subcellular membranes generally follows the same preference for particular lipids in model systems (28). MbSrc4 bound with a partition coefficient (Kp) of 77 μM (Figure 5 and Table 1). Binding was not calcium-dependent, as measurements in the presence of added calcium (500 nM) gave a comparable partition coefficient (Table 1).
Figure 5.
Membrane binding by full-length MbSrc4 and the isolated N-terminus. Large unilamellar vesicles were titrated into 100 nM solutions of CPM-labeled protein. After correcting for dilution and background, the change in fluorescence intensity was plotted as a function of lipid concentration.
Table 1.
Membrane-Binding Experimentsa
| construct | added calcium | Kp(μM) |
|---|---|---|
| full length | no | 77 ± 36 |
| full length | yes (500 nM) | 85 ± 18 |
| N-terminus, His tagged | no | 188 ± 41 |
| N-terminus, untagged | no | 187 ± 38 |
Lipids were in the form of large unilamellar vesicles (0.1 μm) composed of POPC:POPS:POPE (1:1:1). The buffer was 20 mM HEPES, pH 7.4, and 0.16M NaCl.
To test whether the membrane-binding ability of MbSrc4 is derived from the C2 domain, we cloned the N-terminal 136 residues of MbSrc (containing the C2 domain) and expressed the protein as a His-tagged fusion in E. coli. We fluorescently labeled the N-terminus and carried out similar binding studies. The N-terminus bound with a partition coefficient of 188 μM (Figure 5 and Table 1). This suggests that the C2 domain indeed acts as a membrane targeting module but that other elements of MbSrc4 are present that contribute to the tighter binding observed for the full-length protein. To confirm that the N-terminal His tag did not interfere with the binding measurements, we cleaved the His tag with TEV protease and repurified the MbSrc4 N-terminus. Removal of the His tag did not alter the binding properties of the N-terminus (Table 1).
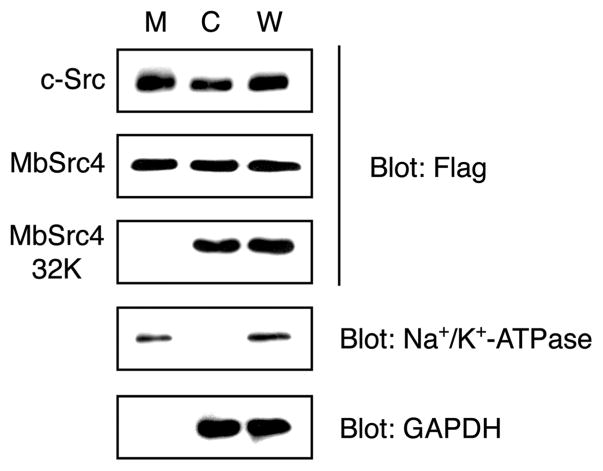
Next, to test the importance of the C2 domain on the membrane localization of MbSrc4 in intact cells, we expressed an N-terminally Flag-tagged version of MbSrc4 in Src/Yes/Fyn triple knockout (SYF) cells, which lack all Src family kinases (14). We conducted subcellular fractionation experiments and visualized MbSrc4 by anti-Flag Western blotting. MbSrc4 and mammalian c-Src were evenly divided between the membrane and cytosolic fractions (Figure 6). In contrast, an MbSrc truncation mutant consisting of the SH3, SH2, and kinase domains but lacking the C2 domain (MbSrc4-32K) was found solely in the cytosol (Figure 6). These results show that the C2 domain mediates membrane targeting in intact cells.
Figure 6.
Subcellular fractionation. SYF cells were transfected with human c-Src, full-length MbSrc4, or MbSrc4-32K. Cells were fractionated as described in Materials and Methods, and membrane and cytosolic fractions (M and C, respectively) and whole cell lysates (W) were analyzed by SDS–PAGE with anti-Flag Western blotting. Equivalent amounts of total protein were analyzed in each lane. To ensure that fractionation was successful, lysates were analyzed by blotting with a membrane marker (Na+/K+-ATPase) and a cytosolic marker (GAPDH); representative blots are shown.
Functional Studies of MbSrc4 in Mammalian Cells
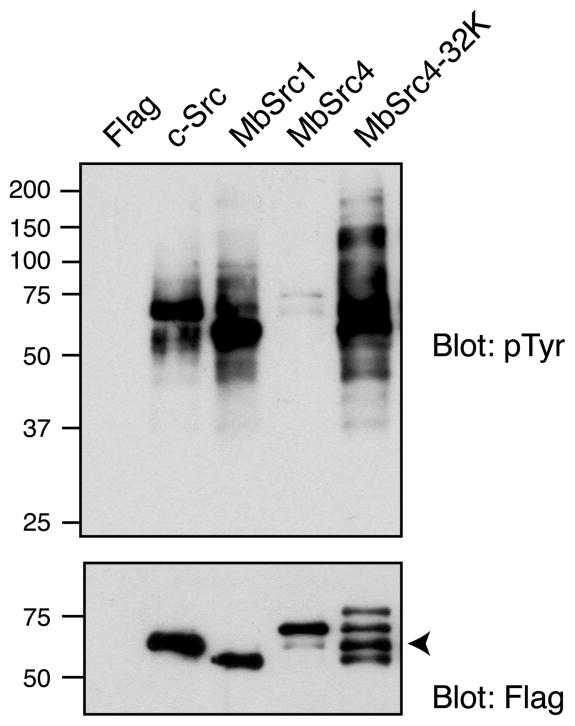
We expressed MbSrc4 in SYF cells and carried out anti-phosphotyrosine Western blotting of SYF lysates as a measure of global MbSrc4 kinase activity. For comparison, we analyzed cells expressing M. brevicollis MbSrc1 or human c-Src. Full-length MbSrc4 displayed much lower levels of activity than c-Src or MbSrc1, suggesting that the enzyme was incapable of recognizing cellular substrates (Figure 7). The single band in the MbSrc4 lysate migrated at a position consistent with autophosphorylated MbSrc4. On the other hand, the MbSrc4-32K construct (lacking the C2 domain) displayed a level of kinase activity that was comparable to that of c-Src or MbSrc1 (Figure 7), suggesting that the C2 domain interfered with phosphorylation of cellular proteins.
Figure 7.
Phosphorylation of mammalian cell substrates. (A) SYF cells were transfected with empty vector (Flag), human c-Src, MbSrc1, full-length MbSrc4, or MbSrc4-32K. Lysates were analyzed by SDS–PAGE and anti-pTyr Western blotting. The blots were reprobed with anti-Flag antibody. The arrowhead indicates the position of MbSrc4-32K.
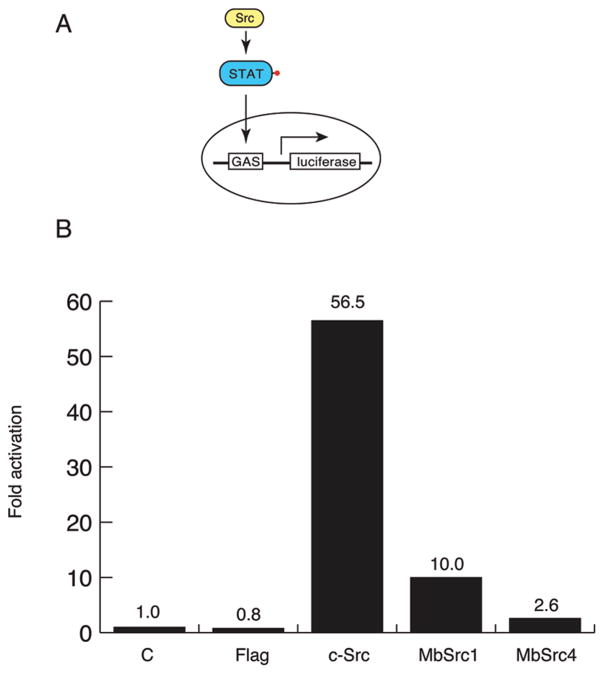
Next, we tested whether MbSrc4 can functionally substitute for mammalian Src using a Src-responsive reporter gene assay. We expressed Flag-tagged MbSrc4, MbSrc1, or c-Src in SYF cells together with a phospho-STAT responsive luciferase reporter construct. As reported previously (9), Src activates transcription in this system by > 50-fold, and MbSrc1 activates by 10-fold (Figure 8). MbSrc4 showed low but reproducible activity in this assay (2.5–3-fold activation). These results are consistent with the Western blotting experiments shown in Figure 7 and suggest that, although MbSrc4 has high kinase activity in vitro, the enzyme has a low level of function in mammalian cells.
Figure 8.
Src reporter assay. (A) Diagram of the STAT-dependent reporter assay. Phosphorylation of STAT drives the transcription of the GAS-luciferase reporter plasmid. (B) Luciferase activity was measured in lysates from SYF cells transfected with the indicated plasmids and the GAS luciferase reporter plasmid. C = a promoter-less renilla luciferase plasmid; Flag = empty vector. Fold activation is presented relative to the control.
DISCUSSION
In the M. brevicollis genome, a majority of the domain combinations involving tyrosine kinase, tyrosine phosphatase, or SH2 domains are not found in any metazoan genome (2, 4). Several nonreceptor tyrosine kinases in M. brevicollis contain modular domains that are not present in tyrosine kinases in metazoans, including C2, FYVE, and PTB domains (5). In contrast, for other signaling systems (Ser/Thr kinases and phosphatases, Ras and Rho signaling), most of the domain combinations in M. brevicollis are similar to metazoans. This suggests a model in which tyrosine kinase signaling emerged in a common ancestor of choanoflagellates and metazoans and diverged significantly after the split (2, 29, 30). The functions of the diverse tyrosine kinases in the unicellular life of M. brevicollis are largely unknown.
One example of a unique domain combination in a M. brevicollis nonreceptor tyrosine kinase is the MbSrc4 protein. The only other tyrosine kinase known to possess a C2 domain is the likely orthologous Src kinase from the choanoflagellate Monosiga ovata (NCBI accession number = BAG55494). Despite their shared genus names, M. brevicollis and M. ovata are evolutionarily remote choanoflagellates; thus, the conservation of the C2 domain increases the likelihood that they are functionally relevant. C2 domains were first described in conventional protein kinase C enzymes, where they act as Ca2+-dependent phospholipid-binding domains (8). Many C2 domains have similar functions, although a subset do not bind Ca2+, and protein targets for some C2 domains have been described. In particular, the C2 domain of protein kinase Cδ has been shown to bind phosphotyrosine-containing peptides and proteins (31). This ability is dependent on a histidine residue in PKCδ to form ring-stacking interactions with phosphotyrosine. The C2 domain of MbSrc4 lacks a histidine residue at the corresponding position and is unlikely to act as a phosphotyrosine-binding module. We demonstrate in this paper that the C2 domain of MbSrc4 is functional as a lipid-targeting module (Figure 5) and that its removal interferes with membrane localization in intact cells (Figure 6). Membrane binding of many C2 domains has been assessed by qualitative methods, but only a few have been quantified using surface plasma resonance, and the Kd values are in the 1–100 nM range, or about 100-fold stronger than the values obtained here (32–34). However, the C2 domains used in those studies were calcium-binding domains, and chelation of calcium ions with phosphoserine headgroups appears to contribute to the strong membrane binding. Since the MbSrc4 C2 domain does not appear to bind calcium, the micromolar membrane affinity reflects a strong protein–lipid interaction and on the order seen for calcium-independent membrane protein domains (35). Our data suggest that MbSrc4, with its alternative membrane-binding mechanism, may represent an example of convergent evolution with other Src kinases (although a recent gene fusion event is also a plausible scenario).
The SH3 and SH2 domains of MbSrc4 are also functional, as demonstrated by their ability to direct phosphorylation of peptide substrates possessing cognate ligands (Figure 4). The substrate targeting ability of the MbSrc4 SH3 and SH2 domains is comparable to that of mammalian Src kinases. Interestingly, the related M. brevicollis kinase MbSrc1 displayed SH3-mediated targeting but not SH2-mediated targeting (9). MbSrc1 has the typical SH3-SH2-kinase architecture found in mammalian SFKs and lacks the N-terminal C2 domain of MbSrc4. The reason for the lack of SH2-mediated targeting in MbSrc1 is not clear but may be related to the divergent linker region between the SH2 and kinase domains of that enzyme. Despite the fact that the C2, SH3, SH2, and kinase domains of MbSrc4 are functional, MbSrc4 had lower activity than mammalian Src or M. brevicollis MbSrc1 in a reporter gene assay (Figure 8). Furthermore, full-length MbSrc4 phosphorylated a lower number of proteins in mammalian cells than c-Src (Figure 7), consistent with the in vitro data showing that the substrate preference of MbSrc4 differs from the mammalian consensus (Figure 2B). The MbSrc4 construct lacking a C2 domain was more active than full length. The latter result suggests either (i) that the C2 domain is involved in an autoinhibitory interaction with the kinase domain or (ii) that, in mammalian cells, the C2 domain targets MbSrc4 to a subcellular location that is incompatible with phosphorylation of protein substrates. In the latter case, deletion of the C2 domain would free MbSrc4 from its membrane tether, permitting the kinase to phosphorylate additional substrates. These issues will be clarified when the roles of the various MbSrc4 domains in M. brevicollis biology are better understood.
Acknowledgments
We thank Nicole King (UC Berkeley) for helpful comments on the manuscript. We thank Nancy Reich and Yan Song (Stony Brook) for reporter constructs and assistance with luciferase experiments and Noriko Yokoyama (Stony Brook) for help with the cell fractionation experiments.
Footnotes
Abbreviations: SH3, Src homology domain 3; SH2, Src homology domain 2; SFKs, Src family kinases; PCR, polymerase chain reaction; Sf9, Spodoptera frugiperda; SYF, Src/Yes/Fyn knockout cells; SDS—PAGE, sodium dodecyl sulfate–polyacrylamide gel electrophoresis; PVDF, polyvinylene difluoride; CPM, 7-diethylamino-3-(4′-maleimidyl-phenyl)-4-methylcoumarin; BCA, bicinchoninic acid; DTT, dithio-threitol; POPE, 1-palmitoyl-2-oleoylphosphatidylethanolamine; POPS, 1-palmitoyl-2-oleoylphosphatidylserine; POPC, 1-palmitoyl-2-oleoylphosphatidylcholine; ATP, adenosine triphosphate; YOP, Yersinia tyrosine phosphatase.
This work was supported by National Institutes of Health Grants CA 58530 to W.T.M. and GM 053132 to S.S.
References
- 1.Steenkamp ET, Wright J, Baldauf SL. The protistan origins of animals and fungi. Mol Biol Evol. 2006;23:93–106. doi: 10.1093/molbev/msj011. [DOI] [PubMed] [Google Scholar]
- 2.King N, Westbrook MJ, Young SL, Kuo A, Abedin M, Chapman J, Fairclough S, Hellsten U, Isogai Y, Letunic I, Marr M, Pincus D, Putnam N, Rokas A, Wright KJ, Zuzow R, Dirks W, Good M, Goodstein D, Lemons D, Li W, Lyons JB, Morris A, Nichols S, Richter DJ, Salamov A, Sequencing JG, Bork P, Lim WA, Manning G, Miller WT, McGinnis W, Shapiro H, Tjian R, Grigoriev IV, Rokhsar D. The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans. Nature. 2008;451:783–788. doi: 10.1038/nature06617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lang BF, O’Kelly C, Nerad T, Gray MW, Burger G. The closest unicellular relatives of animals. Curr Biol. 2002;12:1773–1778. doi: 10.1016/s0960-9822(02)01187-9. [DOI] [PubMed] [Google Scholar]
- 4.Pincus D, Letunic I, Bork P, Lim WA. Evolution of the phospho-tyrosine signaling machinery in premetazoan lineages. Proc Natl Acad Sci USA. 2008;105:9680–9684. doi: 10.1073/pnas.0803161105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Manning G, Young SL, Miller WT, Zhai Y. The protist, Monosiga brevicollis, has a tyrosine kinase signaling network more elaborate and diverse than found in any known metazoan. Proc Natl Acad Sci USA. 2008;105:9674–9679. doi: 10.1073/pnas.0801314105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kamps MP, Buss JE, Sefton BM. Rous sarcoma virus transforming protein lacking myristic acid phosphorylates known polypeptide substrates without inducing transformation. Cell. 1986;45:105–112. doi: 10.1016/0092-8674(86)90542-8. [DOI] [PubMed] [Google Scholar]
- 7.Brown MT, Cooper JA. Regulation, substrates and functions of src. Biochim Biophys Acta. 1996;1287:121–149. doi: 10.1016/0304-419x(96)00003-0. [DOI] [PubMed] [Google Scholar]
- 8.Lemmon MA. Membrane recognition by phospholipid-binding domains. Nat Rev Mol Cell Biol. 2008;9:99–111. doi: 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- 9.Li W, Young SL, King N, Miller WT. Signaling properties of a non-metazoan Src kinase and the evolutionary history of Src negative regulation. J Biol Chem. 2008;283:15491–15501. doi: 10.1074/jbc.M800002200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.King N, Carroll SB. A receptor tyrosine kinase from choanoflagellates: molecular insights into early animal evolution. Proc Natl Acad Sci USA. 2001;98:15032–15037. doi: 10.1073/pnas.261477698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qiu H, Miller WT. Regulation of the nonreceptor tyrosine kinase brk by autophosphorylation and by autoinhibition. J Biol Chem. 2002;277:34634–34641. doi: 10.1074/jbc.M203877200. [DOI] [PubMed] [Google Scholar]
- 12.Casnellie JE. Assay of protein kinases using peptides with basic residues for phosphocellulose binding. Methods Enzymol. 1991;200:115–120. doi: 10.1016/0076-6879(91)00133-h. [DOI] [PubMed] [Google Scholar]
- 13.Garcia P, Shoelson SE, George ST, Hinds DA, Goldberg AR, Miller WT. Phosphorylation of synthetic peptides containing Tyr-Met-X-Met motifs by nonreceptor tyrosine kinases in vitro. J Biol Chem. 1993;268:25146–25151. [PubMed] [Google Scholar]
- 14.Klinghoffer RA, Sachsenmaier C, Cooper JA, Soriano P. Src family kinases are required for integrin but not PDGFR signal transduction. EMBO J. 1999;18:2459–2471. doi: 10.1093/emboj/18.9.2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drin G, Douguet D, Scarlata S. The pleckstrin homology domain of phospholipase Cbeta transmits enzymatic activation through modulation of the membrane-domain orientation. Biochemistry. 2006;45:5712–5724. doi: 10.1021/bi052317n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Philip F, Scarlata S. Influence of membrane components in the binding of proteins to membrane surfaces. Biochemistry. 2004;43:11691–11700. doi: 10.1021/bi049381+. [DOI] [PubMed] [Google Scholar]
- 17.Bjorge JD, Jakymiw A, Fujita DJ. Selected glimpses into the activation and function of Src kinase. Oncogene. 2000;19:5620–5635. doi: 10.1038/sj.onc.1203923. [DOI] [PubMed] [Google Scholar]
- 18.Parsons SJ, Parsons JT. Src family kinases, key regulators of signal transduction. Oncogene. 2004;23:7906–7909. doi: 10.1038/sj.onc.1208160. [DOI] [PubMed] [Google Scholar]
- 19.Songyang Z, Carraway KL, III, Eck MJ, Harrison SC, Feldman RA, Mohammadi M, Schlessinger J, Hubbard SR, Smith DP, Eng C, Lorenzo MJ, Poner BAJ, Mayer BJ, Cantley LC. Catalytic specificity of protein-tyrosine kinases is critical for selective signalling. Nature. 1995;373:536–539. doi: 10.1038/373536a0. [DOI] [PubMed] [Google Scholar]
- 20.LaFevre-Bernt M, Sicheri F, Pico A, Porter M, Kuriyan J, Miller WT. Intramolecular regulatory interactions in the Src family kinase Hck probed by mutagenesis of a conserved tryptophan residue. J Biol Chem. 1998;273:32129–32134. doi: 10.1074/jbc.273.48.32129. [DOI] [PubMed] [Google Scholar]
- 21.Sicheri F, Kuriyan J. Structures of Src-family tyrosine kinases. Curr Opin Struct Biol. 1997;7:777–785. doi: 10.1016/s0959-440x(97)80146-7. [DOI] [PubMed] [Google Scholar]
- 22.Porter M, Schindler T, Kuriyan J, Miller WT. Reciprocal regulation of Hck activity by phosphorylation of Tyr (527) and Tyr(416). Effect of introducing a high affinity intramolecular SH2 ligand. J Biol Chem. 2000;275:2721–2726. doi: 10.1074/jbc.275.4.2721. [DOI] [PubMed] [Google Scholar]
- 23.Miller WT. Determinants of substrate recognition in nonreceptor tyrosine kinases. Acc Chem Res. 2003;36:393–400. doi: 10.1021/ar020116v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pellicena P, Stowell KR, Miller WT. Enhanced phosphorylation of Src family kinase substrates containing SH2 domain binding sites. J Biol Chem. 1998;273:15325–15328. doi: 10.1074/jbc.273.25.15325. [DOI] [PubMed] [Google Scholar]
- 25.Scott MP, Miller WT. A peptide model system for processive phosphorylation by Src family kinases. Biochemistry. 2000;39:14531–14537. doi: 10.1021/bi001850u. [DOI] [PubMed] [Google Scholar]
- 26.Yeagle P. The Structure of Biological Membranes. CRC PressL; Boca Raton, FL: 1992. [Google Scholar]
- 27.Gennis RB. Biomembranes: Molecular Structure and Function. Springer-Verlag; New York: 1989. [Google Scholar]
- 28.Cho W, Stahelin RV. Membrane binding and subcellular targeting of C2 domains. Biochim Biophys Acta. 2006;1761:838–849. doi: 10.1016/j.bbalip.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 29.Suga H, Sasaki G, Kuma K, Nishiyori H, Hirose N, Su ZH, Iwabe N, Miyata T. Ancient divergence of animal protein tyrosine kinase genes demonstrated by a gene family tree including choanoflagellate genes. FEBS Lett s. 2008;582:815–818. doi: 10.1016/j.febslet.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 30.Segawa Y, Suga H, Iwabe N, Oneyama C, Akagi T, Miyata T, Okada M. Functional development of Src tyrosine kinases during evolution from a unicellular ancestor to multicellular animals. Proc Natl Acad Sci USA. 2006;103:12021–12026. doi: 10.1073/pnas.0600021103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Benes CH, Wu N, Elia AE, Dharia T, Cantley LC, Soltoff SP. The C2 domain of PKCdelta is a phosphotyrosine binding domain. Cell. 2005;121:271–280. doi: 10.1016/j.cell.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 32.Ananthanarayanan B, Das S, Rhee SG, Murray D, Cho W. Membrane targeting of C2 domains of phospholipase C-delta isoforms. J Biol Chem. 2002;277:3568–3575. doi: 10.1074/jbc.M109705200. [DOI] [PubMed] [Google Scholar]
- 33.Kulkarni S, Das S, Funk CD, Murray D, Cho W. Molecular basis of the specific subcellular localization of the C2-like domain of 5-lipoxygenase. J Biol Chem. 2002;277:13167–13174. doi: 10.1074/jbc.M112393200. [DOI] [PubMed] [Google Scholar]
- 34.Stahelin RV, Rafter JD, Das S, Cho W. The molecular basis of differential subcellular localization ofC2 domains of protein kinase C-alpha and group IVa cytosolic phospholipase A2. J Biol Chem. 2003;278:12452–12460. doi: 10.1074/jbc.M212864200. [DOI] [PubMed] [Google Scholar]
- 35.Wang T, Pentyala S, Rebecchi MJ, Scarlata S. Differential association of the pleckstrin homology domains of phospholipases C-beta 1, C-beta 2, and C-delta 1 with lipid bilayers and the beta gamma subunits of heterotrimeric G proteins. Biochemistry. 1999;38:1517–1524. doi: 10.1021/bi982008f. [DOI] [PubMed] [Google Scholar]