Abstract
Chromosomal double-strand breaks (DSBs) are considered to be among the most deleterious DNA lesions found in eukaryotic cells due to their propensity to promote genome instability. DSBs occur as a result of exogenous or endogenous DNA damage, and also occur during meiotic recombination. DSBs are often repaired through a process called homologous recombination (HR), which employs the sister chromatid in mitotic cells or the homologous chromosome in meiotic cells, as a template for repair. HR frequently involves the formation and resolution of four-way DNA structures referred to as the Holliday junction (HJ). Despite extensive study, the machinery and mechanisms used to process these structures in eukaryotes have remained poorly understood. Recent work has identified XPG and UvrC/GIY domain-containing structure-specific endonucleases that can symmetrically cleave HJs in vitro in a manner that allows for religation without additional processing, properties that are reminiscent of the classical RuvC HJ resolvase in bacteria. Genetic studies reveal potential roles for these HJ resolvases in repair after DNA damage and during meiosis. The stage is now set for a more comprehensive understanding of the specific roles these enzymes play in the response of cells to DSBs, collapsed replication forks, telomere dysfunction, and meiotic recombination.
Keywords: Holliday junction resolvase, homologous recombination, meiosis, structure-specific endonuclease
Our genomes are constantly under attack by chemical and physical agents in the environment. These agents give rise to various types of damaged DNA, including interstrand and intrastrand DNA cross-links, double-strand breaks (DSBs), and photodimers. In addition, errors in DNA replication contribute to DNA damage in proliferative cell populations. Thus, organismal survival relies heavily on a robust and accurate collection of sensing and repair mechanisms that can respond to a plethora of DNA lesions (Harper and Elledge 2007). Prominent mechanisms include mismatch repair, nucleotide excision repair (NER), nonhomologous end-joining (NHEJ), and homologous recombination (HR), and genes that control these repair processes are often mutated in familial and sporadic cancer predisposition syndromes. In humans, mutations in genes involved in NER such as XPA lead to a genetic condition known as Xeroderma pigmentosum, a human disease characterized by ultraviolet (UV) sensitivity and a predisposition to cancer, particularly cancer of the skin (Hoeijmakers 2009). Likewise, a number of genes involved in HR, including BRCA2 and the BLM helicase, are frequently mutated in cancer (Sung and Klein 2006).
DNA repair often requires the breakage of DNA strands via structure-specific endonucleases (SSEs) (Ciccia et al. 2008). These enzymes recognize and cleave a variety of DNA structures that are formed as intermediates in the repair process. Typically, the products of the action of SSEs are rejoined to complete the repair process. Three major classes of SSEs have been identified in eukaryotes, and linked genetically and biochemically to specific repair pathways; the ERCC4 class typified by XPF and MUS81, the UvrC class typified by SLX1, and the XPG class typified by GEN1 (Ciccia et al. 2008; Klein and Symington 2009; Mimitou and Symington 2009b).
Much of our understanding of SSEs comes from the analysis of their roles in HR, which is used during meiotic recombination to exchange chromosomal information and ensure proper chromosomal segregation, and in mitotic cells to repair DSBs (Sung and Klein 2006; San Filippo et al. 2008). A driving concept in attempts to understand HR are Holliday junctions (HJs). HJs are four-stranded structures involving two homologous DNA duplexes that must be resolved into linear duplexes in order to complete meiosis I (Fig. 1; Duckett et al. 1988; Schwacha and Kleckner 1995). HJs are also featured prominently in the repair of DSBs in mitotic cells produced directly by either damage such as ionizing radiation (IR) or replication fork collapse (Krogh and Symington 2004; San Filippo et al. 2008).
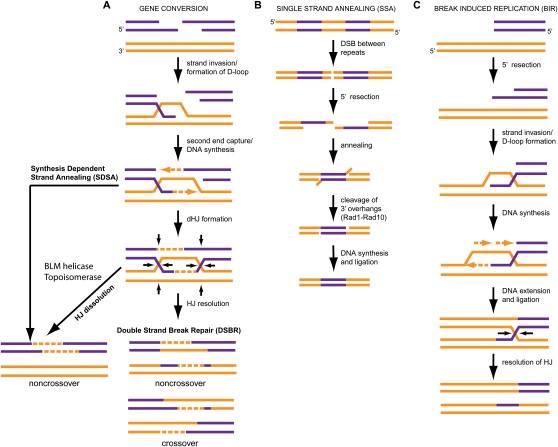
Figure 1.
Pathways for repairing DSBs via HR. (A) GC occurs at DSBs where both ends of the break are available for repair. It is subdivided into three pathways for repair: DSBR, SDSA, and dHJ dissolution. DSBR can generate either crossover or noncrossover products, while SDSA and HJ dissolution generate only noncrossover products. (B) SSA is the primary means of repair when DSBs are in highly repetitive regions. (C) BIR is employed when only one strand of a DSB is available for repair.
HJs were proposed initially by Robin Holliday in 1964 to explain meiotic gene conversion (GC) in fungi (Holliday 1964). Since that time, much has been learned about the structure and biology of HJs, and the enzymatic machinery responsible for their processing. A central question has concerned the mechanisms by which HJs are resolved. Early work in this area identified enzymes in phage and soon after in bacteria that could symmetrically cleave HJs in vitro to produce products that could be religated without further processing (West 1995). These “classical” HJ resolvases have served as a model for understanding HJ processing for the last two decades. However, the identification of the machinery involved in processing HJs in eukaryotes has been more challenging, and only recently have eukaryotic SSEs with properties analogous to “classical” HJ resolvases been identified (for excellent historical perspectives, see Liu and West 2004; West 2009). This is due to multiple factors, including genetic redundancy, the apparent absence of conserved pathways across eukaryotic organisms, and the fact that alternative mechanisms that are independent of SSEs are frequently used to repair DSBs in mitotic cells, thereby complicating the in vivo analysis of candidate SSEs. This review outlines early studies that led to the identification of the first HJ processing enzymes, and summarizes recent work that has culminated in the identification of two new classes of enzymes from multicellular eukaryotes that display properties of a classical HJ resolvase: the GEN1 resolvase (Ip et al. 2008), and the SLX1/SLX4 resolvase, a module within the larger SLX4–endonuclease complex (Andersen et al. 2009; Fekairi et al. 2009; Munoz et al. 2009; Saito et al. 2009; Svendsen et al. 2009). We highlight the relationships between the major SSEs in single and multicellular organisms, and describe the challenges ahead in understanding the context in which these and other SSEs function during meiotic recombination and repair of DSBs.
Homology-driven repair of DSBs
Before describing HJ processing enzymes, it is useful to consider the types of HR intermediates that exist in vivo. HR encompasses three major pathways for repair of DSBs: GC, single-strand annealing (SSA), and break-induced replication (BIR) (Fig. 1; Krogh and Symington 2004; Sung and Klein 2006; Lyndaker and Alani 2009; Pardo et al. 2009; Rouse 2009). The choice of pathways employed depends on the precise type of break. If both ends of the DSB are present, then the damage is repaired by GC using one of three mechanisms: DSB repair (DSBR) involving resolvase-dependent processing of the junction, double HJ (dHJ) dissolution involving helicase-dependent branch migration and topoisomerase-dependent decatenation of the junction, or synthesis-dependent strand annealing (SDSA), which does not involve dHJ structures (Fig. 1). The choice of pathways employed can lead to two major classes of products: crossovers, in which strands are exchanged relative to the initial chromosome organization, and noncrossovers, where no strand exchange occurs. Common steps are used to produce intermediates that are used in all three GC pathways in yeast (relevant vertebrate orthologs are in parentheses): (1) recognition of the DSB by the MRX(N) (Mre11–Rad50–Xrs2[NBS1]) complex; (2) 5′ end resection by the Sgs1(BLM)–Dna2 and the exonuclease Exo1(EXO1) pathways (Gravel et al. 2008; Mimitou and Symington 2008, 2009a; Zhu et al. 2008); (3) homology search and strand exchange, wherein Rad51 forms a nucleofilament on the 3′ ssDNA overhang and, along with Rad52/Rad54, promotes homologous strand invasion to form a D-loop structure (Pardo et al. 2009). In vertebrate cells, BRCA2 is functionally similar to Rad52 in that it interacts with RAD51, has a role in formation of RAD51 foci, and is required for RAD51-mediated HR (Pardo et al. 2009).
In the resolution and dissolution pathways, migration of the D-loop structure results in capture of the second end of the DSB, and ligation to form a dHJ structure (Fig. 1; Krogh and Symington 2004). dHJ dissolution is catalyzed by Sgs1/Top3 helicase–topoisomerase complex (BLM/TOP3a in mammals) and produces noncrossover products (Fig. 1; Ira et al. 2003; Wu and Hickson 2003; Krogh and Symington 2004; Mimitou and Symington 2008, 2009b). In contrast, cutting of dHJ structures produces crossover and/or noncrossover products, depending on the choice of DNA strands that are cut (Fig. 1). SDSA does not involve dHJ formation and results exclusively in noncrossover products (Fig. 1; Allers and Lichten 2001; Hollingsworth and Brill 2004; Krogh and Symington 2004). However, depending on precisely how much DNA is copied, flap endonucleases may be required to remove overhanging DNA sequences during SDSA.
If there is only one DSB end at the site of damage, then BIR is the primary means of HR (Fig. 1), and completion of BIR may also require HJ resolution (McEachern and Haber 2006; Llorente et al. 2008). Breaks that occur in highly repetitive regions such as rDNA loci can be repaired by SSA (Fig. 1), which involves flap endonuclease activity but does not involve a HJ-like intermediate.
The origins of HJ resolution activity
The first evidence of enzymatic machinery capable of resolving HJ structures came with the identification of the bacteriophage T4 endonuclease VII as an enzyme that could symmetrically cut joint plasmids to generate unbranched duplex products that could be religated directly (Mizuuchi et al. 1982). This was soon followed by the demonstration that extracts from yeast and Escherichia coli contain an analogous classical HJ resolving activity (Symington and Kolodner 1985; West and Korner 1985; Parsons and West 1988; Elborough and West 1990). This activity in E. coli is encoded by the RuvC gene (Connolly et al. 1991; Dunderdale et al. 1991; Iwasaki et al. 1991). RuvC acts on HJs as a dimer (Figs. 2A, 3A), and works in conjunction with a branch migration activity composed of the RuvA and RuvB gene products (West 1997; Davies and West 1998; van Gool et al. 1998). RuvA acts as a tetramer to unfold the arms of the HJ, forming a planar conformation (Bennett et al. 1993). The hexameric RuvB complex is brought to the junction via RuvA and employs its ATPase activity to promote branch migration (Yamada et al. 2004). RuvC dimers bind to the junction and cut symmetrically in a sequence-specific manner. Interestingly, the second RuvC cleavage reaction is enhanced compared with the first reaction, suggesting a mechanism by which RuvC may symmetrically cleave HJs that are actively being branch-migrated by RuvAB (Fogg and Lilley 2000). The initial activity observed in budding yeast extracts was ultimately demonstrated to be due to a mitochondrial enzyme, Cce1p, which shares 21% sequence identity with the orthologous enzyme Ydc2 in fission yeast (Kleff et al. 1992; Whitby and Dixon 1997; White and Lilley 1997). Casual inspection of Cce1/Ydc2 and RuvC resolvases reveals little in the way of sequence similarity. Remarkably, however, crystallographic studies of Ydc2 and RuvC reveal extensive structural similarity within major β-sheet and α-helical elements (Fig. 3A,B; Ceschini et al. 2001). Acidic residues implicated in binding to the catalytic Mg+2 ion are also conserved between Ydc2/Cce1 and RuvC (Fig. 3A,B; White and Lilley 1997).
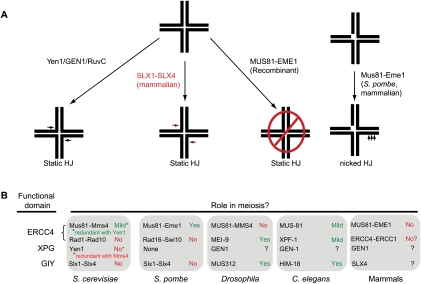
Figure 2.
In vitro and in vivo activities of SSE that process HJs. (A) GEN1/Yen1 and the mammalian SLX1–SLX4 module contain classical HJ resolvase activities and can cleave static HJs symmetrically like the classical HJ resolvase RuvC. Recombinant MUS81–EME1 does not cleave static HJs, but has robust nHJ cleavage activity. (B) During meiosis, HJs are processed to produce crossover products. In budding yeast, Mus81–Mms4 and Yen1 have redundant roles in meiosis, while in fission yeast, Mus81–Eme1 is required for almost all meiotic events. In Drosophila, the SLX4 ortholog MUS312 and the ERCC4/XPF ortholog MEI-9 work in concert to affect meiotic crossovers. In worms, the situation is more complex, with HIM-18, MUS-81, and XPF-1 responsible for subsets of meiotic events. In mammals, it is still unclear what SSEs are required during meiosis, although there is no obvious meiotic phenotype in mice lacking MUS81.
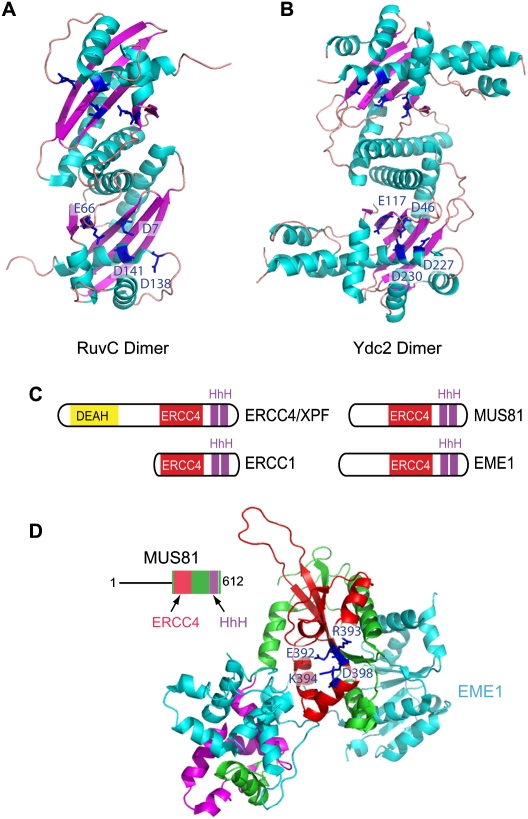
Figure 3.
Structural features of classical HJ resolvases and the ERCC4 family of SSEs. (A,B) Structure of dimeric E. coli RuvC (PDB: 1HJR) and S. pombe Ydc2 (PDB: 1KCF) HJ resolvases. The positions of critical acidic residues important for binding divalent metal ions are shown. (C) Domain structures of members of the ERCC4 family of SSEs. ERCC4/XPF contains an ERCC4 domain that is conserved in ERCC1, MUS81, and EME1. ERCC4/XPF also contains a DEAH helicase domain not found in ERCC1, MUS81, or EME1. The C-terminal regions of these proteins also contain conserved tandem HhH motifs. (D) Crystal structure of a MUS81–EME1 complex (PDB: 2ZIU), with the ERCC4 domain of the MUS81 (zebrafish) subunit shown in red, and the HhH motifs shown in magenta. The EME1 protein (human) is shown in cyan. Catalytic residues (see the text for details) are shown in blue.
The first evidence of HJ resolvase activity in mammalian cells came with the identification of activity in crude calf testes extracts (Elborough and West 1990), and later in extracts from tissue culture cells (Hyde et al. 1994). A feature of the early work on HJ branch migration and resolvase activity was the near universal use of complex plasmid-based substrates (Dunderdale et al. 1991), which greatly limited the ability to search for additional enzymes. The development of oligonucleotide-based HJ substrates made it possible to more systematically search for HJ resolvase activity in purified fractions, facilitating the first purification of mammalian classical HJ resolvase activity in 2001 (Elborough and West 1990). Interestingly, mammalian branch migration and resolvase activities comigrated through several chromatographic steps, and the ability of the resolvase to cleave depended on the branch migration activity (Elborough and West 1990). Thus, the activities of the mammalian enzyme and the bacterial RuvABC enzyme appeared to be highly parallel. Ultimately, this HJ resolvase activity in mammalian cells was referred to as ResA, but several years would elapse before the identity of the protein corresponding to ResA was identified (West 2009).
The MUS81/EME1 complex as a candidate HJ processing enzyme
The first candidate for nuclear HJ processing activity came with the identification of Mus81 and its heterodimeric noncatalytic partner, Eme1, in Schizosaccharomyces pombe and human cells (Boddy et al. 2001; Chen et al. 2001; Ciccia et al. 2003). S. pombe Mus81 was identified initially as a substrate of the Cds1 kinase (Boddy et al. 2000), the S. pombe ortholog of the CHK2 protein kinase in mammals, but genetic analysis revealed a requirement for Mus81 in meiosis, and led the way to its discovery as a HJ processing enzyme (Boddy et al. 2000). Mus81 and Eme1 are related to each other in that they both contain a domain referred to as an ERCC4 domain, named for its presence in the ERCC4/XPF flap endonuclease involved in NER (Fig. 3C), but the ERCC4 domain of Eme1 has diverged significantly from Mus81 and lacks key catalytic residues (Fig. 3D; Nishino et al. 2003; Ciccia et al. 2008). The critical motif in the catalytic center is referred to as the GDXnERKX3D motif (Fig. 3D), which employs its Asp and Glu residues to bind a catalytically important magnesium ion (Nishino et al. 2003; Chang et al. 2008). In addition to the ERCC4 domain, both Mus81 and Eme1 contain C-terminal helix–hairpin–helix (HhH) motifs (Fig. 3C,D), implicated in DNA binding (Ciccia et al. 2008). ERCC4/XPF forms a heterodimer with ERCC1, which, like Eme1, contains ERCC4 and HhH motifs but lacks catalytic activity (Fig. 3C; Ciccia et al. 2008).
S. pombe lacking mus81 or eme1 produce spores that are inviable (<1% survival) due to defects in chromosome segregation during meiosis I (Fig. 2B; Boddy et al. 2000, 2001; Osman et al. 2003). Crossovers are eightfold to 25-fold lower in a mus81 mutant background, while noncrossovers are unaffected (Boddy et al. 2001; Osman et al. 2003). Importantly, meiotic defects could be rescued by expression of a bacterial and phage-derived HJ resolvase called RusA that is unrelated to RuvC, or suppressed by deletion of rec12 (Spo11 ortholog), which promotes meiotic DSBs, suggesting that Mus81 functions downstream from DSB formation and may be involved in HJ processing (Mahdi et al. 1996; Boddy et al. 2001; Osman et al. 2003) . Evidence of a role for Mus81–Eme1 in DNA repair is suggested by sensitivities of fission yeast cells lacking mus81 to various types of DNA damage associated with replication fork progression, including methyl methanesulfonate (MMS), and agents such as camptothecin (CPT), which cause DSBs during replication (Boddy et al. 2000; Doe et al. 2002). Importantly, mus81 mutants are synthetically lethal, with mutations in S. pombe in the RecQ/BLM helicase rqh1, which has important roles in repair of damaged or stalled replication forks (Boddy et al. 2000; Doe et al. 2002).
In contrast to S. pombe, deletion of MUS81 in budding yeast results in only a modest decrease (40%) in meiotic survival, and this defect is not rescued by expression of RusA, suggesting that budding and fission yeast Mus81 proteins may have different functions and/or requirements in meiosis (Fig. 2B; de los Santos et al. 2003). Budding yeast Mus81 is only responsible for a small subset of crossovers during meiosis, and this is dependent on chromosome size (de los Santos et al. 2003). Moreover, mus81Δ cells have a decrease in dHJs, not an increase as one would expect (de los Santos et al. 2003). Recent work suggests that the role of Mus81 during meiotic recombination is to resolve aberrant joint molecules, and that the Sgs1 helicase works alongside Mus81 to ensure that aberrant joint molecules do not persist, suggesting a possible mechanism behind mus81sgs1 synthetic lethality (Jessop and Lichten 2008; Oh et al. 2008). The presence of meiotic crossovers in budding yeast lacking Mus81 appears to reflect a second pathway for resolving meiotic DSBs involving the Msh4–Msh5 complex (de los Santos et al. 2003; Whitby 2005). Fission yeast lacks obvious orthologs of the Msh4–Msh5 complex (Hollingsworth and Brill 2004). This, coupled with the fact that S. pombe also appears to lack an ortholog of the Yen1 HJ resolvase (see below), may explain why meiosis in fission yeast is so dependent on Mus81–Eme1.
In contrast to fungi, there is little evidence of a critical role for mammalian MUS81–EME1 in meiotic recombination or repair of DSBs produced by IR, or in meiotic crossovers in Drosophila (Fig. 2B; McPherson et al. 2004; Dendouga et al. 2005; Hanada et al. 2007; Trowbridge et al. 2007). Loss of EME1 in murine embryonic stem (ES) cells does not produce defects in GC or gene targeting, and EME1−/− ES cells as well as MUS81−/− ES cells are not sensitive to IR (Abraham et al. 2003; Hanada et al. 2007) . MUS81−/− mice are viable and fertile, with no obvious meiotic defects (Fig. 2B; McPherson et al. 2004; Dendouga et al. 2005). This is in contrast to ERCC1−/− mouse embryonic fibroblasts (MEFs), ES cells, and mice, which are sensitive to IR (Ahmad et al. 2008; Al-Minawi et al. 2008). While ERCC1−/− mice are infertile, it is unclear if this is linked to defects in meiotic crossovers or to persistent DNA damage generated by ERCC1 loss (Hsia et al. 2003). In Drosophila, mutations in MUS81 do not affect the extent of crossovers during meiosis, but, in contrast, mutations in MEI-9, the Drosophila XPF ortholog, have severe defects (Fig. 2B; Yildiz et al. 2002; Trowbridge et al. 2007). Thus, Mus81 appears to be more important for meiosis in S. pombe and, to some extent, Saccharomyces cerevisiae than in the multicellular organisms tested. As in S. pombe, budding yeast lacking Mus81 display sensitivity to DNA-damaging agents that produce stalled or collapsed replication forks (Liu et al. 2002; Bastin-Shanower et al. 2003). So it is somewhat surprising that mammalian cells lacking MUS81 have distinct damage sensitivities when compared with yeast, in that MUS81−/− mouse cells are highly sensitive to only DNA cross-linking agents (Abraham et al. 2003; McPherson et al. 2004; Dendouga et al. 2005; Hanada et al. 2007).
Mus81–Eme1 lacks characteristics of a classical HJ resolvase in vitro
Initial experiments examining the activity of S. pombe and human Mus81–Eme1 complexes purified from in vivo sources on model HJ substrates revealed an ability to cleave mobile HJs, but in a manner that is distinct from that of a classical HJ resolvase, such as ResA or RuvC. While the products were linear duplexes, they were not fully symmetrical. Further analysis of the activity and substrate specificity of Mus81–Eme1 from multiple organisms raised further questions concerning whether Mus81–Eme1 acted as a classical HJ resolvase. First, human MUS81–EME1 purified from bacteria was devoid of activity toward static HJs in vitro (Ciccia et al. 2003). Second, MUS81–EME1 displayed robust activity toward replication forks and 3′ flap substrates, raising the question of whether this enzyme might preferentially function as a flap endonuclease (Bastin-Shanower et al. 2003; Ciccia et al. 2003).
A potential resolution to the question of whether MUS81–EME1 functions as a HJ resolvase as opposed to a HJ processing enzyme or flap endonuclease came with the discovery that MUS81–EME1 had potent activity toward nicked HJs (nHJs) (Hollingsworth and Brill 2004). In examining how Mus81–Eme1 promotes crossover products in S. pombe (Fig. 2A), Osman et al. (2003) showed that the enzyme preferentially cleaves D-loop structures that are the precursor to dHJs. Moreover, cleavage of the D-loop occurs on the complimentary strand 3–8 nucleotides (nt) 5′ to the junction, which is the same site specificity observed with replication fork and 3′ flap substrates (Constantinou et al. 2002; Bastin-Shanower et al. 2003; Gaillard et al. 2003; Osman et al. 2003; Fricke et al. 2005; Taylor and McGowan 2008). Second end capture then generates a nHJ that is resolved by Mus81–Eme1 (Fig. 2A; Osman et al. 2003). Cleavage of nHJs by Mus81–Eme1 was 3–6 nt 5′ to the junction on the strand opposite the nicked strand, and, for budding yeast, Mus81–Mms4 cleavage was 4–9 base pairs (bp) 5′ to the junction on the opposite strand, similar to published results for Mus81–Eme1/Mms4 (Gaillard et al. 2003; Osman et al. 2003; Fricke et al. 2005; Taylor and McGowan 2008). Junctions with flap or nick positions both 3′ and 5′ to the branchpoint were resolved by Mus81–Eme1, indicating that this enzyme is capable of acting on a variety of intermediates that may be formed during HJ resolution, as opposed to the fully formed HJ itself (Osman et al. 2003). The discovery of this pathway suggested the likely existence of an additional mechanism by which HJ intermediates could be processed during meiosis. This discovery came at a time when it was presumed that dHJs were the only intermediates resolved during meiosis (Whitby 2005).
Revealing the first classical nuclear eukaryotic HJ resolvases
As MUS81–EME1 clearly did not represent the activity first identified as ResA in mammalian cells (Constantinou et al. 2002; Ciccia et al. 2003), it was evident that classical HJ resolvases awaited discovery. In 2004, Liu et al. (2004) described the purification of a HJ resolvase and branch migration activity from human cells that contained Rad51C and XRCC3, but the specificity was distinct from that of MUS81–EME1. However, several lines of evidence suggested that resolution activity may not be inherent to RAD51C or XRCC3, including the absence of an identifiable nuclease domain in either protein and the absence of activity of recombinant XRCC3–RAD51C complexes (Sharan and Kuznetsov 2007). The identification of the first bona fide classical HJ resolvases functioning in the nucleus of eukaryotic cells came from two parallel and complementary studies performed in budding yeast or extracts from human tissue culture cells (Ip et al. 2008). Both efforts resulted in the identification of orthologous proteins—GEN1 in mammals, and Yen1 in budding yeast—that have a HJ resolvase activity indistinguishable from the ResA activity seen previously in extracts from mammalian cells (Fig. 2A; Ip et al. 2008). In one approach, ResA activity was followed using a synthetic mobile HJ substrate over several chromatographic fractionation steps, ultimately resulting in the identification of the GEN1 protein (Ip et al. 2008). This discovery is remarkable not only because GEN1 is of exceedingly low abundance, but because GEN1 maintains activity even after extraction from SDS-PAGE, which turned out to be a key step in its identification. An alternative approach was employed to identify yeast Yen1 as a HJ resolvase. Taking advantage of a collection of epitope-tagged yeast ORFs, Ip et al. (2008) tested 1100 (of ∼6400) yeast proteins for in vitro HJ cleavage activity, with the collection enriched in low-abundance proteins, ultimately identifying known HJ processing enzymes Mus81–Mms4 and Cce1, as well as the GEN1 ortholog Yen1.
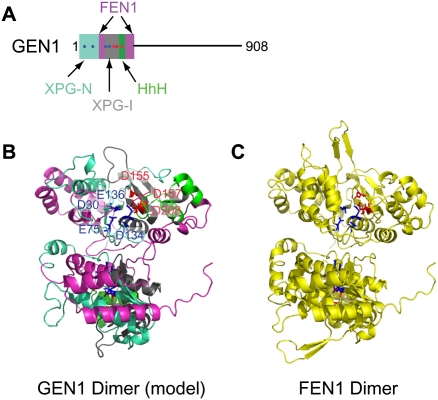
A key to the identification of GEN1 and Yen1 was their homology with proteins of the XPG family (Ip et al. 2008), marking these proteins as potential endonucleases. The XPG gene in mammalian cells encodes a component of a NER complex, and functions to cleave DNA at the single-strand–double-strand junction 3′ to the damaged nucleotide (Scharer 2008). Several other proteins, including the FEN1 flap endonuclease, display homology with the catalytic domain of XPG. Crystallographic analysis of FEN1 (Hosfield et al. 1998) indicates that it forms a homodimer with the catalytic core composed of two clusters of acidic residues that function to bind two magnesium ions important for DNA cleavage (Fig. 4C). The first acidic cluster is formed by four acidic residues embedded within XPG-N and XPG-I motifs, representing N-terminal (N) and internal (I) regions, respectively (Fig. 4C). The second cluster consists of three residues provided by the XPG-I motif and the adjacent HhH-like motif (Fig. 4A,C), analogous to that found in other proteins that interact with DNA, including MUS81 and ERCC4 (Fig. 3C,D). Mutagenesis studies indicate that the deep groove formed by the concave surface of the XPG homology region functions to bind DNA, placing the scissile bond near the catalytic center formed by the acidic clusters (Hosfield et al. 1998). Modeling studies reveal that GEN1 (Fig. 4A,B) and Yen1 (data not shown) are likely to adopt a structure similar to FEN1, and all seven residues that make up the two acidic clusters are conserved in GEN1 and Yen1 (Fig. 4B). Consistent with this, mutation of aspartate or glutamate residues in either acidic cluster in GEN1 abolishes HJ resolvase activity (Ip et al. 2008). Overall, Yen1 and GEN1 display 13% identity (54% similarity) across their length. The central and C-terminal regions of GEN1 and Yen1 lack known protein interaction domains (Fig. 4A), but the extent of conservation is similar to the catalytic domain over these regions, suggesting a conserved function for the central and C-terminal regions of GEN1/Yen1. One speculative hypothesis is that this region interacts with branch migration activity noted previously during the purification of ResA activity from mammalian cells (Constantinou et al. 2001).
Figure 4.
Structural analysis of XPG endonucleases. (A) GEN1 domain structure showing elements within the N-terminal XPG homology region. (B,C) Model of the GEN1 catalytic domain (B) based on the structure of FEN1 (C; PDB: 1B43), a flap endonuclease with an XPG-based catalytic site. The GEN1 model was generated using SWISS-MODEL. Color-coding of motifs within GEN1 is as diagrammed in A. Residues responsible for coordination of the primary and secondary catalytic magnesium ions are in blue and red, respectively.
The substrate specificity of GEN1 and Yen1 has been examined using model DNA substrates in vitro. Both GEN1 and Yen1 symmetrically cleave static HJs 1 nt 3′ to the junction (Fig. 2A), a pattern identical to that observed with ResA (Ip et al. 2008). These products are readily religated, consistent with the identification of GEN1 and Yen1 as classical HJ resolvases. While GEN1 displays the highest specific activity toward HJs, it also is capable of cleaving replication fork and 5′ flap substrates with rates that are 20-fold and sevenfold lower than with HJs, but does not detectably cleave splayed arm or 3′ flap substrates (Ip et al. 2008).
While our understanding of the XPG class of HJ resolvases from a biochemical perspective is well developed, their roles in vivo are not fully understood. In budding yeast, yen1Δ mutants are viable and lack an obvious meiotic phenotype. However, combining yen1Δ and mms4Δ mutants blocks completion of meiosis and results in a failure to resolve meiotic joint molecules, suggesting redundancy between the Mus81–Mms4 endonuclease and the Yen1 HJ resolvase (Ip et al. 2008). Surprisingly, S. pombe lacks an obvious Yen1 ortholog, thereby explaining the strict requirement of Mus81–Eme1 for meiosis in this organism (Ip et al. 2008). Nevertheless, ectopic expression of GEN1 rescues the meiotic phenotype of S. pombe mus81Δ mutants, suggesting that this enzyme can function in HJ resolution in vivo (Lorenz et al. 2009). A definitive analysis of the role of GEN1 in meiotic recombination or DNA repair in multicellular eukaryotes has not been reported. Preliminary RNAi experiments in human tissue culture cells suggest that depletion of GEN1 leads to a mild sensitivity to DNA damage by MMC and CPT (Svendsen et al. 2009). However, further studies using true null alleles are required to fully evaluate the contribution of GEN1 to mitotic DNA repair and meiotic recombination. Additional work also is required to evaluate GEN1’s contribution to cancer. In this regard, two GEN1 mutants that would be predicted to lack HJ resolvase activity have been identified in a cohort of breast cancer patients, suggesting the possibility that loss of GEN1 may contribute to recombination repair defects that are associated with tumorigenesis (Ip et al. 2008).
The mammalian SLX4 complex—a toolkit for DNA repair
Recent work employing multiple experimental strategies and organisms has led to the discovery of a new conserved HJ resolvase in multicellular eukaryotes centered on the SLX4 protein. Based on available data, SLX4 serves a scaffolding function by assembling multiple SSEs and other DNA repair and telomere protection proteins in a unique protein complex that may participate in the repair of DSBs in mitotic and meiotic cells (Andersen et al. 2009; Fekairi et al. 2009; Munoz et al. 2009; Saito et al. 2009; Svendsen et al. 2009).
Yeast Slx4 was identified initially in genetic screens for proteins that are required for viability in the absence of the Sgs1 helicase, an ortholog of the Bloom's helicase (Mullen et al. 2001). The same screen identified Slx1, and biochemical studies revealed that Slx4 and Slx1 interact to form a flap endonuclease in budding yeast and in fission yeast (Fig. 5B; Mullen et al. 2001; Fricke and Brill 2003; Coulon et al. 2004). Slx1–Slx4 is required for S-phase-dependent recombination at rDNA loci in both S. cerevisiae and S. pombe, but slx1Δ single mutants do not have prominent DNA damage sensitivity phenotypes (Fig. 5D; Kaliraman and Brill 2002; Coulon et al. 2004, 2006; Deng et al. 2005; Flott and Rouse 2005). Interestingly, Slx4 has been shown to form an independent complex with Rad1–Rad10, the yeast orthologs of XPF–ERCC1 (Fig. 5B; Flott et al. 2007). The Rad1–Rad10–Slx4 complex is required for SSA (Figs. 1B, 5D; Flott et al. 2007; Li et al. 2008). In addition, slx4Δ mutants (but not slx1Δ mutants) display sensitivity to CPT in the context of a mutation in the TPD1 gene, which encodes a tyrosine phosphodiesterase that reverses CPT–topoisomerase I adducts (Deng et al. 2005). Rad1–Rad10 also plays a role in the repair of CPT-dependent DNA damage, but this role appears to be independent of Slx4 (Deng et al. 2005). The molecular basis for the distinct sensitivities of slx4 and rad1 mutants is unknown.
Figure 5.
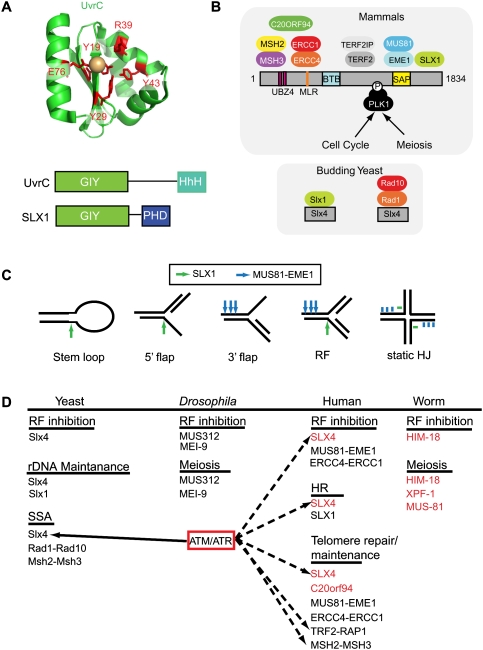
Versatility of the SLX4 complex. (A) The NER endonuclease UvrC as well as SLX1 orthologs contain a GIY motif, also referred to as an Uri domain. The structure of the UvrC domain is shown along with conserved residues (red) involved in binding to a hydrated divalent metal ion (magnesium or manganese, beige sphere). While UvrC has a C-terminal HhH motif, SLX1 contains a PHD-like ring finger motif. (B) Structural anatomy of the SLX4 complex in mammals and its relationship with SLX4 complexes in budding yeast. (C) In vitro cleavage specificity of the SLX4 complex. Bars and arrows represent regions of cleavage. (D) Functions of the yeast, Drosophila, C. elegans, and human SLX4 complexes in vivo.
Slx1 is a member of the UvrC family of endonucleases identified initially in prokaryotes (Fig. 5A). UvrC functions together with UvrAB in NER, and employs an N-terminal “GIY” domain (also called a Uri domain) to make an incision 3–4 nt 3′ to the damage site, a reaction that is analogous to that performed by XPG in mammalian NER. The GIY domain contains several highly conserved residues, including a conserved glutamate (E76 in UvrC from Thermatoga maritima), which is critical for binding the active site divalent cation (magnesium or manganese); three tyrosine or histidine residues (Y19, Y29, and Y43 in UvrC) that form hydrogen bonds with water molecules coordinated to the metal ion; and a fully conserved arginine residue (R39 in UvrC) that may stabilize the negative charge on the phosphate group at the scissile bond (Fig. 5A; Truglio et al. 2005). Unlike UvrC, Slx1 orthologs also contain a C-terminal cysteine-rich domain akin to zinc-binding PHD domains (Fig. 5A; Fricke and Brill 2003; Coulon et al. 2004). Budding yeast Slx1–Slx4 purified from bacteria displays robust endonuclease activity toward 5′ flap structures, splayed arm, and replication fork substrates, but displays much lower levels of activity toward static HJs (Fricke and Brill 2003). Moreover, processing of HJs by both budding and fission yeast Slx1–Slx4 leads to products that cannot be ligated efficiently (Fricke and Brill 2003; Coulon et al. 2004). The activity of S. pombe Slx1–Slx4 in vitro is abolished by mutations in conserved glutamate or arginine residues in the GIY domain (Fig. 5A), and mutation of a conserved cysteine in the C-terminal PHD-like domain of S. cerevisiae Slx1 leads to functional inactivation in vivo (Fricke and Brill 2003; Coulon et al. 2004). Moreover, yeast Slx1 alone is inactive, indicating a requirement for Slx4 in supporting endonucleolytic activity, akin to the requirement of Mus81 for Eme1 and ERCC4 for ERCC1 (Fricke and Brill 2003; Coulon et al. 2004).
While clear orthologs of yeast Slx1 are evident in multicellular eukaryotes, early attempts to identify Slx4 orthologs using conventional sequence comparisons were unsuccessful (Fricke and Brill 2003). Fungal Slx4 proteins contain C-terminal SAP and helix–turn–helix (HtH) domains implicated previously in DNA binding (Coulon et al. 2004). Using more sophisticated PSI-BLAST algorithms, three groups identified the largely unstudied BTBD12 protein as the human ortholog of fungal Slx4 and of the MUS312 protein in Drosophila (Andersen et al. 2009; Fekairi et al. 2009; Munoz et al. 2009). BTBD12 was identified independently as a substrate of the ATM/ATR protein kinase (Matsuoka et al. 2007), and subsequently as a protein that associates with multiple SSEs, including SLX1, using a proteomic approach (Svendsen et al. 2009). Finally, a screen for genes enriched in the germline and required for meiosis in Caenorhabditis elegans identified HIM-18, which displays sequence identity with both fungal Slx4 and mammalian BTBD12 (Saito et al. 2009). Thus, a variety of data indicate that BTBD12 encodes the SLX4 ortholog in multicellular eukaryotes.
Unlike fungal Slx4, which interacts independently with Slx1 and with the Rad1–Rad10 complex, a variety of proteomic and interaction data indicate that mammalian SLX4 interacts with three endonucleases (SLX1, ERCC4[XPF]–ERCC1, and MUS81–EME1); components of the shelterin telomere protection complex (TRF2/RAP1); the MSH2–MSH3 mismatch repair complex; the polo-like kinase PLK1; and a previously unstudied ORF, C20ORF94 (Fig. 5B; Fekairi et al. 2009; Munoz et al. 2009; Svendsen et al. 2009). The various interaction partners are assembled across the length of SLX4, consistent with a scaffolding function (Fig. 5B). SLX1 interacts with sequences at the extreme C terminus of SLX4, which may form an HtH motif (Fekairi et al. 2009; Munoz et al. 2009; Svendsen et al. 2009). This region, and the adjacent SAP domain that may interact with MUS81–EME1, contains the highest degree of sequence similarity across SLX4 orthologs from fungi to mammals (Andersen et al. 2009; Fekairi et al. 2009; Munoz et al. 2009; Svendsen et al. 2009). In contrast to these endonucleases, ERCC4–ERCC1 as well as MSH2–MSH3 and C20ORF94 interact with the N-terminal region of SLX4 (Andersen et al. 2009; Fekairi et al. 2009; Svendsen et al. 2009). Both TRF2–RAP1 and PLK1 associate with the central region of SLX4 between the BTB domain (protein–protein dimerization) and the SAP domain (Svendsen et al. 2009). The interaction of PLK1 with SLX4 appears to require, in part, a candidate Polo-box-interacting sequence (Svendsen et al. 2009). Although not studied in a systematic way, directed yeast two-hybrid studies indicate that Drosophila MUS312 and C. elegans HIM-18 interact with the XPF and SLX1 orthologs (Yildiz et al. 2002; Andersen et al. 2009; Saito et al. 2009). The sequence in MUS312 known to associate with MEI9(XPF)—the MUS312–Mei-9-interacting-like region (MLR)—is conserved among SLX4 orthologs and may serve generally as the binding site for ERCC4, although this has yet to be tested in the vertebrate system (Fig. 5B; Fekairi et al. 2009). However, there are several differences in motifs among the various SLX4 orthologs that are not understood fully. While vertebrate and C. elegans SLX4 orthologs contain recognizable BTB and UBZ4 (ubiquitin-binding zinc finger motif 4) domains, Drosophila and fungal proteins lack such motifs (Fekairi et al. 2009; Munoz et al. 2009; Saito et al. 2009). The BTB domain is known to serve a dimerization function in other contexts (Stogios et al. 2005), but this has not been tested directly in the case of SLX4. Furthermore, zinc-binding UBZ4 motifs are best understood in the context of ubiquitin binding (Bienko et al. 2005; Crosetto et al. 2008), but thus far there is no evidence that SLX4 associates covalently or noncovalently with ubiquitin. Importantly, SLX4 complexes are fully assembled in proliferating cells, and no obvious changes in the composition of the complex is seen upon addition of DNA-damaging agents (Svendsen et al. 2009). Given that SLX4 is phosphorylated by ATM/ATR in response to DNA damage, it may be regulated at a level that is independent of subunit assembly.
SLX1–SLX4 module—a second HJ resolvase in multicellular eukaryotes
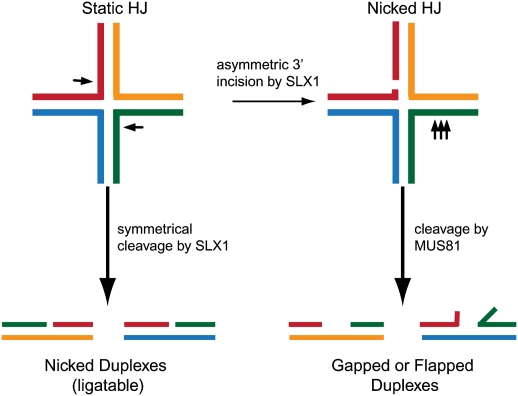
Given the presence of multiple SSEs in association with SLX4, and the previous finding that Slx1–Slx4 complexes from fungi display cleavage activity toward multiple substrates, it is not surprising that SLX4 complexes purified from mammalian cells display activity toward multiple substrates (Fricke and Brill 2003; Coulon et al. 2004; Fekairi et al. 2009; Munoz et al. 2009; Svendsen et al. 2009). However, the activity profile displayed by mammalian SLX4 complexes is unique among previously characterized endonucleases in that SLX4 complexes contain several endonucleases with their representative activities, while ResA (GEN1) complexes contain one endonuclease and its related activity. SLX4 complexes can cleave replication fork and 5′ and 3′ flaps, as well as both static and mobile HJs (Fig. 5C; Fekairi et al. 2009; Munoz et al. 2009; Svendsen et al. 2009). The ability to cleave replication fork and 3′ flap structures appears to reside in the MUS81–EME1–SLX4 module, and the specificity of cleavage is essentially identical to that defined previously for fungal Mus81–Eme1 (Fig. 5C; Bastin-Shanower et al. 2003; Munoz et al. 2009; Svendsen et al. 2009). The ability to cleave HJs resides in the SLX1–SLX4 module (Fekairi et al. 2009; Svendsen et al. 2009). The SLX1–SLX4 module purified from bacteria cleaves static HJs symmetrically 2 nt 3′ to the branchpoint (Figs. 2A, 5C), and this activity requires an intact cation-binding site in the SLX1 catalytic center (Fekairi et al. 2009; Svendsen et al. 2009). The site of cleavage is distinct from GEN1/Yen1, which cleaves 1 nt 3′ to the branchpoint (Ip et al. 2008). In addition, the SLX1–SLX4 module also cleaves mobile HJs, giving rise to duplex products that can be ligated, marking the mammalian SLX1–SLX4 module as a HJ resolvase (Fekairi et al. 2009; Munoz et al. 2009; Svendsen et al. 2009). This activity of mammalian SLX1–SLX4 is somewhat surprising, given that the fungal enzymes do not display this specificity, as purified from S. pombe or bacteria (Fricke and Brill 2003; Coulon et al. 2004). Currently, it is unknown whether the differences in activity seen between the fungal and mammalian enzymes with in vitro substrates represents an intrinsic difference in their catalytic specificities, or differences in the methodology employed for in vitro assay. Moreover, the specificities of Drosophila and C. elegans SLX4 complexes remain to be determined.
When fully intact SLX4 complexes from mammalian cells are exposed to static HJs, a complex series of products are generated that appear to reflect the combined action of SLX1 to cleave 2 nt 3′ to the branchpoint, and MUS81–EME1 to cleave 3–5 nt 5′ to the branchpoint (Figs. 2A, 5C; Svendsen et al. 2009). The products of dual cleavage by SLX1 and MUS81 cannot be ligated without further processing by DNA polymerases, or SSEs on flapped substrates. The simplest explanation for this cleavage pattern is that SLX1 in the context of the MUS81–EME1–SLX4 complex cuts asymmetrically with some frequency, thereby generating a nHJ, the preferred substrate for MUS81–EME1 (Fig. 6). What determines whether the SLX1–SLX4 module cuts symmetrically across the junction is unknown. When SLX1 and MUS81 are not tethered to each other via SLX4, symmetrical cleavage products predominate, suggesting that MUS81 and SLX1 need to be tethered to produce asymmetric cleavage products (Svendsen et al. 2009). An interesting question for the future concerns whether SLX1 and MUS81-dependent activities work independently or in tandem to create distinct types of cleavage intermediates in vivo. One possibility is that, during meiosis, SLX1 makes the initial cut of the first HJ formed after strand invasion, and that MUS81 makes the second cut of the nHJ generated by second end capture (Fig. 7A; Osman et al. 2003). This pathway is advantageous because it generates crossovers, while DSBR can generate crossover and noncrossover products depending on how the dHJ is resolved (Fig. 7B). In addition, SLX4 is required for the activity of SLX1 and has been suggested to enhance the activity of MUS81–EME1 complexes, but the mechanism by which this occurs is unknown (Munoz et al. 2009). The SLX4 protein may contribute structural elements that bind and position the DNA substrate for cleavage by the catalytic SLX1 subunit, much like the Eme1 subunit helps bind and orient DNA substrates on the associated Mus81 endonuclease (Chang et al. 2008). Alternatively, SLX4 may act in an allosteric manner to control SLX1 activity. Finally, given that SLX1 and MUS81 catalytic subunits can be bridged by SLX4, it is conceivable that the previous controversy concerning whether human MUS81–EME1 acts as a HJ resolvase (Constantinou et al. 2002) could have been influenced by small or variable amounts of contaminating SLX1–SLX4 activity.
Figure 6.
Model depicting distinct roles for SLX1 and MUS81–EME1 within the SLX4 complex. SLX4 complexes contain two HJ processing activities. SLX1–SLX4 behaves as a classical HJ resolvase and symmetrically cleaves HJs to generate nicked ligatable DNA duplexes in vitro. Alternatively, SLX1 may cleave HJs asymmetrically to generate a nHJ that is a suitable substrate for MUS81–EME1. This results in flapped and gapped DNA duplex products that cannot be ligated without further processing.
Figure 7.
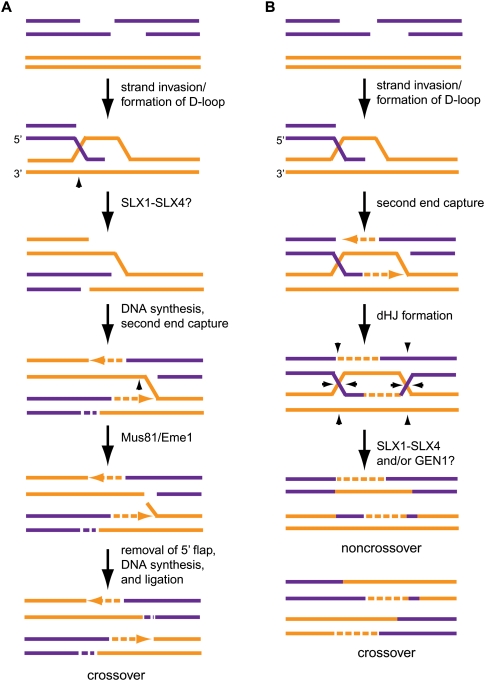
Potential roles for the SLX4 complex during DSBR. (A) In an alternate model for DSBR initially proposed by Osman et al. (2003). SLX1–SLX4 may cleave the HJ-like intermediate formed by strand exchange, and MUS81–EME1 may work downstream to cleave the nHJ generated by second end capture. This mechanism would produce crossover products that are desirable during meiosis. (B) Alternatively, SLX1–SLX4 and/or GEN1 complexes may cleave dHJs during traditional DSBR to generate crossover and/or noncrossover products. Arrowheads represent possible sites of cleavage by SSEs.
Interestingly, the activity of a fragment of SLX4 capable of interacting with MUS81–EME1 and SLX1 but not ERCC4–ERCC1 produces reaction products with HJs, replication forks, and 3′ flaps that are indistinguishable from the full-length SLX4 protein, suggesting that ERCC4–ERCC1 does not play a direct role in cleavage of these substrates in vitro (Svendsen et al. 2009). This is in keeping with the known activity of ERCC4–ERCC1 in cleaving Y structures that mimic intermediates in NER (Ciccia et al. 2008). As described below, cells depleted of SLX4 do not display defects in UV-dependent DNA damage, indicating the absence of an obvious role for SLX4 in NER (Fekairi et al. 2009; Svendsen et al. 2009). Thus, the roles of ERCC4–ERCC1 in the context of the SLX4 complex remain to be determined. However, it seems likely that there will be a role for the ERCC4–ERCC1–SLX4 module in SSA, given the role for the analogous complex in SSA in budding yeast (Li et al. 2008).
SLX4 and the response to DNA damage
The finding that SLX4 is targeted by the ATM/ATR protein kinases (Fig. 5D; Matsuoka et al. 2007) suggests roles in the response to DNA damage and repair. Consistent with this, SLX4 is recruited to sites of DNA damage (Svendsen et al. 2009). The precise signals required for SLX4 localization are unknown. One possibility is that DNA damage-dependent phosphorylation is involved. While SLX4 localizes properly in cells that are mutated for ATM, it has not been determined whether SLX4 localization requires ATR or ATM and ATR in a redundant fashion (Svendsen et al. 2009). Thus, a major question for the future concerns the pathway by which the SLX4 complex is localized to sites of damage. A number of mechanisms are used to mark the sites of DSBs in eukaryotic cells, including phosphorylation, ubiquitination, and sumoylation (Harper and Elledge 2007; Boulton 2009), and each of these signals will need to be interrogated for their involvement in SLX4 recruitment.
Much of our understanding of the role of SLX4 in DNA repair comes from an examination of its role in DNA damage at the replication fork. Early studies indicated that yeast slx4Δ and Drosophila mus312 mutants have increased sensitivity to agents that generate collapsed replication forks, including mitomycin C (MMC) and CPT (Fig. 5D; Yildiz et al. 2002; Deng et al. 2005; Flott and Rouse 2005; Roberts et al. 2006; Flott et al. 2007). Repair of interstrand cross-links generates a DSB as a result of incision on either side of the cross-link, generating a product that must be repaired by HR. CPT leads to the formation of DSBs as the replication fork attempts to pass through a Topoisomerase-1–DNA adduct. Recent work has likewise suggested a role for human SLX4 in repair of damage linked to these types of replication fork abnormalities (Fig. 5D). HeLa, U2OS, or HEK293 cells depleted of SLX4 by RNAi are highly sensitive to cisplatin, MMC, and CPT, and also display sensitivity to MMS, as determined using clonogenic survival or cellular proliferation competition assays (Fekairi et al. 2009; Munoz et al. 2009; Svendsen et al. 2009). Similarly, cells in him-18 embryos are exquisitely sensitive to agents that affect replication fork progression (Saito et al. 2009). In contrast, cells depleted of SLX4 are not sensitive to UV, suggesting the absence of an important role in NER, and consistent with the absence of a role for MUS312 in NER (Fekairi et al. 2009; Svendsen et al. 2009). This is despite the finding that depletion of ERCC4/XPF in parallel experiments leads to major defects in the response to UV, suggesting that ERCC4–ERCC1 functions in NER in a pathway that is independent of its interaction with SLX4 (Svendsen et al. 2009). A major question concerns whether the SLX4 complex is important for repair of DSBs produced outside of DNA replication. While one study has suggested sensitivity of SLX4-depleted cells to IR in HEK293 cells, a second study in U2OS cells indicates little sensitivity to IR under conditions where sensitivity is seen with CPT and MMC, although some sensitivity to IR was seen upon further depletion of SLX4 (Munoz et al. 2009; Svendsen et al. 2009). The impact of depletion of other components of the complex including SLX1 is comparable with that seen with SLX4 in HEK293 cells, but less so in the context of U2OS cells (Munoz et al. 2009; Svendsen et al. 2009). A more complete understanding of the contributions of the mammalian SLX4 complex to DNA repair will likely require analysis of cells genetically engineered to lack these proteins, as the extent of depletion in the various cell lines examined may be responsible for differences in sensitivities seen in various studies. For example, >90% depletion of MUS81, SLX1, or GEN1 has little impact on the sensitivity of cells to multiple types of damaging agents, yet further depletion produces detectable phenotypes (Svendsen et al. 2009). Thus, it is possible that even small amounts of these enzymes are sufficient to catalyze repair, making a definitive analysis by RNAi problematic.
Many DNA damage response components are required not only for repair of exogenously produced damage, but also control the repair of endogenous DNA damage, as can occur during replication. Depletion of SLX4 leads to increased levels of H2AX and 53BP1 phosphorylation, which is indicative of a role in controlling spontaneous genomic instability (Fekairi et al. 2009; Munoz et al. 2009). An inability to repair collapsed replication forks when the SLX4 and SLX1 proteins are absent may underlie activation of the DNA damage signaling system.
A role for SLX4 orthologs during HR in vivo
A central question that is yet to be fully resolved concerns the key HJ resolution activities that contribute to repair of DSBs in both mitotic and meiotic cells. As noted above, S. pombe appears to rely predominantly on Mus81–Eme1, while S. cerevisiae may employ Mus81–Mms4 and Yen1 redundantly, and mammalian cells do not require MUS81–EME1 for meiosis. Ultimately, it will be important to understand not only the identity of the key enzymes involved in individual steps, but also how the pathways for resolution differ with distinct types of DNA damage or meiotic intermediates. Additionally, it will be important to understand the similarities and differences between pathways used to recombine meiotic versus mitotic DSBs. Analysis of HR in mammalian tissue culture cells frequently employs a system wherein restriction enzyme (I-Sce1)-dependent DSB formation leads to repair of a GFP ORF by GC (Xia et al. 2006). Depletion of SLX4 or SLX1 by RNAi resulted in HR defects in this system, although the extent of the defect seen is not as large as is observed with depletion of core components of the repair pathway (XRCC3 and ATR) (Munoz et al. 2009; Svendsen et al. 2009). It is believed that repair in this system reflects primarily SDSA, which, as described above (Fig. 1), does not involve a HJ intermediate. Thus, the available data suggest two possibilities that are not mutually exclusive: Either the extent of repair via SDSA in this system is smaller than thought previously, or the role for SLX4 in this system reflects flap endonuclease activity. For example, removal of the nonhomologous sequences introduced by the I-Sce1 restriction site in the recombination substrate is expected to be required for repair of the DSB by SDSA. Interestingly, work in yeast has found that both the Rad1–Rad10 endonuclease as well as Msh2–Msh3 complex are required to remove nonhomologous regions at DSBs during HR (Paques and Haber 1997; Sugawara et al. 1997; Flott et al. 2007; Li et al. 2008). It is conceivable that the MSH2–MSH3 and ERCC4–ERCC1 modules localized at the N terminus of SLX4 are involved in this process in mammalian cells. A clearer understanding of the role of mammalian SLX4 and SLX1 in mitotic and meiotic HR and HJ resolution in mammals will require analysis of null alleles, as it is possible that incomplete target protein depletion by RNAi does not reveal the full functional potential of SLX1 and SLX4 in DSB repair. It will also be important to understand the extent to which mammalian SLX1–SLX4 and GEN1 display functional overlap in DSB repair in vivo.
In contrast to the mammalian system, there is already strong genetic evidence of a role for Drosophila and C. elegans SLX4 orthologs in HR in vivo. The Drosophila SLX4 ortholog MUS312 is required for formation of meiotic crossovers, and also for repair of interstrand DNA cross-links, a process that requires HR (Fig. 2B; Yildiz et al. 2002). In C. elegans, levels of the SLX4 ortholog HIM-18 are temporally and spatially regulated, and are highest in cells that are engaged in the late stages of HR (premeiotic and late pachytene germline cells) (Saito et al. 2009). him-18 embryos are largely inviable, and of those that survive, the rate of male progeny is 60-fold higher than wild type, suggesting that him-18 gametes have an increased occurrence of X-chromosome nondisjunction reflective of defects in HR (Fig. 2B; Saito et al. 2009). In him-18 meiotic cells, the early steps of DSB repair proceed normally, yet RAD51 foci accumulate, indicative of ongoing defects in DNA repair. HIM-18 functions downstream from both meiotic DSB formation by SPO-11 and strand exchange by MSH-5 (Saito et al. 2009). The accumulation of bivalents suggests that HIM-18 functions after dHJ formation, but before or concurrently with dHJ resolution. However, many of the bivalents are abnormal and represent “fragile” chromosome connections, further implicating HIM-18 in resolution of dHJs. Consistent with this, crossovers within the autosomes and the X chromosome are reduced in him-18 cells (Saito et al. 2009). Taken together with the biochemistry of the mammalian SLX4 complex, it seems likely that the function of SLX4 during meiosis may be to resolve dHJs in a manner that generates crossover products.
While there is strong evidence of a role for SLX4 orthologs in meiotic recombination, the roles of associated catalytic subunits is less clear. Thus far, SLX1 orthologs have not been examined in either Drosophila or C. elegans for their roles in meiosis or mitotic DNA repair, as genetic mutants have not yet been identified. The interaction between MUS312 and MEI-9 (XPF) is required for the generation of crossovers and proper chromosome disjunction during meiosis in Drosophila, and recent data suggest that XPF-1 may also be involved in crossover formation—but not position—in C. elegans (Yildiz et al. 2002; Saito et al. 2009). him-18;xpf-1 double mutants also display a slight increase in the percentage of inviable embryos compared with him-18 mutants alone, suggesting that XPF-1 has meiotic roles that do not fully overlap those of HIM-18 (Saito et al. 2009). Whether or not the major functions of XPF-1 in meiosis involves interaction with HIM-18 in C. elegans remains to be determined. Unlike HIM-18 and XPF-1, MUS-81 is not required for chromosome disjunction in meiosis I, suggesting that, in C. elegans, the MUS-81 HJ processing activity is not required for meiosis. But, as with xpf-1 mutants, mus-81;him-18 double mutants display increase embryonic lethality relative to him-18 mutants alone, suggesting HIM-18-independent roles for MUS-81 (Saito et al. 2009). Thus far, it is not known whether MUS-81 associates with SLX4 orthologs in either Drosophila or C. elegans. An important issue also concerns the extent to which HJ resolution, SDSA, and HJ dissolution contribute to completion of the meiotic program. Recent work has demonstrated that mus312 is synthetic lethal with the Bloom's helicase ortholog mus309, which promotes dHJ dissolution (Fig. 1), and him-18 is likewise synthetic lethal him-6BLM (Andersen et al. 2009; Saito et al. 2009). him-18;him-6 premeiotic and meiotic cells display increased RAD51 foci and chromatin bridges (Saito et al. 2009). These results suggest that HIM-18 and HIM-6 likely function in parallel pathways to dissolve or resolve HJs during meiosis, and possibly during DSB repair in mitotic cells. This is consistent with the notion that only a fraction of SPO-11-induced DSBs result in crossovers during meiosis. Further studies are required to determine to what extent the functions of MUS312/HIM-18 depend on their associated endonucleases in Drosophila and C. elegans, and the mechanisms by which individual endonucleases contribute to processing of HJs.
Regulation of the SLX4 complex by phosphorylation
Murine and human SLX4 (BTBD12) were initially found in screens for targets of the ATM/ATR protein kinases (Matsuoka et al. 2007; Mu et al. 2007). Several candidate Ser/Thr–Gln motifs are present in SLX4, and two of these are conserved throughout vertebrate SLX4 family members, but thus far only a single site of phosphorylation (T1107 in murine SLX4) has been shown to be DNA damage-inducible, and this site is not conserved. Whether and how ATM/ATR-dependent phosphorylation controls SLX4 function is unknown. In addition to SLX4, several other components of the complex (MSH2, MSH3, TRF2, and RAP1) have been identified as potential ATM/ATR substrates, and yeast Slx4 is a known target of Tel1ATM/Mec1ATR in response to MMS, CPT, IR, and hydroxyurea (HU) (Flott and Rouse 2005; Tanaka et al. 2005; Flott et al. 2007; Matsuoka et al. 2007; Dephoure et al. 2008). Phosphorylation of Slx4 by Mec1ATR or Tel1ATM is required for its function in SSA (Fig. 5D; Flott et al. 2007). It is therefore likely that phosphorylation of SLX4 by ATM/ATR has a role in promoting SLX4 function in mammalian cells (Fig. 5D).
The finding that SLX4 also associates reciprocally with PLK1 (Svendsen et al. 2009) provides an alternative mode of regulation by phosphorylation. As mentioned above, the PLK1–SLX4 interaction involves a candidate polo-box-binding site in SLX4, which would be predicted to be targeted for phosphorylation by a cyclin-dependent kinase (Elia et al. 2003). Preliminary studies indicate that SLX4 immune complexes contain an autophosphorylation activity that is inhibited by addition of a specific small molecule inhibitor of PLK1 (Svendsen et al. 2009). Thus, it would appear as though PLK1 may phosphorylate SLX4, at least in vertebrates. In addition to its well-understood role in mitotic cells, PLK1 is also implicated in meiosis in yeast, Drosophila, and mice (Hollingsworth 2008; Sourirajan and Lichten 2008; Xiong et al. 2008; Archambault and Glover 2009). Interestingly, the kinase activity of budding yeast PLK1 ortholog Cdc5 is necessary for HJ resolution during meiosis. Cells lacking Cdc5 or expressing a catalytically dead Cdc5 accumulate dHJs and have decreased crossovers, but normal levels of noncrossovers. This suggests that Cdc5 may control the activity of a resolvase that is critical to formation of crossovers during HJ resolution (Clyne et al. 2003; Sourirajan and Lichten 2008). Future studies will need to address the interplay between PLK1 and SLX4 in mitotic and meiotic cells. For example, it will be important to determine whether the interaction of SLX4 with PLK1 is programmed by the cell division cycle, thereby allowing PLK1 to modulate the endonucleolytic activity of the complex, for example, during mitosis, when aberrant DNA cleavage activity could be detrimental to the cell.
A role for the SLX4 complex at telomeres?
Roles at telomeres have been described for both MUS81 and ERCC4/XPF, and both proteins can be found associated with TRF2 (Zhu et al. 2003; Munoz et al. 2005; Zeng et al. 2009). The endonucleolytic activity of MUS81 is required for recombination-based telomere maintenance and survival in cells that employ the ALT mechanism for telomere maintenance (Zeng et al. 2009). ERCC4/XPF at telomeres can have both positive and negative implications for telomere homeostasis. At unprotected telomeres lacking TRF2, ERCC4/XPF promotes 3′ flap cleavage and NHEJ, but at protected telomeres, ERCC4/XPF has a role in ensuring that D-loops are generated within the telomere (Zhu et al. 2003). The links between MUS81, ERCC4/XPF, and telomeres are interesting in light of the fact that SLX4 associates with TRF2–RAP1 and localizes to a subset of telomeres in HeLa cells (Fig. 5D; Svendsen et al. 2009). Recent work identifying proteins that localize to telomeres in U2OS (ALT) cells not only identified SLX4, but also identified many of the components of the SLX4 complex, including C20orf94, ERCC1, TRF2–RAP1, and MSH2 (Fig. 5D; Dejardin and Kingston 2009). Together, this information suggests that SLX4 localizes to telomeres in hTERT-positive and ALT cells, and raises many interesting questions, including how SLX4 is recruited to telomeres. Does SLX4 gain access to telomeres via the TRF2–RAP1 complex, and, in addition, is the ability of MUS81 to promote recombination at ALT telomeres dependent on its association with SLX4? Given the fact that C20ORF94 is also detected at telomeres, it is possible that it plays a recruitment or maintenance role, although it may also simply be a passenger in the SLX4 complex. Finally, are their roles for SLX1 in telomere recombination either independent of or together with the MUS81–EME1 complex? Clearly, there is still much to be learned about the extent to which SLX4 contributes to telomere biology.
Outlook
The discovery of two new classes of HJ resolvases in multicellular eukaryotes—GEN1 and the SLX4 complex—has reinvigorated studies aimed at understanding the biochemical basis for HJ resolution (Symington and Holloman 2008; Klein and Symington 2009). These enzymes will serve as the focal point for mechanistic and cell biological studies that dissect the individual roles of these enzymes in DSB repair and meiotic recombination. Important questions will include delineation of the types of repair processes that GEN1 and the SLX1–SLX4 module participate in, and a determination of the extent to which these enzymes act redundantly to control cleavage of HJs and possibly other types of DNA intermediates in vivo. Almost nothing is known about how these enzymes are regulated and what determines the choice of which resolvase gets used in different scenarios in vivo. It will be important to identify proteins that interact with GEN1, possibly including branch migration activities, and to establish how GEN1 is integrated into the DNA damage response network (Harper and Elledge 2007). Currently, the SLX4 complex appears to be uniquely poised for important functions at the telomere. Given the critical role that telomeres play in the life of a cell, determining the roles of the SLX4 complex in maintaining or altering telomere structure will be of particular interest. Finally, given the critical roles of other components in the HR machinery, such as BRCA1 and BRCA2 in protecting cells from genomic instability and transformation, it will be interesting to address the question of whether resolvase mutation is linked to human cancer. One also wonders whether additional enzymes capable of processing HJs and related structures remain to be identified. Given that we are approaching the 50th anniversary of the concept of HJs (Holliday 1964), it is satisfying that a mechanistic understanding of how cells resolve these structures is within sight.
Acknowledgments
We thank Alberto Ciccia and John Lydeard for helpful discussions. Work in the Harper laboratory is supported by grants from the National Institutes of Health.
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1903510.
References
- Abraham J, Lemmers B, Hande MP, Moynahan ME, Chahwan C, Ciccia A, Essers J, Hanada K, Chahwan R, Khaw AK, et al. Eme1 is involved in DNA damage processing and maintenance of genomic stability in mammalian cells. EMBO J. 2003;22:6137–6147. doi: 10.1093/emboj/cdg580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad A, Robinson AR, Duensing A, van Drunen E, Beverloo HB, Weisberg DB, Hasty P, Hoeijmakers JH, Niedernhofer LJ. ERCC1–XPF endonuclease facilitates DNA double-strand break repair. Mol Cell Biol. 2008;28:5082–5092. doi: 10.1128/MCB.00293-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allers T, Lichten M. Intermediates of yeast meiotic recombination contain heteroduplex DNA. Mol Cell. 2001;8:225–231. doi: 10.1016/s1097-2765(01)00280-5. [DOI] [PubMed] [Google Scholar]
- Al-Minawi AZ, Saleh-Gohari N, Helleday T. The ERCC1/XPF endonuclease is required for efficient single-strand annealing and gene conversion in mammalian cells. Nucleic Acids Res. 2008;36:1–9. doi: 10.1093/nar/gkm888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen SL, Bergstralh DT, Kohl KP, LaRocque JR, Moore CB, Sekelsky J. Drosophila MUS312 and the vertebrate ortholog BTBD12 interact with DNA structure-specific endonucleases in DNA repair and recombination. Mol Cell. 2009;35:128–135. doi: 10.1016/j.molcel.2009.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archambault V, Glover DM. Polo-like kinases: Conservation and divergence in their functions and regulation. Nat Rev Mol Cell Biol. 2009;10:265–275. doi: 10.1038/nrm2653. [DOI] [PubMed] [Google Scholar]
- Bastin-Shanower SA, Fricke WM, Mullen JR, Brill SJ. The mechanism of Mus81–Mms4 cleavage site selection distinguishes it from the homologous endonuclease Rad1–Rad10. Mol Cell Biol. 2003;23:3487–3496. doi: 10.1128/MCB.23.10.3487-3496.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett RJ, Dunderdale HJ, West SC. Resolution of Holliday junctions by RuvC resolvase: Cleavage specificity and DNA distortion. Cell. 1993;74:1021–1031. doi: 10.1016/0092-8674(93)90724-5. [DOI] [PubMed] [Google Scholar]
- Bienko M, Green CM, Crosetto N, Rudolf F, Zapart G, Coull B, Kannouche P, Wider G, Peter M, Lehmann AR, et al. Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science. 2005;310:1821–1824. doi: 10.1126/science.1120615. [DOI] [PubMed] [Google Scholar]
- Boddy MN, Lopez-Girona A, Shanahan P, Interthal H, Heyer WD, Russell P. Damage tolerance protein Mus81 associates with the FHA1 domain of checkpoint kinase Cds1. Mol Cell Biol. 2000;20:8758–8766. doi: 10.1128/mcb.20.23.8758-8766.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddy MN, Gaillard PH, McDonald WH, Shanahan P, Yates JR, 3rd, Russell P. Mus81–Eme1 are essential components of a Holliday junction resolvase. Cell. 2001;107:537–548. doi: 10.1016/s0092-8674(01)00536-0. [DOI] [PubMed] [Google Scholar]
- Boulton SJ. DNA repair: A heavyweight joins the fray. Nature. 2009;462:857–858. doi: 10.1038/462857a. [DOI] [PubMed] [Google Scholar]
- Ceschini S, Keeley A, McAlister MS, Oram M, Phelan J, Pearl LH, Tsaneva IR, Barrett TE. Crystal structure of the fission yeast mitochondrial Holliday junction resolvase Ydc2. EMBO J. 2001;20:6601–6611. doi: 10.1093/emboj/20.23.6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JH, Kim JJ, Choi JM, Lee JH, Cho Y. Crystal structure of the Mus81–Eme1 complex. Genes & Dev. 2008;22:1093–1106. doi: 10.1101/gad.1618708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XB, Melchionna R, Denis CM, Gaillard PH, Blasina A, Van de Weyer I, Boddy MN, Russell P, Vialard J, McGowan CH. Human Mus81-associated endonuclease cleaves Holliday junctions in vitro. Mol Cell. 2001;8:1117–1127. doi: 10.1016/s1097-2765(01)00375-6. [DOI] [PubMed] [Google Scholar]
- Ciccia A, Constantinou A, West SC. Identification and characterization of the human mus81–eme1 endonuclease. J Biol Chem. 2003;278:25172–25178. doi: 10.1074/jbc.M302882200. [DOI] [PubMed] [Google Scholar]
- Ciccia A, McDonald N, West SC. Structural and functional relationships of the XPF/MUS81 family of proteins. Annu Rev Biochem. 2008;77:259–287. doi: 10.1146/annurev.biochem.77.070306.102408. [DOI] [PubMed] [Google Scholar]
- Clyne RK, Katis VL, Jessop L, Benjamin KR, Herskowitz I, Lichten M, Nasmyth K. Polo-like kinase Cdc5 promotes chiasmata formation and cosegregation of sister centromeres at meiosis I. Nat Cell Biol. 2003;5:480–485. doi: 10.1038/ncb977. [DOI] [PubMed] [Google Scholar]
- Connolly B, Parsons CA, Benson FE, Dunderdale HJ, Sharples GJ, Lloyd RG, West SC. Resolution of Holliday junctions in vitro requires the Escherichia coli ruvC gene product. Proc Natl Acad Sci. 1991;88:6063–6067. doi: 10.1073/pnas.88.14.6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantinou A, Davies AA, West SC. Branch migration and Holliday junction resolution catalyzed by activities from mammalian cells. Cell. 2001;104:259–268. doi: 10.1016/s0092-8674(01)00210-0. [DOI] [PubMed] [Google Scholar]
- Constantinou A, Chen XB, McGowan CH, West SC. Holliday junction resolution in human cells: Two junction endonucleases with distinct substrate specificities. EMBO J. 2002;21:5577–5585. doi: 10.1093/emboj/cdf554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulon S, Gaillard PH, Chahwan C, McDonald WH, Yates JR, 3rd, Russell P. Slx1–Slx4 are subunits of a structure-specific endonuclease that maintains ribosomal DNA in fission yeast. Mol Biol Cell. 2004;15:71–80. doi: 10.1091/mbc.E03-08-0586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulon S, Noguchi E, Noguchi C, Du LL, Nakamura TM, Russell P. Rad22Rad52-dependent repair of ribosomal DNA repeats cleaved by Slx1–Slx4 endonuclease. Mol Biol Cell. 2006;17:2081–2090. doi: 10.1091/mbc.E05-11-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosetto N, Bienko M, Hibbert RG, Perica T, Ambrogio C, Kensche T, Hofmann K, Sixma TK, Dikic I. Human Wrnip1 is localized in replication factories in a ubiquitin-binding zinc finger-dependent manner. J Biol Chem. 2008;283:35173–35185. doi: 10.1074/jbc.M803219200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies AA, West SC. Formation of RuvABC–Holliday junction complexes in vitro. Curr Biol. 1998;8:725–727. doi: 10.1016/s0960-9822(98)70282-9. [DOI] [PubMed] [Google Scholar]
- Dejardin J, Kingston RE. Purification of proteins associated with specific genomic loci. Cell. 2009;136:175–186. doi: 10.1016/j.cell.2008.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de los Santos T, Hunter N, Lee C, Larkin B, Loidl J, Hollingsworth NM. The Mus81/Mms4 endonuclease acts independently of double-Holliday junction resolution to promote a distinct subset of crossovers during meiosis in budding yeast. Genetics. 2003;164:81–94. doi: 10.1093/genetics/164.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dendouga N, Gao H, Moechars D, Janicot M, Vialard J, McGowan CH. Disruption of murine Mus81 increases genomic instability and DNA damage sensitivity but does not promote tumorigenesis. Mol Cell Biol. 2005;25:7569–7579. doi: 10.1128/MCB.25.17.7569-7579.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng C, Brown JA, You D, Brown JM. Multiple endonucleases function to repair covalent topoisomerase I complexes in Saccharomyces cerevisiae. Genetics. 2005;170:591–600. doi: 10.1534/genetics.104.028795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dephoure N, Zhou C, Villen J, Beausoleil SA, Bakalarski CE, Elledge SJ, Gygi SP. A quantitative atlas of mitotic phosphorylation. Proc Natl Acad Sci. 2008;105:10762–10767. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doe CL, Ahn JS, Dixon J, Whitby MC. Mus81–Eme1 and Rqh1 involvement in processing stalled and collapsed replication forks. J Biol Chem. 2002;277:32753–32759. doi: 10.1074/jbc.M202120200. [DOI] [PubMed] [Google Scholar]
- Duckett DR, Murchie AI, Diekmann S, von Kitzing E, Kemper B, Lilley DM. The structure of the Holliday junction, and its resolution. Cell. 1988;55:79–89. doi: 10.1016/0092-8674(88)90011-6. [DOI] [PubMed] [Google Scholar]
- Dunderdale HJ, Benson FE, Parsons CA, Sharples GJ, Lloyd RG, West SC. Formation and resolution of recombination intermediates by E. coli RecA and RuvC proteins. Nature. 1991;354:506–510. doi: 10.1038/354506a0. [DOI] [PubMed] [Google Scholar]
- Elborough KM, West SC. Resolution of synthetic Holliday junctions in DNA by an endonuclease activity from calf thymus. EMBO J. 1990;9:2931–2936. doi: 10.1002/j.1460-2075.1990.tb07484.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia AE, Rellos P, Haire LF, Chao JW, Ivins FJ, Hoepker K, Mohammad D, Cantley LC, Smerdon SJ, Yaffe MB. The molecular basis for phosphodependent substrate targeting and regulation of Plks by the Polo-box domain. Cell. 2003;115:83–95. doi: 10.1016/s0092-8674(03)00725-6. [DOI] [PubMed] [Google Scholar]
- Fekairi S, Scaglione S, Chahwan C, Taylor ER, Tissier A, Coulon S, Dong MQ, Ruse C, Yates JR, 3rd, Russell P, et al. Human SLX4 is a Holliday junction resolvase subunit that binds multiple DNA repair/recombination endonucleases. Cell. 2009;138:78–89. doi: 10.1016/j.cell.2009.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flott S, Rouse J. Slx4 becomes phosphorylated after DNA damage in a Mec1/Tel1-dependent manner and is required for repair of DNA alkylation damage. Biochem J. 2005;391:325–333. doi: 10.1042/BJ20050768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flott S, Alabert C, Toh GW, Toth R, Sugawara N, Campbell DG, Haber JE, Pasero P, Rouse J. Phosphorylation of Slx4 by Mec1 and Tel1 regulates the single-strand annealing mode of DNA repair in budding yeast. Mol Cell Biol. 2007;27:6433–6445. doi: 10.1128/MCB.00135-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogg JM, Lilley DM. Ensuring productive resolution by the junction-resolving enzyme RuvC: Large enhancement of the second-strand cleavage rate. Biochemistry. 2000;39:16125–16134. doi: 10.1021/bi001886m. [DOI] [PubMed] [Google Scholar]
- Fricke WM, Brill SJ. Slx1–Slx4 is a second structure-specific endonuclease functionally redundant with Sgs1–Top3. Genes & Dev. 2003;17:1768–1778. doi: 10.1101/gad.1105203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricke WM, Bastin-Shanower SA, Brill SJ. Substrate specificity of the Saccharomyces cerevisiae Mus81–Mms4 endonuclease. DNA Repair (Amst) 2005;4:243–251. doi: 10.1016/j.dnarep.2004.10.001. [DOI] [PubMed] [Google Scholar]
- Gaillard PH, Noguchi E, Shanahan P, Russell P. The endogenous Mus81–Eme1 complex resolves Holliday junctions by a nick and counternick mechanism. Mol Cell. 2003;12:747–759. doi: 10.1016/s1097-2765(03)00342-3. [DOI] [PubMed] [Google Scholar]
- Gravel S, Chapman JR, Magill C, Jackson SP. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes & Dev. 2008;22:2767–2772. doi: 10.1101/gad.503108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada K, Budzowska M, Davies SL, van Drunen E, Onizawa H, Beverloo HB, Maas A, Essers J, Hickson ID, Kanaar R. The structure-specific endonuclease Mus81 contributes to replication restart by generating double-strand DNA breaks. Nat Struct Mol Biol. 2007;14:1096–1104. doi: 10.1038/nsmb1313. [DOI] [PubMed] [Google Scholar]
- Harper JW, Elledge SJ. The DNA damage response: Ten years after. Mol Cell. 2007;28:739–745. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- Hoeijmakers JH. DNA damage, aging, and cancer. N Engl J Med. 2009;361:1475–1485. doi: 10.1056/NEJMra0804615. [DOI] [PubMed] [Google Scholar]
- Holliday R. A mechanism for gene conversion in fungi. Genet Res. 1964;5:282–304. doi: 10.1017/S0016672308009476. [DOI] [PubMed] [Google Scholar]
- Hollingsworth NM. Deconstructing meiosis one kinase at a time: Polo pushes past pachytene. Genes & Dev. 2008;22:2596–2600. doi: 10.1101/gad.1727508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth NM, Brill SJ. The Mus81 solution to resolution: Generating meiotic crossovers without Holliday junctions. Genes & Dev. 2004;18:117–125. doi: 10.1101/gad.1165904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosfield DJ, Mol CD, Shen B, Tainer JA. Structure of the DNA repair and replication endonuclease and exonuclease FEN-1: Coupling DNA and PCNA binding to FEN-1 activity. Cell. 1998;95:135–146. doi: 10.1016/s0092-8674(00)81789-4. [DOI] [PubMed] [Google Scholar]
- Hsia KT, Millar MR, King S, Selfridge J, Redhead NJ, Melton DW, Saunders PT. DNA repair gene Ercc1 is essential for normal spermatogenesis and oogenesis and for functional integrity of germ cell DNA in the mouse. Development. 2003;130:369–378. doi: 10.1242/dev.00221. [DOI] [PubMed] [Google Scholar]
- Hyde H, Davies AA, Benson FE, West SC. Resolution of recombination intermediates by a mammalian activity functionally analogous to Escherichia coli RuvC resolvase. J Biol Chem. 1994;269:5202–5209. [PubMed] [Google Scholar]
- Ip SC, Rass U, Blanco MG, Flynn HR, Skehel JM, West SC. Identification of Holliday junction resolvases from humans and yeast. Nature. 2008;456:357–361. doi: 10.1038/nature07470. [DOI] [PubMed] [Google Scholar]
- Ira G, Malkova A, Liberi G, Foiani M, Haber JE. Srs2 and Sgs1–Top3 suppress crossovers during double-strand break repair in yeast. Cell. 2003;115:401–411. doi: 10.1016/s0092-8674(03)00886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki H, Takahagi M, Shiba T, Nakata A, Shinagawa H. Escherichia coli RuvC protein is an endonuclease that resolves the Holliday structure. EMBO J. 1991;10:4381–4389. doi: 10.1002/j.1460-2075.1991.tb05016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessop L, Lichten M. Mus81/Mms4 endonuclease and Sgs1 helicase collaborate to ensure proper recombination intermediate metabolism during meiosis. Mol Cell. 2008;31:313–323. doi: 10.1016/j.molcel.2008.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaliraman V, Brill SJ. Role of SGS1 and SLX4 in maintaining rDNA structure in Saccharomyces cerevisiae. Curr Genet. 2002;41:389–400. doi: 10.1007/s00294-002-0319-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleff S, Kemper B, Sternglanz R. Identification and characterization of yeast mutants and the gene for a cruciform cutting endonuclease. EMBO J. 1992;11:699–704. doi: 10.1002/j.1460-2075.1992.tb05102.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein HL, Symington LS. Breaking up just got easier to do. Cell. 2009;138:20–22. doi: 10.1016/j.cell.2009.06.039. [DOI] [PubMed] [Google Scholar]
- Krogh BO, Symington LS. Recombination proteins in yeast. Annu Rev Genet. 2004;38:233–271. doi: 10.1146/annurev.genet.38.072902.091500. [DOI] [PubMed] [Google Scholar]
- Li F, Dong J, Pan X, Oum JH, Boeke JD, Lee SE. Microarray-based genetic screen defines SAW1, a gene required for Rad1/Rad10-dependent processing of recombination intermediates. Mol Cell. 2008;30:325–335. doi: 10.1016/j.molcel.2008.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, West SC. Happy Hollidays: 40th anniversary of the Holliday junction. Nat Rev Mol Cell Biol. 2004;5:937–944. doi: 10.1038/nrm1502. [DOI] [PubMed] [Google Scholar]
- Liu C, Pouliot JJ, Nash HA. Repair of topoisomerase I covalent complexes in the absence of the tyrosyl-DNA phosphodiesterase Tdp1. Proc Natl Acad Sci. 2002;99:14970–14975. doi: 10.1073/pnas.182557199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Masson JY, Shah R, O'Regan P, West SC. RAD51C is required for Holliday junction processing in mammalian cells. Science. 2004;303:243–246. doi: 10.1126/science.1093037. [DOI] [PubMed] [Google Scholar]
- Llorente B, Smith CE, Symington LS. Break-induced replication: What is it and what is it for? Cell Cycle. 2008;7:859–864. doi: 10.4161/cc.7.7.5613. [DOI] [PubMed] [Google Scholar]
- Lorenz A, West SC, Whitby MC. The human Holliday junction resolvase GEN1 rescues the meiotic phenotype of a Schizosaccharomyces pombe mus81 mutant. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkp1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyndaker AM, Alani E. A tale of tails: Insights into the coordination of 3′ end processing during homologous recombination. Bioessays. 2009;31:315–321. doi: 10.1002/bies.200800195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahdi AA, Sharples GJ, Mandal TN, Lloyd RG. Holliday junction resolvases encoded by homologous rusA genes in Escherichia coli K-12 and phage 82. J Mol Biol. 1996;257:561–573. doi: 10.1006/jmbi.1996.0185. [DOI] [PubMed] [Google Scholar]
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- McEachern MJ, Haber JE. Break-induced replication and recombinational telomere elongation in yeast. Annu Rev Biochem. 2006;75:111–135. doi: 10.1146/annurev.biochem.74.082803.133234. [DOI] [PubMed] [Google Scholar]
- McPherson JP, Lemmers B, Chahwan R, Pamidi A, Migon E, Matysiak-Zablocki E, Moynahan ME, Essers J, Hanada K, Poonepalli A, et al. Involvement of mammalian Mus81 in genome integrity and tumor suppression. Science. 2004;304:1822–1826. doi: 10.1126/science.1094557. [DOI] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS. DNA end resection: Many nucleases make light work. DNA Repair (Amst) 2009a;8:983–995. doi: 10.1016/j.dnarep.2009.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS. Nucleases and helicases take center stage in homologous recombination. Trends Biochem Sci. 2009b;34:264–272. doi: 10.1016/j.tibs.2009.01.010. [DOI] [PubMed] [Google Scholar]
- Mizuuchi K, Kemper B, Hays J, Weisberg RA. T4 endonuclease VII cleaves holliday structures. Cell. 1982;29:357–365. doi: 10.1016/0092-8674(82)90152-0. [DOI] [PubMed] [Google Scholar]
- Mu JJ, Wang Y, Luo H, Leng M, Zhang J, Yang T, Besusso D, Jung SY, Qin J. A proteomic analysis of ataxia telangiectasia-mutated (ATM)/ATM–Rad3-related (ATR) substrates identifies the ubiquitin–proteasome system as a regulator for DNA damage checkpoints. J Biol Chem. 2007;282:17330–17334. doi: 10.1074/jbc.C700079200. [DOI] [PubMed] [Google Scholar]
- Mullen JR, Kaliraman V, Ibrahim SS, Brill SJ. Requirement for three novel protein complexes in the absence of the Sgs1 DNA helicase in Saccharomyces cerevisiae. Genetics. 2001;157:103–118. doi: 10.1093/genetics/157.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz P, Blanco R, Flores JM, Blasco MA. XPF nuclease-dependent telomere loss and increased DNA damage in mice overexpressing TRF2 result in premature aging and cancer. Nat Genet. 2005;37:1063–1071. doi: 10.1038/ng1633. [DOI] [PubMed] [Google Scholar]
- Munoz IM, Hain K, Declais AC, Gardiner M, Toh GW, Sanchez-Pulido L, Heuckmann JM, Toth R, Macartney T, Eppink B, et al. Coordination of structure-specific nucleases by human SLX4/BTBD12 is required for DNA repair. Mol Cell. 2009;35:116–127. doi: 10.1016/j.molcel.2009.06.020. [DOI] [PubMed] [Google Scholar]
- Nishino T, Komori K, Ishino Y, Morikawa K. X-ray and biochemical anatomy of an archaeal XPF/Rad1/Mus81 family nuclease: Similarity between its endonuclease domain and restriction enzymes. Structure. 2003;11:445–457. doi: 10.1016/s0969-2126(03)00046-7. [DOI] [PubMed] [Google Scholar]
- Oh SD, Lao JP, Taylor AF, Smith GR, Hunter N. RecQ helicase, Sgs1, and XPF family endonuclease, Mus81–Mms4, resolve aberrant joint molecules during meiotic recombination. Mol Cell. 2008;31:324–336. doi: 10.1016/j.molcel.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman F, Dixon J, Doe CL, Whitby MC. Generating crossovers by resolution of nicked Holliday junctions: A role for Mus81–Eme1 in meiosis. Mol Cell. 2003;12:761–774. doi: 10.1016/s1097-2765(03)00343-5. [DOI] [PubMed] [Google Scholar]
- Paques F, Haber JE. Two pathways for removal of nonhomologous DNA ends during double-strand break repair in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:6765–6771. doi: 10.1128/mcb.17.11.6765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo B, Gomez-Gonzalez B, Aguilera A. DNA repair in mammalian cells: DNA double-strand break repair: How to fix a broken relationship. Cell Mol Life Sci. 2009;66:1039–1056. doi: 10.1007/s00018-009-8740-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons CA, West SC. Resolution of model Holliday junctions by yeast endonuclease is dependent upon homologous DNA sequences. Cell. 1988;52:621–629. doi: 10.1016/0092-8674(88)90474-6. [DOI] [PubMed] [Google Scholar]
- Roberts TM, Kobor MS, Bastin-Shanower SA, Ii M, Horte SA, Gin JW, Emili A, Rine J, Brill SJ, Brown GW. Slx4 regulates DNA damage checkpoint-dependent phosphorylation of the BRCT domain protein Rtt107/Esc4. Mol Biol Cell. 2006;17:539–548. doi: 10.1091/mbc.E05-08-0785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouse J. Control of genome stability by SLX protein complexes. Biochem Soc Trans. 2009;37:495–510. doi: 10.1042/BST0370495. [DOI] [PubMed] [Google Scholar]
- Saito TT, Youds JL, Boulton SJ, Colaiacovo MP. Caenorhabditis elegans HIM-18/SLX-4 interacts with SLX-1 and XPF-1 and maintains genomic integrity in the germline by processing recombination intermediates. PLoS Genet. 2009;5:e1000735. doi: 10.1371/journal.pgen.1000735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- Scharer OD. XPG: Its products and biological roles. Adv Exp Med Biol. 2008;637:83–92. doi: 10.1007/978-0-387-09599-8_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwacha A, Kleckner N. Identification of double Holliday junctions as intermediates in meiotic recombination. Cell. 1995;83:783–791. doi: 10.1016/0092-8674(95)90191-4. [DOI] [PubMed] [Google Scholar]
- Sharan SK, Kuznetsov SG. Resolving RAD51C function in late stages of homologous recombination. Cell Div. 2007;2:15. doi: 10.1186/1747-1028-8-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sourirajan A, Lichten M. Polo-like kinase Cdc5 drives exit from pachytene during budding yeast meiosis. Genes & Dev. 2008;22:2627–2632. doi: 10.1101/gad.1711408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stogios PJ, Downs GS, Jauhal JJ, Nandra SK, Prive GG. Sequence and structural analysis of BTB domain proteins. Genome Biol. 2005;6:R82. doi: 10.1186/gb-2005-6-10-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara N, Paques F, Colaiacovo M, Haber JE. Role of Saccharomyces cerevisiae Msh2 and Msh3 repair proteins in double-strand break-induced recombination. Proc Natl Acad Sci. 1997;94:9214–9219. doi: 10.1073/pnas.94.17.9214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung P, Klein H. Mechanism of homologous recombination: Mediators and helicases take on regulatory functions. Nat Rev Mol Cell Biol. 2006;7:739–750. doi: 10.1038/nrm2008. [DOI] [PubMed] [Google Scholar]
- Svendsen JM, Smogorzewska A, Sowa ME, O'Connell BC, Gygi SP, Elledge SJ, Harper JW. Mammalian BTBD12/SLX4 assembles a Holliday junction resolvase and is required for DNA repair. Cell. 2009;138:63–77. doi: 10.1016/j.cell.2009.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington LS, Holloman WK. Resolving resolvases: The final act? Mol Cell. 2008;32:603–604. doi: 10.1016/j.molcel.2008.11.011. [DOI] [PubMed] [Google Scholar]
- Symington LS, Kolodner R. Partial purification of an enzyme from Saccharomyces cerevisiae that cleaves Holliday junctions. Proc Natl Acad Sci. 1985;82:7247–7251. doi: 10.1073/pnas.82.21.7247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H, Mendonca MS, Bradshaw PS, Hoelz DJ, Malkas LH, Meyn MS, Gilley D. DNA damage-induced phosphorylation of the human telomere-associated protein TRF2. Proc Natl Acad Sci. 2005;102:15539–15544. doi: 10.1073/pnas.0507915102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor ER, McGowan CH. Cleavage mechanism of human Mus81–Eme1 acting on Holliday-junction structures. Proc Natl Acad Sci. 2008;105:3757–3762. doi: 10.1073/pnas.0710291105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trowbridge K, McKim K, Brill SJ, Sekelsky J. Synthetic lethality of Drosophila in the absence of the MUS81 endonuclease and the DmBlm helicase is associated with elevated apoptosis. Genetics. 2007;176:1993–2001. doi: 10.1534/genetics.106.070060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truglio JJ, Rhau B, Croteau DL, Wang L, Skorvaga M, Karakas E, DellaVecchia MJ, Wang H, Van Houten B, Kisker C. Structural insights into the first incision reaction during nucleotide excision repair. EMBO J. 2005;24:885–894. doi: 10.1038/sj.emboj.7600568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gool AJ, Shah R, Mezard C, West SC. Functional interactions between the holliday junction resolvase and the branch migration motor of Escherichia coli. EMBO J. 1998;17:1838–1845. doi: 10.1093/emboj/17.6.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West SC. Holliday junctions cleaved by Rad1? Nature. 1995;373:27–28. doi: 10.1038/373027a0. [DOI] [PubMed] [Google Scholar]
- West SC. Processing of recombination intermediates by the RuvABC proteins. Annu Rev Genet. 1997;31:213–244. doi: 10.1146/annurev.genet.31.1.213. [DOI] [PubMed] [Google Scholar]
- West SC. The search for a human Holliday junction resolvase. Biochem Soc Trans. 2009;37:519–526. doi: 10.1042/BST0370519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West SC, Korner A. Cleavage of cruciform DNA structures by an activity from Saccharomyces cerevisiae. Proc Natl Acad Sci. 1985;82:6445–6449. doi: 10.1073/pnas.82.19.6445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitby MC. Making crossovers during meiosis. Biochem Soc Trans. 2005;33:1451–1455. doi: 10.1042/BST0331451. [DOI] [PubMed] [Google Scholar]
- Whitby MC, Dixon J. A new Holliday junction resolving enzyme from Schizosaccharomyces pombe that is homologous to CCE1 from Saccharomyces cerevisiae. J Mol Biol. 1997;272:509–522. doi: 10.1006/jmbi.1997.1286. [DOI] [PubMed] [Google Scholar]
- White MF, Lilley DM. Characterization of a Holliday junction-resolving enzyme from Schizosaccharomyces pombe. Mol Cell Biol. 1997;17:6465–6471. doi: 10.1128/mcb.17.11.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Hickson ID. The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426:870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- Xia B, Sheng Q, Nakanishi K, Ohashi A, Wu J, Christ N, Liu X, Jasin M, Couch FJ, Livingston DM. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol Cell. 2006;22:719–729. doi: 10.1016/j.molcel.2006.05.022. [DOI] [PubMed] [Google Scholar]
- Xiong B, Sun SC, Lin SL, Li M, Xu BZ, OuYang YC, Hou Y, Chen DY, Sun QY. Involvement of Polo-like kinase 1 in MEK1/2-regulated spindle formation during mouse oocyte meiosis. Cell Cycle. 2008;7:1804–1809. doi: 10.4161/cc.7.12.6019. [DOI] [PubMed] [Google Scholar]
- Yamada K, Ariyoshi M, Morikawa K. Three-dimensional structural views of branch migration and resolution in DNA homologous recombination. Curr Opin Struct Biol. 2004;14:130–137. doi: 10.1016/j.sbi.2004.03.005. [DOI] [PubMed] [Google Scholar]
- Yildiz O, Majumder S, Kramer B, Sekelsky JJ. Drosophila MUS312 interacts with the nucleotide excision repair endonuclease MEI-9 to generate meiotic crossovers. Mol Cell. 2002;10:1503–1509. doi: 10.1016/s1097-2765(02)00782-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng S, Xiang T, Pandita TK, Gonzalez-Suarez I, Gonzalo S, Harris CC, Yang Q. Telomere recombination requires the MUS81 endonuclease. Nat Cell Biol. 2009;11:616–623. doi: 10.1038/ncb1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu XD, Niedernhofer L, Kuster B, Mann M, Hoeijmakers JH, de Lange T. ERCC1/XPF removes the 3′ overhang from uncapped telomeres and represses formation of telomeric DNA-containing double minute chromosomes. Mol Cell. 2003;12:1489–1498. doi: 10.1016/s1097-2765(03)00478-7. [DOI] [PubMed] [Google Scholar]
- Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]