Abstract
Estrogen-related receptor α (ERRα) and proliferator-activated receptor γ coactivator-1α (PGC-1α) play central roles in the transcriptional control of energy homeostasis, but little is known about factors regulating their activity. Here we identified the homeobox protein prospero-related homeobox 1 (Prox1) as one such factor. Prox1 interacts with ERRα and PGC-1α, occupies promoters of metabolic genes on a genome-wide scale, and inhibits the activity of the ERRα/PGC-1α complex. DNA motif analysis suggests that Prox1 interacts with the genome through tethering to ERRα and other factors. Importantly, ablation of Prox1 and ERRα have opposite effects on the respiratory capacity of liver cells, revealing an unexpected role for Prox1 in the control of energy homeostasis.
Keywords: ChIP-on-chip, homeobox, mitochondrial respiration, nuclear receptor, regulon
Regulation of energy homeostasis involves elaborate biochemical pathways that have evolved to react to the metabolic needs of the organism in response to specific physiological states. While homeostatic regulation is generally under hormonal control and achieved through allosteric control and post-translational modifications of metabolic enzymes for immediate needs, organ-specific requirements and lasting adaptation require regulation of metabolic genes at the transcriptional level via the action of diverse classes of transcription factors and coregulatory proteins (Desvergne et al. 2006; Feige and Auwerx 2007). Among those factors, the orphan nuclear receptor estrogen-related receptor α (ERRα, NR3B1) and the coregulator peroxisome proliferator-activated receptor γ coactivator-1α (PGC-1α) have been shown to play a predominant role in controlling several aspects of energy metabolism, most notably mitochondrial biogenesis and oxidative phosphorylation (Oxphos) (Lin et al. 2005; Giguère 2008).
Prospero-related homeobox 1 (Prox1) is a transcription factor essential for the development of numerous tissues, including the liver (Sosa-Pineda et al. 2000; Burke and Oliver 2002; Dudas et al. 2006). In particular, Prox1 plays a critical role in determining the fate of lymphatic endothelial cells and, consequently, Prox1-null embryos are devoid of lymphatic vasculature and die in utero at approximately embryonic day 14.5 (Wigle and Oliver 1999; Johnson et al. 2008). Prox1 haploinsufficient mice also display lymphatic vascular defects that have been proposed to lead to adult-onset obesity via the promotion of adipogenesis and increased fat storage in lymphatic-rich regions (Harvey et al. 2005). Prox1 is also known to regulate the activity of a specific subset of nuclear receptors (Qin et al. 2004; Song et al. 2006; Lee et al. 2009; Yamazaki et al. 2009). Of particular interest, Prox1 was shown to regulate the activity of hepatocyte nuclear factor 4α (HNF4α, NR2A1) and liver receptor homolog-1 (LRH-1, NR5A2) on the CYP7A1 and PCK1 promoters, suggesting a possible role for Prox1 in the regulation of bile acid synthesis and gluconeogenesis in the liver (Qin et al. 2004; Song et al. 2006). Whether Prox1 plays a more comprehensive role in the regulation of energy metabolism is currently unknown.
Results and Discussion
Prox1 interacts with and modulates the activity of the ERRα/PGC-1α complex
An automated yeast two-hybrid interaction screen previously identified fragments of Prox1 as interactors of ERRα (Albers et al. 2005). We first sought to validate the physiological significance of this interaction by performing coimmunoprecipitation experiments with endogenous proteins present in the mouse liver. As observed in Figure 1A, ERRα could be detected in extract immunoprecipitated with a Prox1 antibody, while Prox1 could be detected in liver lysate immunoprecipitated with an ERRα antibody. As expected, but not shown previously, a potent in vivo interaction was observed in the mouse liver between ERRα and PGC-1α (Fig. 1A). Prox1 can be also found in a complex with PGC-1α. Direct interactions were detected between Prox1 and both ERRα and PGC-1α via in vitro GST pull-down experiments (Fig. 1B). Only the N terminus of Prox1 binds to PGC-1α, while both the N terminus and C terminus of Prox1 interact with ERRα. Prox1 interacts with ERRα solely through its DNA-binding domain (DBD) (Fig. 1C). Indeed, an altered Prox1 protein containing inactivation mutations for the two putative LxxLL interaction motifs (NR1/2mut) known to be required for the interaction with LRH1 and HNF4α (Qin et al. 2004; Song et al. 2006) was able to interact physically with ERRα. Finally, Prox1 was found to interact with PGC-1α via a domain comprised of residues 483–631, a domain without a previously assigned function (Fig. 1D). A schematic representation of the potential ERRα/Prox1/PGC-1α trimeric complex is shown in Figure 1E.
Figure 1.
Prox1 interacts with and influences the transcriptional activity of ERRα and PGC-1α. (A) Prox1, ERRα, and PGC-1α interact in vivo. Lysates from mouse liver were subjected to immunoprecipitation and immunoblot analyses with the indicated antibodies. (B) Direct interactions between Prox1, PGC-1α, and ERRα. In vitro translated ERRα and PGC-1α were subjected to pull-down analysis with GST-Prox1 fragments. (N) N terminus; (M) middle; (C) C terminus. (C) Prox1 interacts with the DNA-binding domain (DBD) of ERRα. In vitro translated Prox1 and a LxxLL 1/2 mutant were subjected to pull-down analysis with GST-ERRα fragments. (FL) Full-length; (N) N terminus; (LBD) ligand-binding domain. (D) Prox1 interacts with a new functional domain of PGC-1α. In vitro translated Prox1 with GST-PGC-1α fragments. (E) Schematic representation of a potential trimeric interaction between Prox1, ERRα, and PGC-1α. (AD) Activation domain; (RD) repression domain; (RRM) RNA recognition motif; (AF-2) activation function 2; (white bars) LxxLL motifs. (F) Re-ChIP experiments performed in the mouse liver on the Pdk4 promoter using either anti-ERRα or anti-Prox1 antibodies in a serial manner. (G) Effects of wild-type and mutant Prox1 proteins on the transcriptional activity of ERRα and PGC-1α. The Pdk4 promoter–luciferase reporter gene was cotransfected in HepG2 cells with empty vector (−), ERRα, PGC-1α, or a combination of both expression vectors in the presence or absence of wild-type or mutant Prox1. (H) Same assay as in G using the Cs promoter as the reporter gene.
We next tested whether ERRα and Prox1 could form a complex on chromatin by performing a serial chromatin immunoprecipitation (ChIP) experiment in the liver at the Pdk4 promoter. As shown in Figure 1F, re-ChIP for ERRα generated further enrichment following an initial ChIP for Prox1, while the converse re-ChIP experiment generated even more enrichment for Prox1 at the Pdk4 promoter. Next, the Pdk4 promoter was fused to the luciferase reporter gene, and the construct was cotransfected in HepG2 cells together with expression vectors for ERRα and PGC-1α. As shown in Figure 1G, introduction of Prox1 decreased both the basal and ERRα-induced and/or PGC-1α-induced luciferase activity. In agreement with the physical interaction data, the altered Prox1 protein containing inactivation mutations of the two putative LxxLL interaction motifs (NR1/2mut) still retains a repressive effect, while a Prox1 DBD mutant is no longer functional for ERRα target gene repression. Similar results were obtained when using reporter constructs linked to the Cs, Cycs, and ApoC3/Apoa4 promoters (Fig. 1H; Supplemental Fig. S1).
ChIP-on-chip analyses identify a genomic relationship between ERRα and Prox1
We next performed genome-wide location analyses to assess the extent of the functional interaction between Prox1 and ERRα. To be able to directly relate the binding events to a specific target gene, we performed two distinct ChIP-on-chip experiments using tiled genomic DNA arrays covering the extended promoter regions (−5.5 to +2.5 kb from transcriptional start sites) of ∼17,000 mouse genes and antibodies specific to ERRα and Prox1. Analysis of the ChIP-on-chip data sets identified 2479 and 2266 high-confidence binding sites mapping to the promoters of 2373 and 2069 genes in the mouse liver for ERRα and Prox1, respectively (Fig. 2A; Supplemental Tables S1, S2). Comparison of the data sets from both factors revealed that a total of 937 ERRα target genes are also targets of Prox1 (39.5% of all ERRα targets) (Fig. 2A). Standard ChIP validation and examination of the bound segments revealed four different classes of promoter regions targeted by the two factors (Fig. 2B,C). Of the 937 promoters shared by ERRα and Prox1, 527 contain a common segment bound by both factors (22% of all ERRα targets) (Fig. 2B). Considering that PGC-1α can interact with both Prox1 and ERRα (Fig. 1), we next tested whether PGC-1α associates with chromatin at sites bound specifically by Prox1 or ERRα, or only when both factors are present. Standard ChIP analysis revealed that PGC-1α is recruited at DNA segments recognized by Prox1 or ERRα (Fig. 2B), consistent with the observation that PGC-1α interacts directly with both partners (Fig. 1B).
Figure 2.
Genome-wide promoter occupancy of ERRα and Prox1 in mouse liver. (A) Venn diagrams illustrating the overlap in ERRα (red) and Prox1 (green) direct target genes from ChIP-on-chip analyses in the mouse liver. (B) Standard ChIP validation of a subset of ERRα-enriched (red) and Prox1-enriched (green) segments. Occupancy of PGC-1α on these selected DNA segments bound by ERRα, Prox1, or both factors as assayed by standard ChIP is also shown. (C) Representative binding profiles of ERRα (red line) and Prox1 (green line) on specific or common target extended promoters containing either distinct or overlapping binding sites.
The results of the ChIP-on-chip experiments were then analyzed using motif-finding algorithms. First, we searched for known transcription factor-binding motifs that were enriched in bound segments, and found that, in both ERRα-specific and ERRα/Prox1 shared segments, the most enriched motifs were ERREs (Supplemental Table S3). In agreement with our previous analysis (Dufour et al. 2007), CREB-binding motifs were also enriched in ERRα-specific segments, suggesting that the functional interaction between ERRα and CREB observed in the heart is also operational in the liver. On the other hand, analysis of Prox1-specific segments revealed enrichment of HNF4α- and C/EBPβ-binding sites (Supplemental Table S3). Next, we tested a dictionary of 741 motifs identified by conservation across four mammals (Xie et al. 2005) for enrichment in bound segments using motifADE (Mootha et al. 2004), and again found that the most significantly enriched motifs in both ERRα-specific and ERRα/Prox1 shared segments were ERREs (Table 1). However, while several motifs were enriched in Prox1-specific segments, we were not able to identify with high confidence any homeobox-like motifs.
Table 1.
De novo enriched motifs in ERRα, ERRα/Prox1, or Prox1 target promoters at P < 0.01
Nuclear receptor (NR) includes ERRα (NR3B1), estrogen receptor α (NR3A1), GNCF (NR6A1), thyroid hormone receptor α (NR1A1), RORA (NR1F1), and COUP-TF (NR2F1). (NS) Not significant.
The ERRα regulon
We next evaluated the biological processes associated with genes with promoter regions that are enriched specifically for ERRα, Prox1 or both factors. As expected from previous work (Mootha et al. 2004; Schreiber et al. 2004; Dufour et al. 2007; Sonoda et al. 2007; Deblois et al. 2009), analysis showed ERRα target genes highly enriched for processes linked to metabolism (Fig. 3A; Supplemental Fig. S2). ERRα/Prox1 shared genes were also significantly enriched for the tricarboxylic acid (TCA) cycle as well as pyruvate metabolism. Similarly, Prox1-specific genes were significantly enriched for glycolysis/gluconeogenesis and pyruvate metabolism, and also for bile acid, histidine, and purine metabolism, but were virtually absent for Oxphos and the TCA cycle (Supplemental Fig. S3). Remarkably, precise assignment of ERRα and Prox1 target genes to pathways involved in energy production revealed that ERRα binds to the extended promoter regions of genes encoding enzymes at every step in the glycolytic pathway, pyruvate metabolism, and TCA cycle (Fig. 3B). This cluster of functionally linked genes is subsequently referred to as the ERRα bioenergetic regulon. ERRα-bound segments can also be found in the promoter regions of a large number of genes encoding proteins that constitute the five complexes of the Oxphos pathway (Fig. 3B). The ERRα bioenergetic regulon also includes a significant number of genes whose extended promoter regions are bound by Prox1, most notably genes encoding enzymes at key entry points in energy production pathways such as G6pc, Ldhb, Pdk4, Pcx, Pck1, Cs, and Fh1.
Figure 3.
The ERRα bioenergetic regulon. (A) Enrichment of canonical metabolic pathways in the ChIP-on-chip target genes determined to be common (yellow) or specific to either ERRα (red) or Prox1 (green). (NS) Not significant; (*) P < 0.05. (B) ChIP-on-chip direct target genes specific to ERRα (red) or Prox1 (green) or shared by both factors (yellow) involved in metabolic pathways are shown. All genes involved in glycolysis, pyruvate metabolism, and the TCA cycle are targets of ERRα, a cluster of genes defining the ERRα bioenergetic regulon. Genes labeled in black were not identified as being enriched by either ERRα or Prox1.
Divergent regulation of bioenergetic functions by ERRα and Prox1
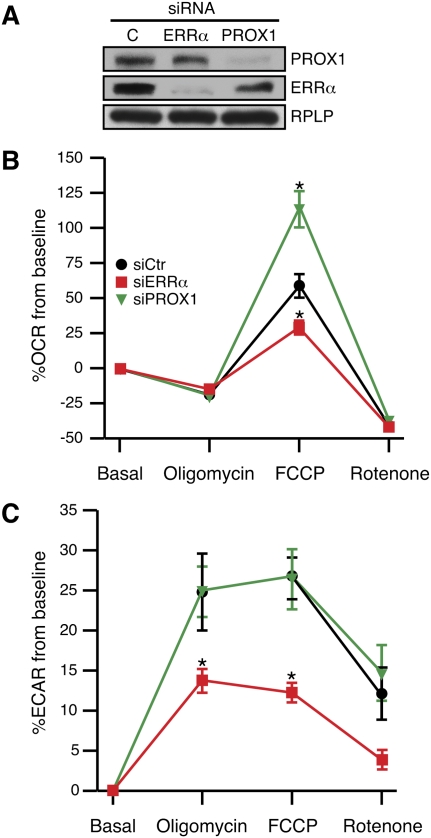
We then examined the role of ERRα and Prox1 in the regulation of bioenergetic functions in HepG2 cells. We first demonstrated that ERRα, Prox1, and PGC-1α are indeed present and can interact with each other in HepG2 cells (Supplemental Fig. S4). We also showed, using siRNAs to silence ERRα and Prox1 expression, that both factors can regulate a subset of metabolic genes indentified as ERRα target genes in the mouse liver (Supplemental Fig. S5). The HepG2 expression data set indicates that ERRα and Prox1 have, in general, contrasting effects on the expression of genes involved in bioenergetic pathways. We next measured in vivo cellular respiration and glycolytic rates in HepG2 cells in the presence or absence of two specific sets of siRNAs against either ERRα or Prox1 (Fig. 4A; Supplemental Fig. S6). HepG2 cells treated with control siRNA displayed an expected cellular respiration rate (OCR) profile that was first inhibited by addition of the ATP synthase (Complex V) inhibitor oligomycin, then enhanced with the uncoupling agent p-trifluoromethoxy carbonyl cyanide phenyl hydrazone (FCCP), and repressed again with the Complex I inhibitor rotenone (Fig. 4B; Supplemental Fig. S6). HepG2 cells lacking ERRα and Prox1 were found to have a decreased and increased response to FCCP relative to control cells, respectively. The FCCP-stimulated OCRs show that HepG2 cells lacking ERRα display impaired mitochondrial function, and that cells lacking Prox1 have a greater cellular respiratory capacity. Our experiment also revealed that cells treated with ERRα siRNA were found to have significantly lower extracellular acidification rates following oligomycin and FCCP addition (Fig. 4C; Supplemental Fig. S6), indicating that the presence of ERRα is indispensable for the ability of HepG2 cells to switch from oxidative to glycolytic metabolism.
Figure 4.
Divergent regulation of mitochondrial functions by ERRα and Prox1. (A) Western blot analysis on lysates prepared from the HepG2 knockdown samples is shown with the respective antibodies as indicated. Detection of RPLP was used a control. (B,C) Cellular oxygen consumption (B) and extracellular acidification rates (C) were measured in intact HepG2 cells treated with either control siRNA or an siRNA against ERRα or Prox1. Rates determined following sequential addition of oligomycin, FCCP, and rotenone were taken from an average of two measurements and are expressed as a percentage of the baseline rates. (*) P < 0.05.
The work presented herein not only extends the repertoire of nuclear receptors with which Prox1 physically interacts to include ERRα, but broadens these functional interactions to a coactivator protein, PGC-1α. Furthermore, the identification of ERRα and Prox1 target genes in the mouse liver establishes a unique relationship between the two factors at both a genomic and functional level. Our study also demonstrates the efficacy of the ChIP-on-chip on promoter array approach to define the regulon of a eukaryotic transcription factor. Indeed, we show that ERRα binds to the extended promoter regions of genes encoding virtually all enzymes involved in glycolysis, pyruvate metabolism, and the TCA cycle. The relevance of the metabolic role of ERRα and Prox1 was further probed by monitoring in vivo cellular respiration and glycolytic rates in HepG2 liver cells upon ERRα and Prox1 knockdown. The results indicate that ERRα and Prox1 have opposite effects on the respiratory capacity of liver cells, and that the presence of ERRα is essential for the switch to glycolysis when mitochondrial Oxphos is unable to meet the energy demands of the cell.
One of the unexpected elements of this study is the identification of the ERRα bioenergetic regulon. In prokaryotes and lower eukaryotes, regulons represent a widespread mechanism to coordinate the concurrent expression of a group of genes by a common transcription factor. In higher eukaryotes, the complexity of gene regulation is often linked to a multitude of extracellular signals, and this may preclude the use of a common factor to regulate all genetic components of an integrated biochemical pathway. Remarkably, ERRα occupies the extended promoter regions of practically all genes encoding enzymes of three well-defined biochemical pathways involved in the generation of energy from glucose. The potential to regulate linked biochemical pathways involved in energy metabolism likely evolved to ensure a coordinated increase in energy output in response to physiological stressors that are known to up-regulate the expression of the ERRα protein ligands PGC-1α and/or PGC-1β.
Prox1 is a homeobox protein, and, as expected, most studies probing its functions focused on embryonic development (Wigle and Oliver 1999; Sosa-Pineda et al. 2000; Kamiya et al. 2008). The results of our study demonstrate a novel and comprehensive role for Prox1 in the direct control of energy homeostasis. Prox1 thus joins HNF1β as a rare example of a homeobox-containing factor involved directly in the transcriptional regulation of metabolism (Desvergne et al. 2006). However, our computational analysis of sequence-specific DNA recognition suggests that Prox1’s sequence-specific binding to the genome in the adult liver is likely through interaction with other factors, most prominently ERRα, C/EBPβ, and HNF4α. Our results thus suggest that Prox1’s main mode of action in this context is that of a corepressor. While the exact molecular mechanism by which Prox1 exerts its repressor effect remains to be determined, preliminary analysis indicates that immunoprecipitated Prox1 is not associated with histone deacetylase (HDAC) activity (Supplemental Fig. S7).
In conclusion, we demonstrated that Prox1 acts as a negative modulator of the ERRα/PGC-1α axis, and, as such, regulates a broad transcriptional program implicated in the control of energy homeostasis in the liver that includes a newly defined bioenergetic regulon controlled by ERRα. Our results also reinforce the concept that the ERR isoforms are essential factors controlling the transition from carbohydrate-based to lipid-based oxidative metabolism (Alaynick et al. 2007; Dufour et al. 2007). The physiological significance of these findings is further highlighted by the recent identification of Prox1 as a genetic locus implicated in fasting glucose homeostasis and increased risk for type II diabetes (Dupuis et al. 2010). Thus, the identification of Prox1 as an important regulator of the ERRα/PGC-1α axis suggests that novel strategies for managing diseases involving long-term energy imbalance can be envisaged.
Materials and methods
Animals
Adult male C57BL/6J mice were housed and fed standard chow in the animal facility at the McGill University Health Center. In all experiments involving mouse livers, mice were sacrificed during the day at ZT (Zeitgeber time) 4.
Cell culture and reporter assays
Luciferase constructs and reporter assays in HepG2 cells are described in the Supplemental Material.
Coimmunoprecipitation, immunoblotting, and GST pull-down assays
Coimmunoprecipitation, Western blot, and GST pull-down assays involving ERRα, PGC-1α, and Prox1 are described in the Supplemental Material.
ChIP, re-ChIP, ChIP-on-chip, ChIP-qPCR, and functional analysis of target genes
Mouse liver ERRα, Prox1, and PGC-1α ChIP, serial ChIP, and genome-wide location analyses performed using Agilent extended promoter arrays are described in the Supplemental Material. Bed files are available on request. Primer sequences used for ChIP-qPCR are shown in Supplemental Table S4. Ingenuity Pathway Analysis software (Ingenuity Systems, http://www.ingenuity.com) was used for functional analysis of target genes (see the Supplemental Material).
Computational motif discovery
Enriched motifs within ERRα and Prox1 ChIP-on-chip targets were identified using MOTIFCLASS (Smith et al. 2006) and motifADE (Mootha et al. 2004) as described in the Supplemental Material.
siRNA, qRT–PCR, and extracellular flux (XF) analysis
ERRα and Prox1 knockdown experiments in HepG2 cells with subsequent qRT–PCR and bioenergetic analysis using a Seahorse Extracellular Flux (XF24) Analyzer are described in the Supplemental Material. Primers used for qRT–PCR are shown in Supplemental Table S5.
Acknowledgments
We thank Carlo Ouellet for skillful technical assistance and Dr. Julie St-Pierre for careful reading of the manuscript. This work was supported by grants from the Canadian Institutes for Health Research (MOP-84227 and MOP-77763) and the American Diabetes Association/Smith Family Foundation.
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1871610.
Supplemental material is available at http://www.genesdev.org.
References
- Alaynick WA, Kondo RP, Xie W, He W, Dufour CR, Downes M, Jonker JW, Giles W, Naviaux RK, Giguère V, et al. ERRγ directs and maintains the transition to oxidative metabolism in the post-natal heart. Cell Metab. 2007;6:16–24. doi: 10.1016/j.cmet.2007.06.007. [DOI] [PubMed] [Google Scholar]
- Albers M, Kranz H, Kober I, Kaiser C, Klink M, Suckow J, Kern R, Koegl M. Automated yeast two-hybrid screening for nuclear receptor-interacting proteins. Mol Cell Proteomics. 2005;4:205–213. doi: 10.1074/mcp.M400169-MCP200. [DOI] [PubMed] [Google Scholar]
- Burke Z, Oliver G. Prox1 is an early specific marker for the developing liver and pancreas in the mammalian foregut endoderm. Mech Dev. 2002;118:147–155. doi: 10.1016/s0925-4773(02)00240-x. [DOI] [PubMed] [Google Scholar]
- Deblois G, Hall JA, Perry MC, Laganiere J, Ghahremani M, Park M, Hallett M, Giguere V. Genome-wide identification of direct target genes implicates estrogen-related receptor α as a determinant of breast cancer heterogeneity. Cancer Res. 2009;69:6149–6157. doi: 10.1158/0008-5472.CAN-09-1251. [DOI] [PubMed] [Google Scholar]
- Desvergne B, Michalik L, Wahli W. Transcriptional regulation of metabolism. Physiol Rev. 2006;86:465–514. doi: 10.1152/physrev.00025.2005. [DOI] [PubMed] [Google Scholar]
- Dudas J, Elmaouhoub A, Mansuroglu T, Batusic D, Tron K, Saile B, Papoutsi M, Pieler T, Wilting J, Ramadori G. Prospero-related homeobox 1 (Prox1) is a stable hepatocyte marker during liver development, injury and regeneration, and is absent from “oval cells”. Histochem Cell Biol. 2006;126:549–562. doi: 10.1007/s00418-006-0191-4. [DOI] [PubMed] [Google Scholar]
- Dufour CR, Wilson BJ, Huss JM, Kelly DP, Alaynick WA, Downes M, Evans RM, Blanchette M, Giguère V. Genone-wide orchestration of cardiac functions by orphan nucler receptors ERRα and γ. Cell Metab. 2007;5:345–356. doi: 10.1016/j.cmet.2007.03.007. [DOI] [PubMed] [Google Scholar]
- Dupuis J, Langenberg C, Prokopenko I, Saxena R, Soranzo N, Jackson AU, Wheeler E, Glazer NL, Bouatia-Naji N, Gloyn AL, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 2010;42:105–113. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feige JN, Auwerx J. Transcriptional coregulators in the control of energy homeostasis. Trends Cell Biol. 2007;17:292–301. doi: 10.1016/j.tcb.2007.04.001. [DOI] [PubMed] [Google Scholar]
- Giguère V. Transcriptional control of energy homeostasis by the estrogen-related receptors. Endocr Rev. 2008;29:677–696. doi: 10.1210/er.2008-0017. [DOI] [PubMed] [Google Scholar]
- Harvey NL, Srinivasan RS, Dillard ME, Johnson NC, Witte MH, Boyd K, Sleeman MW, Oliver G. Lymphatic vascular defects promoted by Prox1 haploinsufficiency cause adult-onset obesity. Nat Genet. 2005;37:1072–1081. doi: 10.1038/ng1642. [DOI] [PubMed] [Google Scholar]
- Johnson NC, Dillard ME, Baluk P, McDonald DM, Harvey NL, Frase SL, Oliver G. Lymphatic endothelial cell identity is reversible and its maintenance requires Prox1 activity. Genes & Dev. 2008;22:3282–3291. doi: 10.1101/gad.1727208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya A, Kakinuma S, Onodera M, Miyajima A, Nakauchi H. Prospero-related homeobox 1 and liver receptor homolog 1 coordinately regulate long-term proliferation of murine fetal hepatoblasts. Hepatology. 2008;48:252–264. doi: 10.1002/hep.22303. [DOI] [PubMed] [Google Scholar]
- Lee S, Kang J, Yoo J, Ganesan SK, Cook SC, Aguilar B, Ramu S, Lee J, Hong YK. Prox1 physically and functionally interacts with COUP-TFII to specify lymphatic endothelial cell fate. Blood. 2009;113:1856–1859. doi: 10.1182/blood-2008-03-145789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Handschin C, Spiegelman BM. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab. 2005;1:361–370. doi: 10.1016/j.cmet.2005.05.004. [DOI] [PubMed] [Google Scholar]
- Mootha VK, Handschin C, Arlow D, Xie X, St Pierre J, Sihag S, Yang W, Altshuler D, Puigserver P, Patterson N, et al. ERRα and GABPAα/β specify PGC-1α-dependent oxidative phosphorylation gene expression that is altered in diabetic muscle. Proc Natl Acad Sci. 2004;101:6570–6575. doi: 10.1073/pnas.0401401101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Gao DM, Jiang QF, Zhou Q, Kong YY, Wang Y, Xie YH. Prospero-related homeobox (Prox1) is a corepressor of human liver receptor homolog-1 and suppresses the transcription of the cholesterol 7-α-hydroxylase gene. Mol Endocrinol. 2004;18:2424–2439. doi: 10.1210/me.2004-0009. [DOI] [PubMed] [Google Scholar]
- Schreiber SN, Emter R, Hock MB, Knutti D, Cardenas J, Podvinec M, Oakeley EJ, Kralli A. The estrogen-related receptor α (ERRα) functions in PPARγ coactivator 1α (PGC-1α)-induced mitochondrial biogenesis. Proc Natl Acad Sci. 2004;101:6472–6477. doi: 10.1073/pnas.0308686101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AD, Sumazin P, Xuan Z, Zhang MQ. DNA motifs in human and mouse proximal promoters predict tissue-specific expression. Proc Natl Acad Sci. 2006;103:6275–6280. doi: 10.1073/pnas.0508169103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song KH, Li T, Chiang JY. A Prospero-related homeodomain protein is a novel co-regulator of hepatocyte nuclear factor 4α that regulates the cholesterol 7α-hydroxylase gene. J Biol Chem. 2006;281:10081–10088. doi: 10.1074/jbc.M513420200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda J, Laganière J, Mehl IR, Barish GD, Chong LW, Li X, Scheffler IE, Mock DC, Bataille AR, Robert F, et al. Nuclear receptor ERRα and coactivator PGC-1β are effectors of IFN-γ induced host defense. Genes & Dev. 2007;21:1909–1920. doi: 10.1101/gad.1553007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosa-Pineda B, Wigle JT, Oliver G. Hepatocyte migration during liver development requires Prox1. Nat Genet. 2000;25:254–255. doi: 10.1038/76996. [DOI] [PubMed] [Google Scholar]
- Wigle JT, Oliver G. Prox1 function is required for the development of the murine lymphatic system. Cell. 1999;98:769–778. doi: 10.1016/s0092-8674(00)81511-1. [DOI] [PubMed] [Google Scholar]
- Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, Lander ES, Kellis M. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki T, Yoshimatsu Y, Morishita Y, Miyazono K, Watabe T. COUP-TFII regulates the functions of Prox1 in lymphatic endothelial cells through direct interaction. Genes Cells. 2009;14:425–434. doi: 10.1111/j.1365-2443.2008.01279.x. [DOI] [PubMed] [Google Scholar]