Abstract
Human pluripotent stem cells, such as embryonic stem cells (hESCs) and induced pluripotent stem cells (iPSCs), have the unique abilities of differentiation into any cell type of the organism (pluripotency) and indefinite self-renewal. Here, we show that the Rem2 GTPase, a suppressor of the p53 pathway, is up-regulated in hESCs and, by loss- and gain-of-function studies, that it is a major player in the maintenance of hESC self-renewal and pluripotency. We show that Rem2 mediates the fibroblastic growth factor 2 (FGF2) signaling pathway to maintain proliferation of hESCs. We demonstrate that Rem2 effects are mediated by suppressing the transcriptional activity of p53 and cyclin D1 to maintain survival of hESCs. Importantly, Rem2 does this by preventing protein degradation during DNA damage. Given that Rem2 maintains hESCs, we also show that it is as efficient as c-Myc by enhancing reprogramming of human somatic cells into iPSCs eightfold. Rem2 does this by accelerating the cell cycle and protecting from apoptosis via its effects on cyclin D1 expression/localization and suppression of p53 transcription. We show that the effects of Rem2 on cyclin D1 are independent of p53 function. These results define the cell cycle and apoptosis as a rate-limiting step during the reprogramming phenomena. Our studies highlight the possibility of reprogramming somatic cells by imposing hESC-specific cell cycle features for making safer iPSCs for cell therapy use.
Keywords: Rem2, cyclin D1, p53, reprogramming, self-renewal
In recent years, the field of pluripotent human embryonic stem cells (hESCs), including the discovery of induced pluripotent stem cells (iPSCs), has moved rapidly in the direction of finding a safe application for clinical use, such as cell replacement therapy and modeling for drug discovery. However, relatively little has been done to advance our mechanistic insights into the properties of self-renewing hESCs, and even less is known about the mechanisms governing iPSC formation. A better understanding of the molecular mechanisms controlling pluripotency and self-renewal would be essential for the clinical translation of hESCs and iPSCs.
hESCs were first derived from the pluripotent cells of the blastocyst inner cell mass and can be maintained in vitro indefinitely with the addition of fibroblastic growth factor 2 (FGF2) and other unknown factors secreted from feeder cell layers (Thomson et al. 1998). The pluripotency of hESCs is regulated by a set of unique transcription factors including Oct4, Sox2, and Nanog (Chambers and Smith 2004). It has been shown that a combination of three or four factors of Oct4, Sox2, and KLf4, with or without Myc, can reprogram somatic cells to generate iPSCs (Takahashi and Yamanaka 2006; Takahashi et al. 2007). Analysis of partially reprogrammed iPSCs reveals temporal and separable contributions of the four factors and indicates that ectopic c-Myc acts earlier than the pluripotency regulators (Sridharan et al. 2009). Indeed, overexpression of Myc is known to regulate cyclin D1, promoting cell cycle progression, although it remains unknown if the cell cycle function of c-Myc plays a separate role to the pluripotency genes (Oct4/sox2/Klf4) in the reprogramming process (Daksis et al. 1994). Recently, it has been shown that loss of p53 function can enhance the efficiency of reprogramming, suggesting that the cell cycle is a rate-limiting step in the reprogramming process (Zhao et al. 2008; Hong et al. 2009; Kawamura et al. 2009; Li et al. 2009; Utikal et al. 2009). Regulators of p53 transcriptional activity in hESC survival or the reprogramming process remain to be defined.
The cell cycle of mouse ESCs and, to a lesser extent, hESCs has been well described; however, defined functional data are lacking (Savatier et al. 2002; Stead et al. 2002; White and Dalton 2005). The core cell cycle regulatory machinery of ESCs—namely, Cyclins A, E, D, and B, and their kinases CDK2, CDK4, and CDK6—are not regulated in a cyclic fashion, with the exception of cyclin B (Savatier et al. 2002; Stead et al. 2002). Cyclin D1 is a unique cyclin in having a destruction box that responds to cellular stress leading to its degradation and cell cycle arrest (Agami and Bernards 2000). Of the core machinery, cyclins A/E/CDK2 are regarded as constitutively on in mouse ESCs, driving an almost nonexistent G1 phase into S phase in which 60%–70% of ESCs are present. Consequently, the rate of ESC proliferation is much faster, with an average cycle lasting just 12 h compared with somatic cells (White and Dalton 2005; Becker et al. 2006). A recent study functionally demonstrated that Cyclin A regulates pluripotency, albeit in mouse ESCs, but is redundant in fibroblasts (Kalaszczynska et al. 2009). In addition to these differences, many of the peripheral genes that control cell cycle, such as p16INK4a, are thought to be inactive, resulting in a different regulation of cell cycle in mouse ESCs (Faast et al. 2004). Given such differences between somatic and pluripotent cells, little is known about the role of the cell cycle in maintaining hESCs or iPSCs in culture. Much less is known about control of apoptosis in hESCs or iPSCs. Recently, the use of a Rho kinase inhibitor has been shown to protect hESCs from apoptosis, and is now used with in vitro culture of hESCs (Watanabe et al. 2007).
We sought to understand the role of the cell cycle and apoptosis in hESCs and formation of iPSCs by investigating the Rem2 GTPase. Rem2 is a recently identified member of the Rem/Rad/Gem/Kir (RGK) family of Ras-related GTPases, which were first described as overexpressed in muscle cells of patients with type II diabetes mediating signal transduction (Reynet and Kahn 1993; Maguire et al. 1994). Recently, Rem2 was identified from a functional genetic screen to bypass a p53-induced senescence to immortalize somatic cells, demonstrating a fundamental role for cell cycle control (Bierings et al. 2008). Here we hypothesize that the immortalization of cells is a feature very similar to self-renewal of hESCs, suggesting a role for Rem2 in self-renewal.
Results
Rem2 GTPase is expressed in hESCs, and is essential for self-renewal and pluripotency
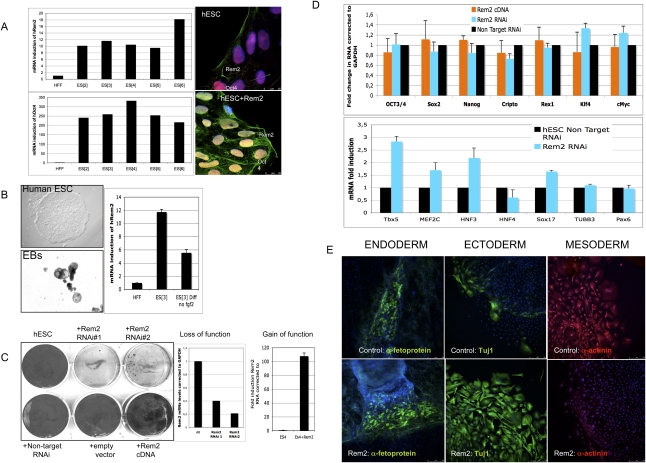
We found that Rem2 GTPase is highly expressed in six independent hESC lines derived, compared with human fibroblasts. Protein expression was found to be located predominantly in the cell membrane; however, when Rem2 was forced, overexpressed protein expression was observed in the cytoplasm and nucleus as well (Fig. 1A). We next tested the expression levels during hESC differentiation and found that Rem2 is down-regulated under general differentiation conditions (Fig. 1B). This suggests that high levels of Rem2 expression are important for maintaining a self-renewing undifferentiated state in hESCs. Given this, we knocked down Rem2 in hESCs using stably expressed shRNAs and found that loss of Rem2 caused loss of hESC self-renewal, as evidenced in colony-formation assays (CFAs) (Fig. 1C). With gain of Rem2 function in hESCs, the differences compared with controls suggested an overgrowth of colony formation, which was overgrown deliberately to show the effect of the Rem2 RNAi (Fig. 1C). Shorter-term CFAs also suggested an increase in growth with Rem2 overexpression (data not shown). To rule out any effects of the virus titer or effect of the virus on the adherence of hESCs to the culture plates following infection, we tested the virus titer and took time-lapse photos following plating and did not observe any significant effect (Supplemental Figs. 1, 2). The karyotype of hESC lines following infection and passaging was found to be normal (Supplemental Fig. 3).
Figure 1.
Rem2 GTPase is expressed in hESCs and is essential for self-renewal. (A, left panels) Real-time PCR of Rem2 and Oct4 RNA levels in hESC lines and endogenous Rem2 or ectopic Rem2 localization by immunostaining. (B, left panels) Photographs of differentiation of hESCs into EBs using general conditions on gelatin (×100). (Right panel) Real-time PCR levels of Rem2 mRNA. (C, left panel) CFA with gain (cDNA) and loss (RNAi) of function of Rem2 in hESCs in undifferentiating conditions. (Middle right panels) Graphs of real-time PCR levels of Rem2 mRNA levels following treatment with two independent Rem2 RNAi hairpins or nontarget RNAi controls and Rem2 cDNA in hESCs. (D, top graph) Graph of real-time PCR of mRNA levels of pluripotency genes with gain and loss of function of Rem2. (Bottom graph) Graph of real-time PCR of mRNA levels of differentiation genes with loss of function of Rem2. (E) Photos of immunohistochemical markers of the three germ layers after 20 d with general in vitro differentiation conditions of hESCs, with or without ectopic Rem2.
The main molecular markers of pluripotency—such as Oct4, Sox2, Nanog, and Klf4—in hESCs did not appear to be affected by Rem2 knockdown or overexpression, including c-Myc, even though the hESCs were dying (Fig. 1D). However, molecular markers of differentiation were affected in undifferentiating in vitro conditions (Fig. 1D). To investigate the role of Rem2 in pluripotency further, we overexpressed Rem2 in hESCs and differentiated them under general conditions (20% fetal calf serum [FCS] on gelatin-coated flasks). We were unable to do the same with loss of function of Rem2 due to the rapid loss of hESC survival (Fig. 1E). Forced expression of Rem2 under general differentiating conditions in vitro pushed ESC fate toward an ectodermal lineage at the expense of mesoderm (Fig. 1E), demonstrating that Rem2 plays a critical role in maintaining a true pluripotent state.
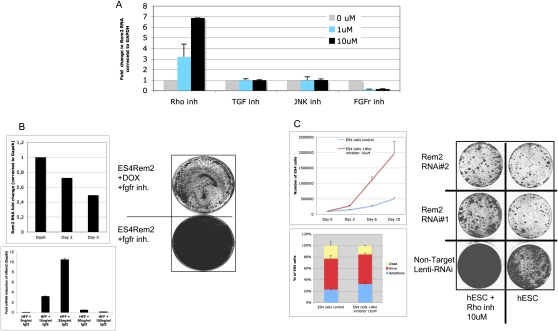
In order to further describe the importance of Rem2 in hESC biology, we tested a panel of established chemical inhibitors known to affect signaling pathways essential for maintenance of hESCs in vitro. We found that chemical inhibition of FGF receptors caused down-regulation of Rem2, and inhibition of the Rho pathway caused up-regulation of Rem2 (Fig. 2A). However, inhibition of the c-Jun N-terminal kinase (JNK) or transforming growth factor (TGF) pathways had no effect on Rem2 expression, suggesting a specific pathway of Rem2 regulation via FGF or Rho pathways (Fig. 2A). To define the role of Rem2 in these pathways further, we overexpressed Rem2 in hESCs and were able to rescue the FGFr inhibitor effects of slowing hESC growth as assessed by CFA (Fig. 2B). We used a dox-regulated lentiviral vector (a kind gift of Professor L. Naldini); the addition of DOX to cell culture reduced expression of Rem2 in 2 d (data not shown). To determine if FGF2 regulated Rem2 further, we added FGF2 to the culture medium of human fibroblasts that express relatively low levels of Rem2 and saw a 10-fold induction of Rem2 RNA with 25 ng/mL (Fig. 2B). Moreover, we removed FGF2 from hESC culture medium and saw a reduction of Rem2 levels over 5 d (Fig. 2B), further supporting that FGF2 regulates Rem2 expression. We also chose to investigate the effects of the Rho inhibitor further, as it has been shown previously to control survival of hESCs (Watanabe et al. 2007). Indeed, loss of Rem2 function by RNAi prevented the ability of the Rho inhibitor to promote survival of hESCs, suggesting that Rem2 antagonizes Rho signaling in hESCs to control survival (Fig. 2C). A role of Rem2 family members in antagonizing Rho signaling has been shown before (Olson 2002). Furthermore, the effects of the Rho inhibitor were to increase the cell cycle of hESCs grown on Matrigel rather than protection against apoptosis, which is in contrast to what has been reported previously (Fig. 2C; Watanabe et al. 2007). Together, these data demonstrate that Rem2 is overexpressed in hESCs compared with fibroblasts, controls self-renewal as well as pluripotency of hESCs, and is regulated by and mediates signaling pathways essential for maintaining hESCs in vitro.
Figure 2.
Rem2 is regulated and mediates FGF2/Rho signaling. (A) Graph of real-time PCR of Rem2 RNA levels in hESCs treated with chemical inhibitors of signaling pathways known to be important in hESC survival: FGFr (SU5402), JNK (SP600125), TGF-b-R1 Kinase-Alk5 (Inhibitor II), and Rho-kinase (Y-27632). (B, top left panel) Graph showing hESCs cultured without FGF2 in the media and Rem2 expression levels measured by real-time PCR. (Bottom left panel) Real-time PCR of Rem2 expression in human fibroblasts treated with different concentrations of FGF2 growth factor. (Right panel) Rescue of effects of FGFr inhibitor (SU5402) by ectopic Rem2. CFA of Rem2-overexpressing hESC growth on Matrigel with and without DOX plus FGFr inhibitor SU5402. (C, top left panel) Graph of effect of Rho kinase inhibitor on hESC proliferation. (Bottom left panel) Graph representing FACs analysis of apoptosis using DilC in hESCs treated with Rho inhibitor. Note that Rho inhibitor does not protect hESCs from apoptosis, but rather increases proliferation of hESCs cultured on Matrigel. (Right panel) Rescue of Rho inhibitor effects on proliferation by Rem2 RNAi in hESCs by CFA.
Rem2 GTPase cell cycle and apoptosis by regulating cyclin D1 expression and localization in hESCs
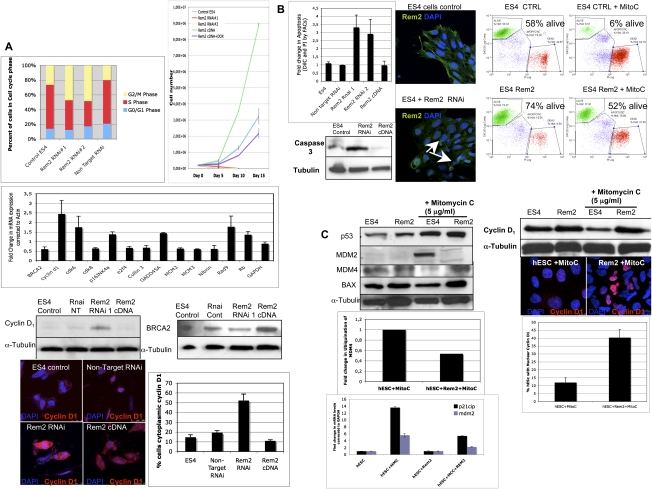
We next sought to understand the mechanisms by which Rem2 regulates hESC self-renewal using the same gain (cDNA) and loss (RNAi) of gene function strategies described above. We first analyzed the effects of Rem2 on the cell cycle of hESCs because we showed previously that Rem2 regulates the cell cycle of endothelial cells via the p53 pathway to immortalize cells (Bierings et al. 2008). Given this, we first examined the effects of Rem2 on p14ARF in hESCs, but did not find any effect (Supplemental Fig. 4). This is not surprising, given that many cell cycle pathways such as p16INK4a–cyclin D are not functional in ESCs (Faast et al. 2004). In ESCs, we found that loss of Rem2 caused a decrease of cells in S phase and an increase in G2/M phase in a short-term experiment (Fig. 3A; Supplemental Figs. 5, 6). With long-term passaging of the cells, we saw that, after 5–7 d, most of the RNAi-treated cells were arrested or dead. Conversely, gain of Rem2 function caused an increase in proliferation of hESCs over time, demonstrating that Rem2 is necessary and sufficient for maintaining the rapid cell cycle of hESCs (Fig. 3A). This supports the initial observation using CFA methodology that overexpression of Rem2 increases growth of hESCs (Fig. 1C).
Figure 3.
Rem2 GTPase controls cell cycle and apoptosis by regulating cyclin D1 and p53. (A, top left panel) Graph of FACs analysis of cell cycle profile of hESCs with loss of function of Rem2. (Top right panel) Proliferation curve of hESCs with loss and gain of Rem2 function. (Middle left panel) Real-time PCR superarray of cell cycle genes (summary of most significant changes) with loss of Rem2 function compared with nontarget RNAi controls in hESCs. (Bottom left panel) Validation of array using Western blot of cyclin D1 protein levels with loss and gain of Rem2 function in hESCs. (Bottom right panel) Validation of array by Western blot of BRCA2 protein levels with loss and gain of Rem2 function in hESCs. (Far bottom panel) Photographs of immunostaining with cyclin D1 with loss and gain of Rem2 function in hESCs. The graph shows quantification of the percent of cells expressing high levels of cyclin D1 in the cytoplasm. (B, top left panel) Graph of FACs analysis of apoptosis using DilC and PI staining of hESCs with loss and gain of Rem2 function. (Top middle panels) Photographs of DAPI/Rem2 immunostaining of hESCs with no infection control or with Rem2 RNAi demonstrating apoptotic nuclei with loss of Rem2 function in undifferentiating conditions. (Top right panels) FACs histograms of apoptosis (assessed by DilC) with and without Rem2 gain of function in hESCs treated with or without 5 μg/mL Mitomycin C. (Bottom left panel) Western blot of activated caspase 3 levels with loss and gain of Rem2 function in hESCs. (C, top left panel) Western blot analysis of p53 pathway with and without Rem2 gain of function in hESCs treated with or without 5 μg/mL Mitomycin C. (Top right panel) Western blot of cyclin D1 protein localization and level of expression with and without Rem2 gain of function in hESCs treated with or without 5 μg/mL Mitomycin C. The graph indicates quantification of the percent of hESCs with cyclin D1 protein localization. (Middle left panel) Quantification of ubiquitination assay for MDM4 with or without Rem2 and with or without Mitomycin C treatment demonstrates reduced ubiquitination of MDM4 with Rem2 overexpression in hESCs treated with Mitomycin C compared with input controls (MDM4). Note: A photo of the blot can be seen in Supplemental Figure 8. (Bottom left panel) Graph of real-time PCR of p53 transcriptional targets MDM2 and p21CIP mRNA levels treated with 5 μg/mL Mitomycin C and with or without Rem2 in hESCs, clearly showing that Rem2 regulates the transcriptional activity of p53 under stressful conditions.
To understand further how loss of Rem2 could be regulating proliferation, we performed a real-time PCR superarray for all cell cycle- and apoptosis-related genes. We found that there were little significant differences of gene expression on the core machinery of cell cycle kinases with loss of Rem2 in hESCs (Fig. 3A). To our surprise, the only exception was up-regulation of cyclin D1/CDK6, which normally promotes cell cycle progression—the opposite of what we observed with hESCs treated with Rem2 RNAi (Figs. 1, 3A). We show that the up-regulation of cyclin D1 with loss of Rem2 is partially cytoplasmic in undifferentiated conditions, and suggests that expression of Rem2 expression regulates cyclin D1 localization to maintain cell cycle and survival of hESCs (Fig. 3A). We also observed a deregulation of DNA damage-controlling genes such as BRCA2 by Rem2, which we validated by Western blot analysis, suggesting further a role for protection of apoptosis pathways (Fig. 3A). The apparent specific regulation of DNA damage-controlling genes by Rem2 suggests a more specific regulation than that known for c-Myc.
Given that loss of Rem2 caused hESC death in vitro, we assessed apoptosis using three approaches. With all three approaches—FACs analysis for DilC, DAPI staining, and Western blot for cleaved caspase 3—an increase in apoptosis with loss of Rem2 was observed (Fig. 3B; Supplemental Fig. 7). Overexpression of Rem2 did not reveal a significant change by FACs; however, by Western blot analysis, we observed a decrease in cleaved caspase 3 with Rem2 cDNA in hESCs (Fig. 3B), suggesting that they were protected from apoptosis with overexpression of Rem2 even under nonstress conditions. To test if overexpression of Rem2 protected hESCs from apoptosis, we treated the hESCs with mitomycin C, an activator of the DNA damage p53/apoptosis pathway. We observed that the hESCs were protected from apoptosis with eightfold more alive cells in Rem2/mitomycin C-treated hESCs compared with controls (Fig. 3C). To dissect the mechanism further, Western blot analyses showed that MDM2 levels, an indirect readout for the transcriptional activity of p53, were not induced in hESC cells with ectopic Rem2 following mitomycin C treatment as compared with the clear p53 transcriptional activation observed in the control, suggesting a role for Rem2 in suppressing p53 activation (Fig. 3C). Moreover, MDM4 protein, which is known to bind directly to p53 to inhibit its activity and needs to be degraded in response to stress to allow for p53 activation, was not degraded with mitomycin C treatment in Rem2 hESCs, further supporting a role for Rem2 in suppression of the p53 pathway (Fig. 3C). We show that Rem2 prevents the ubiquitination and degradation of MDM4 upon stress, suggesting that Rem2 is a novel and active component in the DNA damage–ubiquitination signaling pathway in response to stress (Fig. 3C). To further establish and define that Rem2 suppresses p53 transcriptional activity, we demonstrate that ectopic Rem2 blocks the induction of MDM2 and p21CIP mRNA, two transcriptional targets of p53 observed in control cells in the presence of stress, and that a p53 luciferase reporter construct is suppressed following treatment with mitomycin C with ectopic Rem2 (Fig. 3C; Supplemental Fig. 8). Although the fold activation of the p53 luciferase reporter construct by mitomycin C in controls is small, the obvious suppression by Rem2 in the presence of mitomycin C below basal levels (<1) demonstrates clearly the ability of Rem2 to suppress p53 transcriptional activity under stress (Supplemental Fig. 8).
We observed that the FGF2 inhibitor (SU5402) inhibits Rem2 expression and consequently increases Cyclin D1 mRNA expression levels, consistent with the Rem2 RNAi data (Fig. 3A), implicating the FGF2 pathway in regulation of Cyclin D1 in hESCs (Supplemental Fig. 9). Moreover, Cyclin D1 has been shown previously to be a target of the DNA damage pathway (Agami and Bernards 2000). Given this, we also examined if Rem2 controlled cyclin D1 protein level when hESCs were placed under stress with mitomycin treatment. Whereas cyclin D1 normally is degraded in response to cell stress, which we observed in hESCs by Western blot methodology, we show that ectopic Rem2 protected the degradation of cyclin D1 under stress conditions in hESCs (Fig. 3C). Moreover we observed that stress of Rem2-overexpressing hESCs causes a nuclear localization of cyclin D1, which would enable faster proliferation and survival (Fig. 3C). Therefore, under normal culturing conditions of hESCs, Rem2 maintains cyclin D1 expression and localization, whereas under stressful conditions such as DNA damage, Rem2 maintains cyclin D1 nuclear to promote hESC survival. Taken together, these data define Rem2 as a critical mediator of proliferation and apoptosis in hESCs by suppressing the ability of p53 to transcriptionally induce its targets, as well as the expression/location of cyclin D1 to promote survival and therefore self-renewal of hESCs in vitro.
Rem2 GTPase controls efficiency of reprogramming into iPSCs
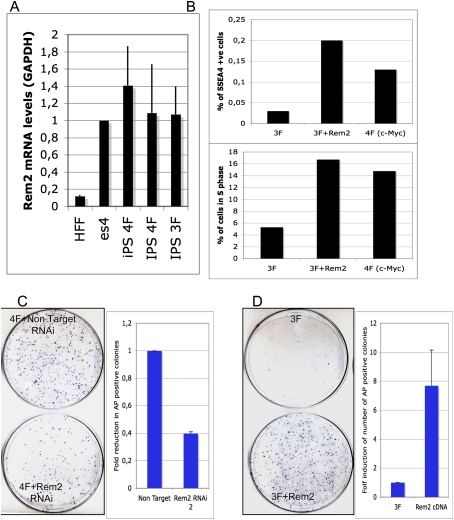
Recent work has demonstrated that ectopic Rem2 regulates the p53 pathway to immortalize primary cultured cells, indicating a possible role with instating self-renewal-like properties in somatic cells (Bierings et al. 2008). Moreover, it has been shown that loss of p53 can enhance reprogramming, although regulators of p53 remain to be defined (Zhao et al. 2008; Hong et al. 2009; Kawamura et al. 2009; Li et al. 2009; Utikal et al. 2009). Given that we showed that Rem2 maintains self-renewal and pluripotency in hESCs, we next asked if Rem2 could enhance reprogramming of somatic cells into iPSCs. To answer these questions, we reprogrammed somatic cells with three factors alone (Oct4/sox2/Klf4) or with either Myc, cyclin D1, or Rem2 (Supplemental Fig. 10).
We first observed that endogenous Rem2 levels increase in reprogrammed iPSCs, similar to levels of Rem2 in hESCs (Fig. 4A). We also found that cyclin D1 protein levels were highly expressed in iPSCs compared with ESCs (Supplemental Fig. 11). Consequently, we investigated if Rem2 could increase the number of pluripotent cells reprogramming with just three factors by staining for the early pluripotency marker SSEA4 (Fig. 4B). The increase in proliferation that we observed was correlated to an eightfold increase in SSEA4-positive cells expressing three factors plus Rem2, which was more than that observed from three factors plus c-Myc (Fig. 4B; Supplemental Fig. 12). Next, we asked if Rem2 expression is essential for the reprogramming process by stably knocking down Rem2 using a lentiviral system to deliver shRNAi in human keratinocytes infected with four factors—Oct4, Sox2, Klf4, and c-Myc. We achieved a 50% knockdown of Rem2 mRNA in human keratinocytes (data not shown), and observed a 60% loss of alkaline phosphatase (AP)-positive-stained colonies (Fig. 4C). This demonstrates that Rem2 expression is an essential component of the reprogramming phenomena of somatic cells to iPSCs. The trend of an increase in SSEA4-positive cells with three factors plus Rem2 correlates to the AP staining, thus validating AP staining as a reliable tool for assessment of reprogramming efficiency. We observed no effect of Rem2 on p14ARF levels in keratinocytes or mouse embryonic fibroblasts (MEFs), ruling out regulation of this pathway during iPSC formation (Supplemental Fig. 4). Finally, we overexpressed Rem2 with just three factors—Oct4, Sox2, and Klf4—to determine if gain of Rem2 function could enhance reprogramming and replace c-Myc. We saw an eightfold increase in efficiency of reprogramming with three factors plus Rem2, which was as efficient as using four factors (Fig. 4C,D).
Figure 4.
Rem2 GTPase controls efficiency of reprogramming into iPSCs. (A) Graph of real-time PCR of Rem2 levels in fibroblasts compared with iPSCs and hESCs. (B, top panel) Graph of SSEA4 staining of early reprogramming cells (day 12) as analyzed by FACs. The trend of an increase in SSEA4-positive cells with 3F plus Rem2 correlates to the AP staining, thus validating AP staining as a reliable tool for assessment of reprogramming efficiency. (Bottom panel) Graph of FACs analysis of the same cells analyzed for SSEA4 and GFP (a polycistronic Oct4/Sox2/Klf4/Myc or Rem2 GFP-tagged expression vector was used for these reprogramming experiments) for percent of early reprogramming cells in S phase (measured by EDU and DAPI). (C, left panel) Photo of colony formation assay of reprogramming with four factors with Rem2 RNAi or nontarget RNAi stained with AP. (Right panel) Graph of quantification of number of AP-positive colonies with four factors with Rem2 RNAi or nontarget RNAi. (D, left panel) Photo of CFA of reprogramming with three (no c-Myc) factors with Rem2 cDNA or controls stained with AP. (Right panel) Graph of quantification of number of AP-positive colonies with three factors with Rem2 cDNA or controls stained with AP.
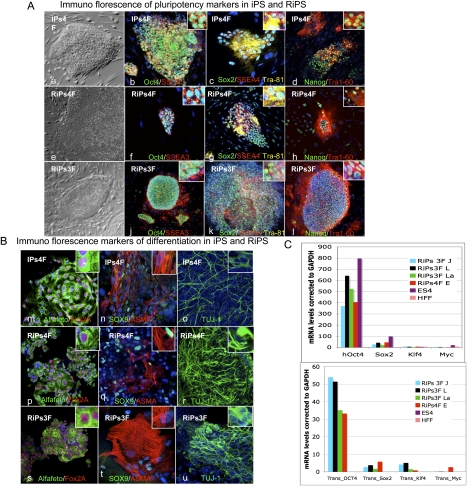
To check that we had made iPSCs, we overexpressed Rem2 with three or four factors and picked colonies for characterization. Rem2 iPSC colonies have been named RiPs (Rem2-induced pluripotent stem cells) and were picked between days 12 and 18 for expansion. We observed that RiPs were threefold more likely to survive picking and expansion than other iPSCs, and that four factor plus Rem2 colonies were more prone to differentiation during expansion than three factors plus Rem2 (data not shown). Further characterization of the RiPs revealed protein expression of ESC markers OCT4, Sox2, Nanog, Tra1-81, Tra1-60, SSEA3, and SSEA4 (Fig. 5A, panels a–l). To determine if RiPs were able to differentiate into the three germ layers, a hallmark of true ESC function, embryoid bodies (EBs) were made using general differentiation conditions, and we show that they were rapidly able to form all three germ cell layers, demonstrating that real iPSCs were made (Fig. 5B, panels m–u). Real-time PCR analysis of the endogenous and transgene expression of the four factors demonstrates that endogenous factors are up-regulated in RiPs and the transgenes are almost all totally silenced (Fig. 5C). We found consistently that the Oct4 transgene shows some residual expression in other studies where we made fully characterized human iPSCs (Aasen et al. 2008). These data demonstrate that Rem2 is able to reprogram human somatic cells into iPSCs with just three factors and is as efficient as c-Myc in reprogramming.
Figure 5.
Characterization of RiPs. (A, panels a–l) Phase contrast and immunostaining of iPSCs (one clone) and RiPs (two clones) for pluripotency markers Oct4, SSEA3, Sox2, SSEA4, Tra-81, Nanog, and Tra-60. (B, panels m–u) Immunostaining of differentiation markers following 3 wk of general differentiation of iPSC and RiP clones (gelatin with 20% FCS medium) for Alfafeto protein, Fox2A, Sox9, Alfa smooth muscle (ASMA), and TUJ1. (C) Real-time PCR of transgenes and pluripotency marker expression in RiPs compared with hESCs and somatic cells.
Rem2 is dependent on p53 and regulates cyclin D1 localization to enhance reprogramming
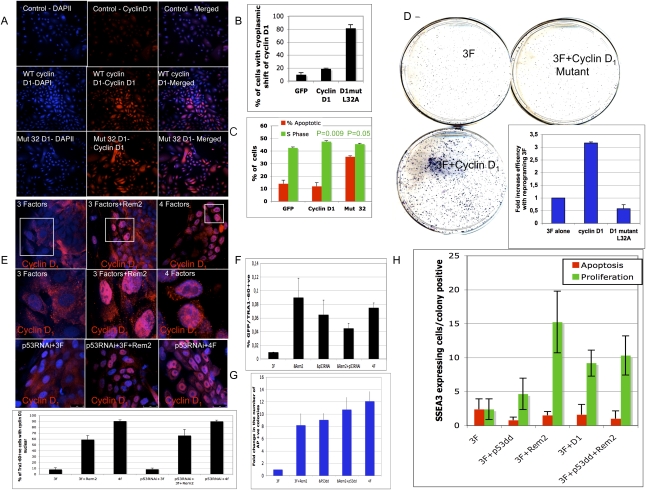
We gleaned from the work in hESCs that Rem2 regulates cyclin D1 expression and localization. Cyclin D1 is a main regulator of the cell cycle and is a target of c-Myc, suggesting further that a change in the cell cycle is required to attain pluripotency during reprogramming. We sought then to understand if the regulation/localization of cyclin D1 is responsible for reprogramming human somatic cells toward iPSCs. We overexpressed wild-type cyclin D1 and a cytoplasmic-expressed cyclin D1 mutant in an attempt to mimic the effects of Rem2 on cyclin D1 regulation and localization in hESCs (Fig. 4A,B). We overexpressed these cyclin D1 constructs with only three factors—Oct4/sox2/Klf4—as Myc is known to regulate cyclin D1 (Daksis et al. 1994). We found that overexpression of wild-type cyclin D1 (a target of c-Myc) increased the efficiency of reprogramming more than threefold, by increasing the number of cells in S phase of the cell cycle (P = 0.009), demonstrating that cyclin D1 up-regulation is sufficient for enhancing reprogramming (Fig. 4C,D). However, the converse was true with overexpression of the cytoplasmic cyclin D1 mutant, which caused apoptosis, loss of S-phase cycling cells (P = 0.05), and loss of reprogramming capacity (Fig. 4C,D). We observed in hESCs that overexpression of Rem2 under stress conditions in vitro protected hESCs to increase survival by maintaining high levels of cyclin D1 protein (Fig. 2). Because reprogramming is thought to evoke high levels of cellular stress, we measured Cyclin D1 protein levels and location by immunostaining in early reprogramming colonies (at 12 d) that were made with three factors alone (Oct4/Sox2/Klf4) with either Rem2 or c-Myc as well. We show that cyclin D1 is mainly cytoplasmic, where it is inactive, when reprogramming with three factors, which may explain why it is so inefficient compared with four factors. However, there is a clear up-regulation of cyclin D1 protein levels and a shift to the nucleus with three factors plus Rem2, where it is active to phosphorylate Retinoblastoma Protein (pRb) and promote cell cycle progression (Fig. 4E). This was also the case for three factors plus c-Myc, where most early reprogramming cells have nuclear-located cyclin D1 expression (Fig. 4E). Next, we asked whether localization of cyclin D1 was independent of p53, and found that, in early developing human iPSCs (RNAi for p53) or mouse iPSCs (using p53-null MEFs), loss of p53 did not affect the localization of cyclin D1 by Rem2 or c-Myc, suggesting that it is independent of p53 (Fig. 4E). Taken together, the data demonstrate that the Rem2 control of the localization of cyclin D1 is critical to the reprogramming phenomena.
We next asked if the role of Rem2 was independent of the p53 pathway during reprogramming, and if their effects during reprogramming are via proliferation and/or apoptosis pathways. We infected human keratinocytes with Rem2, with and without a p53 dominant-negative construct (p53dd), a p53 RNAi construct we published previously (Kawamura et al. 2009), and expression was checked by Western blot analysis (Supplemental Fig. 12). Infected cells were split into CFAs to assess efficiency (on feeder layers) or into slide flasks for assessment of apoptosis (tunnel) and proliferation (phospho-H3) of early reprogramming colonies (day 12) by selecting only SSEA3-positive colonies (Supplemental Fig. 11). We found that loss of p53 with three factors increased efficiency of reprogramming eightfold compared with three factors alone, and this was due to reducing apoptosis and increasing the number of proliferating cells, suggesting that both the apoptotic and cell cycle functions of p53 are critical for the reprogramming process (Fig. 4F,G). In the absence of an active p53, overexpression of Rem2 enhanced reprogramming to ∼10-fold; similar to three factors with p53dd alone, demonstrating that Rem2 cannot enhance reprogramming in the absence of an active p53 (Fig. 4F,G). Overexpression of Rem2 or cyclin D1 caused a large increase in the number of SSEA3-positive proliferating cells in early reprogramming colonies, demonstrating that control of cell cycle during reprogramming is essential for the process (Fig. 4H). Taken together, these data demonstrate that a cell cycle element is essential for the early reprogramming process, and that Rem2 increases efficiency of early reprogramming events by accelerating proliferation and protection of cells via its regulation of cyclin D1 localization and suppression of p53 (Fig. 4).
Discussion
In order to develop further our understanding of human pluripotent ESCs, we investigated the role of Rem2 GTPase because it is known to regulate p53 to immortalize somatic cells, a feature similar to self-renewal of both hESCs and hiPSCs. This study identified Rem2 GTPase as a gene highly expressed in hESCs and iPSCs compared with fibroblasts to promote survival and therefore self-renewal of hESCs in vitro. Furthermore, we make the novel discovery that Rem2 regulates the ability of hESCs to differentiate into all three germ layers, suggesting a role in pluripotency. By manipulating the expressed levels of Rem2, we can force hESCs toward an ectodermal lineage under general differentiating conditions. Rem2 has been identified in neuronal developments such as control neuron synapse formation (Paradis et al. 2007) and has enriched expression in the central extended amygdala (Becker et al. 2008). Recently, two microarray-based experiments identified Rem2 as overexpressed in the formation of two different adult stem cells: those making pancreas, and those making dopamine neurons (Lee et al. 2006; Treff et al. 2006). Further work to define the role of Rem2 and its cell cycle targets toward a specific cell fate is warranted.
The control of apoptosis and cell cycle are two essential functions for ESCs to maintain their self-renewal capacity. In hESCs we show that Rem2 regulates cyclin D1 expression and p53 transcriptional activation controlling cell cycle and apoptosis of hESCs. The regulation of DNA damage genes such as p53 and the cell cycle kinase cyclin D1 suggests that Rem2 is an important regulator of apoptosis and maintaining an ESC-like cell cycle profile. The ability of Rem2 to regulate the localization of cyclin D1 and to suppress p53 transcriptional activity during stress underscores a pivotal functional role for Rem2 to maintain survival and a rapid rate of cell cycle, and therefore self-renewal of hESCs. Indeed, assessment of SSEA4-, SSEA3-, and TRA1-60-positive cells with three factors (Oct4/Sox2/Klf4) plus Rem2 during reprogramming reveals that Rem2 can increase the number of pluripotent cells during the early reprogramming process (Fig. 6H). By using either p53RNAi or p53dd cells for reprogramming, and FACs sorting for SSEA3- or TRA1-60-positive cells, Rem2 cannot enhance reprogramming. This suggests that Rem2's main mechanism of action is via the p53 pathway (Fig. 6F,H). We also showed that overexpression of cyclin D1, which is also a target of c-Myc, increases the number of SSEA4-positive cells in S phase (Fig. 4B), demonstrating for the first time that the cell cycle functions of c-Myc plays an important role in the efficiency of reprogramming. Taken together, the data clearly define that the core machinery of the cell cycle—namely, cyclin D1—is a rate-determining step in the reprogramming phenomena. We propose that imposing a hESC-like cell cycle profile in somatic cells with Rem2 or cyclin D1, in addition to the pluripotency module (Oct4/Sox2/Klf4), may be a safer and more efficient way to make iPSCs. It remains to be defined if other cell cycle genes overlap with pluripotency or not, which future gain- and loss-of-function studies of other cell cycle genes would reveal.
Figure 6.
Rem2 is dependent on p53 and regulates cyclin D1 localization to enhance reprogramming. (A) Photographs of immunostaining with cyclin D1 in somatic cells before reprogramming, overexpressing GFP (control), cyclin D1, and cyclin D1 mutant (L32A). Magnification, 200× . (B) Graph of the quantification of the number of somatic cells before reprogramming expressing cyclin D1 in the cytoplasm, demonstrating that the expression of the mutant cyclin D1 is more cytoplasmic. (C) Graph summarizing FACs analysis of the percent of cells in S phase or apoptotic cells by DilC plus PI staining for overexpressing GFP (control), cyclin D1, and cyclin D1 mutant (L32A) in somatic cells before reprogramming, demonstrating that the mutant causes apoptosis and reduced proliferation. P-values given by a Student's t-test. (D) Photo of CFA of reprogramming colonies at day 21with three factors, three factors plus cyclin D1, or three factors plus cyclin D1 mutant (L32A) stained with AP. (Insert) The number of colonies was quantified by image analysis software and is presented as a graph, demonstrating loss of reprogramming efficiency with expression of mutant cyclin D1. (E) Photos of localization of cyclin D1 during early reprogramming (at day 12), demonstrating a nuclear localization with ectopic Rem2 plus three factors (Oct4/Sox2/Klf4) and three factors plus c-Myc (four factors). P53 RNAi-treated cells were also reprogrammed, and demonstrate nuclear localization of cyclin D1. (E) Graph of quantification of percent of cells with nuclear-located cyclin D1. (F) Assessment of Tra1-60-positive cells in early reprogramming colonies with p53 RNAi-treated cells, Rem2 and c-Myc (4F) assessed by FACs analysis. (G) Assessment of AP-positive colonies in early reprogramming colonies with p53dd-treated cells. The number of colonies was quantified by image analysis software and is presented as a graph. (H) Quantification of proliferation and apoptosis in early developing reprogramming colonies with three factors (Oct4/Sox2/Klf4) plus Rem2 or cyclin D1 or p53dd at day 12 by immunostaining for tunnel (red bars), phospho-H3 (green bars), and SSEA3 (to assess specific reprogramming colonies) for different gene treatments as described in the figure.
When we stressed hESCs with a DNA-damaging agent, we observed that cyclin D1 is degraded, as expected. With overexpression of Rem2 in the presence of stress, cyclin D1 is not degraded in hESCs and moves nuclear, relative to hESCs without Rem2 but equally stressed, suggesting that localization of cyclin D1 is critical for maintaining self-renewal and pluripotency. Indeed, overexpression of Rem2 during the early reprogramming stages up-regulates and maintains cyclin D1 exclusively nuclear, further supporting that cyclin D1 localization is an important rate-determining step during acquisition of pluripotency (Fig. 6E). Moreover, overexpression of a mutant cyclin D1 that is cytoplasmically expressed (to mimic the effects of loss of Rem2 on cyclin D1 in hESCs) caused an increase in apoptosis of somatic cells and loss of reprogramming efficiency, consistent with the idea that maintenance of proper cyclin D1 location in human pluripotent cells is essential for self-renewal and pluripotency. This critical function of Rem2 to regulate Cyclin D1 under stressful conditions (which reprogramming presents to a cell), and to maintain it nuclear during early reprogramming, increases the efficiency of reprogramming by accelerating the cell cycle. The phosphorylation of Rb has been described as a ground state of the hESC cell cycle profile, suggesting that Rem2 can instate a hESC cell cycle profile during reprogramming. The novel discovery that the localization and overexpression of cyclin D1 controls reprogramming defines a critical role for the cell cycle during the reprogramming process. How Rem2 controls cyclin D1 localization during reprogramming to achieve pluripotency remains to be defined, although modification of the ubiquitination and protein transport pathways seem likely pathways involved. Rem2 GTPase is a unique GTPase in having an extended N-terminal protein tail with many signal transduction-binding sites, and we did not rule out the GTPase activity of Rem2 in regulating cell survival decisions.
Indeed, the process of reprogramming is thought to evoke huge cellular stresses caused by the introduction of the four factors and the viral or transfection procedures themselves. It follows then that the introduction of genes such as Rem2 in somatic cells, which can control the effects of cellular stresses, increases survival and therefore the efficiency of reprogramming. Here we show that Rem2 GTPase can enhance the reprogramming efficiency of somatic cells into iPSCs eightfold (Fig. 4). This supports the hypothesis that loss of p53 signaling is an important step in making reprogramming more efficient, and suggests that signaling proteins upstream of p53 are important contributors to reprogramming by monitoring cellular stress levels.
In conclusion, the identification of Rem2 function in hESCs and reprogramming will help with understanding the molecular mechanisms of survival and proliferation that are essential for self-renewal and pluripotency of hESCs. Our studies highlight the possibility of reprogramming somatic cells by imposing hESC-specific cell cycle features—rather than relying on oncogenes such as c-Myc—for making safer iPSCs for cell therapy use.
Materials and methods
Culture of hESCs
hESCs were derived and fully characterized at the CMR[B] (Raya et al. 2008). They were maintained on either human feeder layers or on Matrigel-coated plates with HUES medium, consisting of KO-DMEM (Invitrogen) supplemented with 10% KO-Serum Replacement (Invitrogen), 0.5% human albumin (Grifols), 2 mM Glutamax (Invitrogen), 50 μM 2-mercaptoethanol (Invitrogen), nonessential amino acids (Cambrex), and 10 ng/mL bFGF (Peprotech). Cultures were maintained at 37°C, 5% CO2, with media changes every other day. hESCs were routinely tested for normal karyotype. For hESC lines adapted to Matrigel-coated plates, HUES-conditioned medium from irradiated MEFs was used instead. MEFs were cultured using 10% FCS with Dulbecco's modified Eagle's medium (DMEM).
Constructs and lentivirus production
Rem2 cDNA made from RNA extracted from hESCs was cloned into the pCCL TET-off-inducible lenti-vector. Rem2 RNAi constructs and nontarget RNAi controls were purchased from RZPD and were cloned into the pLKO.1puro lenti-vector (Sigma-Aldrich). All lentiviruses were made using a third-generation approach. Briefly, MDL (6.5 μg), VSV (4 μg), REV (2.5 μg), and lentiviral constructs (10 μg) were transfected into 293T cells using Lipofectamine2000 overnight. 293T cells (American Type Culture Collection no. CRL-12103) were used for the production of lentiviruses. These cell lines were grown in DMEM (GIBCO) supplemented with 10% fetal bovine serum (FBS; Biowhitaker). The next day, medium was refreshed, and the following day virus was harvested, tested for titer using an HIV-1 p24 ELISA kit (Perkin Elmer), and stored at −80°C in 1-mL aliquots. hESCs (200,000) were infected in suspension with 1 mL of viral supernatant for 1 h at 37°C and then plated in six-well Matrigel-coated plates with a further 1 mL of conditioned media. The next day, media was changed, cells were allowed to recover, and experiments were performed. For proliferation curves, equal cells were plated and then counted every 5 d.
Constructs and retroviral production
Cyclin D1 pBabe puro cDNA constructs were a kind gift from Reuven Agami (Agami and Bernards 2000). cDNAs for Rem2, Oct4, and Sox2 were amplified from ES[4] total RNA by RT–PCR. Klf-4 was amplified from IMAGE clone 5111134. c-Myc T58A mutant cDNA was amplified from DNA kindly provided by Dr. Luciano DiCroce. The amplified cDNAs were cloned into the EcoRI/ClaI sites of a modified pMSCVpuro vector, which allows the expression of N-terminal Flag-tagged proteins. The single polycistronic vector containing three factors (Oct4, Sox2, and Klf4) or four factors plus a GFP tag has been described elsewhere (Gonzalez et al. 2009). Retroviruses for the four factors were produced independently after transfecting the cell line Phoenix Amphotropic using Fugene 6 reagent (Roche) according to the manufacturer's directions. After 24 h, the DMEM was replaced, cells were incubated at 32°C, and the viral supernatant was harvested after 24 h and 48 h.
Generation of iPSCs
The generation of iPSCs using human primary keratinocytes has been described before (Aasen et al. 2008). Briefly, cells were isolated from juvenile foreskins (2–16 yr old) using dispase to remove the dermis from the epidermis followed by trypsinization and culture in serum-free low-calcium medium (Epilife; Invitrogen) at 37°C, 5% CO2, 5% O2, and used between two and four passages. For reprogramming experiments, ∼50,000 or 100,000 cells were seeded per well of a six-well plate and infected with a virus from a polycistronic vector (Oct4, Sox2, and Klf4 plus Rem2) or with a 1:1:1:1 mix of retroviral supernatants of Flag-tagged Rem2, OCT4, SOX2, KLF4, and c-MYCT58A in the presence of 1 mg/mL polybrene. Infection consisted of a 45-min spinfection at 750g, washed with PBS and with keratinocyte medium replaced. Two rounds of infections on consecutive days were performed. Two days after beginning the last round of infection, cells were trypsinized and seeded onto feeder layers of irradiated MEFs in the same culture medium. The medium was changed upon plating to hESC medium, consisting of KO-DMEM (Invitrogen) supplemented with 10% KO-Serum Replacement (Invitrogen), 0.5% human albumin (Grifols), 2 mM Glutamax (Invitrogen), 50 μM 2-mercaptoethanol (Invitrogen), nonessential amino acids (Cambrex), and 10 ng/mL bFGF (Peprotech). Cultures were maintained at 37°C, 5% CO2, with media changes every other day. IPSCs were picked, expanded, and characterized. For CFA experiments, plates were fixed with 50% methanol/10% acetic acid/40% water for 5 min, stained with 0.1% Coomassie blue for 5 min, and washed with water, and an electronically scanned photo was produced.
Immunofluorescence and AP analysis
Cells were grown on plastic coverslide chambers and fixed with 4% paraformaldehyde (PFA). The antibodies used have been described before (Aasen et al. 2008). Rem2 antibody has been described before (Bierings et al. 2008). Cyclin D1 was used at 1:100 (Santa Cruz Biotechnologies). The secondary antibodies used were all the Alexa Fluor series from Invitrogen (all 1:500). Images were taken using a Leica SP5 confocal microscope. Direct AP activity was analyzed using an Alkaline Phosphatase Blue/Red Membrane Substrate solution kit according to the manufacturer's guidelines (Sigma).
Chemical inhibitors
Inhibitors were used at the concentration and time described in Figure 2 or with vehicle alone. FGFr (SU5402), JNK (SP600125), TGF-b-R1 Kinase-Alk5 (Inhibitor II), and Rho-kinase (Y-27632) were all purchased from Calbiochem.
In vitro differentiation
Differentiation toward endoderm, mesoderm, and neuroectoderm was carried out by plating EBs on gelatin and DMEM medium with 20% FCS changed every second day for 2–3 wk. Cells were then stained for appropriate markers as described in the figures.
Flow cytometry analyses
All analyses were performed on a MoFlo cell sorter (DakoCytomation) running Summit software. For measuring apoptosis and proliferation, we used the commercial kits from Invitrogen—the “MitoProbe DilC1(5) Assay kit” and the “Click-iT EdU AlexaFluor647 Flow Cytometry Assay kit,” respectively—following the manufacturer's instructions. For the proliferation assay using the click-IT kit, instead of using the supplied DNA dyes, we used a homemade DNA DAPI-staining solution (0.1 M Tris Base at pH 7.4, 0.9% or 150 mM NaCl, 1 mM CaCl2, 0.5 mM MgCl2, 0.2% BSA, 0.1% Nonidet NP-40, 10 mg/mL DAPI) at 0.5 mL per test (2 h at room temperature or overnight at 4°C).
Western blot
Western blot analyses were performed as described previously, using extracts of cells collected by centrifugation, washed twice in PBS, lysed in 1× lysis buffer (50 nM Tris-HCl, 70 mM 2-mercaptoethanol, 2% sodium dodecylsulfate [SDS]), then boiled for 5 min and subjected to 12% polyacrylamide SDS gel electrophoresis. After electrophoresis, proteins were transferred to a nitrocellulose membrane using a submerged transfer apparatus (Bio-Rad) filled with 25 mM Tris Base, 200 mM glycine, and 20% methanol. After blocking with 5% nonfat dried milk in TBS-T (50 mM Tris-HCl at pH 8.0, 150 mM NaCl, 0.1% Tween 20), the membrane was incubated with the primary antibodies diluted in TBS-T and washed extensively. Primary antibodies were anti-Flag (Sigma), BRCA-1 (Santa Cruz Biotechnologies), p53 (Santa Cruz Biotechnologies, D01), MDM2 (4B2) (Chen et al. 1993), MDM4 (Affinity, bioreagents PA1-24307), Bax (Santa Cruz Biotechnologies), tubulin, and cleaved Caspase 3 (Cell Signaling, 5A1). The membrane was washed three times with TBS-T and then incubated with the appropriate horseradish peroxidase-linked secondary antibody (Amersham). The detection was performed with the Western Breeze Immunodetection kit (Invitrogen). The concentration of protein was measured by the Bradford assay. For the ubiquitination assay for MDM4, we used a previously described protocol (Xirodimas et al. 2001). Briefly, hESCs were infected with ubiquitin lentivirus, and stably expressed Ub-hESCs were then infected again with Rem2. Cells were then treated with Mitomycin C, and the ubiquitination assay for MDM4 was performed.
Real-time PCR
Total mRNA was isolated using Trizol and 1 μg was used to synthesize cDNA using the Invitrogen Cloned AMV First-Strand cDNA synthesis kit. One microliter to 2 μL of the reaction were used to quantify gene expression by quantitative PCR for transgenes, endogenous pluripotent genes, as described previously (Aasen et al. 2008). For analysis of cell cycle and apoptosis-related genes, we used a real-time PCR superarray for human cell cycle genes. For Rem2, MDM2, and cyclin D1, we used the following primers: hRem2: FOR, AGATGCCACGCTACTAAAGAAG, and REV, GCCCAAGGAGTCAGACGAGCCA; hCyclin D1: FOR, GATCAAGTGTGACCCGGACT, and REV, TCCTCCTCCTCTTCCTCCTC; hMDM2: FOR, TGTTGGTGCACAAAAAGACA, and REV, CACGCCAAACAAATCTCCTA; p21: FOR, ACCTGTCACTGTCTTGTACCCTTGT, and REV, GTTTGGAGTGGTAGAAATCTGTCATG.
Real-time PCR superarrays for cell cycle genes were purchased, and the manufacturer's instructions were followed (SA Biosciences).
Quantification using computer-aided analysis software
Analysis of the number of colonies in CFAs was done using computer-assisted video analysis using Metamorph software. Briefly, a region of interest was set in a photograph of the CFA, a threshold for color was set (blue), and the number of colonies was exported directly to Excel for analysis. All data were analyzed using Excel spreadsheet software for mean, standard deviation, and Student's t-test.
Acknowledgments
We are indebted to José Miguel Andrés Vaquero for assistance with flow cytometry; Meritxell Carrió for expert assistance with cell culture techniques; Esther Melo, Lola Mulero Pérez, and Mercé Gaudes Martí for bioimaging assistance; and Reuven Agami for the cyclin D1 constructs. Cristina Menchon is partly supported by a predoctoral grant from MICINN. This work was supported by grants from the Fondo de Investigaciones Sanitarias, MICINN, RETICS, CIBER, Fundacion Cellex, and The G. Harold and Leila Y. Mathers Charitable Foundation.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1876710.
Supplemental material is available at http://www.genesdev.org.
References
- Aasen T, Raya A, Barrero MJ, Garreta E, Consiglio A, Gonzalez F, Vassena R, Bilic J, Pekarik V, Tiscornia G, et al. Efficient and rapid generation of induced pluripotent stem cells from human keratinocytes. Nat Biotechnol. 2008;26:1276–1284. doi: 10.1038/nbt.1503. [DOI] [PubMed] [Google Scholar]
- Agami R, Bernards R. Distinct initiation and maintenance mechanisms cooperate to induce G1 cell cycle arrest in response to DNA damage. Cell. 2000;102:55–66. doi: 10.1016/s0092-8674(00)00010-6. [DOI] [PubMed] [Google Scholar]
- Becker KA, Ghule PN, Therrien JA, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Self-renewal of human embryonic stem cells is supported by a shortened G1 cell cycle phase. J Cell Physiol. 2006;209:883–893. doi: 10.1002/jcp.20776. [DOI] [PubMed] [Google Scholar]
- Becker JA, Befort K, Blad C, Filliol D, Ghate A, Dembele D, Thibault C, Koch M, Muller J, Lardenois A, et al. Transcriptome analysis identifies genes with enriched expression in the mouse central extended amygdala. Neuroscience. 2008;156:950–965. doi: 10.1016/j.neuroscience.2008.07.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierings R, Beato M, Edel MJ. An endothelial cell genetic screen identifies the GTPase Rem2 as a suppressor of p19ARF expression that promotes endothelial cell proliferation and angiogenesis. J Biol Chem. 2008;283:4408–4416. doi: 10.1074/jbc.M707438200. [DOI] [PubMed] [Google Scholar]
- Chambers I, Smith A. Self-renewal of teratocarcinoma and embryonic stem cells. Oncogene. 2004;23:7150–7160. doi: 10.1038/sj.onc.1207930. [DOI] [PubMed] [Google Scholar]
- Chen J, Marechal V, Levine AJ. Mapping of the p53 and mdm-2 interaction domains. Mol Cell Biol. 1993;13:4107–4114. doi: 10.1128/mcb.13.7.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daksis JI, Lu RY, Facchini LM, Marhin WW, Penn LJ. Myc induces cyclin D1 expression in the absence of de novo protein synthesis and links mitogen-stimulated signal transduction to the cell cycle. Oncogene. 1994;9:3635–3645. [PubMed] [Google Scholar]
- Faast R, White J, Cartwright P, Crocker L, Sarcevic B, Dalton S. Cdk6-cyclin D3 activity in murine ES cells is resistant to inhibition by p16(INK4a) Oncogene. 2004;23:491–502. doi: 10.1038/sj.onc.1207133. [DOI] [PubMed] [Google Scholar]
- Gonzalez F, Barragan Monasterio M, Tiscornia G, Montserrat Pulido N, Vassena R, Batlle Morera L, Rodriguez Piza I, Izpisua Belmonte JC. Generation of mouse-induced pluripotent stem cells by transient expression of a single nonviral polycistronic vector. Proc Natl Acad Sci. 2009;106:8918–8922. doi: 10.1073/pnas.0901471106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong H, Takahashi K, Ichisaka T, Aoi T, Kanagawa O, Nakagawa M, Okita K, Yamanaka S. Suppression of induced pluripotent stem cell generation by the p53–p21 pathway. Nature. 2009;460:1132–1135. doi: 10.1038/nature08235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalaszczynska I, Geng Y, Iino T, Mizuno S, Choi Y, Kondratiuk I, Silver DP, Wolgemuth DJ, Akashi K, Sicinski P. Cyclin A is redundant in fibroblasts but essential in hematopoietic and embryonic stem cells. Cell. 2009;138:352–365. doi: 10.1016/j.cell.2009.04.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura T, Suzuki J, Wang YV, Menendez S, Morera LB, Raya A, Wahl GM, Belmonte JC. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature. 2009;460:1140–1144. doi: 10.1038/nature08311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MS, Jun DH, Hwang CI, Park SS, Kang JJ, Park HS, Kim J, Kim JH, Seo JS, Park WY. Selection of neural differentiation-specific genes by comparing profiles of random differentiation. Stem Cells. 2006;24:1946–1955. doi: 10.1634/stemcells.2005-0325. [DOI] [PubMed] [Google Scholar]
- Li H, Collado M, Villasante A, Strati K, Ortega S, Canamero M, Blasco MA, Serrano M. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature. 2009;460:1136–1139. doi: 10.1038/nature08290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire J, Santoro T, Jensen P, Siebenlist U, Yewdell J, Kelly K. Gem: An induced, immediate early protein belonging to the Ras family. Science. 1994;265:241–244. doi: 10.1126/science.7912851. [DOI] [PubMed] [Google Scholar]
- Olson MF. Gem GTPase: Between a ROCK and a hard place. Curr Biol. 2002;12:R496–R498. doi: 10.1016/S0960-9822(02)00968-5. [DOI] [PubMed] [Google Scholar]
- Paradis S, Harrar DB, Lin Y, Koon AC, Hauser JL, Griffith EC, Zhu L, Brass LF, Chen C, Greenberg ME. An RNAi-based approach identifies molecules required for glutamatergic and GABAergic synapse development. Neuron. 2007;53:217–232. doi: 10.1016/j.neuron.2006.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raya A, Rodriguez-Piza I, Aran B, Consiglio A, Barri PN, Veiga A, Izpisua Belmonte JC. Generation of cardiomyocytes from new human embryonic stem cell lines derived from poor-quality blastocysts. Cold Spring Harb Symp Quant Biol. 2008;73:127–135. doi: 10.1101/sqb.2008.73.038. [DOI] [PubMed] [Google Scholar]
- Reynet C, Kahn CR. Rad: A member of the Ras family overexpressed in muscle of type II diabetic humans. Science. 1993;262:1441–1444. doi: 10.1126/science.8248782. [DOI] [PubMed] [Google Scholar]
- Savatier P, Lapillonne H, Jirmanova L, Vitelli L, Samarut J. Analysis of the cell cycle in mouse embryonic stem cells. Methods Mol Biol. 2002;185:27–33. doi: 10.1385/1-59259-241-4:27. [DOI] [PubMed] [Google Scholar]
- Sridharan R, Tchieu J, Mason MJ, Yachechko R, Kuoy E, Horvath S, Zhou Q, Plath K. Role of the murine reprogramming factors in the induction of pluripotency. Cell. 2009;136:364–377. doi: 10.1016/j.cell.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stead E, White J, Faast R, Conn S, Goldstone S, Rathjen J, Dhingra U, Rathjen P, Walker D, Dalton S. Pluripotent cell division cycles are driven by ectopic Cdk2, cyclin A/E and E2F activities. Oncogene. 2002;21:8320–8333. doi: 10.1038/sj.onc.1206015. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Treff NR, Vincent RK, Budde ML, Browning VL, Magliocca JF, Kapur V, Odorico JS. Differentiation of embryonic stem cells conditionally expressing neurogenin 3. Stem Cells. 2006;24:2529–2537. doi: 10.1634/stemcells.2006-0082. [DOI] [PubMed] [Google Scholar]
- Utikal J, Polo JM, Stadtfeld M, Maherali N, Kulalert W, Walsh RM, Khalil A, Rheinwald JG, Hochedlinger K. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature. 2009;460:1145–1148. doi: 10.1038/nature08285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Ueno M, Kamiya D, Nishiyama A, Matsumura M, Wataya T, Takahashi JB, Nishikawa S, Nishikawa S, Muguruma K, et al. A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nat Biotechnol. 2007;25:681–686. doi: 10.1038/nbt1310. [DOI] [PubMed] [Google Scholar]
- White J, Dalton S. Cell cycle control of embryonic stem cells. Stem Cell Rev. 2005;1:131–138. doi: 10.1385/SCR:1:2:131. [DOI] [PubMed] [Google Scholar]
- Xirodimas D, Saville MK, Edling C, Lane DP, Lain S. Different effects of p14ARF on the levels of ubiquitinated p53 and Mdm2 in vivo. Oncogene. 2001;20:4972–4983. doi: 10.1038/sj.onc.1204656. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Yin X, Qin H, Zhu F, Liu H, Yang W, Zhang Q, Xiang C, Hou P, Song Z, et al. Two supporting factors greatly improve the efficiency of human iPSC generation. Cell Stem Cell. 2008;3:475–479. doi: 10.1016/j.stem.2008.10.002. [DOI] [PubMed] [Google Scholar]