Abstract
Eukaryotic chromosomal DNA replication requires cyclin-dependent kinase (CDK) activity. CDK phosphorylates two yeast replication proteins, Sld3 and Sld2, both of which bind to Dpb11 when phosphorylated. These phosphorylation-dependent interactions are essential and are the minimal requirements for CDK-dependent activation of DNA replication. However, how these interactions activate DNA replication has not been elucidated. Here, we show that CDK promotes the formation of a newly identified fragile complex, the preloading complex (pre-LC) containing DNA polymerase ɛ (Pol ɛ), GINS, Sld2, and Dpb11. Formation of the pre-LC requires phosphorylation of Sld2 by CDK, but is independent of DNA replication, protein association with replication origins, and Dbf4-dependent Cdc7 kinase, which is also essential for the activation of DNA replication. We also demonstrate that Pol ɛ, GINS, Dpb11, and CDK-phosphorylated Sld2 form a complex in vitro. The genetic interactions between Pol ɛ, GINS, Sld2, and Dpb11 suggest further that they form an essential complex in cells. We propose that CDK regulates the initiation of DNA replication in budding yeast through formation of the pre-LC.
Keywords: DNA replication, cell cycle, CDK, GINS, Pol ɛ, yeast
Eukaryotic chromosome DNA replicates from multiple origins only once per cell cycle. This is regulated mainly at the initiation steps by two protein kinases, cyclin-dependent kinase (CDK) and Dbf4-dependent Cdc7 kinase (DDK), both of which are activated at the G1/S boundary of the cell cycle. These kinases cause many replication proteins—including replicative DNA polymerases α, δ, and ɛ (Pol α, Pol δ, and Pol ɛ)—to associate at origins with the prereplicative complex (pre-RC). The pre-RC is formed from late M phase to G1 phase when CDK activity is low by loading of the Mcm2–7 helicase to the replication origins that are bound by the six-subunit origin recognition complex (ORC) throughout the cell cycle in budding yeast. This loading also requires Cdt1 and Cdc6 (Bell and Dutta 2002; Schwob and Labib 2006; Sivaprasad et al. 2006; Walter and Araki 2006).
CDK phosphorylates two replication proteins, Sld3 and Sld2, both of which bind to Dpb11 when phosphorylated. Dpb11 has two pairs of tandem BRCT domains, known as a phosphopeptide-binding domain (Glover et al. 2004). The N-terminal pair of the BRCT domains binds to phosphorylated Sld3, and the C-terminal pair binds to phosphorylated Sld2. These phosphorylation-dependent interactions are essential, and represent the minimal requirement for CDK-dependent activation of DNA replication in budding yeast (Masumoto et al. 2002; Tanaka et al. 2007; Zegerman and Diffley 2007). DDK phosphorylates Mcm, and this phosphorylation is thought to enhance the interaction between Mcm and other replication proteins (Masai et al. 2006; Sheu and Stillman 2006) by alleviating an inhibitory activity in Mcm4 (Sheu and Stillman 2010).
Sld3, Sld2, and Dpb11 interact with many replication proteins. Sld3 and Cdc45 form a complex that associates with the pre-RC at the early firing origins even in G1 phase and with the late-firing origins in S phase, in a mutually dependent and Mcm-dependent manner (Kamimura et al. 2001; Kanemaki and Labib 2006). Dpb11 and Sld2 interact with Pol ɛ genetically and physically. Increasing the dosage of DPB11 or SLD2 suppresses the growth defect caused by mutations occurring in Pol ɛ (Araki et al. 1995; Kamimura et al. 1998). Moreover, combined mutations in Pol ɛ, SLD2, and DPB11 cause inviability (Araki et al. 1995; Kamimura et al. 1998; Wang and Elledge 1999). In a two-hybrid assay, Sld2 and Dpb11 interact with the C-terminal portion of Pol2 (see below), the catalytic subunit of Pol ɛ (Edwards et al. 2003). Cross-linking experiments indicate that Dpb11 and Pol ɛ coprecipitate mainly in S phase and associate with replication origins in a mutually dependent manner (Masumoto et al. 2000).
Pol ɛ comprises four subunits—Pol2, Dpb2, Dpb3, and Dpb4—and forms a globular domain connected to a more extended tail-like structure (Asturias et al. 2006; Pursell and Kunkel 2008). Pol2, the largest and a catalytic subunit of Pol ɛ, has a DNA polymerase domain in the N-terminal portion followed by a long C-terminal stretch. Surprisingly, the DNA polymerase domain is not essential for cell growth and DNA replication, whereas deletion of the C-terminal portion of Pol2 confers lethality (Dua et al. 1999; Kesti et al. 1999; Feng and D'Urso 2001). Thus, whereas Pol ɛ normally synthesizes the leading strand at the replication forks (Pursell et al. 2007; Kunkel and Burgers 2008; Burgers 2009), we and others have proposed that the C-terminal portion of Pol2 has an essential function as a scaffold for other replication proteins (Masumoto et al. 2000; Feng and D'Urso 2001). Moreover, Lou et al. (2008) reported recently that Pol2 interacts with Mrc1, a checkpoint mediator, and functions in checkpoint control. In addition, two nonessential small subunits, Dpb3 and Dpb4, both of which have a histone fold, form a subassembly that interacts with histones and functions in transcriptional silencing caused by chromatin structures (Iida and Araki 2004; Tackett et al. 2005; Tsubota et al. 2006). Therefore, Pol ɛ itself seems to be an important regulator of chromosome dynamics.
Dpb11 also interacts with GINS, which comprises Sld5, Psf1, Psf2, and Psf3, and participates in the initiation and elongation steps of chromosomal DNA replication (Kanemaki et al. 2003; Takayama et al. 2003; Labib and Gambus 2007). Dpb11 and GINS associate with origins in a mutually dependent manner. GINS is one of the replication proteins found at the replication forks and forms a complex with Cdc45 and Mcm, called the CMG complex in Drosophila embryo extracts (Moyer et al. 2006), the Unwindsome in Xenopus egg extracts (Pacek et al. 2006), and the replisome progression complex (RPC) in budding yeast (Gambus et al. 2006). The CMG complex purified to homogeneity shows a higher DNA helicase activity than Mcm alone (Ilves et al. 2010), suggesting that it works as a replicative DNA helicase. The RPC contains other factors that regulate fork progression in addition to components of the CMG complex. Thus, the CMG complex seems to comprise the minimal form of the replicative DNA helicase.
Although phosphorylation-dependent interactions between Dpb11, Sld2, and Sld3 are essential for CDK-dependent activation of DNA replication (Masumoto et al. 2002; Tanaka et al. 2007; Zegerman and Diffley 2007), how these interactions promote the initiation of DNA replication has not been elucidated. To investigate the implication of these interactions in DNA replication, we tried to identify the protein complexes that form in a CDK-dependent manner. Using a cross-linking reagent, we identified a fragile complex called the preloading complex (pre-LC), which contains Pol ɛ, GINS, Sld2, and Dpb11. The pre-LC forms before any association with origins in a CDK-dependent and DDK-independent manner. Pol ɛ, GINS, Sld2, and Dpb11 can form a complex in vitro, and their genetic interactions indicate the importance of the complex formation in vivo. Based on these findings, we propose that CDK activity regulates the initiation of DNA replication through formation of the pre-LC.
Results
Fragile DNA replication complexes are detected in the cross-linked cell extracts
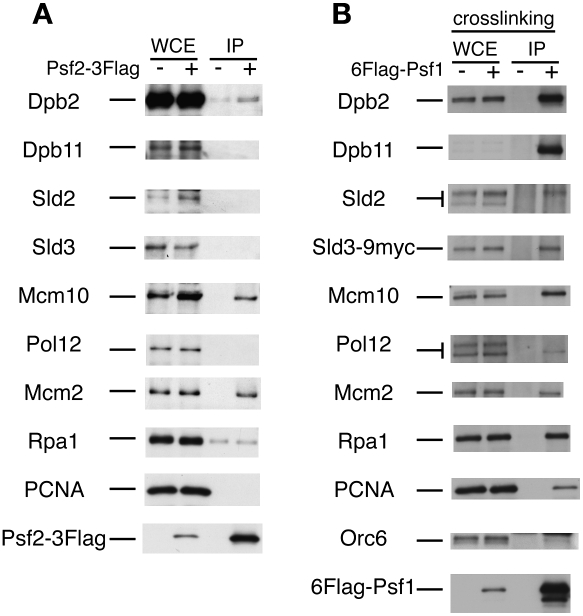
To analyze the CDK-dependent formation of complexes containing replication proteins, we first precipitated Flag-tagged Psf2, a subunit of GINS, with anti-Flag antibody from S-phase cells in which CDK is activated. We then examined the coprecipitates using antibodies against various replication proteins and detected Dpb2 (the second largest subunit of Pol ɛ), Mcm10, and Mcm2 (Fig. 1A). (We did not examine whether Cdc45 coprecipitates with GINS because of the lack of strong antibodies against Cdc45.) The genetic analysis and two-hybrid assay showed that GINS interacts with several replication proteins other than the coprecipitated proteins (Takayama et al. 2003). The lack of these replication proteins in this precipitate may relate to fragility of the complexes of these proteins. We then used a cross-linking reagent, formaldehyde, to stabilize the fragile complex, and identified Dpb11, Sld2, Sld3, Pol12 (the second largest subunit of Pol α), Mcm2, Rpa1 (the largest subunit of an ssDNA-binding protein, RPA), and PCNA (a sliding clamp for Pol δ and Pol ɛ) (Fig. 1B). After formaldehyde treatment, anti-Dpb11 antibodies hardly reacted with Dpb11 in whole-cell extracts, although they detected the precipitated Dpb11, and anti-Sld2 antibodies reacted with Sld2 very weakly. This coprecipitation occurred only with specific proteins. Orc6, a subunit of Orc, did not coprecipitate, and only the slow-migrating form of Sld2 and the fast-migrating form of Pol12 coprecipitated. Sld2 is phosphorylated by S-phase-specific CDK (S-CDK), and its slow-migrating form appears from the G1/S boundary (Masumoto et al. 2002), whereas Pol12 is phosphorylated by M-phase-specific CDK (M-CDK), and the slow-migrating form appears from the S/G2 boundary (Foiani et al. 1995). These results suggest that GINS forms complexes with Sld2 and Pol α at the G1/S boundary and in S-phase cells, which have active S-CDK.
Figure 1.
Coprecipitation of replication proteins with GINS in the absence or presence of cross-linking agent. (A) YNIG20 (PSF2-3Flag-1HA) cells were arrested with hydroxyurea (HU) (0.2 M, 2 h), and cell extracts were prepared. Proteins were immunoprecipitated with the anti-Flag antibody. After the proteins were separated by SDS-PAGE, Western blotting was performed with the antibodies indicated. The samples used as whole-cell extracts (WCEs) corresponded to 2% of proteins used for immunoprecipitation. (B) Asynchronous YYK61 (6FlagPSF1 SLD3-9myc) cells were cross-linked with formaldehyde. The cell extracts were prepared using a bead shocker (Yasui Kikai) and immunoprecipitated with the anti-Flag antibody. Whole-cell extracts corresponded to 1% of the proteins used for immunoprecipitation except for 5% for Orc6 and Sld2. The Dpb11 protein was hardly detected in the formaldehyde-fixed whole-cell extracts.
Complex formation is cell cycle-dependent
To elucidate the complex formation further, we synchronized the cells in which the Psf1 subunit of GINS was tagged with Flag, and CDC6 was expressed under the control of galactose. Cdc6 is required for the pre-RC formation, which is essential for further association of replication proteins with origins. This was confirmed by no association of Sld2 with the origin (Supplemental Fig. 1). Thus, the Cdc6-depleted cells do not initiate DNA replication. Cells synchronized in M phase by nocodazole were released into the medium containing α-factor and glucose so that they completed mitosis and were arrested in the subsequent G1 phase in the absence of expression of Cdc6 and formation of the pre-RC. The cells were then released from α-factor, treated with formaldehyde, and subjected to immunoprecipitation with anti-Flag antibody. As a control, cells with CDC6 expressed from the wild-type promoter were treated in the same manner.
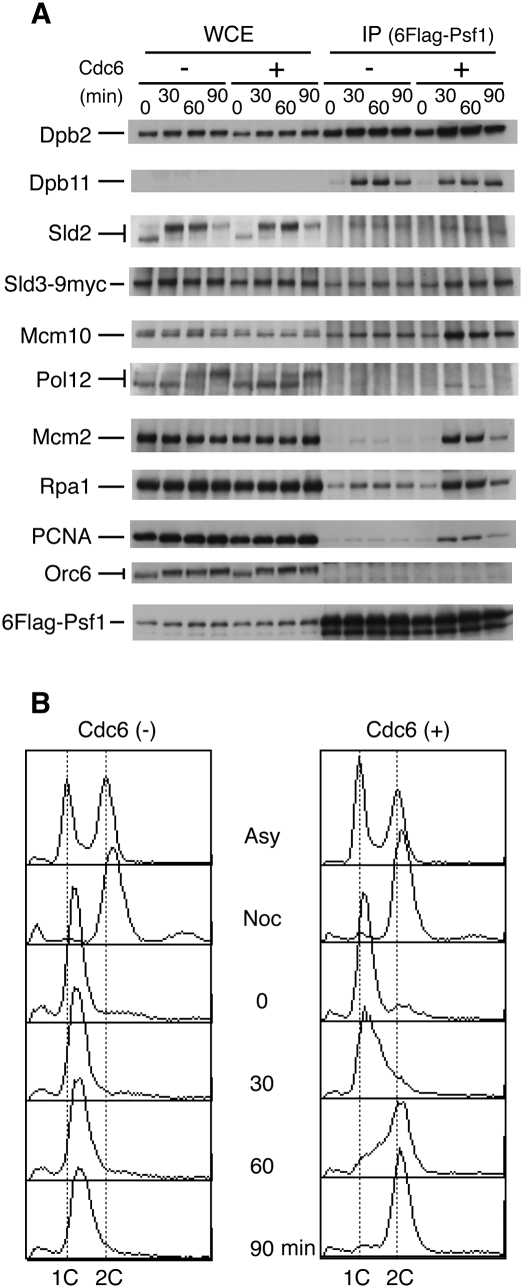
At first, we noticed that the proteins we examined could be classified into four groups (Fig. 2A): Orc6, the smallest subunit of Orc, did not associate with Psf1 at any point; Dpb2 associated with Psf1 throughout the cell cycle; Dpb11 and Sld2 associated with Psf1 only when the cells were released from G1 block; and Sld3, Mcm10, Pol12, Mcm2, Rpa1, and PCNA associated with Psf1, and their association was enhanced during DNA replication (Fig. 2A,B). Sld3 and Mcm10 coprecipitated slightly with Psf1 even in the absence of Cdc6, and their associations increased when replication started. Since the purified Sld3 and Mcm10 proteins bind to various proteins weakly (our unpublished results), we concluded that these proteins associate with Psf1 during DNA replication. Thus, the proteins of this group seem to associate mainly with the replication forks. We discuss the function of Sld3 further in the Discussion. Instead, Psf1, Dpb2, Dpb11, and Sld2 formed a complex irrespective of DNA replication (Fig. 2A, Cdc6 −). Because the subunits of GINS and Pol ɛ hardly dissociate from the complexes, this result indicates that GINS, Pol ɛ, Dpb11, and Sld2 form a complex in the absence of their association with replication origins.
Figure 2.
Coprecipitation of replication proteins with GINS during the cell cycle. (A) YYK61 (6Flag-PSF1 SLD3-9myc) (+) and YYK62 (6Flag-PSF1 SLD3-9myc GAL1p-CDC6) (−) cells were arrested at G2/M phase with nocodazole (5 μg/mL) for 2.5 h in YPAGal (galactose as a carbon source), transferred to YPDA (glucose as a carbon source) containing nocodazole, released in YPDA containing α-factor (30 ng/mL), and incubated for 2.5 h at 24°C. Aliquots of the cells released from α-factor were withdrawn at the times indicated, cross-linked with formaldehyde, and immunoprecipitated with anti-Flag antibodies. Note that the Sld2 protein level fluctuated during the cell cycle and reached the lowest level at G1 (0 min). The Sld2-Flag protein level does not fluctuate significantly during the cell cycle (Masumoto et al. 2002), probably because the Flag-tagged C-terminal stabilizes the fusion protein. The amount of proteins applied as whole-cell extracts corresponds to 2% of that used for immunoprecipitation. (B) The cells were also subjected to flow cytometry analysis.
The pre-LC contains Pol ɛ, GINS, Sld2, and Dpb11
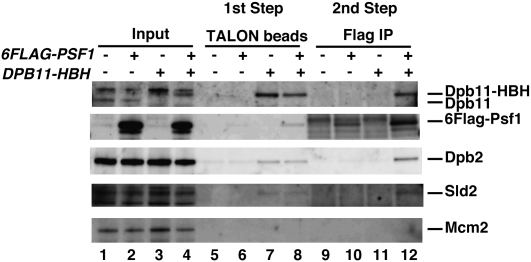
As shown above, GINS forms a complex with Pol ɛ, Sld2, and Dpb11. To determine whether they are included in the same complex, we purified the putative Pol ɛ–Dpb11–Sld2–GINS complex by a two-step precipitation from the cells in which CDC6 was expressed under the control of galactose, and Psf1 and Dpb11 were tagged with Flag and HBH (Tagwerker et al. 2006), respectively.
Synchronized cells that did not express Cdc6 were fixed 40 min after release from G1 block. After disrupting the cells, we first precipitated Dpb11-HBH with TALON beads and eluted the bound proteins with imidazole. Psf1, Dpb2, and Sld2 coeluted with Dpb11 (Fig. 3, lanes 6,7). We then precipitated 6Flag-Psf1 from the material using anti-Flag antibody beads, and found Sld2, Dpb2, and Dpb11, but not Mcm2, in this precipitate (Fig. 3, lane 12). Without the Flag tag on Psf1, they did not coprecipitate on the beads (Fig. 3, lane 11). These results suggest the presence of a Pol ɛ–Dpb11–Sld2–GINS complex or a mixture of Sld2–Dpb11–GINS and Pol ɛ–Dpb11–GINS complexes. Because an efficient association between Dpb11 and GINS requires Pol ɛ and Sld2 (see below), the Sld2–Dpb11–GINS and Pol ɛ–Dpb11–GINS complexes hardly exist in vivo. Therefore, it seems likely that Pol ɛ, Dpb11, Sld2, and GINS are involved in the same complex, which we call the pre-LC.
Figure 3.
Two-step coprecipitation of Sld2, Dpb11, Pol ɛ, and GINS. YYK60 (Galp-CDC6), YYK62(Galp-CDC6 6Flag-PSF1), YNIG231(Galp-CDC6 DPB11-HBH) and YNIG217(Galp-CDC6 6Flag-PSF1 DPB11-HBH) cells were synchronized at 24°C in the condition that depletes Cdc6, as described in the legend for Figure 2. At 40 min after release from the G1 block, the cells were fixed with formaldehyde and disrupted. The resultant cell extracts were mixed with Dynabeads TALON (Dynal). The proteins were eluted from these beads with imidazole and precipitated with Dynabeads Protein A (Dynal) conjugated to anti-Flag (M2). The bound proteins were denatured by heat and subjected to SDS-PAGE, followed by Western blotting.
Formation of the pre-LC requires CDK but not DDK
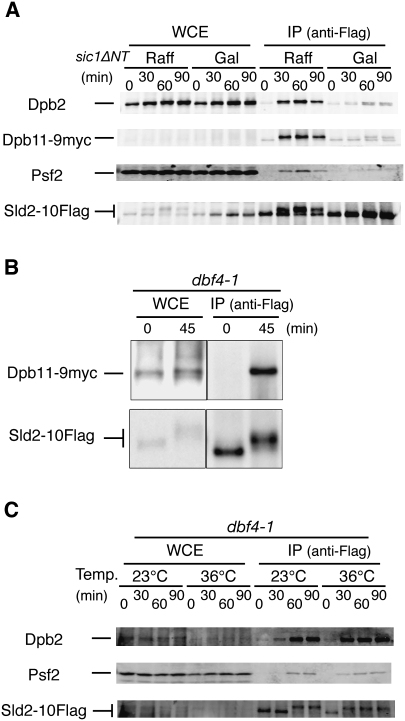
Two protein kinases, CDK and DDK, are essential for the initiation of DNA replication. To examine the requirement for CDK activity in the formation of the pre-LC, we arrested cells in G1 phase with α-factor and expressed Sic1ΔNT, a stable inhibitor of S-CDK and M-CDK that is under the control of galactose, and then released them from α-factor. Expression of Sic1ΔNT inhibits S-CDK and M-CDK, and consequently arrested the cells at the G1/S boundary. As expected, the slow-migrating form of Sld2, a hyperphosphorylated form dependent on CDK, appeared in the absence of Sic1ΔNT (Fig. 4A, Raff) but not in the cells expressing Sic1ΔNT (Fig. 4A, Gal). Although Dpb2, Dpb11, and Psf2 precipitated with Sld2-10Flag after release from the G1 block in the absence of Sic1ΔNT, their coprecipitation decreased when the cells expressed Sic1ΔNT (Fig. 4A), indicating that their association depends on CDK activity.
Figure 4.
CDK-dependent formation of the pre-LC. (A) YNIG193 (SLD2-10Flag GAL1p-sic1ΔNT DPB11-9myc) cells were arrested at G1 phase with α-factor (30 ng/mL) in YPRaff for 1.5 h at 25°C. The culture was then split into two, and galactose (2%) was added to one half. After 45 min, the cells were released from α-factor in YPAGal (0 min). The other half was released from α-factor in YPRaff. Aliquots of cells released from α-factor were withdrawn at the times indicated, cross-linked with formaldehyde, and immunoprecipitated with the anti-Flag antibody as described in the legend for Figure 1. Western blotting was performed with the antibodies indicated. Dpb11-9myc and Sld2-10Flag were detected using anti-myc antibody 9E10 and anti-Flag antibody M2, respectively. (Raff) Raffinose; (Gal) galactose. (B) YS136 cells (dbf4-1 SLD2-10Flag DPB11-9myc) arrested with α-factor for 2.5 h at 25°C were shifted to 36°C and incubated further for 30 min at 36°C. The cells were then released (0 min) and cultivated for 45 min at 36°C. The cells were lysed with Lyticase (Sigma), and the proteins were precipitated with anti-Flag antibody M2 as described (Takayama et al. 2003). (C) YS134 (dbf4-1 SLD2-10Flag) cells were arrested at G1 phase with α-factor (30 ng/mL) in YPDA for 1.5 h at 23°C. The culture was split into two, and one half was shifted up to 36°C. After 30 min, cells were released from α-factor (0 min). Aliquots of cells were withdrawn at the times indicated, cross-linked with formaldehyde, and immunoprecipitated with anti-Flag antibody, as described in the legend for Figure 1. Western blotting was then performed with the antibodies indicated. The FACS patterns of the synchronized cells are shown in Supplemental Figure 5.
Sld2-P1 and Dpb11-C proteins, truncated versions of Sld2 and Dpb11, form a complex in vitro when Sld2-P1 is phosphorylated by CDK (Tak et al. 2006), suggesting that DDK is not required for this complex formation. In the cells that are arrested in G1 by α-factor and released at nonpermissive temperature for dbf4-1—a temperature-sensitive allele of DBF4 that encodes a regulatory subunit of DDK—Sld2 and Dpb11 coprecipitated without applying a cross-linking reagent (Fig. 4B). When the cells were cross-linked with formaldehyde, Dpb2 and Psf2 also precipitated with Sld2-10Flag after release from G1 block without DDK activity (Fig. 4C). Thus, the pre-LC formation appears to depend on CDK but not on DDK.
Pol ɛ, GINS, Sld2, and Dpb11 form a complex in vitro
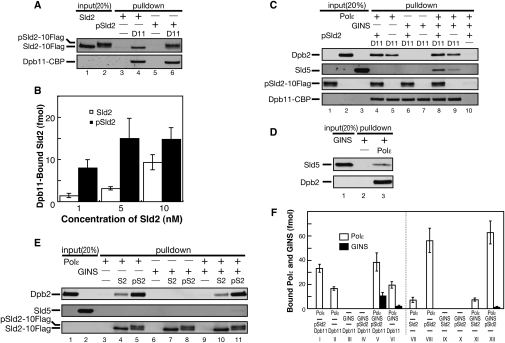
Coimmunoprecipitation of Pol ɛ, GINS, Sld2, and Dpb11 suggested that all are components of the pre-LC complex. However, we did not know whether Pol ɛ, GINS, Sld2, and Dpb11 can form a complex, or whether other missing components are required for complex formation. We next tried to form a complex from purified Pol ɛ, Sld2, Dpb11, and GINS proteins in the absence of formaldehyde (Supplemental Fig. 2).
We first mixed various amounts of Sld2 with 0.25 pmol of Dpb11 immobilized on the beads. Sld2 bound to Dpb11 specifically because it did not bind to Cdc45 or Mcm4 (Supplemental Fig. 3). Moreover, phosphorylation of Sld2 by CDK enhanced this binding by threefold to fivefold (Fig. 5A,B). Next, we mixed 0.5 pmol (10 nM) each of Pol ɛ and GINS with the pSld2–Dpb11 complex formed on beads (∼0.1 pmol of phosphorylated Sld2 [pSld2] bound to the immobilized Dpb11 [0.5 pmol]). In this reaction, Pol ɛ and GINS associated with pSld2 and Dpb11 on the beads (Fig. 5C, lane 8), indicating that they had formed a complex.
Figure 5.
In vitro complex formation from purified proteins. (A) Dpb11–CBP immobilized to beads (0.250 pmol of Dpb11) was incubated with 0.125 pmol (5 nM) of Sld2-10Flag and CDK-phosphorylated Sld2-10Flag for 10 min at 4°C. The Sld2 and Dpb11 were detected by antibodies against Flag and CBP tagged to Sld2 and Dpb11, respectively. (B) Dpb11-immobilized beads (0.5 pmol of Dpb11) were mixed with various concentration of Sld2, and the amounts of phosphorylated Sld2 and bound Sld2 were measured. The results are the average of three independent experiments and standard deviation. (C) Dpb11-immobilized beads with phosphorylated Sld2 (0.5 pmol of Dpb11 and 0.1 pmol of phosphorylated Sld2) were incubated with 0.5 pmol (10 nM) of Pol ɛ and GINS for 1 h at 4°C. The Pol ɛ and GINS bound to the beads were detected by antibodies against the Dpb2 subunit of Pol ɛ and the Sld5 subunit of GINS. (D) Pol ɛ (0.5 pmol) immobilized to beads was incubated with 0.5 pmol (10 nM) of GINS for 1 h at 4°C. (E) Sld2 or CDK-phosphorylated Sld2 immobilized to beads (0.5 pmol) was incubated with 0.5 pmol (10 nM) of Pol ɛ and GINS for 1 h at 4°C. (F) The amounts of Pol ɛ and GINS bound to beads in C and E were estimated from Western blotting, and are shown as the average of three independent experiments and standard deviation.
We then examined the requirements of the complex formation in vitro. When we did not add Sld2, Pol ɛ binding to Dpb11 was reduced to half (Fig. 5C [lanes 4,5,8,9], F [columns I,II,V,VI]), indicating an important role of Sld2 in Pol ɛ binding. Moreover, Pol ɛ was required for GINS to bind Dpb11 (Fig. 5C, lanes 6–9). Consistent with this observation, Pol ɛ and GINS formed a complex in vitro (Fig. 5D).
Interestingly, the ratio of GINS/Pol ɛ was higher in the GINS–Polɛ–Dpb11 complex (∼12% [2.2 fmol/19 fmol]) (Fig. 5C [lane 9], F [column VI]) than in the complex formed between them (∼6.0% [30 fmol/500 fmol]) (Fig. 5D). The interaction between Dpb11 and the Psf1 subunit of GINS in a two-hybrid assay (Takayama et al. 2003) suggests that Dpb11 stabilizes the association of GINS with Pol ɛ through the weak Dpb11–Psf1 interaction. The ratio of GINS/Pol ɛ was increased more by the addition of pSld2 (29% [11 fmol/38 fmol]) (Fig. 5C [lane 8], F [column V]), suggesting that pSld2 or the pSld2–Dpb11 complex further stabilizes the association between GINS and Pol ɛ.
To understand how Sld2 enhances the interaction between Dpb11, Pol ɛ, and GINS, we also examined the association between Sld2, Pol ɛ, and GINS. Sld2 (0.5 pmol) was immobilized on the beads using Flag tag and anti-Flag antibody, and was mixed with 0.5 pmol (10 nM) each of Pol ɛ and GINS. We did not add Dpb11 to this reaction because Dpb11 binds to the beads nonspecifically. Pol ɛ bound to the pSld2 beads more efficiently than to the Dpb11 beads. GINS did not bind to Sld2 in the absence of Pol ɛ (Fig. 5E, lanes 8,11), indicating that GINS bound to pSld2 through Pol ɛ. Unlike Dpb11, the ratio of GINS/Pol ɛ in the GINS–Polɛ–Sld2 complex (∼1.7% [1.1 fmol/63 fmol]) (Fig. 5F) was less than that observed for the complex formed between them (∼6.0%), indicating that pSld2 binds efficiently to Pol ɛ but does not stabilize the association between GINS and Pol ɛ. This result suggests that the increase in the ratio of GINS/Pol ɛ by the addition of pSld2 seems to be caused by formation of the pSld2–Dpb11 complex. These results also suggest that Dpb11, Sld2, Pol ɛ, and GINS can form a complex without the aid of other proteins, and that Sld2 and Pol ɛ play roles in the binding of GINS to Dpb11.
Phosphorylation of Sld2 enhanced the binding to Pol ɛ (Fig. 5E,F). We surmise that this phosphorylation-dependent enhancement increases the efficiency of the initiation of DNA replication but may not be essential. This is because alanine substitutions of serine/threonine at all of the CDK phosphorylation motifs except Thr84 in Sld2 do not inhibit the cell growth, and because phospho-Thr84 is required for Dpb11 binding (Tak et al. 2006). Moreover, substitution of alanine for Thr84 did not reduce its binding to Pol ɛ in vitro (Supplemental Fig. 4).
CDK-dependent pre-LC formation requires Sld2
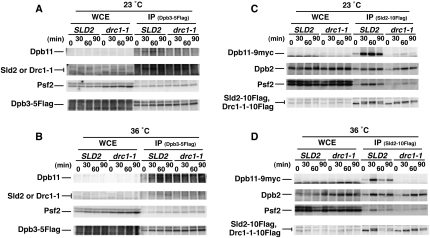
The in vitro complex formation suggested that Sld2 enhances the interaction between Pol ɛ, GINS, and Dpb11. This could explain the CDK dependency of their interactions in vivo (Fig. 3A), because CDK phosphorylates Sld2. We then examined the role of Sld2 in the formation of the pre-LC in vivo. For this purpose, we used a temperature-sensitive allele of SLD2, drc1-1 (Wang and Elledge 1999), which reduces the interaction between Sld2 and Dpb11 (Masumoto et al. 2002). The Drc1-1 has four amino acid alterations (K59R, P85S, G282C, and R358Q) in the Sld2 protein (our unpublished results), in which P85S is responsible for temperature-sensitive growth and interaction with Dpb11 as observed in Sld2-6 (Kamimura et al. 1998).
As shown in Figure 6, A and B, Dpb3-5Flag always precipitated with Psf2 in drc1-1 cells, as observed in the wild-type cells at 23°C and at 36°C. Sld2 also coprecipitated with Dpb3-5Flag in drc1-1 cells when phosphorylated, as observed in the wild-type cells. In contrast, Dpb11 coprecipitated poorly even at 23°C (Fig. 6A), and this coprecipitation was diminished at 36°C (Fig. 6B). We also precipitated Sld2-10Flag of drc1-1 cells. Dpb2 coprecipitated at 23°C and at 36°C, as observed in the wild-type cells (Fig. 6C,D). This coprecipitation was enhanced when the cells were released from the G1 block and Sld2 was phosphorylated, which was consistent with the in vitro result (Fig. 5E). However, Psf2 and Dpb11 coprecipitated poorly at 23°C (Fig. 6C), and their coprecipitation was diminished at 36°C (Fig. 6D). These results indicate that Pol ɛ and GINS form a complex in the absence of Sld2 function, whereas they require Sld2 to interact with Dpb11 in vivo.
Figure 6.
Sld2 function is required for pre-LC formation. (A,B) YNIG14 (Galp-CDC6 DPB3-5Flag) and YNIG224 (Galp-CDC6 drc1-1 DPB3-5Flag) cells were synchronized in G1 phase at 23°C in the condition that depletes Cdc6, as described in the legend for Figure 2. The culture was split into two, and one half was shifted up to 36°C. After 30 min, cells were released from G1 block, withdrawn at 30 min intervals, cross-linked, and immunoprecipitated with anti-Flag antibody M2. (C,D) YNIG225 (Galp-CDC6 SLD2-10Flag DPB11-9myc) and YNIG226 (Galp-CDC6 drc1-1-10Flag DPB119myc) were treated as described in A and B.
Genetic interactions are consistent with the formation of the pre-LC
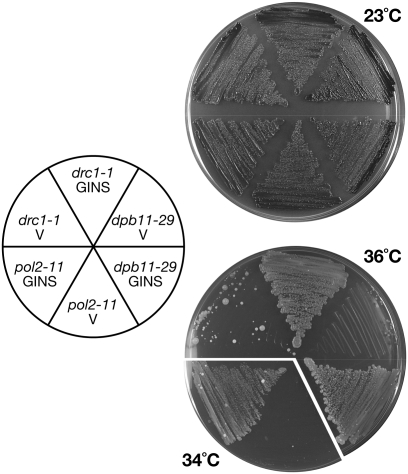
A mutation occurring in one subunit of a complex is often suppressed by increasing the dosage of other subunits. This is true for the Sld2–Dpb11 (Kamimura et al. 1998; Wang and Elledge 1999), GINS (Takayama et al. 2003), and Pol ɛ (Araki et al. 1995; Iida and Araki 2004) complexes. We extended this high-copy suppression analysis to GINS. We cloned the genes encoding the four subunits of GINS on high-copy plasmids and introduced them into pol2-11, dpb11-29, and drc1-1 mutant cells defective in Pol ɛ, Dpb11, and Sld2, respectively. Whereas the cells harboring one of those temperature-sensitive mutations could not grow at the restrictive temperature, simultaneous introduction of the high-copy plasmids bearing all four genes of GINS restored the cell growth (Fig. 7). This suppression was specific because the growth defect of sld3 or cdc45 cells was not suppressed by the same plasmid (data not shown). Temperature-sensitive growth of pol2-11 mutant cells defective in a catalytic subunit of Pol ɛ is suppressed by high copy of DPB11 and SLD2 (Araki et al. 1995; Kamimura et al. 1998; Wang and Elledge 1999), suggesting that Pol ɛ interacts with Sld2 and Dpb11, and that the pol2-11 mutation reduces its interaction. The dpb11-29 mutation, which occurs in the C-terminal half of Dpb11, and the drc1-1 mutation reduce the interaction between Dpb11 and Sld2 (Wang and Elledge 1999; Masumoto et al. 2002). Because GINS requires the Dpb11–Sld2 complex and Pol ɛ for efficient interaction between GINS and Dpb11 (Fig. 5), the suppression of pol2-11, dpb11-29, and drc1-1 mutations by increasing the dosage of GINS suggests that GINS interacts with Pol ɛ, Dpb11, and Sld2 specifically, and that the formation of the pre-LC complex is essential.
Figure 7.
Suppression of thermosensitive growth of pol2-11, dpb11-29, and drc1-1 (sld2) by increasing dosages of GINS. YHA211 (pol2-11), YNIG61 (dpb11-29), and Y799 (drc1-1) cells carrying both YEp195 and YEp181 (V) or both YEp195SLD5-PSF1 and YEp181PSF2-PSF3 (GINS) were streaked onto YPD plates and were incubated for 4 d at the temperatures indicated.
Discussion
CDK-catalyzed phosphorylation of Sld2 and Sld3, and their subsequent association with Dpb11, is essential for the initiation of chromosomal DNA replication in budding yeast (Tanaka et al. 2007; Zegerman and Diffley 2007). In this study, we identified a complex, the pre-LC, that contains Pol ɛ, GINS, Sld2, and Dpb11. We argue that the formation of the pre-LC is part of the CDK-dependent process mediated by the interaction between Sld2 and Dpb11.
The pre-LC complex is essential for the initiation of chromosomal DNA replication
Although the pre-LC was detected only in formaldehyde-fixed cell extracts, several lines of evidence strongly suggest that the formation of the pre-LC occurs in vivo. First, coimmunoprecipitation of proteins after formaldehyde treatment was still specific (Figs. 1, 2). For example, Orc did not precipitate with components of the pre-LC. Second, we demonstrated that purified Pol ɛ, GINS, Sld2, and Dpb11 proteins form a complex without a cross-linker (Fig. 5). Thus, Pol ɛ, GINS, Sld2, and Dpb11 proteins are the minimal components constituting the pre-LC. Third, the genetic interactions observed previously (Araki et al. 1995; Kamimura et al. 1998; Takayama et al. 2003) and in this study are consistent with formation of the pre-LC. We thus propose that the pre-LC forms in cells, and that this formation is mediated by the interaction between Sld2 and Dpb11. Because a small amount of the pre-LC was recovered from the formaldehyde-treated cells, the pre-LC appears to be formed transiently.
Pol ɛ, GINS, Sld2, and Dpb11 proteins, the components of the pre-LC, are all essential for the initiation of chromosomal DNA replication, and associate with replication origins in a mutually dependent manner (Masumoto et al. 2000; Kamimura et al. 2001; Takayama et al. 2003). The dpb2-1 and dpb11-1 mutations reduce the interaction between Pol ɛ and Dpb11 (Masumoto et al. 2000) and the interaction between Dpb11 and Sld2 (Kamimura et al. 1998), respectively, indicating that these mutations reduce formation of the pre-LC. This study also showed that the drc1-1 mutation defective in Sld2 reduces the formation of the pre-LC (Fig. 6). Thus, reducing the pre-LC formation seems to confer a defect in DNA replication. In contrast, increasing the dosage of DPB11, SLD2, or GINS suppresses dpb2-1, drc1-1, and dpb11-1 (Araki et al. 1995; Kamimura et al. 1998; Takayama et al. 2003), probably by stabilizing the pre-LC complex. Taken together, these data suggest that formation of the pre-LC is essential for DNA replication, especially for the initiation step.
How is the pre-LC loaded onto origins?
We argue that the pre-LC complex is formed before its components are loaded properly onto origins. The pre-LC was formed in the absence of the pre-RC (Fig. 2); in this condition, the components of the pre-LC do not associate with origin DNA, and formation of the pre-LC was resistant to DNase I (our unpublished results). Moreover, Dpb11, Sld2, Pol ɛ, and GINS associate with replication origins in a mutually dependent manner (Masumoto et al. 2000; Takayama et al. 2003). Thus, the components of the pre-LC seem to associate with origins only when they form the pre-LC complex.
How does the pre-LC associate with replication origins? We speculate that the pre-LC forms first and then binds preferentially to phosphorylated Sld3 on replication origins because Sld3, together with Cdc45, associates with origins before and independently of the pre-LC (Kamimura et al. 2001), and because phosphorylated Sld3 binds to the N-terminal tandem BRCT domain of Dpb11 (Tanaka et al. 2007; Zegerman and Diffley 2007). A trace amount of Sld3 precipitated with GINS throughout the cell cycle, and no significant increase in the coprecipitation after release from the G1 block was observed in the Cdc6-depleted and formaldehyde-fixed cells. In contrast, the coprecipitation of Sld3 and GINS increased slightly at the G1/S boundary and in the S phase of wild-type cells in which Sld3 associates with origins (Fig. 2A). In addition, protein–protein interactions between subunits of the pre-RC, GINS, and Pol ɛ may stabilize the complex of the pre-LC and pre-RC on origins. In a two-hybrid assay, the Psf1 subunit of GINS interacts with Sld3 (Takayama et al. 2003), and Sld3 interacts with Mcm, a component of the pre-RC (our unpublished results). A recent proteomic study showed that Dpb2, the second largest subunit of Pol ɛ coprecipitates with the Orc1 and Orc4 subunits of Orc (Krogan et al. 2006). This suggests that the pre-LC but not the complex lacking GINS or Pol ɛ associates with replication origins.
Pol ɛ is important for pre-LC formation
Pol ɛ is essential for DNA replication. However, the catalytic domain located in Pol2, the largest subunit of Pol ɛ, is dispensable, whereas the Pol2 C-terminal portion is essential for DNA replication (Dua et al. 1999; Kesti et al. 1999; Feng and D'Urso 2001). How the essential C-terminal portion of Pol2 works in DNA replication is a long-standing question. This study suggests that one of the essential functions of the C-terminal portion of Pol2 in budding yeast is formation of the pre-LC because it is required for GINS to bind to the Sld2–Dpb11 complex. The C-terminal portion of Pol2 interacts with Dpb11 and Sld2 as well as the Dpb2 subunit of Pol ɛ (Edwards et al. 2003). Moreover, Dpb2 interacts with the Psf1 subunit of GINS (Takayama et al. 2003). Thus, the C-terminal portion of Pol2 connects the Sld2–Dpb11 complex to GINS via Dpb2. This connection is also supported by the genetic interaction; that is, the pol2-11 mutation occurring in the C terminus is suppressed by increasing dosages of Dpb11, Sld2, and GINS (Fig. 7; Araki et al. 1995; Kamimura et al. 1998). Dpb2 is also indispensable for cell growth, although the degraded catalytic subunit of Pol ɛ alone shows a polymerizing activity indistinguishable from that of four-subunit Pol ɛ (Hamatake et al. 1990). One of the essential functions of this subunit may also be attributed to the pre-LC formation.
The essential function of Pol ɛ in the initiation of DNA replication may be conserved in higher eukaryotes. Pol ɛ is dispensable for SV40 virus DNA replication and does not associate with SV40 DNA (Zlotkin et al. 1996), whereas it is indispensable for chromosomal DNA replication (Pospiech et al. 1999; Waga et al. 2001). T-antigen encoded by SV40 recognizes its origin and functions as a replicative helicase instead of Orc and CMG (Waga and Stillman 1998). It is conceivable that Pol ɛ in mammalian cells also aids the proper association of replication proteins, such as GINS, at replication origins, and thus SV40 DNA replicates in the absence of Pol ɛ. The essential function of mammalian Pol ɛ in the initiation step is also consistent with localization of Pol ɛ in S phase, because it colocalizes with PCNA, a sliding clamp at the replication forks, only in late but not in early S phase (Fuss and Linn 2002). If Pol ɛ functions in the proper association of replication proteins with origins, it may associate transiently with origins, and thus it may not colocalize with PCNA. In late S phase, the heterochromatic region is replicated. Two nonessential subunits of Pol ɛ, Dpb3 and Dpb4, have histone fold motifs and function in the silencing of gene expression in heterochromatin-like regions of budding yeast (Iida and Araki 2004; Tackett et al. 2005). Thus, it is possible that, in mammalian cells, Pol ɛ also works with PCNA to regulate heterochromatin during late S phase.
The results obtained from Xenopus egg extracts contradict our idea apparently. Depletion of either Pol ɛ or Pol δ from the extracts reduces but does not abolish DNA replication, while depletion of both diminishes almost all DNA replication (Waga et al. 2001; Fukui et al. 2004). This result suggests that two DNA polymerases, Pol ɛ and Pol δ, work for DNA replication and partially compensate each other. Moreover, even in the absence of Pol ɛ, Cdc45 associates with chromatin (Fukui et al. 2004). It is thus suggested that Pol ɛ does not participate in the initiation step that occurs before chromatin association of Cdc45. Egg extracts contain an extraordinary amount of DNA replication proteins, which support several rounds of replication. In this condition, GINS may associate with replication origins in the absence of the specific machinery, like the pre-LC. However, further study is required to reveal whether the initiation step occurs properly in the absence of Pol ɛ in the extracts.
Role of the pre-LC complex in the initiation step of chromosomal DNA replication
Recent studies identified the complex comprising Cdc45, Mcm, and GINS (CMG complex) from various organisms (Gambus et al. 2006; Moyer et al. 2006; Pacek et al. 2006). Moreover, the CMG complex from Drosophila embryo cells shows robust helicase activity (Ilves et al. 2010), suggesting that it works as a replicative helicase at replication forks. Cdc45 associates with the Mcm-bound replication origins in an Sld3-dependent manner, and GINS joins this complex as a component of the pre-LC. According to this viewpoint, the pre-LC is a carrier of GINS to the pre-RC to form a CMG complex and to activate the helicase activity of Mcm. The association between Dpb11 and GINS is enhanced mostly by the CDK-catalyzed phosphorylation-dependent interaction between Dpb11 and Sld2 (Fig. 5F). Thus, CDK might promote the initiation of DNA replication by enhancing GINS recruitment to origins through the formation of the pre-LC. Once DNA replication starts, Sld2, Sld3, and Dpb11 do not move with the replication forks, whereas Cdc45 and GINS form an active helicase with MCM and work as major components at the replication forks. In this scenario, the pre-LC must be disrupted for the initiation of DNA replication. The fragile characteristic of the pre-LC is suitable for this mechanism.
We do not know whether the pre-LC or related complex is formed in other organisms. However, BRCT-containing proteins (Dpb11-related proteins) seem to play important roles in recruiting GINS to replication origins. In fission yeast, Sld3 associates with replication origins in a DDK-dependent and Cdc45-independent manner. GINS and Cut5, a counterpart of Dpb11, associate with origins in a mutually dependent manner, and their association is a prerequisite for the association between Cdc45 and the origin (Yabuuchi et al. 2006). This step requires CDK activity, which enhances the interaction between Cut5 and Drc1, a counterpart of Sld2 (Noguchi et al. 2002). Although the association order of replication proteins with origins differs from budding yeast, the BRCT-containing protein Cut5 seems to play a role in recruiting GINS to origins. This is also true in Xenopus egg extracts. Chromatin association of Cut5/TopBP1, a counterpart of Dpb11, comprises two distinct modes: one CDK-independent and the other CDK-dependent. CDK-independent Cut5 association is enough for further associations of replication proteins. When CDK is activated, Cdc45 and GINS associate with chromatin in a mutually dependent and Cut5-dependent manner (Hashimoto and Takisawa 2003).
Materials and methods
Yeast strains and plasmids
All of the yeast strains used in this study originated from W303 and are listed in Supplemental Table 1. Plasmids harboring two of four genes encoding GINS proteins were constructed as follows: The XbaI–KpnI fragment bearing SLD5 obtained from YCp22SLD5 (Takayama et al. 2003) was inserted into the XbaI–KpnI-cleaved YEp195PSF1 (Takayama et al. 2003), and the resultant plasmid was named YEp195(SLD5 + PSF1). The Tth111I–NcoI fragment bearing PSF3 was amplified by PCR and was cloned directly into the SmaI site of YEplac195 (Gietz and Sugino 1988) for YEp195PSF3 (Takayama et al. 2003). The EcoRI–SalI and EcoRI–BamHI fragments of YEp195PSF3 bearing PSF3 were ligated with EcoRI- and SalI-cleaved YEplac181 (Gietz and Sugino 1988) for YEp181PSF3, and with EcoRI and BamHI-cleaved YCplac22 (Gietz and Sugino 1988) for YCp22PSF3 (Takayama et al. 2003), respectively. The SalI–SphI fragment bearing PSF2 was obtained from YEp195PSF2 (Takayama et al. 2003), and was cloned into the SalI–SphI gap of YEp181PSF3. The resultant plasmid was named YEp181(PSF2 + PSF3).
Immunoprecipitation of formaldehyde-treated cell extracts
Immunoprecipitation of formaldehyde-treated cell extracts was performed essentially the same way as described in the chromatin immunoprecipitation assay (Takayama et al. 2003). The detailed procedure is described in the Supplemental Material.
For two-step coprecipitation, the cell extracts were mixed with Dynabeads TALON (Dynal). The proteins eluted from these beads with 150 mM imidazole were precipitated with Dynabeads Protein A (Dynal)-conjugated anti-Flag (M2).
In vitro pull-down assay
All in vitro analyses were performed at 4°C with the reaction buffer (50 mM HEPES-KOH at pH 7.5, 200 mM potassium acetate, 2 mM magnesium acetate, 10% glycerol, 0.1% Tween 20, 0.01% Nonidet P40, 1 mg/mL bovine serum albumin, partially hydrolyzed casein [10 mg/mL for Sld2–Dpb11 reactions; 2.5 mg/mL for other reactions]). The method for purifying proteins is described in the Supplemental Material.
Dpb11 was immobilized to anti-CBP antibody (Upstate Biotechnologies)-conjugated Dynabeads M-270 Epoxy (Dynal) prepared according to the supplier's instructions with post-treatment with ethanolamine. For the Dpb11–Sld2 interaction assay, ∼5 × 106 beads with 0.25 pmol of Dpb11–CBP were incubated with 25 μL of the reaction buffer containing various amounts of Sld2-10Flag for 10 min. For interactions among Dpb11, Sld2, Pol ɛ, and GINS, the Dpb11 beads were first incubated with phosphorylated or unphosphorylated Sld2-10Flag for 20 min in the reaction buffer. After washing, the Dpb11–Sld2 beads (0.5 pmol of Dpb11 and 0.1 pmol of Sld2 on 1 × 107 beads) were mixed with 50 μL of the reaction buffer containing 0.5 pmol of Pol ɛ and GINS, and were incubated for 1 h.
Phosphorylated or unphosphorylated Sld2-10Flag was immobilized onto anti-Flag M2 antibody (Sigma-Aldrich)-conjugated Dynabeads M-270 Epoxy (Dynal) and was used for the Sld2–Pol ɛ–GINS interaction assay. For the Pol ɛ–GINS interaction assay, Pol ɛ bearing Dpb3-Flag was immobilized onto anti-Flag M2 antibody-conjugated Dynabeads M-270 Epoxy.
After the reactions, beads were washed twice with 1 mL of the reaction buffer, and the bound proteins were separated by 5%–20% gradient SDS-PAGE and analyzed by Western blotting. Proteins were visualized and quantified using Odyssey Infrared Imaging System (LI-COR).
Phosphorylation of Sld2
Sld2 protein was phosphorylated using recombinant Cdc28–Clb5 (Tak et al. 2006) in phosphorylation buffer (50 mM HEPES-KOH at pH 7.5, 2 mM MgCl2, 0.2 mM ATP, 0.25 mg/mL partially hydrolyzed casein) for 90 min at 30°C. For the unphosphorylated control, Sld2 was incubated in the buffer without kinase.
Acknowledgments
We thank B. Stillman, M. Foiani, and Y. Kawasaki for antibodies; S. Elledge and S. Maki for yeast strains; P. Kaiser for HBH plasmid; N. Shimamoto for advice on in vitro experiments; and K. Labib, H. Masukata, H. Takisawa, and S. Tanaka for critical reading of the manuscript. S.M. and Y.K. examined coprecipitation of proteins from cross-linked cells, and S.M. also examined high-copy suppressions of mutants. K.H. performed in vitro analysis of complex formation. T.Y.-S. examined Sld2–Dpb11 coprecipitation in dbf4 cells. H.A. organized this project and wrote the paper. This study is supported partly by CREST and by Grants-in-Aid from Ministry of Education, Culture, Sports, Science, and Technology to H.A.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1883410.
Supplemental material is available at http://www.genesdev.org.
References
- Araki H, Leem SH, Phongdara A, Sugino A. Dpb11, which interacts with DNA polymerase II (ɛ) in Saccharomyces cerevisiae, has a dual role in S-phase progression and at a cell cycle checkpoint. Proc Natl Acad Sci. 1995;92:11791–11795. doi: 10.1073/pnas.92.25.11791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asturias FJ, Cheung IK, Sabouri N, Chilkova O, Wepplo D, Johansson E. Structure of Saccharomyces cerevisiae DNA polymerase ɛ by cryo-electron microscopy. Nat Struct Mol Biol. 2006;13:35–43. doi: 10.1038/nsmb1040. [DOI] [PubMed] [Google Scholar]
- Bell SP, Dutta A. DNA replication in eukaryotic cells. Annu Rev Biochem. 2002;71:333–374. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- Burgers PM. Polymerase dynamics at the eukaryotic DNA replication fork. J Biol Chem. 2009;284:4041–4045. doi: 10.1074/jbc.R800062200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dua R, Levy DL, Campbell JL. Analysis of the essential functions of the C-terminal protein/protein interaction domain of Saccharomyces cerevisiae pol ɛ and its unexpected ability to support growth in the absence of the DNA polymerase domain. J Biol Chem. 1999;274:22283–22288. doi: 10.1074/jbc.274.32.22283. [DOI] [PubMed] [Google Scholar]
- Edwards S, Li CM, Levy DL, Brown J, Snow PM, Campbell JL. Saccharomyces cerevisiae DNA polymerase ɛ and polymerase σ interact physically and functionally, suggesting a role for polymerase ɛ in sister chromatid cohesion. Mol Cell Biol. 2003;23:2733–2748. doi: 10.1128/MCB.23.8.2733-2748.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W, D'Urso G. Schizosaccharomyces pombe cells lacking the amino-terminal catalytic domains of DNA polymerase ɛ are viable but require the DNA damage checkpoint control. Mol Cell Biol. 2001;21:4495–4504. doi: 10.1128/MCB.21.14.4495-4504.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foiani M, Liberi G, Lucchini G, Plevani P. Cell cycle-dependent phosphorylation and dephosphorylation of the yeast DNA polymerase α-primase B subunit. Mol Cell Biol. 1995;15:883–891. doi: 10.1128/mcb.15.2.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui T, Yamauchi K, Muroya T, Akiyama M, Maki H, Sugino A, Waga S. Distinct roles of DNA polymerases δ and ɛ at the replication fork in Xenopus egg extracts. Genes Cells. 2004;9:179–191. doi: 10.1111/j.1356-9597.2004.00716.x. [DOI] [PubMed] [Google Scholar]
- Fuss J, Linn S. Human DNA polymerase ɛ colocalizes with proliferating cell nuclear antigen and DNA replication late, but not early, in S phase. J Biol Chem. 2002;277:8658–8666. doi: 10.1074/jbc.M110615200. [DOI] [PubMed] [Google Scholar]
- Gambus A, Jones RC, Sanchez-Diaz A, Kanemaki M, van Deursen F, Edmondson RD, Labib K. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat Cell Biol. 2006;8:358–366. doi: 10.1038/ncb1382. [DOI] [PubMed] [Google Scholar]
- Gietz RD, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Glover JN, Williams RS, Lee MS. Interactions between BRCT repeats and phosphoproteins: Tangled up in two. Trends Biochem Sci. 2004;29:579–585. doi: 10.1016/j.tibs.2004.09.010. [DOI] [PubMed] [Google Scholar]
- Hamatake RK, Hasegawa H, Clark AB, Bebenek K, Kunkel TA, Sugino A. Purification and characterization of DNA polymerase II from the yeast Saccharomyces cerevisiae. Identification of the catalytic core and a possible holoenzyme form of the enzyme. J Biol Chem. 1990;265:4072–4083. [PubMed] [Google Scholar]
- Hashimoto Y, Takisawa H. Xenopus Cut5 is essential for a CDK-dependent process in the initiation of DNA replication. EMBO J. 2003;22:2526–2535. doi: 10.1093/emboj/cdg238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida T, Araki H. Noncompetitive counteractions of DNA polymerase ɛ and ISW2/yCHRAC for epigenetic inheritance of telomere position effect in Saccharomyces cerevisiae. Mol Cell Biol. 2004;24:217–227. doi: 10.1128/MCB.24.1.217-227.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilves I, Petojevic T, Pesavento JJ, Botchan MR. Activation of the MCM2–7 helicase by association with Cdc45 and GINS proteins. Mol Cell. 2010;37:247–258. doi: 10.1016/j.molcel.2009.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamimura Y, Masumoto H, Sugino A, Araki H. Sld2, which interacts with Dpb11 in Saccharomyces cerevisiae, is required for chromosomal DNA replication. Mol Cell Biol. 1998;18:6102–6109. doi: 10.1128/mcb.18.10.6102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamimura Y, Tak YS, Sugino A, Araki H. Sld3, which interacts with Cdc45 (Sld4), functions for chromosomal DNA replication in Saccharomyces cerevisiae. EMBO J. 2001;20:2097–2107. doi: 10.1093/emboj/20.8.2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanemaki M, Labib K. Distinct roles for Sld3 and GINS during establishment and progression of eukaryotic DNA replication forks. EMBO J. 2006;25:1753–1763. doi: 10.1038/sj.emboj.7601063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanemaki M, Sanchez-Diaz A, Gambus A, Labib K. Functional proteomic identification of DNA replication proteins by induced proteolysis in vivo. Nature. 2003;423:720–724. doi: 10.1038/nature01692. [DOI] [PubMed] [Google Scholar]
- Kesti T, Flick K, Keranen S, Syvaoja JE, Wittenberg C. DNA polymerase ɛ catalytic domains are dispensable for DNA replication, DNA repair, and cell viability. Mol Cell. 1999;3:679–685. doi: 10.1016/s1097-2765(00)80361-5. [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- Kunkel TA, Burgers PM. Dividing the workload at a eukaryotic replication fork. Trends Cell Biol. 2008;18:521–527. doi: 10.1016/j.tcb.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labib K, Gambus A. A key role for the GINS complex at DNA replication forks. Trends Cell Biol. 2007;17:271–278. doi: 10.1016/j.tcb.2007.04.002. [DOI] [PubMed] [Google Scholar]
- Lou H, Komata M, Katou Y, Guan Z, Reis CC, Budd M, Shirahige K, Campbell JL. Mrc1 and DNA polymerase ɛ function together in linking DNA replication and the S phase checkpoint. Mol Cell. 2008;32:106–117. doi: 10.1016/j.molcel.2008.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masai H, Taniyama C, Ogino K, Matsui E, Kakusho N, Matsumoto S, Kim JM, Ishii A, Tanaka T, Kobayashi T, et al. Phosphorylation of MCM4 by Cdc7 kinase facilitates its interaction with Cdc45 on the chromatin. J Biol Chem. 2006;281:39249–39261. doi: 10.1074/jbc.M608935200. [DOI] [PubMed] [Google Scholar]
- Masumoto H, Sugino A, Araki H. Dpb11 controls the association between DNA polymerases α and ɛ and the autonomously replicating sequence region of budding yeast. Mol Cell Biol. 2000;20:2809–2817. doi: 10.1128/mcb.20.8.2809-2817.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masumoto H, Muramatsu S, Kamimura Y, Araki H. S-Cdk-dependent phosphorylation of Sld2 essential for chromosomal DNA replication in budding yeast. Nature. 2002;415:651–655. doi: 10.1038/nature713. [DOI] [PubMed] [Google Scholar]
- Moyer SE, Lewis PW, Botchan MR. Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc Natl Acad Sci. 2006;103:10236–10241. doi: 10.1073/pnas.0602400103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi E, Shanahan P, Noguchi C, Russell P. CDK phosphorylation of Drc1 regulates DNA replication in fission yeast. Curr Biol. 2002;12:599–605. doi: 10.1016/s0960-9822(02)00739-x. [DOI] [PubMed] [Google Scholar]
- Pacek M, Tutter AV, Kubota Y, Takisawa H, Walter JC. Localization of MCM2-7, Cdc45, and GINS to the site of DNA unwinding during eukaryotic DNA replication. Mol Cell. 2006;21:581–587. doi: 10.1016/j.molcel.2006.01.030. [DOI] [PubMed] [Google Scholar]
- Pospiech H, Kursula I, Abdel-Aziz W, Malkas L, Uitto L, Kastelli M, Vihinen-Ranta M, Eskelinen S, Syvaoja JE. A neutralizing antibody against human DNA polymerase ɛ inhibits cellular but not SV40 DNA replication. Nucleic Acids Res. 1999;27:3799–3804. doi: 10.1093/nar/27.19.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pursell ZF, Kunkel TA. DNA polymerase ɛ: A polymerase of unusual size (and complexity) Prog Nucleic Acid Res Mol Biol. 2008;82:101–145. doi: 10.1016/S0079-6603(08)00004-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pursell ZF, Isoz I, Lundstrom EB, Johansson E, Kunkel TA. Yeast DNA polymerase ɛ participates in leading-strand DNA replication. Science. 2007;317:127–130. doi: 10.1126/science.1144067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwob E, Labib K. Regulating initiation events in yeasts. In: DePamphilis ML, editor. DNA replication and human disease. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2006. pp. 295–311. [Google Scholar]
- Sheu YJ, Stillman B. Cdc7-Dbf4 phosphorylates MCM proteins via a docking site-mediated mechanism to promote S phase progression. Mol Cell. 2006;24:101–113. doi: 10.1016/j.molcel.2006.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheu YJ, Stillman B. The Dbf4-Cdc7 kinase promotes S phase by alleviating an inhibitory activity in Mcm4. Nature. 2010;463:113–117. doi: 10.1038/nature08647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivaprasad U, Dutta A, Bell SP. Assembly of pre-replication complexes. In: DePamphilis ML, editor. DNA replication and human disease. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2006. pp. 63–88. [Google Scholar]
- Tackett AJ, Dilworth DJ, Davey MJ, O'Donnell M, Aitchison JD, Rout MP, Chait BT. Proteomic and genomic characterization of chromatin complexes at a boundary. J Cell Biol. 2005;169:35–47. doi: 10.1083/jcb.200502104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagwerker C, Zhang H, Wang X, Larsen LS, Lathrop RH, Hatfield GW, Auer B, Huang L, Kaiser P. HB tag modules for PCR-based gene tagging and tandem affinity purification in Saccharomyces cerevisiae. Yeast. 2006;23:623–632. doi: 10.1002/yea.1380. [DOI] [PubMed] [Google Scholar]
- Tak YS, Tanaka Y, Endo S, Kamimura Y, Araki H. A CDK-catalysed regulatory phosphorylation for formation of the DNA replication complex Sld2-Dpb11. EMBO J. 2006;25:1987–1996. doi: 10.1038/sj.emboj.7601075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama Y, Kamimura Y, Okawa M, Muramatsu S, Sugino A, Araki H. GINS, a novel multiprotein complex required for chromosomal DNA replication in budding yeast. Genes & Dev. 2003;17:1153–1165. doi: 10.1101/gad.1065903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Umemori T, Hirai K, Muramatsu S, Kamimura Y, Araki H. CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature. 2007;445:328–332. doi: 10.1038/nature05465. [DOI] [PubMed] [Google Scholar]
- Tsubota T, Tajima R, Ode K, Kubota H, Fukuhara N, Kawabata T, Maki S, Maki H. Double-stranded DNA binding, an unusual property of DNA polymerase ɛ, promotes epigenetic silencing in Saccharomyces cerevisiae. J Biol Chem. 2006;281:32898–32908. doi: 10.1074/jbc.M606637200. [DOI] [PubMed] [Google Scholar]
- Waga S, Stillman B. The DNA replication fork in eukaryotic cells. Annu Rev Biochem. 1998;67:721–751. doi: 10.1146/annurev.biochem.67.1.721. [DOI] [PubMed] [Google Scholar]
- Waga S, Masuda T, Takisawa H, Sugino A. DNA polymerase ɛ is required for coordinated and efficient chromosomal DNA replication in Xenopus egg extracts. Proc Natl Acad Sci. 2001;98:4978–4983. doi: 10.1073/pnas.081088798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter JC, Araki H. Activation of pre-replication complexes. In: DePamphilis ML, editor. DNA replication and human disease. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2006. pp. 89–104. [Google Scholar]
- Wang H, Elledge SJ. DRC1, DNA replication and checkpoint protein 1, functions with DPB11 to control DNA replication and the S-phase checkpoint in Saccharomyces cerevisiae. Proc Natl Acad Sci. 1999;96:3824–3829. doi: 10.1073/pnas.96.7.3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabuuchi H, Yamada Y, Uchida T, Sunathvanichkul T, Nakagawa T, Masukata H. Ordered assembly of Sld3, GINS and Cdc45 is distinctly regulated by DDK and CDK for activation of replication origins. EMBO J. 2006;25:4663–4674. doi: 10.1038/sj.emboj.7601347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegerman P, Diffley JF. Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature. 2007;445:281–285. doi: 10.1038/nature05432. [DOI] [PubMed] [Google Scholar]
- Zlotkin T, Kaufmann G, Jiang Y, Lee MY, Uitto L, Syvaoja J, Dornreiter I, Fanning E, Nethanel T. DNA polymerase ɛ may be dispensable for SV40- but not cellular-DNA replication. EMBO J. 1996;15:2298–2305. [PMC free article] [PubMed] [Google Scholar]