Abstract
Electronic cell sorting for isolation and culture of dinoflagellates and other marine eukaryotic phytoplankton was compared to the traditional method of manually picking cells using a micropipette. Trauma to electronically sorted cells was not a limiting factor, as fragile dinoflagellates, such as Karenia brevis (Dinophyceae), survived electronic cell sorting to yield viable cells. The rate of successful isolation of large-scale (> 4 litres) cultures was higher for manual picking than for electronic cell sorting (2% vs 0.5%, respectively). However, manual picking of cells is more labor intensive and time consuming. Most manually isolated cells required repicking, as the cultures were determined not to be unialgal after a single round of isolation; whereas, no cultures obtained in this study from electronic single-cell sorting required resorting. A broad flow cytometric gating logic was employed to enhance species diversity. The percentages of unique genotypes produced by manual picking or electronic cell sorting were similar (57% vs 54%, respectively), and each approach produced a variety of dinoflagellate or raphidophyte genera. Alternatively, a highly restrictive gating logic was successfully used to target K. brevis from a natural bloom sample. Direct electronic single-cell sorting was more successful than utilizing a pre-enrichment sort followed by electronic single-cell sorting. The appropriate recovery medium may enhance the rate of successful isolations. Seventy percent of isolated cells were recovered in a new medium (RE) reported here, which was optimized for axenic dinoflagellate cultures. The greatest limiting factor to the throughput of electronic cell sorting is the need for manual postsort culture maintenance and assessment of the large number of isolated cells. However, when combined with newly developed automated methods for growth screening, electronic single-cell sorting has the potential to accelerate the discovery of new algal strains.
Keywords: Dinoflagellate, Cell sorting, Flow cytometry
INTRODUCTION
Dinoflagellates are unicellular, flagellated, often photosynthetic marine and freshwater protists. Current methods for the establishment of unialgal cultures of dinoflagellates and other marine protists typically involve either serial dilution or microscopic observation, combined with manual picking of single cells of a specific morphological type from complex microbial assemblages (Brand 1990). The latter approach is time consuming and requires a high level of experimenter expertise. Additionally, the rates of successful culture establishment can be as low as 10% even with the most careful attention to culture conditions (Brand 1981). These obstacles are an impediment to the use of marine phytoplankton for biotechnological applications such as drug discovery and gene mining (Shimizu & Li 2006). Moreover, these obstacles slow studies of the ecology, physiology and population genetics of many dinoflagellate species.
Flow cytometry (FCM) was first applied to environmental science for the study of phytoplankton (Veldhuis & Kraay 2000). This remains the most common environmental application of FCM. Phytoplankton vary in size (0.8- 200 μm) and pigmentation (due to variations in chlorophyll a, b, and c; phycoerythrin; and phycocyanin cell quotas). These properties have historically favored their cytometric discrimination (Dubelaar & Jonker 2000). High-speed flow cytometers facilitate the rapid analysis of many cells when compared to epifluorescence microscopy.
Electronic cell sorting may be used in conjunction with FCM to deposit single cells (`clone sorting') or to collect a large number of cells with similar properties (`enrichment'). Broad sort-gate logic may be used to collect a general class of cells, or highly restrictive sort-gate logic may be employed to target cells of specific genera or of a desired morphology. The potential applications of flow cytometric cell sorting for the isolation and culture of dinoflagellates have not been widely explored, as many dinoflagellates are particularly fragile and problematical to isolate. However, flow cytometric cell sorting has recently been utilized successfully for the isolation of several types of microalgae both from mixed culture assemblages and directly from natural samples (Sieracki et al. 2005).
The study reported here had two aims. First, this study evaluated the potential of viable electronic clone sorting and electronic enrichment sorting for the generation of clonal cultures of marine phytoplankton, particularly dinoflagellates and raphidophytes, from natural samples. Second, the efficiency and success of this automated method was compared to traditional handpicking techniques. This work was done in conjunction with a larger project screening natural marine samples for new toxic microalgal strains and new bioactive compounds. This project presented a need for rapid high-throughput isolation of new algal strains. This study resulted in the successful establishment of 57 unialgal dinoflagellate and raphidophyte cultures. The phylogenetic analysis and characterization of these isolates have been reported elsewhere (Scorzetti et al. 2009), and the characterization of bioactive compounds from these strains is currently ongoing. This study provides an example of the potential of automated cell sorting for use in marine phytoplankton isolation and bioprospecting.
MATERIAL AND METHODS
Sample collection
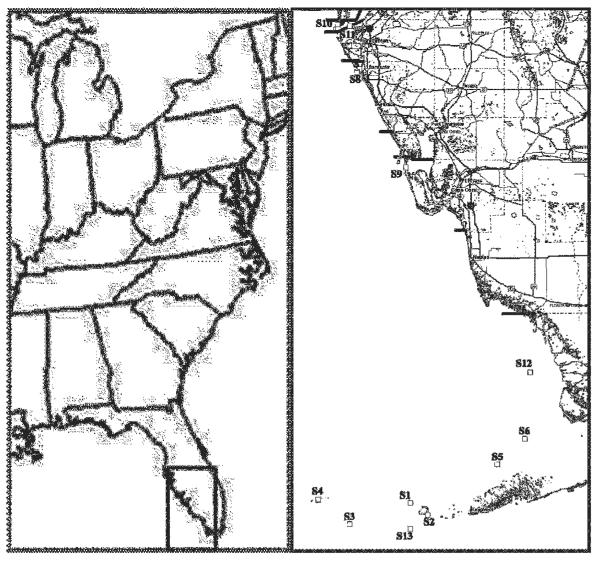
Nearshore surface water samples (2 litres) were collected using a sterile bucket at sites 7, 10 and 11. The remaining sites were sampled by plankton net tows (1.4 m3 of seawater) using a 30-μm mesh plankton net (General Oceanics) (Table 1, Fig. 1). The sample site Global Positioning System coordinates are based on the WGS84 datum. Coordinates were collected by both shipboard and handheld GPS instruments (various models). Sampling locations are listed in Table 1.
Table 1.
Collection site and date for each isolated strain
| Site no. | GPS coordinate (N) | GPS coordinate (W) | Date | Strain no. (CCMP no.) |
|---|---|---|---|---|
| S1 | 24°37'0.12" | 82°13'24.96" | 21 May 2004 | 1, 2 (2980), 3, 4, 5 (2770) |
| 4 Jan. 2005 | 11 (3174) | |||
| S2 | 24°32'30.12" | 82°5'60.00" | 19 Oct. 2004 | 6 (2933), 7, 8, 9 |
| 4 Jan. 2005 | 10 (2982), 12, 13, 25 (2976), 27 (2794), 28 (2981), 29 (3077), 30 (2810), 31, 34 (2796), 35 (2818), 38 (3113), 40 (2804) | |||
| S3 | 24°29'6.00" | 82°39'12.24" | 19 Oct. 2004 | 32 (2774), 33 (2813) 39 |
| S4 | 24°38'12.48" | 82°52'37.20" | 4 Jan. 2005 | 14 (2819), 15 (2795), 16 (2797), 17 (2932), 18 (2771) |
| S5 | 24°51'31.68" | 81°36'38.88" | 4 Jan. 2005 | 19 (3031) |
| S6 | 25°1'0.12" | 81°24'57.24" | 4 Jan. 2005 | 20 (2776) |
| S7 | 27°20'0.60" | 82°34'58.80" | 8 Feb. 2005 | 21 (2812), 22 (3122), 23 (2772), 24, 45 (2820) |
| 24 Aug. 2005 | 52, 53 (2775), 55 (2811) | |||
| S8 | 27°18'43.92" | 82°36'3.60" | 28 Feb. 2005 | 26 (2979), (3080), 37 (3099), 41 (2778) |
| S9 | 26°43'11.28" | 82°15'18.00" | 28 Feb. 2005 | 49 (3029) |
| S10 | 27°36'45.36" | 82°44'16.08" | 8 Jul. 2005 | 42 (2793), 43 (2821), 44 (2816), 46 (2822) |
| S11 | 27°36'16.20" | 82°39'0.00" | 8 Jul. 2005 | 47 (2814), 48 (2931) |
| 24 Aug. 2005 | 50 (2815), 51 (2817), 54 (2780) | |||
| S12 | 25°25'57.00" | 81°22'40.08" | 19 Oct. 2004 | 56 (2934) |
| S13 | 27°27'14.40" | 82°13'26.40" | 19 Oct. 2004 | 57 (3103) |
Fig. 1.
Sampling sites. Strains isolated from each site are listed in Table 1.
Twenty-millilitre subsamples from the plankton tows were diluted 1 : 1 with either RE, L1 or K (Guillard 1975) medium in clear 50-ml polystyrene culture flasks (BD Biosciences). Samples were maintained under Grow-Lux (Sylvania) wide-spectrum lamps at room temperature and transported back to the laboratory within 72 h of collection.
Flow cytometry
Flow cytometry was performed using a MoFlo-MLS (Dakocytomation) equipped with a water-cooled tunable Argon laser (Coherent InnovaTM I90C-A6). Analyses were conducted with a 488-nm laser line at 100 mW. Optical alignment employed 6-μm Flowcheck beads (Beckman Coulter) that yielded coefficients of variation of ,2%. Forward scatter (FSC) signal was collected by a forward scatter diode. Side scatter (SSC) and red (FL4, 670/30 nm) or green fluorescence (FL1, 530/40 nm) were collected by photomultiplier tubes (PMT). All parameters were collected with logarithmic amplification.
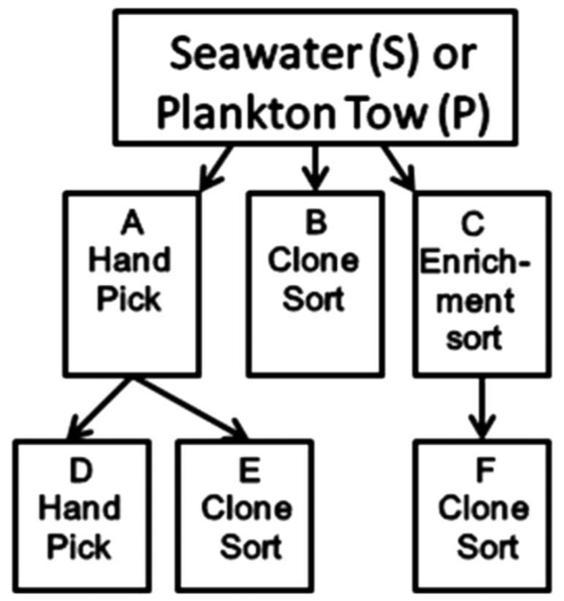
Samples were sorted for enrichment of dinoflagellate cells into clear 50-ml polystyrene culture flasks (Corning) containing 25 ml of receiving medium (10,000 cells sorted per flask), and clones were sorted (one cell per well) into 48-well microtitre plates (Corning) containing 1 ml of media or subjected to manual picking of dinoflagellate cells into 48-well microtitre plates (Fig. 2). Recovery media included RE, L1-Si, f/2-Si and K medium. Multiple enrichment sorts were incubated for four weeks (with periodic media refreshment) and resorted using single-cell sorting into microwell plates containing the same type of media. Sorting conditions consisted of sheath fluid composed of seawater (collected from the sampling sites and triple filtered through 0.2-micron Millipore capsule filters) at a sheath pressure of 30 psi, with a sheath/sample pressure differential of 6.89 × 103 Pa, laser excitation of 100 mW at 488 nm, sort mode setting of `sort-single-1-drop', triggered by SSC.
Fig. 2.
Flow diagram of combined cell sorting and handpicking strategies for isolation of dinoflagellates. The letters in the flow diagram correspond to the isolation methods listed in Table 2.
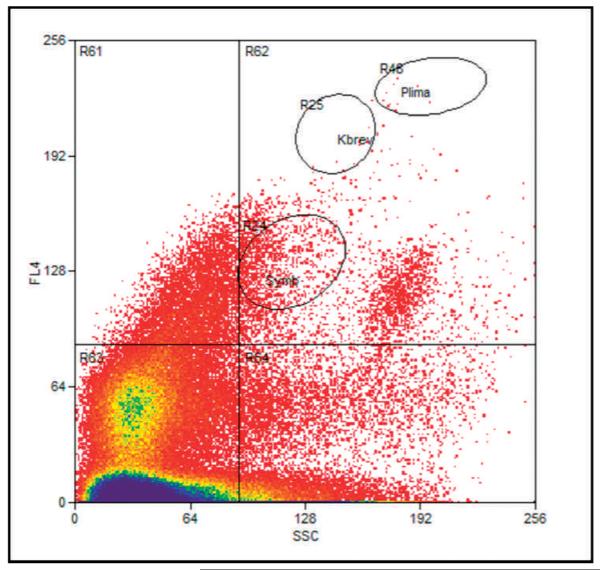
Using laboratory cultures of several genera of dinoflagellates, including Symbiodinium sp., Amphidinium sp., Karenia brevis (Davis) G. Hansen & Moestrup comb. nov. (formerly Gymnodinium breve Davis) and Prorocentrum lima (Ehrenberg) Dodge, a broad gating logic of SSC vs FL4 (FL4 = chlorophyll fluorescence, emission filter 670/30 nm) was developed. This was anticipated to recover a wide range of dinoflagellate cell types (gate R62; Figs. 3, 4). The smaller oval sort gates labeled R24, R25 and R26 are for orientation of relative size and fluorescent intensity. These gates correspond to the population location that would be seen with unialgal laboratory cultures of Symbiodium sp., Karenia sp. and P. lima, respectively. Growth in microwell plates and culture flasks was monitored by microscopic observation using an Axiovert microscope with an AxioCam digital imaging system (Carl Zeiss AG). Wells that appeared to contain multiple algal species were resorted by single-cell sorting until they appeared unialgal by microscopic inspection.
Fig. 3.
Example of broad sort-gate logic (R62) used for enrichment and clone sorting of a diversity of dinoflagellates from a natural water sample. Ovals R24, R25 and R46 represent locations of Symbiodinium, Karenia and Prorocentrum cells, respectively.
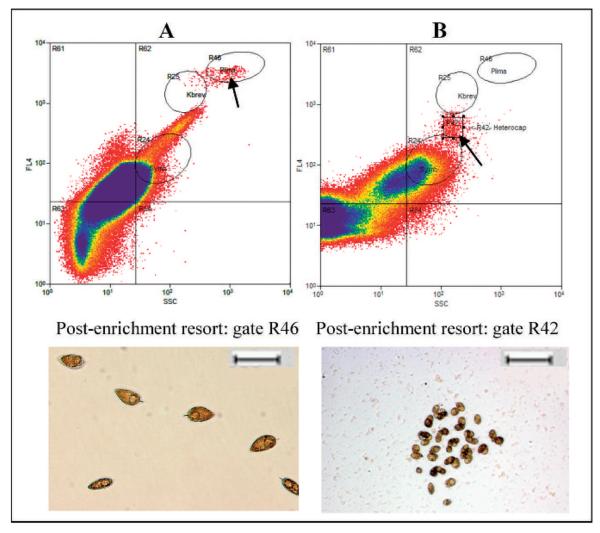
Fig. 4.
Example of group-specific sort-gate logic for single-cell resorting of enrichment cultures. Cells were initially enrichment sorted using the upper-right quadrant sort gate labeled R62 (Fig. 3). Cells were later resorted by single-cell sorting into microwell plates using the Prorocentrum-specific gate R24 (A) or the square custom gate R42 (B). Sequence analyses identified the resulting isolated unialgal cultures as Prorocentrum micans (27) and Heterocapsa sp. (13). Scale bar = 100 μm.
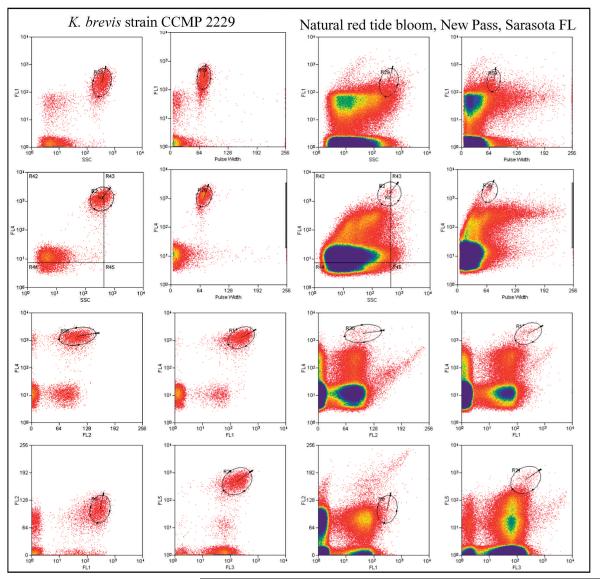
For Karenia-specific sorting direct from natural algal blooms, target cells had to simultaneously satisfy a multi-parametric combination of eight specific sort gates (Fig. 5). These included gating regions of eight bivariate plots for SSC vs FL1, PW vs FL1, SSC vs FL4, PW vs FL4, FL2 vs FL4, FL1 vs FL4, FL1 vs FL2 and FL3 vs FL5, where, PW = pulse width and emission filters FL1 = 530/40 nm, FL2 = 580/30 nm, FL3 = 630/30 nm and FL5 = 740 nm long pass, respectively.
Fig. 5.
Multiparameter flow cytometry logic gates were defined using CCMP2229 (left) and used to sort K. brevis cells from a natural Karenia bloom (right).
Culture conditions
Cultures were maintained in a variety of media - RE, K, f/2-Si and L-Si (Guillard 1975) - at 20-22°C under constant illumination from either Cool White (GE) or Grow-Lux (Sylvania) wide-spectrum lamps. RE medium is a novel dinoflagellate enrichment medium that we report here. It is based on previously described media such as ASP-M (McLachlan 1964) and L1 (Guillard 1995) but differs from most media by the inclusion of higher concentrations of biotin and vitamin B12, both known to be essential for many algae. CaCl2 is sterilized separately to avoid calcium precipitation. The RE medium consists of nine separately sterilized solutions (autoclaved unless otherwise noted): (1) 1 litre of artificial seawater containing, per litre, 23.38 g NaCl, 4.93 g MgSO4•7H2O, 4.06 g MgCl2•6H2O, 0.75 g KCl, 0.2 g KBr and 12.4 mg H3BO3; (2) 1 ml of RE trace metals containing, per litre, 200 mg MnSO4•H2O, 200 mg Na2MoO4•2H2O, 100 mg ZnSO4•7H2O, 20 mg Cu-SO4•5H2O, 20 mg CoCl2•6H2O, 20 mg NiSO4•6H2O, 1.84 mg Na3VO4 and 1.73 mg Na2SeO3; (3) 1 ml of RE vitamins, filter sterilized, containing, per litre, 200 mg vitamin B12 (cyanocobalamine), 200 mg thiamine-HCl and 100 mg biotin; (4) 1 ml of NaNO3 (75 g l-1; (5) 1 ml of NaH2PO4•H2O (5 g l-1); (6) 10 ml of CaCl2•2H2O (147 g l-1); (7) 5 ml of Trizma pH 8.1-8.2 (279.6 g l-1); (8) 2 ml of NaHCO3 (84 g l-1; and (9) 1 ml of FeSO4•7H2O (3.06 g l-1 freshly prepared and filter sterilized). Cells were transferred to 10-ml test tubes containing 2 ml of media at cell concentrations ranging from 500 to 1500 cells ml-1.
RESULTS
Seventeen seawater and plankton tow samples were collected from 13 sites on seven sampling trips (Table 1; Fig. 1). Samples were subjected to a combination of traditional manual picking of cells, electronic cell sorting for enrichment and electronic single-cell sorting for isolation. A minimum of five 48-well plates were sorted per sample (240 wells). However, the number of single-cell isolation attempts varied from site to site. During the course of this study, it was determined that some sites (such as sites S1, S2, S7 and S11) consistently yielded a high diversity of species. Therefore, the number of single-cell isolation attempts from these sites was doubled with samples from subsequent trips. On average, 288 cells were sorted per sample by electronic cell sorting, for approximately 5000 single-cell isolation attempts. Ninety-six single-cell isolation attempts were made per sample by manual picking of cells, for a total of 1632 isolation attempts. This resulted in the successful establishment of 57 unialgal dinoflagellate and raphidophyte cultures (Table 2).
Table 2.
Isolated unialgal cultures.
| Isolate nos.1 | Nearest GenBank sequence match or nearest area of identification | Isolation methods and no. of strains2 |
|---|---|---|
| 34, 56 | DQ779987 Akashiwo sanguinea | D |
| 2 | AJ417899 and AJ417899 Amphidinium eilatiensis and A. carterae | D |
| 12, 14, 15 | AY460587 Amphidinium gibbosum | B |
| 32, 33 | AY55670 and AF260381 Amphidinium massartii and A. klebsii | D |
| 1, 3, 4, 5 | Heterocapsal Cachonina area | B |
| 10 | Heterocapsal Cachonina area | B |
| 13 | AY371082 Heterocapsa sp. | F |
| 31 | Heterocapsal Cachonina area | D |
| 42-44, 46-48 | AF409126 Chatonella subsalsa | A |
| 39 | U92258 and AF244942 Coolia monotis and C. malayense | D |
| 49 | AF033868 Fragilidium subglubosum | D |
| 52 | AF318226 Gymnodinium corii | C |
| 57 | U94907 Gymnodinium sp. | A |
| 11, 28, 29 | EF613355 Heterocapsa triquetra | D |
| 24, 50, 51 | AF409124 Heterosigma akashiwo | A (1), F(1) |
| 45 | U92248 Karenia brevis | C |
| 41 | AY245692 Karlodinium micrum | B |
| 22 | AY833516 andAY833515 Prorocentrum donghaiense and P. dentatum | B |
| 40 | AY027907 Prorocentrum hoffmanianum | D |
| 27, 23 | AF260377 Prorocentrum micans and AY259165 P. gracile | A (1), F (1) |
| 21, 54, 55 | AF042813 Prorocentrum minimum | F (1), C (2) |
| 6, 7, 8, 9 | AY259167 Prorocentrum rhathymum | E |
| 25 | AY259167 Prorocentrum rhathymum | D |
| 19, 20, 38 | AY027907 Protoceratium reticulatum | A (1), B (2) |
| 16, 17, 18 | AY628427 Scripsiella trochoidea (differs by 6 base pairs) | A |
| 26, 36, 37 | AY628427 Scripsiella trochoidea (identical) | A |
| 30 | Scripsiella area | A |
| 35 | Scripsiella area | A |
| 53 | Scripsiella area | C |
Strains in the same row are identical genotypes. Strains shown in bold are potential new species.
Strains were isolated according to Fig. 2.
More than two-thirds of the strains (n = 38 of 57) were isolated from plankton tows as opposed to unconcentrated water samples. Thirty-one of the 57 strains were isolated by manual picking of cells alone (path A or D; Fig. 2). Manual picking, followed by subsequent electronic single-cell sorting, was successful in isolating four species (path E; Fig. 2). More than half the handpicked cells had to be either handpicked a second time or single-cell sorted (path D or E; Fig. 2) because the initial cultures were determined not to be unialgal by microscopic examination. By applying a broad-range sort-gate logic (gate R62 in Figs 3, 4), 20 strains were isolated by electronic single-cell sorting (path B; Fig. 2). The use of the same broad sort-gate logic was used for preliminary enrichment. The resulting mixed culture was single-cell sorted into microwell plates after 4-8 weeks (path F; Fig. 2), ultimately yielding two species. Overall recovery rates of large-scale cultures were ~0.5% for electronic cell sorting and ~2% for manual picking.
Manual picking yielded 20 different genotypes, as determined by a comparison of the D1D2 domain of the large-subunit rRNA (Scorzetti et al. 2009). Twelve different genotypes were isolated by electronic single-cell sorting. The percentages of unique genotypes from manual picking or electronic cell sorting were similar (57% vs 54%, respectively). Manual picking provided 11 unique genera (35%), whereas electronic cell sorting isolated nine (41%) unique genera. There appeared to be little difference in genera isolated by each technique. Seven genera were common to both the manually picked and the electronically sorted groups. Four genera were isolated exclusively by picking cells (Chatonella, Akashiwo, Coolia and Fragilidinium), while two were unique to electronic sorting (Karenia and Karlodinium). Traditional handpicking for isolation may be more effective for larger and more fragile dinoflagellates or those with asymmetrical or elongated protrusions, such as Coolia or Fragilidinium, as these were not isolated by electronic cell sorting.
Using three K. brevis control cultures (CCMP718, CCMP2228 and CCMP2229; Provasoli-Guillard National Center for the Culture of Marine Phytoplankton), a highly targeted gating logic was developed for high-purity sorting of K. brevis cells (Fig. 5). This logic involved the simultaneous gating of eight bivariate parameters (FSC, SSC, pulse width and fluorescence from 488-nm excitation at emissions of 530, 580, 630, 670 and 740 nm and above).
Using these gating parameters, 192 single-cell isolation attempts were made from laboratory cultures of CCMP2229. Simultaneously, 192 isolation attempts were made by manual picking of cells. Growth was monitored by microscopic examination (Table 3). Four electronically sorted cells survived beyond 3 months to provide cultures of high cell concentration, while only one manually picked cell survived beyond 3 months. The application of this simultaneous multiparameter sort-gate logic was effective at directly isolating Karenia type cells from a highly complex microbial assemblage of an early-stage natural red tide bloom sampled on 8 February 2005 from site S7 (Figs 5, 6).
Table 3.
Comparison of K. brevis culture recovery after handpicking or electronic single-cell sorting
| Single-cell isolation approach for CCMP2229 | Electronic sorting | Handpicking |
|---|---|---|
| No. of single-cell isolation attempts | 192 | 192 |
| Isolations surviving 2 generations (4 cells) | 39 | 43 |
| Isolations surviving 5 generations (32 cells) | 6 | 4 |
| Isolations surviving 10 generations (1024 cells) | 6 | 2 |
| Isolations surviving minimum of 3 months and generating large scale (> 4 1) cultures | 4 | 1 |
| % Successful isolation recovery | ~ 2% | ~ 0.5% |
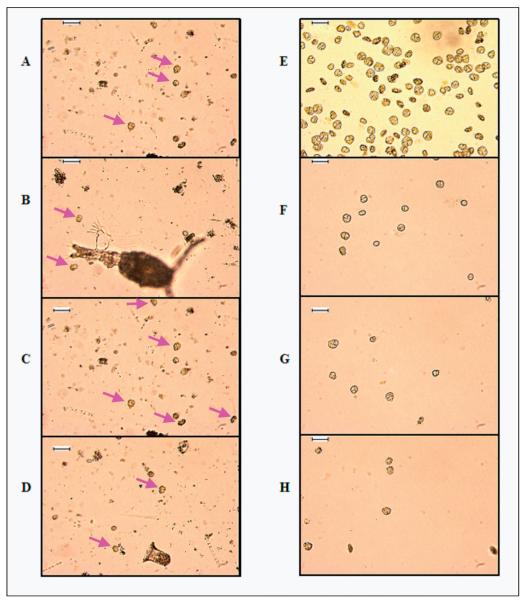
Fig. 6.
Panels A-D are four independent micrographs of the same sample, showing the complex microbial assemblage of an early-stage Karenia bloom, with only a few Karenia-type cells. The arrows highlight some cells typical of Karenia-type morphology in various orientations. Panels E-H are four independent micrographs after sorting, using the gate logic shown in Fig. 5. Scale bar = 100 μm.
DISCUSSION
Flow cytometric cell sorting represents a useful addition to the phycological toolbox. Both electronic cell sorting and handpicking have strengths and limitations (Andersen & Kawachi 2005; Sieracki et al. 2005). Handpicking is a more gentle approach, maximizing survival of large and/or delicate cells but is limited by throughput and requires a high degree of morphological expertise and manual dexterity. Electronic cell sorting requires expensive instrumentation and a skilled instrument operator. Electronic sorting may also subject the cells to more trauma than handpicking. It has the advantage of higher throughput, which can be crucial since culture recovery from any of these approaches is low. Therefore, electronic cell sorting may be used to accelerate the rate of culture isolation of dinoflagellates, raphidophytes, prymesiophytes and other algae of interest.
Dinoflagellates may be armored or unarmored (with or without cellulose theca, respectively). Such structural differences might have an effect on their fragility and on our ability to successfully sort and culture individual cells in the laboratory. During the sorting process in high-speed cell sorters, cells can undergo a variety of traumas. First, elevated pressures are required to produce the desired jet velocity. Second, cells are queried briefly (< 1 × 10-6 s) with laser light at an intensity of over 3.2 × 1010 W m22. By comparison, full sunlight intensity on Florida coastal surface waters is about 4 × 10-3 W m-2. Electronic sorting may also cause electrodamage, by the field pulse applied for deflection purposes (Reckermann 2000). Additional cell trauma may be caused by the expulsion of the sorted cells at speeds in excess of 14 m s-1 into the collection vessels and their impact into the receiving medium. Successful viable cell sorting requires minimization of these deleterious effects and optimization of culture recovery conditions.
Despite these challenges, flow cytometry has been shown to have minimal impacts on long-term growth of many fragile phytoplankton, and electronic sorting effects on cell viability tend to be temporary (Haugen et al. 1987). The greatest instrumental impact on phytoplankton appears to be due to laser exposure. Rivkin et al. (1986) showed a loss of photosynthetic capability of phytoplankton cells immediately after exposure to laser light during sorting, but there was apparently no long-term effect from the sorting process. Prior studies with microalgal strains, including Isochrysis, Thalassiosira, Rhodomonas, Micromonas, Heterocapsa and Synechococcus, have shown that, overall, the viability of flow cytometrically sorted single-cell isolates was slightly less or comparable to those expected by traditional micropipette handpicking techniques (Sieracki et al. 2005). The results we report here for dinoflagellate and raphidophyte isolation also support this observation.
Our initial efforts focused on determining if fragile and unarmored dinoflagellates could survive the flow cytometry sorting process to provide large-scale cultures by the application of the technique to laboratory cultures of K. brevis (CCMP2229). Comparisons of culture recovery between manual picking and single-cell electronic sorting (Table 3) indicate that recovery of this fragile dinoflagellate after isolation by flow cytometry is comparable to recovery by traditional handpicking, using identical microwell plates and the same medium and culture conditions.
Two different approaches may be employed to generate dinoflagellate cultures by electronic cell sorting. The first approach used a broad gating logic that selects algal cells based on size and chlorophyll content. This approach may be used to select a variety of species. The second approach used multiple narrow gating criteria to find a specific species or subgroups. Using the former approach, 22 unialgal cultures were established from plankton tows and seawater samples. Highly restrictive gating proved effective at isolating and enriching K. brevis cells from a red tide bloom (Fig. 5, right side). At the time of sampling, fish kills were observed in local waters. However, the histograms (Fig. 5, right side) and micrographs (Fig. 6, left side) indicate that K. brevis cells made up a minority of the complex algal assemblage during this stage of the bloom. Despite their low abundance, the sort-gating strategy utilized in Fig. 5 was effective in concentrating and sorting K. brevis type cells from this assemblage. While this sort logic specifically isolated Karenia-type cells from a natural bloom, it could not discriminate between species of Karenia.
In our hands, electronic single-cell sorting using a broad gate logic was slightly less effective in terms of recovery of viable cultures and comparable or slightly superior in species diversity when compared to manual picking of cells. Preliminary enrichment sorting using the same broad gate logic, followed by electronic single-cell sorting several weeks later, resulted in the recovery of only two species: Prorocentrum micans Ehrenberg (27) and Heterocapsa (13). Given that this particular sample (site 2, 4 January 2005; Table 1) yielded more strains (12 strains or 11 unique genotypes by manual picking and electronic single-cell sorting) than any other sample, preliminary enrichment sorting appears to be the least successful strategy.
Our rates of successful isolations by handpicking or electronic single-cell sorting were lower than rates reported earlier (Brand 1981; Sieracki et al. 2005). Dinoflagellate cultures proved to be most sensitive immediately after picking or electronic single-cell sorting. In our own work with dinoflagellates, we have found that recovery cultures are very unstable until they reach concentrations of 103-104cells ml-1 (depending on strain and other culture conditions) regardless of the isolation technique. A positive correlation between cell concentration and recovery after electronic sorting has also been observed for several types of microalgae (Sieracki et al. 2005). Studies on viable microalgae recovery after electronic cell sorting have indicated that culture conditions and recovery vessel format may have more impact than instrument sorting conditions on viable recovery of many microalgae. In particular for cell sorting into microwell plates, the size of the wells appears to affect the recovery. Sieracki et al. (2005) reported no recovery of viable Heterocapsa triquetra Stein when electronically single-cell sorted into 96-well plates but did report 30% and 60% recovery when sorted into larger wells of 24-well plates, using f/20 and f/2 medium, respectively. The reasons for this striking difference in recovery with different receptacle formats is not entirely clear but may be influenced by the surface-to-volume ratio of the well. Smaller volumes of water fluctuate more rapidly in temperature and salinity, increasing stress on the cells. The larger surface-to-volume ratio of vessel to media exposes the cells to greater potential contamination from the culture container. Both sorted and handpicked cells were isolated into plastic containers, which may leach plasticizer, as has been suggested by previous studies (Sieracki et al. 2005). The higher survival rate reported by Brand (1981) is probably the result of two factors. Cultures were developed in 10-ml glass test tubes, while ours developed in 1.6-ml well plates. Brand (1981) used rigorous cleaning techniques to reduce chemical contamination of this culture media and test tubes. All media and culture vessels used in this study were sterilized, but no special effort was made to purify the culture media or vessels of possible plasticizer resins or other potentially inhibiting compounds.
While four different media (RE, K, f/2-Si and L-Si) were used equally in our isolation efforts, RE medium proved the most successful recovery medium. Thirty-seven (69%) of the established cultures were sorted into this medium. RE medium was initially developed in an effort to isolate and maintain axenic algal cultures, and it contains increased concentrations of vitamins that were previously provided by bacteria. Sensen et al. (1983) found that up to 20% of cultures of algae established by electronic cell sorting were axenic, even when sorting from highly contaminated samples, and we anticipated that this medium would enhance the survival rate of electronically sorted cells. RE is an artificial seawater medium based on previously described media, primarily ASPM (Guillard 1975) and L1 (Guillard & Hargraves 1993). The trace element and vitamin compositions of bacteriological media (Guirard & Snell 1981) were also consulted. It contains increased concentrations of trace metals that serve as enzyme prosthetic groups. RE also contains vitamins, biotin, thiamine and cyanocobalamin, for which K. brevis and other algae are commonly deficient (Provasoli 1958).
High-throughput electronic cell sorting is a rapid technique allowing for thousands of single-cell isolation attempts in an afternoon. The time spent performing electronic cell sorting (including instrument setup and postsort cleaning) was roughly 10% of the time spent handpicking cells. The necessity for two or three iterations increased the amount of time and labor required for manual picking. This observation is consistent with previous reports attributing the lack of successful isolations of dinoflagellate cultures to domination by contaminating species (Brand 1981). By comparison, none of the single-cell isolations by electronic cell sorting (path B) required resorting. Therefore, the yield per unit of time is higher with electronic cell sorting than manual picking.
A major challenge facing high-throughput electronic cell sorting is the need for manual postsort culture maintenance and assessment of the large number of isolated cultures for growth. However, there are a number of possibilities for automating this stage as well, thus improving the real throughput of this approach. Sieracki et al. (2005) demonstrated the use of high-sensitivity fluorescent micro-plate reading to rapidly screen for growth on a routine basis for large numbers of isolation cultures in microwell plates. This approach used automated microplate readers to measure increases in chlorophyll autofluorescence from cultures in microwell plates over time. Procentrum micans Ehrenberg and H. triquetra were detected in microwell plates using a Turner fluorometer at cell concentrations as low as 40 and 100 cells well-1, respectively. Thus, automated single-cell sorting, combined with automated fluorescent screening of culture growth, could allow the initial assessment of large numbers of isolation attempts. Purity and identity of the isolated cultures would need to be established by manual methods. Nonetheless, this combined approach could improve the throughput of isolation efforts over that of current manual techniques.
This study involved primarily the isolation of single cells of particulate celled strains. Viable electronic cell sorting methods might be more problematical for large colonial and filamentous species such as Gymnodinium catenatum Graham or for cyanobacterial filaments of genera such as Anabaena or Oscillatoria. Isolating single cells of such colonial types by electronic sorting may not be feasible since single cells cannot readily be dissociated from the filaments during the sorting process. However, we have utilized combinations of flow cytometric parameters in conjunction with pulse width to isolate single short filaments of Lyngbya, Anabaena and Cylindrospermopsis from mixed assemblages of laboratory cultures (data not shown). Nonetheless, routine sorting of filamentous strains still poses a significant challenge to electronic cell sorting at this time.
Electronic flow cytometric cell sorting for generating clonal, unialgal cultures from natural samples has the potential to facilitate screening for toxic algae and toxins, drug discovery and understanding ecosystem population dynamics and characteristics of harmful algal blooms yet remains underexplored. The ability to electronically sort live cells of dinoflagellates, raphidophytes and prymesiophytes without a significant impact on their viability is the first step towards the routine use of flow sorting for establishing unialgal cultures. Previous studies have successfully utilized flow sorting for the isolation and culturing of microalgae and cyanobacteria belonging to genera such as Cyanophora, Haematococcus, Monomastix, Scherffelia, Spermatozopsis, Synechococcus, Microcystis, Hemiselmis, Teleaulax, Rhodomonas, Thalassiosira and Prochlorococcus as well as other unidentified eukaryotic picoplankton (Sensen et al. 1993; Moore et al. 1998; Reckermann 2000; Crosbie et al. 2003). We show here that flow cytometric sorting can also be used to directly isolate a variety of dinoflagellates and raphidophytes, including fragile unarmored species such as K. brevis from field samples. However, the condition of the cells being isolated and especially the care and attention to culture and incubation conditions after cell sorting remain a critical factor in the successful isolation of new unialgal cultures.
ACKNOWLEDGEMENTS
This work was supported by the National Institute of Environmental Health Sciences (NIEHS) grant S11 ES11181. Grant support was also provided by the NSF-NIEHS Oceans and Human Health Center Program (National Science Foundation grant 0432368 and NIEHS grant P50 ES12736-01). Work by K.R. and J.W. was also supported by USDA grant NRICGP 2002-35201-11671. The authors thank Terri Teshiba and Marcela Yaafar for expert technical assistance with algal cultures.
REFERENCES
- Andersen RA, Kawachi M. Traditional microalgae isolation techniques. In: Andersen RA, editor. Algal culturing techniques. Elsevier Academic Press; New York: 2005. pp. 101–116. [Google Scholar]
- Brand LE. Genetic variability in reproduction rates in marine phytoplankton populations. Evolution. 1981;35:1117–1127. doi: 10.1111/j.1558-5646.1981.tb04981.x. [DOI] [PubMed] [Google Scholar]
- Brand LE. The isolation and culture of microalgae for biotechnological applications. In: Labeda DP, editor. Isolation of biotechnological organisms from nature. McGraw-Hill; New York: 1990. pp. 81–115. [Google Scholar]
- Crosbie N, Pökle M, Weiss T. Rapid establishment of clonal isolates of freshwater autotrophic picoplankton by single-cell and single-colony sorting. Journal of Microbiological Methods. 2003;55:361–370. doi: 10.1016/s0167-7012(03)00167-2. [DOI] [PubMed] [Google Scholar]
- Dubelaar GBJ, Jonker RR. Flow cytometry as a tool for the study of phytoplankton. Scientia Marina. 2000;64:135–156. [Google Scholar]
- Guillard RRL. Culture of phytoplankton for feeding marine invertebrates. In: Smith WL, Chanley MH, editors. Culture of marine invertrebrate animals. Plenum Press; New York: 1975. pp. 29–60. [Google Scholar]
- Guillard RRL. Culture methods. In: Hallegraeff GM, Anderson DM, Cembella AD, editors. Manual on harmful marine microalgae. UNESCO Publishing; New York: 1995. pp. 45–62. [Google Scholar]
- Guillard RRL, Hargraves PE. Stichochrysis immobilis is a diatom, not a chrysophyte. Phycologia. 1993;32:234–236. [Google Scholar]
- Guirard BM, Snell EE. Biochemical factors in growth. In: Gerhardt P, editor. Manual of methods in general microbiology. ASM Press; Washington, DC: 1981. pp. 79–111. [Google Scholar]
- Haugen EM, Cucci TL, Yentsch CM, Shapiro LP. Effects of flow cytometric analysis on morphology and viability of fragile phytoplankton. Applied and Environmental Microbiology. 1987;53:2677–2679. doi: 10.1128/aem.53.11.2677-2679.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mclachlan J. Some considerations of the growth of marine algae in artificial media. Canadian Journal of Microbiology. 1964;10:769–782. doi: 10.1139/m64-098. [DOI] [PubMed] [Google Scholar]
- Moore LR, Rocap G, Chisholm SW. Physiology and molecular phylogeny of coexisting Prochlorococcus ecotypes. Nature. 1998;393:464–467. doi: 10.1038/30965. [DOI] [PubMed] [Google Scholar]
- Provasoli L. Nutrition and ecology of protozoa and algae. Annual Review of Microbiology. 1958;12:279–308. doi: 10.1146/annurev.mi.12.100158.001431. [DOI] [PubMed] [Google Scholar]
- Reckermann M. Reckermann M, Colijn F, editors. Flow sorting in aquatic ecology. Aquatic flow cytometry: achievements and prospects. 2000;64:235–246. Scientia Marina. [Google Scholar]
- Rivkin RB, Phinney DA, Yentsch CM. Effects of flow cytometric analysis and cell sorting on photosynthetic carbon uptake by phytoplankton in cultures and from natural populations. Applied and Environmental Microbiology. 1986;52:935–938. doi: 10.1128/aem.52.4.935-938.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scorzetti G, Brand LE, Hitchcock GL, Rein KS, Sinigalliano C, Fell JW. Multiple simultaneous detection of harmful algal blooms (HABs) through a high throughput bead array technology, with potential use in phytoplankton community analysis. Harmful Algae. 2009;8:196–211. doi: 10.1016/j.hal.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sensen CW, Heimann K, Melkonian M. The production of clonal and axenic cultures of microalgae using fluorescence-activated cell sorting. European Journal of Phycology. 1993;28:93–97. [Google Scholar]
- Shimizu Y, Li B. Microalgae as a source of bioactive molecules: special problems and methodology. In: Proksch P, Muller WEG, editors. Frontiers in marine biotechnology. Horizon Bioscience; Wymondham, UK: 2006. pp. 145–174. [Google Scholar]
- Sieracki M, Poulton N, Crosbie N. Automated isolation techniques for microalgae. In: Anderson RA, editor. Algal culturing techniques. Elsevier Academic Press; New York: 2005. pp. 101–116. [Google Scholar]
- Veldhuis MJW, Kraay GW. Reckermann M, Colijn F, editors. Application of flow cytometry in marine phytoplankton research: current applications and future perspectives. Aquatic flow cytometry: achievements and prospects. 2000;64:121–134. Scientia Marina. [Google Scholar]